Abstract
Previous studies have shown that hippocampal subfields may be differentially affected by Alzheimer’s disease (AD). This study used an automated analysis technique and two large cohorts to (1) investigate patterns of subfield volume loss in mild cognitive impairment (MCI) and AD, (2) determine the pattern of subfield volume loss due to age, gender, education, APOE ε4 genotype, and neuropsychological test scores, (3) compare combined subfield volumes to hippocampal volume alone at discriminating between AD and healthy controls (HC), and predicting future MCI conversion to AD at 12 months. 1,069 subjects were selected from the AddNeuroMed and Alzheimer’s disease neuroimaging initiative (ADNI) cohorts. Freesurfer was used for automated segmentation of the hippocampus and hippocampal subfields. Orthogonal partial least squares to latent structures (OPLS) was used to train models on AD and HC subjects using one cohort for training and the other for testing and the combined cohort was used to predict MCI conversion. MANCOVA and linear regression analyses showed multiple subfield volumes including Cornu Ammonis 1 (CA1), subiculum and presubiculum were atrophied in AD and MCI and were related to age, gender, education, APOE ε4 genotype, and neuropsychological test scores. For classifying AD from HC, combined subfield volumes achieved comparable classification accuracy (81.7 %) to total hippocampal (80.7 %), subiculum (81.2 %) and presubiculum (80.6 %) volume. For predicting MCI conversion to AD combined subfield volumes and presubiculum volume were more accurate (81.1 %) than total hippocampal volume. (76.7 %).
Keywords: Alzheimer’s disease, MCI conversion, MRI, Freesurfer, OPLS
Introduction
In recent years, research efforts in Alzheimer’s disease (AD) have focused upon the discovery of clinically meaningful and non-invasive biomarkers that can reliably monitor disease progression and predict future conversion to the disease. Several groups including our own have proposed the use of magnetic resonance imaging (MRI) based tools to aid in the diagnosis of AD (Desikan et al. 2009; Liu et al. 2009) and predict future conversion from the prodromal stage of disease often referred to as mild cognitive impairment (MCI) (McEvoy et al. 2009; Westman et al. 2011a).
Hippocampal atrophy has been frequently observed in AD (Jack et al. 1992; Fox et al. 1996) and has been demonstrated in MCI subjects (Devanand et al. 2007), with increased risk of future conversion to AD in subjects with smaller hippocampal volumes (Apostolova et al. 2006; Csernansky et al. 2005). Hippocampal volumetry has been a useful marker of AD pathology but seems to be insufficiently sensitive for distinguishing between MCI subjects bearing a high risk of AD conversion from those who remain clinically stable (Mueller et al. 2010). Post-mortem studies have also demonstrated that hippocampal atrophy in AD is non-uniform, with Cornu Ammonis 1 (CA1) and subicular atrophy reported in early AD (Braak and Braak 1991; West et al. 2004).
So far, only a few studies have attempted to measure regional atrophic changes within the hippocampus using manual delineation and 3D surface mapping (Mueller and Weiner 2009; Apostolova et al. 2010; Costafreda et al. 2011). Manual delineation of the subfield boundaries is both a time consuming and labour intensive process which limits its widespread applicability in practice. However, recent developments in image acquisition have made it possible to segment the hippocampus into its subfields in a fully automated fashion and this method has now been validated using ultra high resolution MRI (Van Leemput et al. 2009). A recent small study applied this technique to 15 MCI subjects using conventional 3D T1 weighted volume imaging and demonstrated that segmenting subfields increased sensitivity in diagnosing MCI (Hanseeuw et al. 2011). The current study uses an extensive dataset created by combining two large cohorts from the AddNeuroMed and the Alzheimer Disease Neuroimaging Initiative (ADNI) studies to build on and extend this earlier work.
In this study we aimed to (1) investigate the differences in hippocampal subfields between subject groups at baseline in a cohort of 1,069 subjects, (2) determine patterns of subfield volume loss in relation to age, gender, education, APOE ε4 genotype, and neuropsychological tests from mini mental state exam (MMSE) and Alzheimer disease Assessment Score-1 (ADAS-1) scores, and (3) compare combined subfield volumes using orthogonal partial least squares (OPLS) multivariate analysis to hippocampal volume alone for discriminating between AD and healthy control (HC) subjects and predicting future conversion from MCI to AD at 12 months.
Materials and Methods
Study Data and Inclusion and Diagnostic Criteria
The data used in this study were derived from two large multicentre cohorts, the AddNeuroMed and ADNI cohorts. The AddNeuroMed study is an integrated project funded by the European Union Sixth Framework Program and aims to establish and validate novel biomarkers of disease and treatment based upon in vitro and in vivo human and animal models of AD. Data was collected from six participating sites across Europe: University of Kuopio, Finland; University of Perugia, Italy; Aristotle University of Thessaloniki, Greece; King’s College London, United Kingdom; University of Lodz, Poland; and University of Toulouse, France (Lovestone et al. 2009; Simmons et al. 2009, 2011).
Data from the ADNI study was downloaded from the ADNI at the LONI website (www.loni.ucla.edu/ADNI, PI Michael M. Weiner). The initiative was launched in 2003 by the National Institute on Ageing, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration, private pharmaceutical companies and non-profit organisations, as a 5 years public–private partnership. The primary goal of ADNI has been to test whether MRI, positron emission tomography (PET), and other biological markers are useful in clinical trials of MCI and early AD. Subjects aged 55–90 from over 50 sites across the U.S and Canada participated in the research, and imaging, clinical, and biological samples were collected at multiple time points (Jack et al. 2008). A detailed description of the inclusion criteria for the study can be found on its webpage (http://www.adni-info.org/scientists/aboutADNI.aspx#).
A total of 1,069 subjects were included in this study (AD = 291, MCI = 447, HC = 331). The demographics of the cohorts are given in Table 1. Of the 447 MCI subjects in our whole cohort, 90 converted to an AD diagnosis (MCI converters) at 12 months.
Table 1.
Demographic, clinical and neuropsychological data in AD, MCI converters, stable MCI, and control subjects
| AD (n = 291) | MCI converters (n = 90) | Stable MCI (n = 357) | HC (n = 331) | p* | |
|---|---|---|---|---|---|
| Gender (male/female) | 131/160a | 54/36 | 216/141b | 166/165 | 0.001 |
| Age | 75.4 ± 7.0 | 74.1 ± 6.6 | 75.1 ± 7.0 | 75.0 ± 5.7 | 0.439 |
| Years of education | 12.1 ± 4.7a,b,c | 14.0 ± 4.1 | 14.3 ± 4.4 | 14.3 ± 4.4 | <0.001 |
| MMSE | 22.4 ± 3.4a,b,c | 26.5 ± 1.8b | 27.1 ± 1.7b | 29.1 ± 1.1 | <0.001 |
| ADAS-1 | 6.3 ± 1.5a,b,c | 5.3 ± 1.3a,b | 4.6 ± 1.4b,c | 3.1 ± 1.3 | <0.001 |
| CDR | 0.9 ± 0.4a,b,c | 0.5b | 0.5b | 0 | <0.001 |
| APOE ε4 genotype (+ive/-ive) | 183/108 | 57/33 | 171/186 | 93/238 | <0.001 |
Data are represented as mean ± and standard deviation. Chi square was used for gender and APOE ε4 genotype comparison. ANOVA with Bonferroni post hoc test was used for age, education, and neuropsychological scores
aSignificant compared to stable MCI
bSignificant compared to healthy controls (HC)
cSignificant compared to MCI converters
* p values corrected for multiple comparisons using the Bonferroni method
For the AddNeuroMed cohort, subjects were patients who attended local memory clinics and received a diagnosis of MCI while HC subjects were recruited from non-related members of the patient’s families, caregiver relatives, and social centres for the elderly or General Practitioner (GP) surgeries. Informed consent was obtained for all subjects and the study was approved by the ethical review boards of each participating country. The general inclusion and exclusion criteria were as follows.
AD
(1) diagnosis established by National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association (NINCDS-ADRDA) and Diagnostic and Statistical Manual of Mental Disorders IV (DSM IV) criteria, (2) MMSE score ranged from 12 to 28. Subjects were excluded from the study if any psychiatric or neurological illness other than AD was present, and if subjects presented with a systemic illness or signs of organ failure.
MCI
(1) subjects had MMSE scores between 24 and 30, (2) subjective memory complaint with preserved activities of daily living, (3) Clinical Dementia Rating (CDR) score of 0.5, (4) Geriatric depression scale score less than or equal to 5, (5) absence of dementia in accordance with NINCDS-ARDA criteria. A 12 months follow up was used to determine whether MCI subjects converted to AD (MCI converters) or remained clinically stable (stable MCI).
HC
(1) MMSE scores between 24 and 30, (2) CDR score of 0, (3) no presence of neurological or psychiatric illness, and non-demented.
MMSE, CDR, and the Consortium to Establish a Registry for Alzheimer’s Disease (CERAD) cognitive battery were assessed for each subject. The CERAD cognitive battery was replaced with the Alzheimer’s disease assessment scale for AD subjects in AddNeuroMed. The CERAD battery employs the same 10 word recall task as the Alzheimer’s assessment scale, only the scoring is inverted. Therefore, the mean number of words not recalled in the CERAD word list task was calculated in order to obtain comparable measures of memory for all diagnostic groups. This revised cognitive parameter was named ADAS-1 corresponding to the first subtest of the Alzheimer’s disease assessment scale.
MRI Acquisition
Standardized MRI data acquisition techniques were in place for AddNeuroMed and ADNI to ensure homogeneity across data acquisition sites. A detailed description of the ADNI data acquisition protocol can be found at www.loni.ucla.edu/ADNI/research/Cores/index.shtml. The imaging protocol included a 1.5T high resolution T1 weighted sagittal 3D MP-RAGE volumes (voxel size 1.1 × 1.1 × 1.2 mm3), and axial proton density with T2 weighted fast spin echo images. A comprehensive quality control procedure was carried out on all MR images according to the AddNeuroMed quality control framework (Simmons et al. 2009, 2011).
Hippocampal Subfield Segmentation
Image analysis was carried out using the Freesurfer image analysis pipeline (version 5.1.0). These procedures have been described in detail in previous publications (Dale et al. 1999; Fischl et al. 2002; Ségonne et al. 2004; Fischl et al. 2004). Initially volumetric segmentation involved the removal of non-brain tissue using a hybrid watershed/surface deformation procedure (Ségonne et al. 2004), automated Talairach transformation, segmentation of the subcortical white matter and deep grey matter volumetric structures (Fischl et al. 2004).
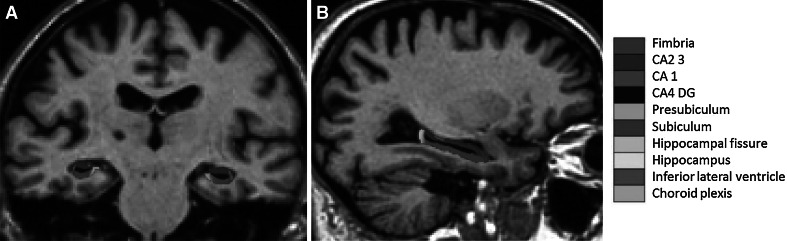
Automated segmentation of the hippocampus was performed to define anatomical subfield labels using a Bayesian modelling approach and a computational model of the areas surrounding the hippocampus. An atlas mesh had previously been built and validated from manual delineations in ultra-high resolution MRI scans of 10 individuals (Van Leemput et al. 2009). These delineations include the fimbria, presubiculum, subiculum, CA1, CA2/3, and CA4-DG subfields as well as the hippocampal fissure. Figure 1 illustrates the delineations made to define the different subfields of the hippocampus. For more details about this technique and the borders used to define the different subfields, see Van Leemput et al. (2009).
Fig. 1.
a Coronal and b sagittal views of the hippocampus
All subfield measures were normalised by the subject’s intracranial volume derived from Freesurfer using the following formula: volumenorm = volumeraw × 1,000/ICV in cm3 (Westman et al. 2013). This automated segmentation approach has been recently applied to a small group of MCI subjects (Hanseeuw et al. 2011).
Statistical Analysis
Statistical analysis was conducted using PASW Statistics (Version 17. 0; SPSS inc., USA). Categorical variables were inspected using the Chi square test while continuous variables were tested using ANOVA with Bonferroni post hoc comparisons. Hippocampal subfield volumes were first analysed using MANCOVA utilising Bonferroni correction by adopting a general linear model procedure, adjusting for age, gender, education, and APOE ε4 genotype as covariates. Bonferroni pairwise comparisons were performed to inspect subfield volume differences between the groups.
Multiple regression analyses were conducted in R version 2.15.2 using the lm function from the R stats package and Bonferroni correction for multiple comparisons. Patterns of subfield volume loss were tested in relation to the effects of age, gender, education, APOE ε4 genotype, and neuropsychological test scores from MMSE and ADAS-1. In this step, all subfield measures were tested as dependent variables by disease group (AD, MCI converters, stable MCI, and HC) as a whole. Age, gender, years of education, APOE ε4 genotype, and neuropsychological scores from MMSE and ADAS-1 tests was treated as independent variables for identifying subfield specific effects. 10 fold cross validation was performed by fitting linear regression models to the data, excluding 1/10th of the data in each fold and using the fitted model for prediction on data that was excluded from the fold.
Hippocampal subfields were subsequently analysed using Orthogonal Partial Least Squares (OPLS) (Wiklund et al. 2008; Trygg and Wold 2002), a supervised multivariate data analysis method included in the software package SIMCA (Umetrics AB, Umea, Sweden). Al1 14 variables (left and right subfields) were used for OPLS analysis. Classification models were created for distinguishing between AD and HC subjects at baseline. The AD versus HC models were subsequently treated as classifiers to investigate how well the hippocampal subfields could predict future MCI conversion to AD at 12 months follow up. Seven-fold cross validation was used for all models. Using this approach we created 4 OPLS models; 2 for the total hippocampus and 2 for the combination of subfield volumes. The first model for each region comprised the AddNeuroMed cohort and the second model comprised the ADNI cohort. To further validate the models created the AddNeuroMed cohort was used as the training set and the ADNI cohort as a test set (and vice versa) to see how well the models could predict new and unseen data. The combined ADNI and AddNeuroMed cohort from the AD versus HC comparison was used as a classifier to investigate the reliability of predicting MCI conversion to AD at 12 months. This OPLS classification approach has been extensively validated (Bylesjo et al. 2006; Wiklund et al. 2008; Westman et al. 2011c) and applied to several biomarker discovery studies in AD (Mangialasche et al. 2010; Westman et al. 2011a, 2012; Spulber et al. 2013).
Sensitivity and specificity were calculated from the cross-validated prediction values of the OPLS models. The positive and negative likelihood ratios (LR+ = sensitivity/(100−specificity) and LR− = (100−sensitivity)/specificity)) were determined. A positive likelihood ratio between 5 and 10 or a negative likelihood ratio between 0.1 and 0.2 increases the diagnostic value in a moderate way, while a value above 10 or below 0.1 significantly increases the diagnostic value of the test.
Receiver operating characteristic (ROC) curves were calculated for the individual subfield volume models using the ROCR library (version 2.1) in R. ROC curves provide a graphical means to interpret the quality of separation and are created by plotting the true positive rate (sensitivity) versus the false positive rate (1−specificity) for various thresholds. The discriminant value of the corresponding ROC curve can be obtained by calculating the area under the curve (AUC). AUC values range from 0.5 (random discriminations no better than chance) to 1.0 (perfect discrimination). The pROC (Receiver Operating Characteristic) package (version 1.5.4) (Robin et al. 2011) in R was used to perform area under the curve (AUC) statistical comparisons between the combined subfield and total hippocampal volume models in the AD vs. HC and MCI converter vs MCI non-converter models.
Results
Demographics, Neuropsychological, and Global Clinical Measurements
1,069 subjects were included in the current study (AD = 291, MCI = 447, HC = 331) from the AddNeuroMed and ADNI cohorts. Results from global, clinical and cognitive assessments revealed that scores on MMSE, CDR, and ADAS-1 were poorest amongst AD and best amongst control subjects as expected (Table 1).
Hippocampal Subfields
Hippocampal subfield volumes from the left and the right hemisphere were used to determine the pattern of subfield atrophy in AD, MCI-converter, MCI stable and HC subjects. Comparisons of the bilateral CA1, CA2-3, CA4-DG, subiculum, and presubiculum were significant across all groups (<0.0001) after correction for multiple comparisons and demonstrated similar results in pairwise comparisons (Table 2 and Fig. 2). No significant volume differences were found for the left and right hippocampal fissure between these groups.
Table 2.
Hippocampal subfield differences in AD, MCI converters, stable MCI, and healthy control subjects
| AD (n = 291) | MCI converters (n− = 90) | Stable MCI (n = 357) | HC (n = 331) | p | |
|---|---|---|---|---|---|
| Left presubiculum | 309.2 ± 63.5a,b | 323.7 ± 64.6a,b | 363.2 ± 70.9b,c | 409.5 ± 62.963.1 | <0.0001 |
| Left subiculum | 449.0 ± 86.7a,b | 465.9 ± 84.1a,b | 520.9 ± 91.0b,c | 579.5 ± 76.2 | <0.0001 |
| Right presubiculum | 307.9 ± 64.0a,b | 318.7 ± 63.5a,b | 359.2 ± 70.8b,c | 399.0 ± 64.3 | <0.0001 |
| Right subiculum | 449.9 ± 90.4a,b | 467.1 ± 92.7a,b | 523.5 ± 96.4b,c | 575.5 ± 77.5 | <0.0001 |
| Left CA4-DG | 399.2 ± 73.5a,b | 416.2 ± 67.6a,b | 453.1 ± 79.0b,c | 494.2 ± 67.7 | <0.0001 |
| Left CA2-3 | 716.4 ± 134.2a,b | 747.6 ± 127.1a,b | 804.6 ± 139.8b,c | 877.9 ± 121.0 | <0.0001 |
| Right CA4-DG | 418.3 ± 78.0a,b | 436.1 ± 80.6a,b | 476.9 ± 83.2b,c | 511.6 ± 69.8 | <0.0001 |
| Right CA2-3 | 765.4 ± 143.4a,b | 793.5 ± 146.4a,b | 858.8 ± 146.0b,c | 920.7 ± 127.7 | <0.0001 |
| Left fimbria | 29.9 ± 21.5a,b | 33.6 ± 25.6b | 37.9 ± 23.3b | 49.1 ± 23.1 | <0.0001 |
| Left CA1 | 286.8 ± 55.5a,b | 298.2 ± 49.1b | 308.1 ± 50.1b | 322.4 ± 45.0 | <0.0001 |
| Right CA1 | 293.8 ± 57.0a,b | 301.5 ± 53.2b | 315.7 ± 54.2b,c | 328.5 ± 45.8 | <0.0001 |
| Right fimbria | 26.9 ± 18.8a,b | 31.8 ± 21.5b | 32.3 ± 18.8b | 41.5 ± 19.9 | <0.0001 |
| Right hippocampal fissure | 44.8 ± 26.2 | 46.2 ± 23.8 | 45.1 ± 23.8 | 47.7 ± 24.1 | 2.144 |
| Left hippocampal fissure | 39.8 ± 22.7 | 37.6 ± 22.8 | 40.7 ± 21.6 | 41.4 ± 21.1 | 3.276 |
Absolute Hippocampal subfields are presented (mm3). However, normalised hippocampal subfields (absolute hippocampal subfield/intracranial volume) were used in MANCOVA with Bonferroni pairwise comparisons. Age, gender, education, and APOE ε4 genotype were used as covariates. p values were corrected for multiple comparisons using the Bonferroni method
aSignificant compared to stable MCI
bSignificant compared to healthy controls (HC)
cSignificant compared to MCI converters
Fig. 2.

Bar plot of subfield volumes of AD (n = 291), MCI converters (n = 90), MCI stable (n = 357), and healthy control (HC) subjects (n = 331). Error bars represent SEM = SD/√n. Subfield volumes are represented in mm3. R = subfield volumes from the right hemisphere, L = subfield volumes from the left hemisphere
In the left hippocampus, presubiculum (F = 144.5, p < 0.0001) and subiculum (F = 144.3, p < 0.0001) volumes were most significantly reduced in AD, MCI converter and MCI stable subjects compared to healthy controls. The same pattern of subfield atrophy was also observed in the right hippocampus for these groups for both presubiculum (F = 122.1, p < 0.0001) and subiculum (F = 120.0, p < 0.0001) relative to healthy controls. MCI-converters displayed significant subfield volume losses in the bilateral subiculum (right = p < 0.0001, left = p < 0.0001), subiculum (right = p < 0.0001, left = p < 0.0001), CA4-DG (right = p < 0.0001, left = p < 0.0001), and CA2-3 (right = p < 0.0001, left = p < 0.0001) relative to stable MCI subjects. However, no significant differences in any of the subfield volume measures were observed between AD and MCI-converter subjects.
Relationship Between Neuropsychological Test Scores and Hippocampal Subfields
A significant positive effect for MMSE was found in relation to all hippocampal subfield volumes except bilateral hippocampal fissure, indicating that subjects with lower MMSE scores had reduced subfield volumes (Table 3). On the other hand, a significant negative effect was observed for ADAS-1 scores across all subfield volumes except the bilateral hippocampal fissure, indicating that subjects with higher ADAS-1 scores (mean number of words not recalled) had lower subfield volumes (Table 3).
Table 3.
MMSE and ADAS-1 effect on hippocampal subfield volumes in the combined cohort
| MMSE score | ADAS-1 score | |||||
|---|---|---|---|---|---|---|
| Hippocampus | β | t | p* | β | t | p* |
| Left CA1 | 0.24 | 8.41 | <0.001 | −0.19 | −6.68 | <0.001 |
| Right CA1 | 0.23 | 8.15 | <0.001 | −0.16 | −5.47 | <0.001 |
| Left CA2-3 | 0.34 | 12.29 | <0.001 | −0.34 | −11.93 | <0.001 |
| Right CA2-3 | 0.35 | 12.40 | <0.001 | −0.28 | −9.67 | <0.001 |
| Left CA4-DG | 0.37 | 13.47 | <0.001 | −0.36 | −12.51 | <0.001 |
| Right CA4-DG | 0.37 | 13.31 | <0.001 | −0.31 | −10.65 | <0.001 |
| Left Fimbria | 0.23 | 8.08 | <0.001 | −0.23 | −8.07 | <0.001 |
| Right Fimbria | 0.19 | 6.51 | <0.001 | −0.18 | −6.19 | <0.001 |
| Left Presubiculum | 0.43 | 15.51 | <0.001 | −0.42 | −15.19 | <0.001 |
| Right Presubiculum | 0.40 | 14.11 | <0.001 | −0.39 | −13.60 | <0.001 |
| Left Subiculum | 0.43 | 15.57 | <0.001 | −0.42 | −15.06 | <0.001 |
| Right Subiculum | 0.40 | 14.49 | <0.001 | −0.36 | −12.67 | <0.001 |
Age, gender, education and APOE ε4 genotype were introduced as covariates in these models
* p values from the regression models were corrected for multiple comparisons using the Bonferroni method
Relationship Between Age, Education, APOE ε4 Genotype, and Hippocampal Subfields
A significant negative effect of age was observed in relation to all subfield volumes indicating that older subjects had lower hippocampal subfield volumes. In particular, the strongest effects of age were found in the right presubiculum (β = −0.32, p < 0.001), and left presubiculum areas, the right fimbria (β = −0.31, p < 0.001), and the right subiculum and left subiculum areas (β = −0.28, p < 0.001).
Linear regression models were also created to test for the effect of gender on subfield volume differences in the male (n = 567) and female (n = 502) subjects. A significant positive effect of gender was found in the right fimbria and left fimbria areas, and in the right and left CA4-DG subfield volumes (Table 4).
Table 4.
Age and Gender effect on hippocampal subfields in the combined cohort
| Hippocampus | Agea | Genderb | ||||||
|---|---|---|---|---|---|---|---|---|
| β | t | PRESS | p value* | β | t | PRESS | p value* | |
| Left CA1 | −0.1 | −3.31 | 42.8 | 0.012 | 0.04 | 1.16 | 0.239 | 2.964 |
| Right CA1 | −0.09 | −3.01 | 42.7 | 0.036 | 0.04 | 1.3 | 0.239 | 2.304 |
| Left CA2-3 | −0.24 | −7.75 | 41.0 | <0.001 | 0.09 | 2.93 | 0.237 | 0.036 |
| Right CA2-3 | −0.23 | −7.62 | 40.8 | <0.001 | 0.1 | 3.2 | 0.237 | 0.012 |
| Left CA4-DG | −0.24 | −7.92 | 40.6 | <0.001 | 0.1 | 3.19 | 0.237 | 0.012 |
| Right CA4-DG | −0.25 | −8.2 | 40.7 | <0.001 | 0.12 | 3.74 | 0.236 | <0.001 |
| Left Fimbria | −0.25 | −8.42 | 39.6 | <0.001 | 0.12 | 3.74 | 0.236 | <0.001 |
| Right Fimbria | −0.29 | −9.72 | 40.5 | <0.001 | 0.13 | 4.24 | 0.235 | <0.001 |
| Left Presubiculum | −0.31 | −10.28 | 40.0 | <0.001 | 0.09 | 2.71 | 0.238 | 0.084 |
| Right Presubiculum | −0.32 | −10.91 | 40.1 | <0.001 | 0.09 | 2.89 | 0.237 | 0.048 |
| Left Subiculum | −0.28 | −9.09 | 38.9 | <0.001 | 0.12 | 3.85 | 0.236 | 0.012 |
| Right Subiculum | −0.28 | −9.25 | 39.3 | <0.001 | 0.12 | 3.38 | 0.236 | 0.012 |
aGender, years of education, and APOE E4 genotype were used as covariates
bSubject age, years of education, and APOE E4 genotype were used as covariates
* p values from each regression model were corrected for multiple comparisons using the Bonferroni method
A significant negative effect of education was only found in the right CA1 (β = −0.95, p = 0.024).
The analysis was repeated for subjects that were carriers and non-carriers of the APOE ε4 allele. APOE E4 genotype was negatively related to all subfield volumes suggesting that subjects with an APOE E4 allele had smaller subfield volumes (Table 5).
Table 5.
Years of education and APOE genotype effect on hippocampal subfields in the combined cohort
| Years of educationa | APOE E4 genotypeb | |||||
|---|---|---|---|---|---|---|
| β | t | p* | β | t | p* | |
| Left CA1 | −0.59 | −1.92 | 0.66 | 0.22 | −7.29 | 0.012 |
| Right CA1 | −0.95 | −3.11 | 0.024 | 0.17 | −5.74 | 0.012 |
| Left CA2-3 | −0.004 | −0.14 | 10.668 | 0.24 | −7.89 | <0.001 |
| Right CA2-3 | −0.01 | −0.45 | 7.86 | 0.26 | −8.46 | <0.001 |
| Left CA4-DG | −0.002 | −0.06 | 11.46 | 0.253 | −8.31 | <0.001 |
| Right CA4-DG | −0.009 | −0.27 | 9.408 | 0.27 | −8.8 | <0.001 |
| Left Fimbria | 0.028 | 0.88 | 4.548 | 0.1 | −3.05 | 0.024 |
| Right Fimbria | 0.08 | 2.39 | 0.204 | 0.07 | −2.28 | 0.276 |
| Left Presubiculum | 0.03 | 0.81 | 5.004 | 0.24 | −7.8 | <0.001 |
| Right Presubiculum | 0.05 | 1.57 | 1.404 | 0.23 | −7.28 | <0.001 |
| Left Subiculum | 0.03 | 1.06 | 3.48 | 0.27 | −8.84 | <0.001 |
| Right Subiculum | 0.02 | 0.65 | 6.216 | 0.27 | −8.66 | <0.001 |
aSubject age, gender, and APOE E4 genotype were used as covariates
b† Subject age, gender and years of education were used as covariates
* p values from each regression model were corrected for multiple comparisons using the Bonferroni method
AD and HC Classification for the Combined AddNeuroMed and ADNI Cohort
For the joint AddNeuroMed and ADNI AD versus HC model, combining the subfield volumes resulted in an accuracy of 81.7 % (sensitivity = 80.4 %, specificity = 82.8 %, AUC = 0.895) compared to 80.7 % for total hippocampal volume (sensitivity = 79.2 %, specificity = 82.8 %, AUC = 0.887) (Table 6). These result were statistically significantly different in terms of the observed AUC differences between the two models (AUC difference = 0.008, p = 0.001).
Table 6.
Comparison of performance for the different cohort models in the AD vs. HC classification
| Total Hippocampus | Hippocampal subfields | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ACC (%) | SENS (%) | SPE (%) | AUC | Q2 (Y) | ACC (%) | SENS (%) | SPE (%) | AUC | Q2 (Y) | |
| AddNeuroMed (cv) | 80.7 (74.8–85.4) | 76.2 (67.2–83.3) | 85.1 (77.1–90.6) | 0.897 | 0.439 | 80.2 (74.3–85.0) | 78.1 (69.3–84.9) | 82.2 (73.9–88.3) | 0.90 | 0.441 |
| ADNI (cv) | 81.5 (77.4–84.9) | 81.2 (75.0–86.2) | 81.7 (76.1–86.2) | 0.884 | 0.404 | 82.0 (77.9–85.6) | 81.2 (75.0–86.2) | 82.6 (77.1–87.0) | 0.892 | 0.433 |
| Combined (cv)* | 80.7 (77.4–83.6) | 79.2 (74.1–83.5) | 82.8 (78.3–86.5) | 0.887 | – | 81.7 (78.4–84.5) | 80.4 (75.5–84.6) | 82.8 (78.3–86.5) | 0.895 | – |
| AddNeuroMeda | 81.1 (75.3–85.8) | 75.2 (66.2–82.5) | 86.9 (79.2–92.0) | 0.897 | – | 82.1 (76.4–86.7) | 77.1 (68.2–84.1) | 86.9 (79.2–92.0) | 0.90 | – |
| ADNIb | 80.2 (76.1–83.8) | 84.4 (78.5–88.9) | 76.8 (70.8–81.8) | 0.884 | – | 80.7 (76.6–84.3) | 88.2 (82.7–92.1) | 74.6 (68.5–79.8) | 0.881 | – |
aAddNeuroMed dataset used as the test set and ADNI data as the training set
bADNI data used as the test set and AddNeuroMed dataset as the training set
* AddNeuroMed and ADNI cohorts used as the combined cohort model, confidence intervals presented within parenthesis
CV cross validation, AUC area under the curve
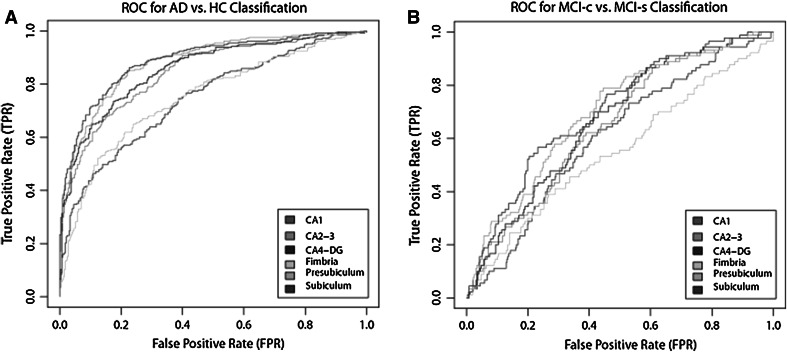
Combining subfield volumes resulted in similar classification accuracy to the subiculum (accuracy = 81.2 %, sensitivity = 83.5 %, specificity = 79.2 %, AUC = 0.887) and presubiculum alone (accuracy = 80.6 %, sensitivity = 83.2 %, specificity = 78.3 %, AUC = 0.882), but higher accuracies than the other individual subfield volume measures (Table 7). Figure 3 illustrates ROC curves for the corresponding individual subfield volumes for distinguishing AD and HC subjects.
Table 7.
Comparison of performance for OPLS AD vs. HC classification models
| ACC (%) | SEN (%) | SPE (%) | AUC | PPV (%) | NPV (%) | LR+ | LR− | |
|---|---|---|---|---|---|---|---|---|
| CA1 | 66.0 (62.2–69.6) | 68.0 (62.5–73.1) | 65.9 (60.6–70.8) | 0.749 | 63.7 | 70.1 | 1.99 (1.68–2.36) | 0.49 (0.40–0.58) |
| CA2-3 | 75.4 (71.9–78.6) | 77.0 (71.8–81.4) | 74.0 (69.0–78.5) | 0.843 | 72.3 | 78.5 | 2.96 (2.44–3.59) | 0.31 (0.25–0.39) |
| CA4-DG | 76.9 (73.4–80.0) | 79.7 (74.7–83.9) | 74.3 (69.4–78.7) | 0.853 | 73.2 | 80.7 | 3.11 (2.56–3.76) | 0.27 (0.22–0.35) |
| Fimbria | 68.2 (64.4–71.7) | 69.8 (64.3–74.8) | 66.8 (61.5–71.6) | 0.745 | 64.9 | 71.5 | 2.10 (1.77–2.49) | 0.45 (0.37–0.55) |
| Presubiculum | 80.6 (74.9–83.4) | 83.2 (78.4–87.0) | 78.3 (73.5–82.4) | 0.882 | 77.1 | 84.1 | 3.08 (3.10–4.72) | 0.22 (0.17–0.28) |
| Subiculum | 81.2 (77.9–84.1) | 83.5 (78.8–87.3) | 79.2 (74.5–83.2) | 0.887 | 77.9 | 84.5 | 4.01 (3.23–4.97) | 0.21 (0.16–0.27) |
Confidence intervals are presented within parenthesis. AUC area under the curve, PPV positive predictive value, NPV negative predictive value, LR+ positive likelihood ratio, LR− negative likelihood ratio
Fig. 3.
a ROC curve for AD versus HC classification using individual subfield measures, b ROC curve for MCI-c and MCI-s classification using individual subfield measures. The curve is calculated with a 95 % probability assurance. ROC receiver operating characteristic, AD Alzheimer’s disease, HC healthy control, MCI-c MCI-converter, MCI-s stable MCI
Model Validation for AD and HC Classification
Seven fold cross validation was used to determine the robustness of all the models. In this study models were validated using an external test set. The ADNI model was used as a training set and predictions were made using the AddNeuroMed cohort as the external test set and vice versa. The results are similar to those obtained by cross validation (Table 6). For the combination of hippocampal subfields, using the AddNeuroMed cohort as the test set and the ADNI cohort as the training set resulted in a similar classification accuracy, 82.1 % (sensitivity = 77.1 %, specificity = 86.9 %, AUC = 0.90) compared to 81.1 % for total hippocampal volume (sensitivity = 75.2 %, specificity = 86.9 %, AUC = 0.897). Similar results for the combination of hippocampal subfields and total hippocampal volume were obtained when using the ADNI cohort as the test set and the AddNeuroMed cohort as the training set (Table 5). For further validation, we compared if subjects were classified differently between the different models for the combination of hippocampal subfields (for example classified as AD in one model and HC in another model). We compared the single cohort cross validated models with the combined cohort model and the single cohort models using the train/test approach. The results demonstrate that classification agreement for the different comparisons were high, lying between 89.5–98.8 % (Table 8).
Table 8.
Comparison of subject classification between cohort models
| ANM and ANM/ADNI | ADNI and ANM/ADNI | ANM and ANMonADNI | ADNI on ADNIonANM | |
|---|---|---|---|---|
| Total n | 212 | 410 | 212 | 410 |
| Same classification (n) | 197 | 405 | 196 | 367 |
| Same classification (%) | 92.9 | 98.8 | 92.5 | 89.5 |
| Different classification (n) | 15 | 5 | 16 | 43 |
| Different classification (%) | 7.1 | 1.2 | 7.5 | 10.5 |
ANM AddNeuroMed cohort model, ANM/ADNI combined AddNeuroMed and ADNI cohort model, ANMonADNI AddNeuroMed cohort test set and ADNI training set, ADNIonANM ADNI cohort test set and AddNeuroMed cohort training set, total n AD and HC subjects, Same classification number of subjects predicted alike, % same classification percentage of subjects predicted alike, Different classification number of subjects predicted differently, Different classification (%) percentage of subjects predicted differently
Predicting MCI Conversion
Models previously constructed using AD and HC subjects from the combined cohort were applied to our large external test set of MCI subjects (n = 447) to predict future conversion to AD. These classifiers subsequently identified MCI subjects with an AD like brain structure (percentage classified as AD-like) or a healthy control-like brain structure (percentage classified as HC-like). During the 12 month follow up interval, 90 MCI subjects from the AddNeuroMed and ADNI cohorts met the clinical criteria for AD, and 357 MCI subjects remained clinically stable.
The combined subfield volumes classifier correctly identified 81.1 % of MCI converters from baseline images, with the presubiculum also correctly identifying 81.1 % of MCI-c with an AD-like pattern of subfield atrophy. In comparison, total hippocampal volume identified 76.7 % of MCI-c correctly. The predictive accuracies from the classifiers ranged between 56.7–81.1 % for MCI-c predictions (Table 9).
Table 9.
MCI predictions using the baseline OPLS AD versus HC classifiers
| MCI-c classification (n = 90)b | MCI-s classification (n = 357) | |||
|---|---|---|---|---|
| AD like (%)* | HC like (%) ** | AD like (%)* | HC like (%) ** | |
| CA1 | 65.6 (59) | 34.4 (31) | 48.2 (172) | 51.8 (185) |
| CA2-3 | 75.6 (68) | 24.4 (22) | 52.1 (186) | 47.9 (171) |
| CA4-DG | 74.4 (67) | 25.6 (23) | 51.8 (185) | 48.2 (172) |
| Fimbria | 56.7 (51) | 43.3 (39) | 53.2 (190) | 46.8 (167) |
| Presubiculum | 81.1 (73) | 18.9 (17) | 51.0 (182) | 49.0 (175) |
| Subiculum | 77.8 (70) | 22.2 (20) | 51.3 (183) | 48.7 (174) |
| Combineda | 81.1 (73) | 18.9 (17) | 51.3 (183) | 48.7 (174) |
| Total Hippocampal volume | 76.7 (69) | 23.3 (21) | 49.9 (178) | 50.1 (179) |
AD Alzheimer’s disease, MCI mild cognitive impairment, MCIc MCI converter, MCI-s MCI stable, HC healthy control
* Sensitivity at each time point is the percentage of MCIc subjects correctly classified as AD in bold
** Specificity at each time point is the percentage of MCI-s subjects correctly classified as HC in bold
aThe combined model used the combination of subfields for classification
bOnly includes subjects that underwent conversion from MCI to AD at 12 months follow up
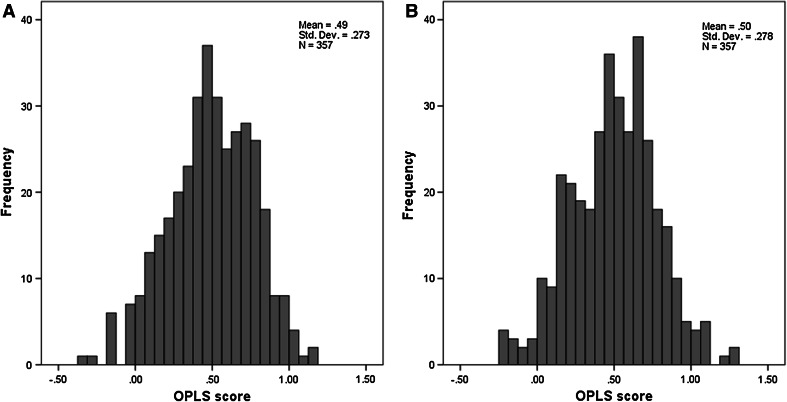
However, a considerable number of MCI-s subjects were also predicted with an AD-like pattern of atrophy despite their clinically stable condition at 12 months follow up. For instance, the combined subfield volumes classifier, which was the most robust for MCI-c prediction, identified only 48.7 % of MCI-s with a HC-like subfield structure. A similar result was observed for the total hippocampal volume classifier which only identified 50.1 % of MCI-s correctly. Mean OPLS scores from the combined subfield volumes classifier and total hippocampal volume classifier were 0.50 ± 028 and 0.49 ± 0.27 (mean ± standard deviation) respectively (Fig. 4). As a result, differences in OPLS scores between the two classifiers were not statistically significant (Wilcoxon signed rank sum test, Z = −0.725, p = 0.469) despite the difference in AD-like MCI-c predictions. HC-like predictive accuracies in MCI-s predictions only ranged between 46.8–51.8 % which is because many of these MCI-s subjects will convert to AD at a future stage and already demonstrate an Alzheimer like pattern of hippocampal subfield atrophy.
Fig. 4.
a OPLS scores from the total hippocampal volume classifier for MCI-s predictions, b OPLS scores from the combined subfields volume classifier for MCI-s predictions
Discussion
Using an automated image analysis pipeline to explore the subfields of the hippocampus, we found that AD and MCI converters displayed a widespread pattern of subfield atrophy, including the bilateral subiculum, presubiculum and CA1 which have been reported in previous studies. Using the same image analysis approach, Hanseeuw et al. (2011) previously reported significant volume losses in the subiculum and CA2-3 region of the hippocampus in a small group of 15 amnestic MCI subjects and 15 healthy controls. We have extended this preliminary work to data from two large studies which together contain a more heterogeneous group of AD and MCI subjects that more accurately reflect the population of MCI and AD. The pattern of hippocampal volume loss that was found was wider than previous reports which have used either manual delineation techniques hippocampal subfield segmentation (Mueller and Weiner 2009; Mueller et al. 2010), 3D surface mapping (Apostolova et al. 2010) or shape analysis techniques (Csernansky et al. 2005; Costafreda et al. 2011) A similar pattern of subfield atrophy was observed for AD subjects and MCI converters suggesting that MCI converters may represent an imaging profile more similar to AD subjects than stable MCI. The pattern of hippocampal subfield loss, though wider than previously reported is in agreement with previous neuropathological studies reporting early neuronal loss in the subiculum, CA4-DG, and CA1 (West et al. 2004). Larger datasets are more likely to contain subjects with different types of atrophy which could explain the widespread pattern of subfield volume losses reported in the present study.
Relationship Between Neuropsychological Test Scores and Hippocampal Subfields
A significant positive effect for both MMSE and ADAS-1 was found in relation to all hippocampal subfield volumes, indicating that subjects with lower MMSE scores and higher ADAS-1 scores had lower subfield volumes. This confirms the relationship between diffuse hippocampal volume loss and poorer neuropsychological test scores (Scheltens et al. 1992; Liu et al. 2009).
Relationship Between Age, Gender, Education, APOE ε4 Genotype, and Hippocampal Subfields
Previous studies investigating the influence of age on hippocampal subfields have found significant negative effects associated with CA1 and CA2-3 subfield volumes (Mueller and Weiner 2009). Consistent with this previous work, using a larger dataset we also found a significant negative effect of age but in relation to all subfield volumes. However, years of education was only significantly associated with the right CA1 and right fimbria. Gender specific differences in the pattern of subfield volume loss were found, with female subjects demonstrating lower bilateral CA2-3, CA4-DG, fimbria, presubiculum and subiculum volumes. Previous work with AD patients suggests that gender specific differences in the rate of hippocampal volume loss are not entirely clear. For example, a previous study has reported a higher prevalence and incidence of AD in females (Barnes et al. 2005), whereas sex hormone differences have been suggested as an explanation of any gender divergence (Gouras et al. 2000). On the other hand, our findings suggest that carriers of the ε4 allele had smaller subfield volumes. which is in agreement with previous studies that have demonstrated a strong neuroanatomic effect of APOE ε4 genotype on the entire hippocampal region. (Jack et al. 1998; Reiman et al. 1998).
AD and HC Classification
In this study we used the multivariate OPLS technique to distinguish between AD and control subjects. This method has previously been used for distinguishing between AD and control subjects (Westman et al. 2011a, b, c, d), as well as predicting conversion from MCI to AD using MRI regional measures and a combination of MRI regional measures and magnetic resonance spectroscopy (MRS) measures (Westman et al. 2010, 2011a, b, c, d). Hippocampal subfields have not been studied using this approach but several other studies have used alternative multivariate techniques including support vector machines, principal components analysis, and partial least squares to latent structures and linear discriminant analysis to analyse multiple MRI regional measures (Fan et al. 2008; McEvoy et al. 2009; Klöppel et al. 2008; Vemuri et al. 2008; Plant et al. 2010).
Studies that have attempted to distinguish between AD and control subjects have often done so using medial temporal structures such as the hippocampus and entorhinal cortex and reported accuracies of 80–90 % (Fox et al. 1996; Jack et al. 1992). Although prior studies have reported accuracies of up to 100 % in discriminating between AD and control subjects(Fan et al. 2008; Lerch et al. 2008), some studies used smaller samples, included more severely impaired AD patients or failed to cross-validate their findings. Here, using two large multicentre studies we segmented the hippocampus into its different subfields to examine whether subfield volumes could improve the sensitivity of MRI in detecting AD. The results suggest that the OPLS technique with fully automated hippocampal subfield volumes performs as accurately as total hippocampal volume, presubiculum volume and subiculum volume in distinguishing between AD and control subjects. The OPLS method which combined hippocampal subfield measures produced a classification accuracy of 81.7 % (sensitivity = 80.4 %, specificity = 82.8 %, AUC = 0.895), while total automated hippocampal volume produced an accuracy of 80.7 % (sensitivity 79.2 %, specificity 82.8 %, AUC = 0.887). Although significantly different, the magnitude of the difference is small and does not offer a particular advantage over hippocampal volume. Recent work has also found that the visual rating assessment of the medial temporal lobe produces accuracies that are comparable to that of manual hippocampal volume in distinguishing between AD and controls (Westman et al. 2011d).
Although our study is the first to use multivariate analysis of automated hippocampal subfields, previous research has examined the combination of automated regional cortical thicknesses and regional volumes in distinguishing between AD and control subjects using support vector machines and linear discriminant analysis (Vemuri et al. 2008; McEvoy et al. 2009).
Predicting MCI Conversion
Building robust classification models on new and unseen data is of great importance for accurately predicting future MCI conversion to AD. MCI predictions were performed using AD vs. HC models in the combined ADNI and AddNeuroMed cohorts as classifiers and MCI subjects as our external validation test set. This approach has been applied previously and demonstrates how larger training sets can be used to assess MCI predictions that are more balanced in terms of sensitivity and specificity (Westman et al. 2011a). Previous studies in the neuroimaging literature utilising advanced methods of high dimensional pattern classification (Fan et al. 2008; Misra et al. 2009), and whole brain structural MRI (Karas et al. 2008; Davatzikos et al. 2010) have demonstrated the complexity of differential atrophy patterns observed in MCI-c and MCI-s subjects. Moreover, studies including our own have also shown heterogeneous patterns of brain atrophy exist in MCI subjects that convert to AD and those who remain clinically stable (Westman et al. 2011c; McEvoy et al. 2009). Consequently, hippocampal subfields were of interest following a small pilot study in MCI subjects (Hanseeuw et al. 2011).
Using a large external validation test set (n = 447) we sought to identify MCI subjects based on the similarity of their hippocampal subfield pattern to AD patients (% AD-like) or healthy control subjects (% HC-like). Unlike some previous studies, the large number of MCI subjects in our study served to more accurately represent the heterogeneity of MCI subjects and included both amnestic and non-amnestic subtypes.
The results demonstrated that the combination of subfield volumes and the presubiculum were the most robust classifiers, identifying 81.1 % of MCI-c correctly, and were better than using total hippocampal volume alone. However, a considerable number of MCI-s subjects were also predicted with AD-like patterns of atrophy despite having a clinically stable MCI condition at 12 months follow up. Although beyond the scope of the current study we intend in future to study longitudinal change in hippocampal subfield measures over longer follow up times in the ADNI cohort. The utility of structural MRI plays a key role in this domain and represents one of the 3 main biomarkers for AD diagnosis (Dubois et al. 2007; Frisoni et al. 2010). However, more focus needs to be addressed towards the standardisation of acquisition and analysis methods in order to facilitate the integration of findings across studies. Recently there has been much interest in exploring the combination of different MRI imaging techniques (i.e. Tensor based morphometry, cortical thicknesses and volumes) with cerebrospinal fluid (CSF) biomarkers, 18F-fluorodeoxyglucose PET, and clinical examination for classifying AD and predicting MCI conversion to AD (Wolz et al. 2011; Vemuri et al. 2009; Zhang et al. 2011; Furney et al. 2011). In regards to our study, a longer follow up time would be helpful to refine our estimates of model specificity for MCI-s predictions. A more robust algorithm that could potentially predict future MCI time to AD conversion would be of future interest to validate our findings in this present study.
Conclusion
Hippocampal subfield volume loss in AD is widespread affecting regions such as the CA-1, subiculum, and presubiculum. Using an automated hippocampal subfield measurement technique we found prominent subfield volume losses in MCI converters and AD. Each of the subfield measures was related to both clinical predictors of AD (Age, gender, years of education, APOE E4 genotype) and cognitive scores (MMSE and ADAS-1 tests). Combined subfield volumes using the OPLS technique produced a similar classification accuracy to total hippocampal volume, presubiculum volume and subiculum volume in distinguishing between AD and HC subjects, but were more accurate than total hippocampal volume measurements at predicting MCI conversion to AD at 12 months.
Acknowledgments
This study was supported by InnoMed, (Innovative Medicines in Europe) an Integrated Project funded by the European Union of the Sixth Framework program priority FP6-2004-LIFESCIHEALTH-5, Life Sciences, Genomics and Biotechnology for Health. Data collection and sharing for this project was funded by the ADNI (National Institutes of Health Grant U01 AG024904). Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. Also, thanks to the Foundation Gamla Tjänarinnor, the Swedish Alzheimer’s Association and Swedish Brain Power, Health Research Council of Academy of Finland and Stockholm Medical Image Laboratory and Education (SMILE). AS, EW, J-SM and SL were supported by funds from NIHR Biomedical Research Centre for Mental Health at the South London and Maudsley NHS Foundation Trust and Institute of Psychiatry, King’s College London.
Footnotes
Data used in preparation of this article were obtained from the Alzheimer’s disease Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.ucla.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Wasim Khan and Eric Westman have contributed equally to the manuscript.
References
- Apostolova LG, et al. Conversion of mild cognitive impairment to Alzheimer disease predicted by hippocampal atrophy maps. Arch Neurol. 2006;63(5):693–699. doi: 10.1001/archneur.63.5.693. [DOI] [PubMed] [Google Scholar]
- Apostolova LG, et al. 3D comparison of low, intermediate, and advanced hippocampal atrophy in MCI. Hum Brain Mapp. 2010;31(5):786–797. doi: 10.1002/hbm.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes LL, et al. Sex differences in the clinical manifestations of Alzheimer disease pathology. Arch Gen Psychiatry. 2005;62(6):685–691. doi: 10.1001/archpsyc.62.6.685. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Bylesjo M, et al. OPLS discriminant analysis : combining the strengths of PLS-DA and SIMCA classification y. J Chemom. 2006;20:341–351. doi: 10.1002/cem.1006. [DOI] [Google Scholar]
- Costafreda SG, et al. Automated hippocampal shape analysis predicts the onset of dementia in mild cognitive impairment. Neuroimage. 2011;56(1):212–219. doi: 10.1016/j.neuroimage.2011.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csernansky JG, et al. Preclinical detection of Alzheimer’s disease: hippocampal shape and volume predict dementia onset in the elderly. Neuroimage. 2005;25(3):783–792. doi: 10.1016/j.neuroimage.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, et al. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol Aging. 2010;32(12):2322e19–2322e27. doi: 10.1016/j.neurobiolaging.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, et al. Automated MRI measures identify individuals with mild cognitive impairment and Alzheimer’s disease. Brain. 2009;132(8):2048–2057. doi: 10.1093/brain/awp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanand DP, et al. Hippocampal and entorhinal atrophy in mild cognitive impairment: prediction of Alzheimer disease. Neurology. 2007;68(11):828–836. doi: 10.1212/01.wnl.0000256697.20968.d7. [DOI] [PubMed] [Google Scholar]
- Dubois B, et al. Research criteria for the diagnosis of Alzheimer’s disease: revising the NINCDS-ADRDA criteria. Lancet Neurol. 2007;6(8):734–746. doi: 10.1016/S1474-4422(07)70178-3. [DOI] [PubMed] [Google Scholar]
- Fan Y, et al. Spatial patterns of brain atrophy in MCI patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline. Neuroimage. 2008;39(4):1731–1743. doi: 10.1016/j.neuroimage.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/S0896-6273(02)00569-X. [DOI] [PubMed] [Google Scholar]
- Fischl B, et al. Sequence-independent segmentation of magnetic resonance images. Neuroimage. 2004;23(Suppl 1):S69–S84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Fox NC, et al. Presymptomatic hippocampal atrophy in Alzheimer’s disease. A longitudinal MRI study. Brain. 1996;119(Pt 6):2001–2007. doi: 10.1093/brain/119.6.2001. [DOI] [PubMed] [Google Scholar]
- Frisoni GB, et al. The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol. 2010;6(2):67–77. doi: 10.1038/nrneurol.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furney SJ, et al. Combinatorial markers of mild cognitive impairment conversion to Alzheimer’s disease–cytokines and MRI measures together predict disease progression. J Alzheimers Dis. 2011;26(Suppl 3(3)):395–405. doi: 10.3233/JAD-2011-0044. [DOI] [PubMed] [Google Scholar]
- Gouras GK, et al. Testosterone reduces neuronal secretion of Alzheimer’s β-amyloid peptides. Proc Natl Acad Sci USA. 2000;97(3):1202–1205. doi: 10.1073/pnas.97.3.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanseeuw BJ, et al. Mild cognitive impairment: differential atrophy in the hippocampal subfields. AJNR Am J Neuroradiol. 2011;32(9):1658–1661. doi: 10.3174/ajnr.A2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, et al. MR-based hippocampal volumetry in the diagnosis of Alzheimer’s disease. Neurology. 1992;42(1):183–188. doi: 10.1212/WNL.42.1.183. [DOI] [PubMed] [Google Scholar]
- Jack CR, et al. Hippocampal atrophy and apolipoprotein E genotype are independently associated with Alzheimer’s disease. Ann Neurol. 1998;43(3):303–310. doi: 10.1002/ana.410430307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, et al. The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27(4):685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karas G, et al. Amnestic mild cognitive impairment: structural MR imaging findings predictive of conversion to Alzheimer disease. AJNR Am J Neuroradiol. 2008;29(5):944–949. doi: 10.3174/ajnr.A0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöppel S, et al. Automatic classification of MR scans in Alzheimer’s disease. Brain. 2008;131(Pt 3):681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JP, et al. Automated cortical thickness measurements from MRI can accurately separate Alzheimer’s patients from normal elderly controls. Neurobiol Aging. 2008;29(1):23–30. doi: 10.1016/j.neurobiolaging.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Liu Y, et al. Combination analysis of neuropsychological tests and structural MRI measures in differentiating AD, MCI and control groups-The AddNeuroMed study. Neurobiol Aging. 2009;32(7):1198–1206. doi: 10.1016/j.neurobiolaging.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Lovestone S, et al. AddNeuroMed–the European collaboration for the discovery of novel biomarkers for Alzheimer’s disease. Ann N Y Acad Sci. 2009;1180:36–46. doi: 10.1111/j.1749-6632.2009.05064.x. [DOI] [PubMed] [Google Scholar]
- Mangialasche F, et al. High plasma levels of vitamin E forms and reduced Alzheimer’s disease risk in advanced age. J Alzheimers Dis. 2010;20(4):1029–1037. doi: 10.3233/JAD-2010-091450. [DOI] [PubMed] [Google Scholar]
- McEvoy LK, et al. Alzheimer disease: quantitative structural neuroimaging for detection and prediction of clinical and structural changes in mild cognitive impairment. Radiology. 2009;251(1):195–205. doi: 10.1148/radiol.2511080924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra C, Fan Y, Davatzikos C. Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: results from ADNI. Neuroimage. 2009;44(4):1415–1422. doi: 10.1016/j.neuroimage.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW. Selective effect of age, Apo e4, and Alzheimer’s disease on hippocampal subfields. Hippocampus. 2009;19(6):558–564. doi: 10.1002/hipo.20614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, et al. Hippocampal atrophy patterns in mild cognitive impairment and Alzheimer’s disease. Hum Brain Mapp. 2010;31(9):1339–1347. doi: 10.1002/hbm.20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant C, et al. Automated detection of brain atrophy patterns based on MRI for the prediction of Alzheimer’s disease. Neuroimage. 2010;50(1):162–174. doi: 10.1016/j.neuroimage.2009.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, et al. Hippocampal volumes in cognitively normal persons at genetic risk for Alzheimer’s disease. Ann Neurol. 1998;44(2):288–291. doi: 10.1002/ana.410440226. [DOI] [PubMed] [Google Scholar]
- Robin X, et al. pROC: an open-source package for R and S + to analyze and compare ROC curves. Bioinformatics. 2011;12(1):77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheltens P, et al. Atrophy of medial temporal lobes on MRI in “probable” Alzheimer’s disease and normal ageing: diagnostic value and neuropsychological correlates. J Neurol Neurosurg Psychiatry. 1992;55(10):967–972. doi: 10.1136/jnnp.55.10.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ségonne F, et al. A hybrid approach to the skull stripping problem in MRI. Neuroimage. 2004;22(3):1060–1075. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Simmons A, et al. MRI measures of Alzheimer’s disease and the AddNeuroMed study. Ann N Y Acad Sci. 2009;1180:47–55. doi: 10.1111/j.1749-6632.2009.05063.x. [DOI] [PubMed] [Google Scholar]
- Simmons A, et al. The AddNeuroMed framework for multi-centre MRI assessment of Alzheimer’s disease: experience from the first 24 months. Int J Geriatr Psychiatry. 2011;26(1):75–82. doi: 10.1002/gps.2491. [DOI] [PubMed] [Google Scholar]
- Spulber G, et al. An MRI-based index to measure the severity of Alzheimer’s disease-like structural pattern in subjects with mild cognitive impairment. J Intern Med. 2013;273(4):396–409. doi: 10.1111/joim.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS) J Chemom. 2002;16(3):119–128. doi: 10.1002/cem.695. [DOI] [Google Scholar]
- Van Leemput K, et al. Automated segmentation of hippocampal subfields from ultra-high resolution in vivo MRI. Hippocampus. 2009;19(6):549–557. doi: 10.1002/hipo.20615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri P, et al. Alzheimer’s disease diagnosis in individual subjects using structural MR images: validation studies. Neuroimage. 2008;39(3):1186–1197. doi: 10.1016/j.neuroimage.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri P, et al. MRI and CSF biomarkers in normal, MCI, and AD subjects: predicting future clinical change. Neurology. 2009;73(4):294–301. doi: 10.1212/WNL.0b013e3181af79fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MJ, et al. Hippocampal neurons in pre-clinical Alzheimer’s disease. Neurobiol Aging. 2004;25(9):1205–1212. doi: 10.1016/j.neurobiolaging.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Westman E, Wahlund LO, Foy C et al (2010) Combining MRI and MRS to distinguish between Alzheimer’s disease and healthy controls. J Alzheimers Dis 22(1):171–181 [DOI] [PubMed]
- Westman E, Simmons A, Muehlboeck J-S, et al. AddNeuroMed and ADNI: similar patterns of Alzheimer’s atrophy and automated MRI classification accuracy in Europe and North America. Neuroimage. 2011;58(3):818–828. doi: 10.1016/j.neuroimage.2011.06.065. [DOI] [PubMed] [Google Scholar]
- Westman E, Wahlund L, Foy C, et al. Magnetic Resonance Imaging and Magnetic Resonance Spectroscopy for Detection of Early Alzheimer’ s Disease. J Alzheimers Dis. 2011;26:307–319. doi: 10.3233/JAD-2011-0028. [DOI] [PubMed] [Google Scholar]
- Westman E, Simmons A, Zhang Y, et al. Multivariate analysis of MRI data for Alzheimer’s disease, mild cognitive impairment and healthy controls. Neuroimage. 2011;54(2):1178–1187. doi: 10.1016/j.neuroimage.2010.08.044. [DOI] [PubMed] [Google Scholar]
- Westman E, Cavallin L, Muehlboeck J-S, et al. Sensitivity and Specificity of Medial Temporal Lobe Visual Ratings and Multivariate Regional MRI Classification in Alzheimer’s Disease J. Laks, ed. PLoS One. 2011;6(7):9. doi: 10.1371/journal.pone.0022506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westman E, Muehlboeck J-S, Simmons A. Combining MRI and CSF measures for classification of Alzheimer’s disease and prediction of mild cognitive impairment conversion. Neuroimage. 2012;62(1):229–238. doi: 10.1016/j.neuroimage.2012.04.056. [DOI] [PubMed] [Google Scholar]
- Westman E, et al. Regional magnetic resonance imaging measures for multivariate analysis in Alzheimer’s disease and mild cognitive impairment. Brain Topogr. 2013;26(1):9–23. doi: 10.1007/s10548-012-0246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiklund S, et al. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem. 2008;80(1):115–122. doi: 10.1021/ac0713510. [DOI] [PubMed] [Google Scholar]
- Wolz R, et al. Multi-method analysis of MRI images in early diagnostics of Alzheimer’s disease. PLoS One. 2011;6(10):e25446. doi: 10.1371/journal.pone.0025446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, et al. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. Neuroimage. 2011;55(3):856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]