Abstract
There are many complex interactions between transposable elements (TEs) and host genomes. Environmental changes that induce stressful conditions help to contribute for increasing complexity of these interactions. The transposon mariner-Mos1 increases its mobilization under mild heat stress. It has putative heat shock elements (HSEs), which are probably activated by heat shock factors (HSFs). Ultraviolet radiation (UVC) is a stressor that has been suggested as able to activate heat shock protein genes (Hsp). In this study, we test the hypothesis that if UVC induces Hsp expression, as heat does, it could also promote mariner-Mos1 transposition and mobilization. The Drosophila simulans white-peach is a mutant lineage that indicates the mariner-Mos1 transposition phenotypically through the formation of mosaic eyes. This lineage was exposed to UVC or mild heat stress (28 °C) in order to evaluate the induction of mariner-Mos1 expression by RT-qPCR, as well as the mariner-Mos1 mobilization activity based on the count number of red spots in the eyes. The effects of both treatments on the developmental time of flies and cell cycle progression were also investigated. Both the analysis of eyes and mariner-Mos1 gene expression indicate that UVC radiation has no effect in mariner-Mos1 transposition, although heat increases the expression and mobilization of this TE soon after the treatment. However, the expression of Hsp70 gene increased after 24 h of UVC exposure, suggesting different pathway of activation. These results showed that heat promotes mariner-Mos1 mobilization, although UVC does not induce the expression or mobilization of this TE.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-015-0611-2) contains supplementary material, which is available to authorized users.
Keywords: Transposable elements, Mechanisms of transposition, Drosophila simulans white-peach, Temperature, UV radiation
Introduction
Transposable elements (TEs) are DNA sequences with the ability to move from one chromosomal location to another in the genome. They are widely distributed among the majority of studied organisms and can correspond to a greater portion of their genomes, as much as 45 % in humans or 50 to 90 % in certain grass genomes (Feschotte and Pritham 2007; Pritham 2009). TEs show huge sequence diversity and are classified in a taxonomic system based on their transposition mechanism (Wicker et al. 2007).
The biological consequence of TE mobilization is their mutagenic effect, ranging from small nucleotide changes to chromosome rearrangements and epigenetic modifications. In general, TE mobilization is detrimental, but from an evolutionary perspective, they are a formidable resource of genetic variability to feed evolution (Hua-Van et al. 2011). To minimize the unfavorable effects of TEs, their “host” genomes developed mechanisms such as RNAi and epigenetic silencing through DNA methylation or heterochromatinization (Yamanaka et al. 2013; Creasey et al. 2014). Many TEs mobilize only in germ cells, and for a long time, this was the main focus of TE research. However, evidence on the importance of somatic mobilization is now accumulating (Kazazian 2011), indicating its involvement in genomic instability related to cancer (Helman et al. 2014), aging (De Cecco et al. 2013), and neurodegenerative diseases (Li et al. 2013).
Stressors act as activators of transposition of many TEs (Capy et al. 2000; Guerreiro 2011). The mariner-Mos1 transposon is a good genetic tool to investigate how a transposition mechanism is induced under stressful conditions (Guerreiro 2011). Chakrani et al. (1993) have shown the effect of increased temperature upon mariner-Mos1 mobilization experimentally. This study also compared the 5′ terminal inverted repeats (TIR) sequence of mariner-Mos1 with the promoter sequences of four heat shock protein (Hsp) genes, finding homology among them. A homology of 57 % was found between a 14-bp stretch of mariner-Mos1 and the Hsp70 gene. These observations suggest that mariner-Mos1 TIR contains functional heat shock elements (HSEs) activated by heat shock factors (HSFs). The heat shock proteins (HSPs) are the best characterized and conserved set of polypeptides that respond to thermal stress (Lindquist and Craig 1988). The HSPs are also implicated in the cell cycle regulation, in resistance to stress-induced apoptosis or necrotic cell death, and in antioxidative defense (Helmbrecht et al. 2000; Takayama et al. 2003; Mosser et al. 1997; Buzzard et al. 1998).
UV radiation is a stressor able to increase Hsp gene expression in human skin cells and fish tissues during embryonic stages (Trautinger 2001; Vehniäinen et al. 2012). Furthermore, UVC promoted transposition and excision of the Tc1/mariner superfamily of fungus Aspergillus oryzae (Ogasawara et al. 2009). UVC radiation damages DNA molecules, which is the major cellular chromophore of UVC light. This absorption generates lesions known as DNA photoproducts, in which the most common are cyclobutane pyrimidine dimers (CPDs) and (6–4) pyrimidine-pyrimidone photoproducts (6–4PPs) (Ravanat et al. 2001; Schuch et al. 2013). The presence of these lesions drastically alters the metabolic processes in DNA, since they represent a physical block to both the replication and transcription machinery (De Santis et al. 2002; Costa et al. 2003). As a result, a cell cycle arrest in the G1 phase is observed because the cells are unable to progress through S phase, thus triggering cell death (Ortolan and Menck 2013).
Considering that mariner-Mos1 element (i) is activated by heat and (ii) has putative HSE and that the UVC radiation can induce cell stress, in this study, we tested the hypotheses that heat and UVC would increase mariner-Mos1 expression and that UVC could induce mariner-Mos1 transposition, as heat does. For testing these hypotheses, we used Drosophila simulans strains containing a specific mutation called white-peach.
The D. simulans white-peach strain is an interesting model for investigating the transposition mechanism because it is an excellent system allowing to quantify the somatic mobilization and is easily manipulated in vivo (Medhora et al. 1991). The white-peach strain has a defective copy of mariner-Mos1 inserted in the promoter region of the white gene. The white gene encodes an enzyme involved in the production of red pigmentation in the eyes of wild flies. This insertion results in the eye color becoming white-peach instead of red (Jacobson and Hartl 1985; Jacobson et al. 1986). Furthermore, this defective copy of mariner-Mos1 (nonautonomous) can be mobilized in trans by the transposase enzyme synthesized from an autonomous copy of mariner-Mos1. This transposition occurs during the eyes’ development, and the mariner-Mos1 mobilization generates a new phenotype: white-peach eyes with red spots (mosaic) (Capy et al. 1992).
Therefore, we investigated whether UVC and the mild heat stress would induce Hsp gene expression in the lineage D. simulans white-peach. Moreover, if UVC can induce Hsp genes, it is possible that it could also promote mariner-Mos1 activation, transcription, and mobilization, since this element has a putative promoter sequence homologous to the Hsp promoter.
Materials and methods
Drosophila strains
In this study, we used a D. simulans white-peach isoline called Dswp test, which has active copies of the mariner-Mos1 element. This isoline was produced by crossing a D. simulans white-peach female (which has no active mariner-Mos1) (Bryan et al. 1990) with wild-type D. simulans males collected in Brasília, Brazil (which has active mariner-Mos1 elements). Furthermore, in the F2 generation, an isoline was established expressing the mosaic eye phenotype. This isoline presents a basal rate of mariner-Mos1 activity, which was confirmed by the presence of red spots in the eyes and one active copy of mariner-Mos1 in the genome, as estimated by qPCR. For the experimental procedures performed in this work, we chose the second larvae instar since the first is very sensitive to manipulation (influencing larval survival) and the third is too late to evaluate the transposition activity with reliable fidelity.
Estimation of transposition rate
To estimate the mariner-Mos1 transposition rate under variable stress conditions, we quantified the red spots in the mosaic eyes of adult flies that were submitted to stress during the second larval stage. Each spot is interpreted as one transposition event of mariner-Mos1, and the individuals were classified in different levels related to the number of observed spots: level 0 = without spots, level 1 = one to four spots, level 2 = five to ten spots, and level 3 = more than ten spots (Chakrani et al. 1993; Jardim and Loreto 2011). Twenty-five larvae were submitted to each stress treatment, in three replicates, for three independent experiments. The same number of control larvae was maintained at 20 °C, and the adults were quantified for spots in their eyes.
Exposure to UVC radiation
In order to estimate a sub-lethal dose and perform the other procedures with UVC, first, a larvae group was submitted to different UVC doses one under in which the survival rate was similar to control. The larvae were irradiated with 10, 25, 50, 75, and 100 J/m2 generated by a UVC germicide lamp (Sanyo G-light, 15 W). The UVC measurements were performed with a portable radiometer (EKO UV Monitor MS-211-1, Japan). For this procedure, groups of 25 second-instar larvae were collected manually and exposed to UVC light in Petri dishes containing 200 μl of phosphate-buffered saline (1× PBS).
With the purpose of evaluating if UVC is able to activate mariner-Mos1 transposase transcription or induce mariner-Mos1 mobilization, groups of larvae were irradiated with the chosen UVC sub-lethal dose and were submitted to different manipulations and analyses (three independent experiments): (i) a larvae group was maintained in culture medium until becoming adult flies in order to analyze the developmental time and carry out the phenotypic analysis; (ii) the total messenger RNA (mRNA) was extracted from other larvae groups, 6 and 24 h after stress, to analyze the expression of mariner-Mos1, Hsp70, and reference genes, by RT-qPCR; and (iii) the third group had the cells dissociated out of larvae tissues and submitted to flow cytometry, 48 h after the UVC irradiation. The control group samples to all procedures were manipulated in the same manner, but the UV lamps were not turned on. These results were statistically discriminated by one-way ANOVA followed by Dunnett’s test.
Mild heat stress
The temperature of 28 °C was used as a mild heat stress. This temperature was used as a stressor agent in a previous work (Jardim and Loreto 2011). A group of 25 second-instar larvae was maintained in culture medium while another group was kept at 28 °C, until becoming adult flies in the sense of analyzing the developmental time and carrying out phenotypic analysis. For gene expression analyses, other larvae groups were submitted at 28 °C for different periods of time: 6 or 24 h. The molecular procedures, mRNA extraction, were performed soon after treatment. Twenty-five larvae were collected for both treatments (three independent experiments). For flow cytometry, other larvae groups were maintained at 28 °C for 48 h and the cells dissociated out of larvae tissues. The control group samples to all procedures were manipulated in the same manner, but the temperature was maintained at 20 °C. These results were statistically discriminated by one-way ANOVA followed by Dunnett’s test.
Estimation of mariner-Mos1 copy number
For the estimation of mariner-Mos1 copy number in the Dswp-test strain, qPCR of a unique copy reference gene (Ribosomal protein L17-RPL17) was performed and compared with the mariner-Mos1 amplification. DNA from 20 flies was extracted individually using the protocol described in Oliveira et al. (2009), quantified in a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA), and diluted to a concentration of 3 ng/μl. Each qPCR reaction was performed in a 20-μl volume containing 20 pmol of each primer, 5 mM of dNTPs, 1× PCR buffer, 2.5 mM MgCl2, 1× SYBR Green (Molecular Probes, USA), 0.5 U of Platinum Taq DNA polymerase (Invitrogen, USA), and 15 ng of DNA. The primers used are described in Supplementary Table 1. The mariner-Mos1 copy number was estimated by the 2−∆∆Ct method (Livak and Schmittgen 2001) using the Ct values obtained in the ECO™ Real-Time PCR System (Illumina, USA). The estimated mariner-Mos1 copy number in the Dswp-test strain was 1.84 (±0.83) copies. As all individuals have a copy of the inactive peach mariner-Mos1 in the white gene, we can conclude that in this strain, the majority of individuals have one copy of an active element and a few have either none or two copies.
RNA extraction and cDNA synthesis
Total RNA was extracted from larvae with TRIzol® reagent (Invitrogen, CA, USA). The quality of the RNA samples was assessed by 1 % agarose gel electrophoresis and quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). Afterward, the samples were treated with DNaseI (Promega, USA) to eliminate possible DNA contamination. The complementary DNA (cDNA) synthesis was performed with M-MLV reverse transcriptase enzyme (Invitrogen, CA, USA) and oligo-dT primers.
mariner-Mos1 gene expression
To evaluate the putative activation of transcriptions of mariner-Mos1 transposase gene by stress, the Hsp70 gene was used as a positive marker, since this gene is expressed under different stressful conditions (Sørensen et al. 2005; Trautinger 2001). We used the RT-qPCR method, performed on a StepOnePlus™ Real-Time PCR System (Applied Biosystems, USA). The reaction was carried out in a final volume of 20 μl containing 10 μl of diluted cDNA (1:100), 0.25 U Platinum Taq DNA Polymerase (Invitrogen, CA, USA), 1× PCR reaction buffer, 3 mM MgCl2, 25 μM dNTPs, 0.2 μM of each reverse and forward primer, and 1× SYBR Green (Molecular Probes, USA).
The RT-qPCR amplification parameters were as follows: 95 °C for 7 min, followed by 40 cycles of 94 °C for 15 s, 60 °C for 15 s, and 72 °C for 20 s. Normalization input was performed with reference genes (GPDH, RPL17q2, and EF1). The primer sequences are described in Supplementary Table 1. The Hsp70 primers amplify only the Hsp70 genes Aa and Ba and do not anneal in their cognates. The relative gene expression was based on 2−ΔΔcq (Livak and Schmittgen 2001) using the Cq values. Furthermore, the efficiency of the PCR reactions was calculated using LinRegPCR (Ruijter et al. 2009) software and varied between 1.85 and 1.91 (±0.03). The relative expression indices were compared at each treatment time (6 and 24 h) with the respective control using one-way ANOVA followed by Dunnett’s test.
Cell cycle analyses
To carry out the cell cycle analysis, the larvae of second instar were treated with UVC light (25 J/m2) and maintained for 48 h at 20 °C or heat-shocked (28 °C for 48 h). As control, other larvae were maintained for the whole time at 20 °C. After each treatment, the samples were washed with 10 % bleach for 1 min and rinsed three times with 1× PBS. Then, 30 larvae were stretched out using tweezers and their internal contents were removed and maintained in 1× PBS solution supplemented with 10 % fetal bovine serum (FBS). Next, the tissue was broken up as much as possible by pipetting it up and down for 5 min. Afterward, the samples were centrifuged for 10 min at 1500 rpm. The supernatant was discarded, and 100 μl of trypsin/EDTA solution 250 mg/l was added for 5 min. Subsequently, 1 ml of 1× PBS (10 % FBS) solution was added to inactivate the trypsin and the suspension was centrifuged again. The cell pellet was fixed with 70 % ethanol and maintained at −20 °C. The dissociated cells were resuspended in 500 μl of 1× PBS solution, and after 5 min of decantation, the supernatant was transferred to a new tube (this step is required to clean the sample in order to avoid obstruction of the cytometer). Then, 200 μl of a solution containing 200 μg/ml of RNase A (Macherey-Nagel, Germany), 20 μg/ml of propidium iodide, 0.1 % Triton X-100, and 1× PBS was added in the supernatant. The samples were submitted to flow cytometry analysis in a BD Accuri C6 cytometer (BD Biosciences USA). Statistical analysis was performed by one-way ANOVA followed by Dunnett’s test.
Results
Determination of sub-lethal UVC dose to be applied
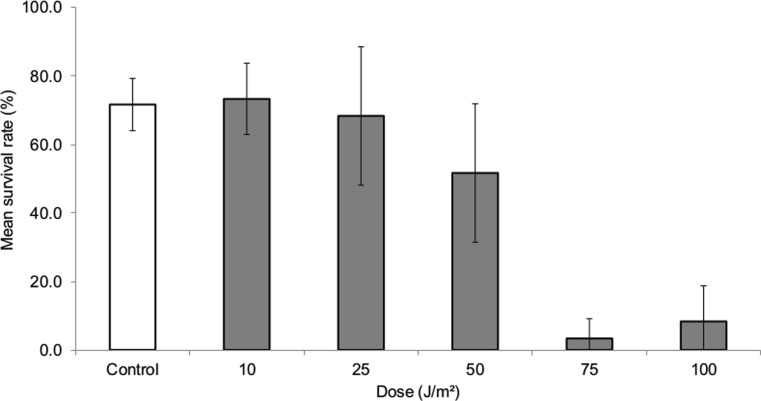
To evaluate if the damage caused by UVC promotes transposition of the mariner-Mos1 element, we firstly determined a sub-lethal dose to be applied in all other procedures performed in this work. As can be seen in Fig. 1, a UVC dose of 25 J/m2 showed a mean survival rate similar to that observed in control and with a dose of 10 J/m2. However, exposure to higher UVC doses (50, 75, and 100 J/m2) decreased the survival rate considerably, as expected.
Fig. 1.
Survival rate of larvae after UVC treatments. Average survival rate (percentage) and standard deviation from three independent experiments. The larvae of second instar were treated with different doses of UVC. The dose of 25 J/m2 presented a survival superior to 50 % and was chosen to perform the other procedures with UVC
mariner-Mos1 gene expression under mild heat and UVC stress
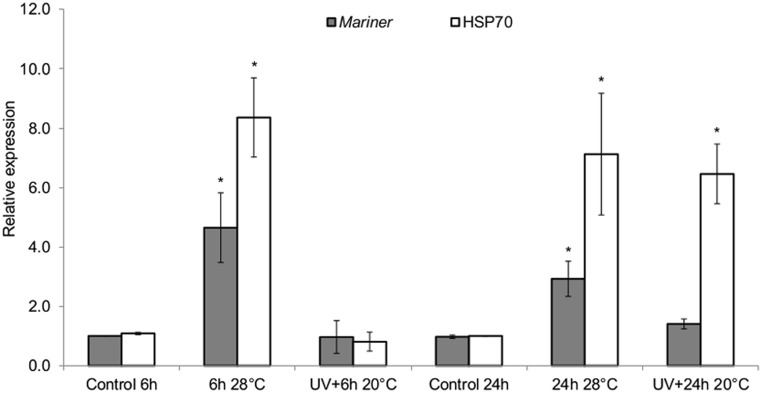
Here, we tested if UVC and mild heat stress could increase the transcriptions of mariner-Mos1. A significant increase in the expression of Hsp70 occurred when the larvae were exposed to 28 °C for 6 and 24 h (Fig. 2). This corresponded to about an eightfold increase in comparison to the control larvae (p < 0.05). Curiously, for the UV treatment, the increased expression of Hsp70 only occurred 24 h after irradiation (p < 0.05). With regard to the mariner-Mos1 transposase gene, this result suggests that 28 °C induces a rapid increase in mariner-Mos1 expression, since its expression level after 6 h of incubation at 28 °C was higher in comparison with control and remains high for the next 24 h. The relative expression of mariner-Mos1 at 28 °C was still about threefold higher than that observed in the control samples (20 °C) (p < 0.05). On the other hand, after the UV treatment, the mariner-Mos1 expression was basal and similar to that observed in the control, regardless of the time of analysis. This result clearly shows that UVC did not induce transcription of the mariner-Mos1 transposase gene, although it can activate expression of the Hsp70 gene as a late response.
Fig. 2.
Relative expression of mariner-Mos1 and Hsp70 genes after heat and UVC treatments. Average and standard deviation from three independent experiments. The input normalization was performed with the reference genes GPDH, RPL17q2, and EF1 genes. The larvae (second instar) were maintained at 28 °C or irradiated (25 J/m2) and maintained at 20 °C for 6 or 24 h. After RNA extraction, cDNA and expression protocols were performed. One-way ANOVA followed by Dunnett’s test, which compare the treatments with the respective control (p < 0.05)
mariner-Mos1 transposition rate under mild heat and UVC stress
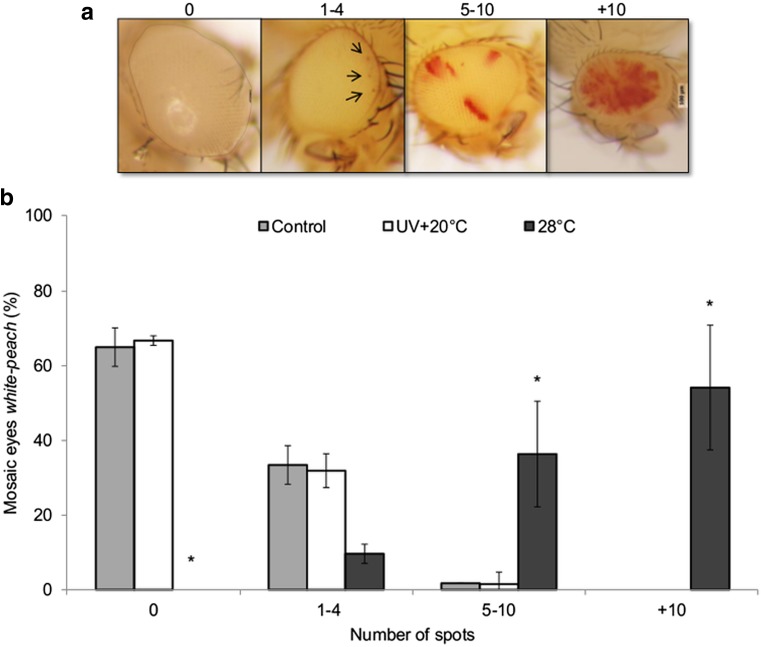
Phenotypic analysis of the adult flies’ eyes was performed after the larvae treatment with UV dose of 25 J/m2 and maintained at 20 °C or by keeping the larvae at 28 °C, until becoming adult flies. The estimation of mariner-Mos1 transposition frequency was achieved by quantifying the red spots in the eyes (Fig. 3). UVC-treated flies showed similar numbers of red spots as observed in nonirradiated control samples (0 and 1–4, respectively), suggesting that UVC does not induce mariner-Mos1 transposition, because the mosaic phenotype could not be observed. However, when the larvae were submitted to the mild heat treatment at 28 °C, the result was the opposite. Most of the individuals were distributed between 5–10 (32 %) and +10 (60 %) spot levels. These data showed that heat was able to increase the mariner-Mos1 mobilization frequency in the Dswp-test strain, despite UVC failed to activate it.
Fig. 3.
Frequency of red spots in the eyes of adult flies after treatments. a Eyes with and without spots were classified into levels related to the number of red spots: level 0 = without spots, level 1 = one to four spots, level 2 = five to ten spots, and level 3 = more than ten spots. b Average percentage of each level of mosaic eyes and standard deviation from three independent experiments. The larvae (second instar) were maintained at 28 °C or irradiated (25 J/m2) and maintained at 20 °C until becoming adult flies. One-way ANOVA followed by Dunnett’s test, which compare the treatments with the respective control (p < 0.05)
Developmental time and cell cycle progression after stress conditions
During the phenotypic analysis, it was observed that the developmental time of treated samples was different in relation to the control ones. Then, we registered the days that second-instar larvae took to become adult flies. The larvae that were maintained at 20 °C required 10–14 days to complete their development, whereas those kept at 28 °C completed it in only 8 days. In contrast, UVC treatment resulted in delayed development, since UV-exposed larvae took 15 to 17 days to become adult flies.
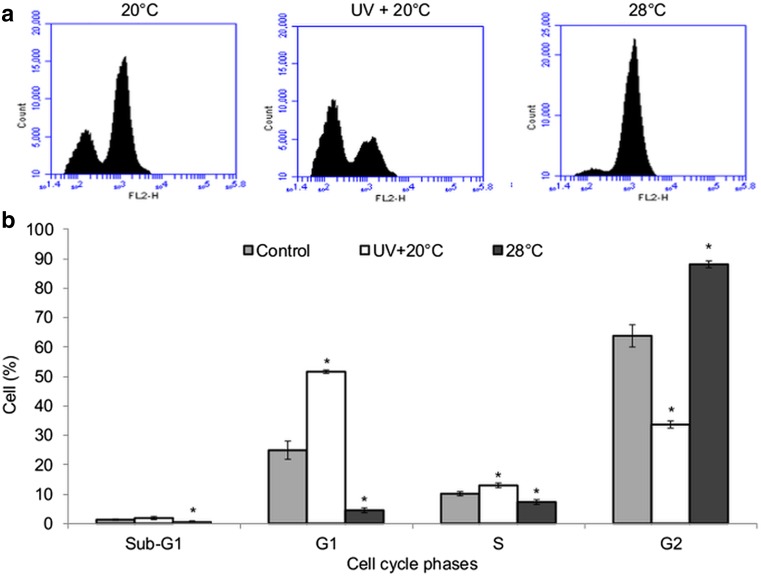
Flow cytometry analyses were then performed to investigate if the effects caused by these stressors on the developmental time could be related to cell cycle progression. For this purpose, larvae that were previously treated with UVC or mild heat stress for 48 h had their cells separated to be submitted to flow cytometry. The proportion of cells in each phase of cell cycle is shown in Fig. 4. The cell proportion in the UV + 20 °C was bigger in the phases G1/S than control sample, indicating an arrest as consequence of UV-induced DNA damage. On the other hand, the mild heat stress (28 °C) resulted in an accumulation of cells in the G2/M.
Fig. 4.
Effect of mild heat and UVC radiation on cell cycle progression. a Histogram generated by a BD Accuri C6 cytometer. b Average percentage and standard deviation from three independent experiments showing the proportion of cells in the sub-G1, G1, S, and G2 phases after each treatment. One-way ANOVA followed by Dunnett’s test, which compare the treatments with the respective control (p < 0.05). The larvae (second instar) were maintained at 28 °C or irradiated (25 J/m2) and maintained at 20 °C for 48 h; after, the cell dissociation protocol was performed and the cells were analyzed
Discussion
As TEs represent a substantial portion of many genomes, with many having potentially active elements, an open question that remains unanswered in functional genomics is how the mechanisms of silencing and activation of TEs work (Hua-Van et al. 2011). Activation by stress factors possesses a special relevance, mainly in somatic mobilization of TEs, since it can be source of deleterious mutations related to cancer, aging, and neurodegenerative diseases (Kazazian 2011). However, not only detrimental effects have been suggested for somatic mobilization of TEs. Some evidence suggests that this process could be involved in phenotypic plasticity (Micale et al. 2012; Iyengar et al. 2014). In the present study, our main focus was to investigate the action of two different stressors on the activation mechanism for somatic mobilization of the mariner-Mos1 element.
In vivo testing using strains with autonomous TEs is sometimes problematic due to the mobile nature of these sequences, which can increase or decrease the copy number during the experimental period, thus interfering with the interpretation of the results. Therefore, we constructed a strain containing only one copy of the active element. As suggested by our qPCR copy number estimation, our strain presents many individuals with a single active copy of mariner-Mos1, but some polymorphism is still maintained. However, this polymorphism did not interfere with the results since the variations observed between replicates were small.
It is already known that heat shock activates the transposition of copia-like retrotransposons and 412 in Drosophila genomes (Junakovic et al. 1986; Strand and McDonald 1985; Ratner et al. 1992; Vasilyeva et al. 1999). More recently, it was reported that the retrotransposon MAGGY was induced by heat shock in Magnaporthe grisea and Magnaporthe oryzae (Chadha and Sharma 2014; Ikeda et al. 2001). For the mariner-Mos1 element, Giraud and Capy (1996) have shown that temperature is involved in the regulation of somatic transposition in natural populations of D. simulans, and Chakrani et al. (1993) showed similar results in a controlled laboratory experiment. In addition, it was demonstrated that mariner-Mos1 has a sequence homologous to the hsp promoters, thus suggesting a possible co-activation of both mariner-Mos1 and hsp genes possibly by HSF.
Some studies have indicated that UV radiation can mobilize TEs (Kuan et al. 1991; Eichenbaum and Livneh 1998; Qüesta et al. 2010, 2013; Myakishev et al. 2008; Morales et al 2003). The only reported mobilization in the Tc1/mariner superfamily is that of Ogasawara et al. (2009) for the fungus A. oryzae. In addition, it has already been demonstrated that UV is able to increase Hsp expression in human skin cells (Trautinger 2001), in fish (Vehniäinen et al. 2012), and in sea urchin (Bonaventura et al. 2006), thereby suggesting a possible activation of mariner-Mos1 mobilization as well.
In this work, we tested the hypothesis that mariner-Mos1 could be co-activated with Hsp70 genes as a result of the stress caused by mild heat and UVC radiation. Corroborating the previous studies, the results presented in this work reinforce the activation of mariner-Mos1 by mild heat, showing that the transposase is upregulated at 28 °C (Fig. 2). In addition, the Hsp70 gene is also upregulated at 28 °C (Fig. 2) and the frequency of mariner-Mos1 mobilization is high under this condition (Fig. 3). Hsp genes have conserved cis sequences called heat shock elements (HSEs). We have reanalyzed the 5′UTR region of mariner-Mos1 element and found a new HSE in addition to that previously described by Chakrani et al. (1993) (Supplementary Figs. 1S and 2S). Together, these results suggest that mariner can be co-activated with Hsp genes, possibly due to the action of heat shock factor (HSFs).
On the other hand, although UVC activated the Hsp70 gene, it did not induce activation of the mariner-Mos1 transposase (Fig. 2). Furthermore, flies subjected to UVC treatment showed similar numbers of red spots in the eyes of adult flies as observed in the nonirradiated control (Fig. 3), confirming that it indeed did not induce mariner-Mos1 transposition. One aspect that should be highlighted is that the pattern of Hsp70 activation was different between the mild heat stress and UVC exposure. With mild heat stress, the activation was rapid (6 h) after the treatment, whereas it occurred much later (24 h) after UVC exposition. It is possible that UV-treated cells need to arrive at this time point before activation of the Hsp70 gene can occur, using a different pathway of activation from the one that is used by mariner-Mos1. Although mammals and plants have four different genes for heat shock factors (HSFs), invertebrates have only one (Åkerfelt et al. 2010). However, even having only one HSF, in invertebrates, this factor can interact with wide range of other biotic and abiotic factors. So, heat and UV could induce differently HSF to activate hsp70 and mariner-Mos1.
Additionally, it is already known that mild heat accelerates cell cycle progression, thus facilitating cell growth and differentiation (Park et al. 2005). In contrast, UVC induces a blockage of the DNA replication fork and causes an arrest in cell cycle progression (Song 2005). As shown by the cell cycle and developmental time analyses, UV promotes an arrest in the cell cycle in the G1 phase (Fig. 4), possibly due to the generation of DNA lesions that need to be repaired and the developmental time that is delayed. On the other hand, 28 °C resulted in an accumulation of cells in the G2 phase and it accelerated the developmental time of the treated flies.
We can conclude that mild heat promotes somatic mobilization of the mariner-Mos1 transposon, increasing transcription of the transposase gene, while UV radiation promotes neither this transcription nor this mobilization. Both stressors activate the Hsp70 gene, but with different patterns.
Electronic supplementary material
(PDF 146 kb)
Acknowledgments
The authors would like to thank the members of the LabDros laboratory group at the Universidade Federal de Santa Maria for comments and discussion relevant to the current study, Daniela Bitencourt Rosa Leal, PhD., and Marcela Zart Arend, MSc., for assistance with the flow cytometry equipment, and Ana Paula Christoff, MSc., for valuable knowledge about RT-qPCR and CNPq and CAPES for financial support.
References
- Åkerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11:545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventura R, Poma V, Russo R, et al. Effects of UV-B radiation on development and hsp70 expression in sea urchin cleavage embryos. Mar Biol. 2006;149(1):79–86. doi: 10.1007/s00227-005-0213-0. [DOI] [Google Scholar]
- Bryan G, Garza D, Hartl D. Insertion and excision of the transposable element mariner in Drosophila. Genetics. 1990;125:103–114. doi: 10.1093/genetics/125.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzard KA, Giaccia AJ, Killender M, et al. Heat shock protein 72 modulates pathways of stress-induced apoptosis. J Biol Chem. 1998;273(27):17147–17153. doi: 10.1074/jbc.273.27.17147. [DOI] [PubMed] [Google Scholar]
- Capy P, Koga A, David JR, et al. Sequence analysis of active mariner elements in natural populations of Drosophila simulans. Genetics. 1992;130:499–506. doi: 10.1093/genetics/130.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capy P, Gasperi G, Biémont C, et al. Stress and transposable elements: co-evolution or useful parasites? Heredity. 2000;85:101–106. doi: 10.1046/j.1365-2540.2000.00751.x. [DOI] [PubMed] [Google Scholar]
- Chadha S, Sharma M. Transposable elements as stress adaptive capacitors induce genomic instability in fungal pathogen Magnaporthe oryzae. PLoS ONE. 2014;9(4):e94415. doi: 10.1371/journal.pone.0094415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrani F, Capy P, David JR. Developmental temperature and somatic excision rate of mariner transposable element in three natural populations of Drosophila simulans. Genet Sel Evol. 1993;25:121–132. doi: 10.1186/1297-9686-25-2-121. [DOI] [Google Scholar]
- Costa RMA, Chiganças V, Galhardo RS, et al. The eukaryotic nucleotide excision repair pathway. Biochimie. 2003;85:1083–1099. doi: 10.1016/j.biochi.2003.10.017. [DOI] [PubMed] [Google Scholar]
- Creasey KM, Zhai J, Borges F, et al. miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature. 2014;508:411–415. doi: 10.1038/nature13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco M, Criscione SW, Peterson AL, et al. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging. 2013;5(12):867–883. doi: 10.18632/aging.100621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santis LP, Garcia CL, Balajeeb AS, et al. Transcription coupled repair efficiency determines the cell cycle progression and apoptosis after UV exposure in hamster cells. DNA Repair. 2002;1:209–223. doi: 10.1016/S1568-7864(01)00017-9. [DOI] [PubMed] [Google Scholar]
- Eichenbaum Z, Livneh Z. UV light induces IS10 transposition in Escherichia coli. Genetics. 1998;149:1173–1181. doi: 10.1093/genetics/149.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud T, Capy P. Somatic activity of mariner transposable element in natural populations of Drosophila simulans. Proc R Soc Lond B Biol Sci. 1996;263:1481–1486. doi: 10.1098/rspb.1996.0216. [DOI] [PubMed] [Google Scholar]
- Guerreiro MP. What makes transposable elements move in the Drosophila genome? Heredity. 2011;108:461–468. doi: 10.1038/hdy.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helman E, Lawrence MS, Stewart C, et al. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014;24:1053–1063. doi: 10.1101/gr.163659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmbrecht K, Zeise E, Rensing L. Chaperones in cell cycle regulation and mitogenic signal transduction: a review. Cell Prolif. 2000;33:341–365. doi: 10.1046/j.1365-2184.2000.00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua-Van A, Le Rouzic A, Boutin TS, et al. The struggle for life of the genome’s selfish architects. Biol Direct. 2011;6:19. doi: 10.1186/1745-6150-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Nakayashiki H, Takagi M, et al. Heat shock, copper sulphate and oxidative stress activate the retrotransposon MAGGY resident in the plant pathogenic fungus Magnaporthe grisea. Mol Genet Genomics. 2001;266:318–325. doi: 10.1007/s004380100560. [DOI] [PubMed] [Google Scholar]
- Iyengar BR, Choudhary A, Sarangdhar MA, et al. Non-coding RNA interact to regulate neuronal development and function. Front Cell Neurosci. 2014;8:47. doi: 10.3389/fncel.2014.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson JW, Hartl DL. Coupled instability of two linked genes in Drosophila mauritiana: germinal e somatic mutability. Genetics. 1985;111:57–65. doi: 10.1093/genetics/111.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson JW, Medhora MM, Hartl DL. Molecular structure of a somatically unstable transposable element in Drosophila. Proc Natl Acad Sci U S A. 1986;83:8684–8688. doi: 10.1073/pnas.83.22.8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardim SS, Loreto ELS. Comparative methodologies for estimating mariner activity using white-peach assay in Drosophila simulans. Drosoph Inf Serv. 2011;94:129–132. [Google Scholar]
- Junakovic N, Di Franco C, Barsanti P, Palumbo G. Transposition of copia-like nomadic elements can be induced by heat- shock. J Mol Evol. 1986;24:89–92. doi: 10.1007/BF02099955. [DOI] [Google Scholar]
- Kazazian HH., Jr Mobile DNA transposition in somatic cells. BMC Biol. 2011;9:62. doi: 10.1186/1741-7007-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan C-T, Liu S-K, Tessman I. Excision and transposition of Tn5 as an SOS activity in Escherichia coli. Genetics. 1991;28:45–57. doi: 10.1093/genetics/128.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Prazak L, Chatterjee N, et al. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat Neurosci. 2013 doi: 10.1038/nn.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Medhora M, Maruyama K, Hart DL. Molecular and functional analysis of the mariner mutator element Mos1 in Drosophila. Genetics. 1991;128:311–318. doi: 10.1093/genetics/128.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micale L, Loviglio MN, Manzoni M, et al. A fish-specific transposable element shapes the repertoire of p53 target genes in zebrafish. PLoS ONE. 2012;7(10) doi: 10.1371/journal.pone.0046642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales JF, Snow ET, Murnane JP. Environmental factors affecting transcription of the human L1 retrotransposon. II Stressors. Mutagenesis. 2003;18(2):151–158. doi: 10.1093/mutage/18.2.151. [DOI] [PubMed] [Google Scholar]
- Mosser DD, Caron AW, Bourget L, et al. Role of the human heat shock protein hsp70 in protection against stress-induced apoptosis. Mol Cell Biol. 1997;17:5317–5327. doi: 10.1128/mcb.17.9.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myakishev M, Polesskaya O, Kulichkova V, et al. PCR-based detection of Pol III-transcribed transposons and its application to the rodent model of ultraviolet response. Cell Stress Chaperones. 2008;13:111–116. doi: 10.1007/s12192-008-0010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara H, Obata H, Hata Y, et al. Crawler, a novel Tc1/mariner-type transposable element in Aspergillus oryzae transposes under stress conditions. Fungal Genet Biol. 2009;46:441–449. doi: 10.1016/j.fgb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Oliveira LFV, Wallau GL, Loreto ELS. Isolation of high quality DNA: a protocol combining “rennet” and glass milk. Electron J Biotechnol. 2009;12:1–6. doi: 10.2225/vol12-issue2-fulltext-4. [DOI] [Google Scholar]
- Ortolan TG, Menck CFM. UVB-induced cell death signaling is associated with G1-S progression and transcription inhibition in primary human fibroblasts. PLoS ONE. 2013;8(10) doi: 10.1371/journal.pone.0076936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HG, Han SI, Oh SY, et al. Cellular responses to mild heat stress. Cell Mol Life Sci. 2005;62:10–23. doi: 10.1007/s00018-004-4208-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritham EJ. Transposable elements and factors influencing their success in eukaryotes. J Hered. 2009;100(5):648–655. doi: 10.1093/jhered/esp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qüesta JI, Walbot V, Casati P. Mutator transposon activation after UV-B involves chromatin remodeling. Epigenetics. 2010;5:352–363. doi: 10.4161/epi.5.4.11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qüesta JI, Walbot V, Casati P. UV-B radiation induces Mu element somatic transposition in maize. Mol Plant. 2013;6:2004–2007. doi: 10.1093/mp/sst112. [DOI] [PubMed] [Google Scholar]
- Ratner VA, Zabanov SA, Kolesnikova OV, et al. Induction of the mobile element Dm-412 transpositions in the Drosophila genome by heat shock treatment. Proc Natl Acad Sci U S A. 1992;89:5650–5654. doi: 10.1073/pnas.89.12.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanat J-L, Douki T, Cadet J. Direct and indirect effects of UV radiation on DNA and its components. J Photochem Photobiol B. 2001;63:88–102. doi: 10.1016/S1011-1344(01)00206-8. [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, et al. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuch AP, Garcia CCM, Makita K, Menck CFM. DNA damage as a biological sensor for environmental sunlight. Photochem Photobiol Sci. 2013 doi: 10.1039/c3pp00004d. [DOI] [PubMed] [Google Scholar]
- Song Y-H. Drosophila melanogaster: a model for the study of DNA damage checkpoint response. Mol Cells. 2005;19(2):167–179. [PubMed] [Google Scholar]
- Sørensen JG, Nielsen MM, Kruhøffer M, et al. Full genome gene expression analysis of the heat stress response in Drosophila melanogaster. Cell Stress Chaperones. 2005;10(4):312–328. doi: 10.1379/CSC-128R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand DJ, McDonald JF. Copia is a transcriptionally responsive to environmental stress. Nucleic Acids Res. 1985;13:4401–4410. doi: 10.1093/nar/13.12.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Reed JC, Homma S. Heat-shock proteins as regulators of apoptosis. Oncogene. 2003;322:9041–9047. doi: 10.1038/sj.onc.1207114. [DOI] [PubMed] [Google Scholar]
- Trautinger F. Heat shock proteins in the photobiology of human skin. J. Photochem Photobiol. 2001;63:70–77. doi: 10.1016/S1011-1344(01)00203-2. [DOI] [PubMed] [Google Scholar]
- Vasilyeva LA, Bubenshchikova EV, Ratner VA. Heavy heat shock induced retrotransposon transposition in Drosophila. Genet Res. 1999;74:111–119. doi: 10.1017/S0016672399003973. [DOI] [PubMed] [Google Scholar]
- Vehniäinen E-R, Vähäkangas K, Oikari A. UV-B exposure causes DNA damage and changes in protein expression in northern pike (Esox lucius) posthatched embryos. Photochem Photobiol. 2012;88:363–370. doi: 10.1111/j.1751-1097.2011.01058.x. [DOI] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Mehta S, Reyes-Turcu FE, Zhuang F, et al. RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature. 2013;493:557–560. doi: 10.1038/nature11716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 146 kb)