Summary
Repair of DNA interstrand crosslinks requires action of multiple DNA repair pathways, including homologous recombination. Here, we report a de novo heterozygous T131P mutation in RAD51/FANCR, the key recombinase essential for homologous recombination, in a patient with Fanconi anemia-like phenotype. In vitro, RAD51-T131P displays DNA-independent ATPase activity, no DNA pairing capacity and a co-dominant negative effect on RAD51 recombinase function. However, the patient cells are homologous recombination proficient due to the low ratio of mutant to wildtype RAD51 in cells. Instead, patient cells are sensitive to crosslinking agents and display hyperphosphorylation of Replication Protein A due to increased activity of DNA2 and WRN at the DNA interstrand crosslinks. Thus, proper RAD51 function is important during DNA interstrand crosslink repair outside of homologous recombination. Our study provides a molecular basis for how RAD51 and its associated factors may operate in a homologous recombination-independent manner to maintain genomic integrity.
Keywords: Fanconi anemia, homologous recombination, DNA interstrand crosslink repair, RAD51, FANCR, DNA2, WRN
Introduction
The maintenance of genomic stability requires the accurate repair of DNA damage that accrues as a result of endogenous metabolism, environmental exposures or chemotherapy. DNA interstrand crosslinks (ICLs) are a particularly serious form of DNA damage that can be formed by anti-cancer alkylating agents or endogenous compounds including aldehydes (Garaycoechea et al., 2012; Langevin et al., 2011; reviewed in Clauson et al., 2013; Kottemann and Smogorzewska, 2013). Inaccurate repair of ICLs impacts organ function, as exemplified by karyomegalic interstitial nephritis (Zhou et al., 2012), a kidney failure predisposition syndrome, and Fanconi anemia (FA), a disorder characterized by developmental abnormalities, bone marrow failure, cancer predisposition, and profound sensitivity to crosslinking agents (Auerbach, 2009).
The 18 genes mutated in FA patients encode proteins implicated in a common pathway that coordinates multiple repair processes and checkpoint signaling events necessary for the accurate removal of ICL lesions (Bogliolo et al., 2013; Hira et al., 2015; Kashiyama et al., 2013; Kottemann and Smogorzewska, 2013; Rickman et al., 2015; Sawyer et al., 2015; Wang and Smogorzewska, 2015). ICL repair occurs predominantly during S-phase following replication fork stalling at the ICL that triggers monoubiquitination of FANCD2 and FANCI, the central event of the FA pathway (Akkari et al., 2000; Garcia-Higuera et al., 2001; Knipscheer et al., 2009; Smogorzewska et al., 2007). Incisions around the lesion allow unhooking of ICLs from one of the two strands and result in DNA strand breaks at the fork (Klein Douwel et al., 2014; Räschle et al., 2008; reviewed in Sengerova et al., 2011; Zhang and Walter, 2014). Following translesion synthesis to restore the DNA template (Räschle et al., 2008), the homologous recombination (HR) machinery utilizes the processed ends of broken duplex DNA to complete the repair (Howlett et al., 2002; Long et al., 2011; Reid et al., 2007; Xia et al., 2007).
Besides being important for the completion of ICL repair, HR is widely used to ensure genome stability when a homologous template is available for repair. In the case of double-strand DNA break (DSB) repair, broken DNA ends are nucleolytically processed by multiple factors including the MRE11-RAD50-XRS2/NBS1 complex, DNA2, EXO1 and CTIP (Nimonkar et al., 2011; reviewed in Symington and Gautier, 2011), exposing ssDNA that is immediately coated by ssDNA binding protein Replication Protein-A (RPA). Recombination is initiated when RAD51 recombinase actively displaces RPA to form the presynaptic filament that searches for and invades the homologous template (Sugiyama et al., 1997; Sung and Robberson, 1995). The displacement process is assisted by a number of recombination mediators, including the breast cancer susceptibility genes, BRCA2, and RAD51 paralogs (Jensen et al., 2010; Liu et al., 2010; Pierce et al., 1999; Takata et al., 2000). Humans with biallelic mutations in HR factors such as BRCA2/FANCD1, have 97% cumulative risk of any malignancy by age 5.2 years (Alter et al., 2007), stressing the importance of HR in assuring genome stability and tumor suppression.
The requirement of RAD51 for proper ICL repair was demonstrated in Xenopus egg extract repair assays (Long et al., 2011), and its involvement was thought to be strictly dependent on its recombinase function in resolving DSBs at the late stage of ICL repair. However, surprisingly, RAD51 appeared to bind ICL-induced stalled replication forks early during repair, preceding the arrival of the activated FANCD2/FANCI complex and DNA incisions that would result in DSBs (Long et al., 2011). This observation suggests an early role for RAD51 at ICLs. RAD51 has been implicated in both maintaining replication fork stability and facilitating replication fork restart following treatment with other replication fork stalling agents, including hydroxyurea (HU) (Hashimoto et al., 2010; Petermann et al., 2010; Schlacher et al., 2011; Schlacher et al., 2012; Zellweger et al., 2015). RAD51 foci formation and recruitment to chromatin were observed in cells following HU treatment (Petermann et al., 2010). In the absence of RAD51, ssDNA gaps accumulate behind replication forks as a result of MRE11-mediated degradation of nascent DNA (Hashimoto et al., 2010). MRE11-dependent nascent strand degradation following HU treatment is particularly pronounced in cells defective in BRCA2 or the Fanconi proteins. In that genetic setting, RAD51 was shown to have a protective role at the HU-stalled forks (Schlacher et al., 2011; Schlacher et al., 2012; Ying et al., 2012). Most recently, it was demonstrated that in response to a range of DNA damaging agents, RAD51 is involved in mediating replication fork reversal (Zellweger et al., 2015), likely to facilitate replication fork restart (Petermann et al., 2010). How RAD51 is recruited to and functions at sites of stalled replication forks and whether these functions of RAD51 at the fork are dependent on its strand exchange activity remain poorly understood. Furthermore, given that DSBs can arise if replication forks collapse following HU treatment it has been difficult to distinguish the fork protection role of RAD51 from its canonical function in HR. Here, we report a patient-derived mutation in RAD51 that is consistent with a role for RAD51 in protection of DNA during the course of ICL repair that is independent of its DNA strand exchange activity.
Results
RAD51 mutation in a patient with FA-like phenotype
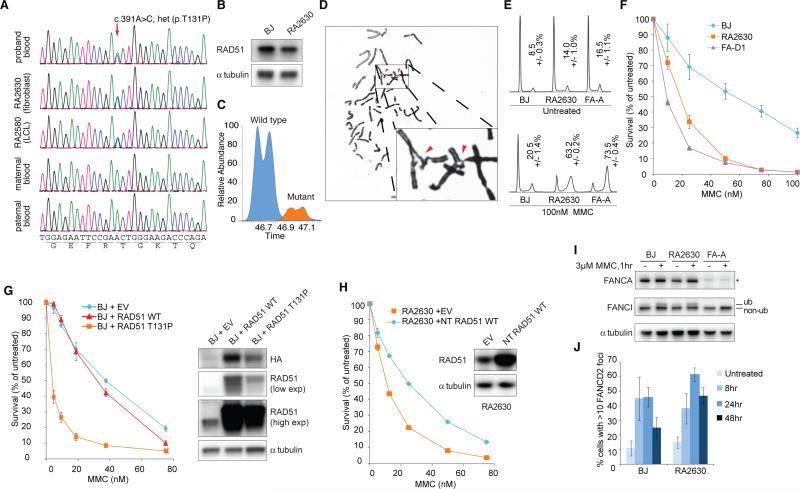
A one-year-old girl born with radial dysplasia, absent right thumb, pelvic left kidney and increased DNA damage sensitivity, including the appearance of radial chromosomes in peripheral blood lymphoblasts and skin fibroblasts after treatment with diepoxybutane (DEB) and mitomycin C (MMC) (See Table S1 for a full description of the phenotype) was entered into the International Fanconi Anemia Registry. No mutation in known FA genes was identified in this subject. Whole exome sequencing of DNA from the lymphoblastoid cell line (RA2580) revealed a heterozygous mutation in RAD51 (c.391A>C) that was likely de novo since it was absent from either of the parents (Figure 1A; also see Supplemental Experimental Procedures for other variants identified in the subject). The same mutation was present in whole blood as well as in the primary fibroblasts (RA2630) from this subject (Figure 1A) suggesting that the subject is not mosaic for the mutation. The mutation results in a single amino acid residue change (p.T131P) within the Walker A domain important for ATP binding and hydrolysis (reviewed in San Filippo et al., 2008) and this residue is completely evolutionarily conserved down to yeasts. Total levels of RAD51 in the RA2630 cell lines are comparable to those in the wild type BJ fibroblasts (Figure 1B). The mutant allele is expressed at the mRNA and protein levels (Figure S1A-D), although the mutant is much less abundant than the WT RAD51 on the protein level (Figure 1C, S1C-D). We were able to quantify the level of the wild type and mutant protein by performing an immunoprecipitation with anti-RAD51 antibody and assessing amounts of the wild type and mutant peptides present in the pull down by mass spectrometry. Based on area under curve for the wild type and mutant peptides (Figure 1C) and selected fragment ions (not shown) a mutant-to-wild type ratio of 1-to-5 was consistently measured (0.21, s.d. 0.017, n=3). Mutant and wild type RAD51 were also detected in the patient LCL sample.
Figure 1.
RAD51 mutation in a Fanconi anemia-like patient. (A) Sequencing traces of genomic DNA identifying the variant (arrow) in RAD51 in the proband but not in the parents. (B) RAD51 protein expression levels in patient cells (RA2630). (C) Assessment of the relative amounts of the WT and mutant RAD51 protein by measurement of mutant and the wild type peptides of RAD51 in fibroblast sample. Extracted ion chromatograms based on the monoisotopic ion corresponding to the mutant (orange) and wild type (blue) peptides. Measured area suggests a ~5-fold difference between the wild type and mutant peptides. Also see Figure S1B-D. (D) Representative metaphase of RA2630 following mitomycin C (MMC) treatment. Arrows mark sites of aberration. Also see Table S2 for quantification of chromosomal breakage levels. (E) Cell cycle profiles with or without MMC treatment. FA-A: FANCA patient cells. (F) MMC sensitivity of indicated fibroblasts. BJ: wild type fibroblasts; FA-D1: BRCA2/FANCD1 mutant patient cells. Error bars: s.d. (n=3) (G) Sensitization of BJ cells to MMC by RAD51 T131P mutant overexpression. Also see Table S3 for quantification of chromosomal breakage levels upon overexpression of WT and mutant RAD51. (H) Suppression of RA2630 MMC sensitivity by overexpression of non-tagged (NT) RAD51. EV: empty vector. (I) FANCI monoubiquitination 24 hours after one-hour 3μM MMC treatment. (J) FANCD2 foci formation post MMC treatment. Error bars: s.e.m. (n=3). Also see Figure S1.
Cells expressing the RAD51 T131P allele are hypersensitive to cross-linking agents
All tested cells derived from the subject showed moderately increased levels of chromosomal breakage in response to DEB and MMC that, while increased compared to controls, are significantly lower than those seen in typical FA-A cell lines with biallelic mutations in the FANCA gene (Figure 1D; Table S2). We observed an increased accumulation of cells in the late S/G2 phases of the cell cycle after MMC treatment (Figure 1E) and both fibroblasts and lymphoblasts derived from the subject were sensitive to cross-linking agents including MMC, DEB, and cisplatin in survival assays (Figure 1F, Figure S1F-J). Collectively, these results show that ICL repair fails in cells expressing the T131P RAD51 allele. However, the chromosome breakage phenotype of cells derived from the subject is less severe than classical FA cells and is more akin to the phenotype of cells with RAD51C mutations (Vaz et al., 2010).
We observed increased MMC sensitivity (Figure 1G) and crosslinking-induced chromosomal aberrations (Table S3) when HA-FLAG tagged RAD51 T131P mutant, but not WT RAD51, was overexpressed in wild type fibroblasts (BJ cells). Conversely, overexpression of non-tagged (NT) WT RAD51 significantly improved the MMC sensitivity of RA2630 cells (Figure 1H, see also Figure S3C). These results suggest that the T131P RAD51 may act as a dominant negative allele in the patient cells, disrupting proper function of WT RAD51 during ICL repair.
To determine whether the RAD51 mutation affects the activation of the FA pathway, we examined the ubiquitination of FANCI and FANCD2 foci formation following MMC treatment of RA2630 cells. FANCI ubiquitination and FANCD2 foci formation are normal in response to MMC in RA2630 cells (Figure 1I, J), indicating the FA pathway activation is intact. However, FANCD2 foci (Figure 1J) and CHK1 phosphorylation (Figure S1K) persist in MMC-treated patient cells suggesting a defect in the completion of DNA repair.
Cells expressing T131P allele are homologous recombination proficient
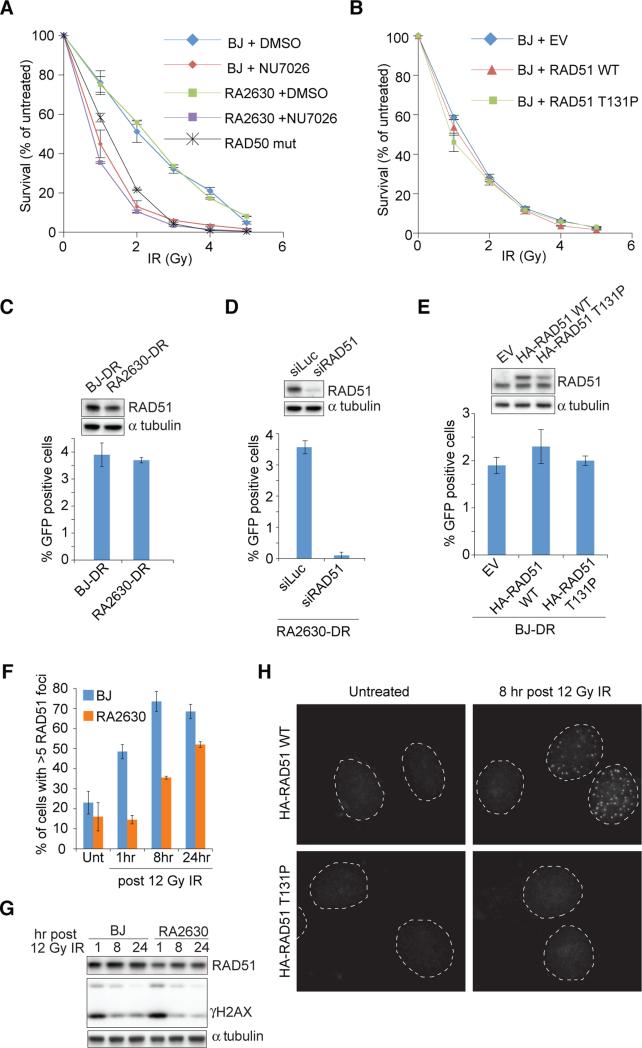
Given the importance of RAD51 in HR-dependent DSB repair, we examined the sensitivity of RA2630 to ionizing radiation (IR) that requires HR for repair. Surprisingly, the patient cells were not more sensitive to IR than the BJ fibroblast, even when an additional DSB repair pathway, non-homologous end-joining was inhibited by the DNA-PKcs inhibitor, NU7026 (Figure 2A). Cells mutated in RAD50 were used as a control (Waltes et al., 2009) and showed increased sensitivity to IR (Figure 2A). In response to camptothecin, patient cells mutated in BRCA2 (FAD1) showed higher sensitivity than RA2630 (Figure S2A), suggesting that the RAD51 patient mutation results in a defect distinct from a more severe HR defect associated with biallelic BRCA2 mutations. However, RA2630 were considerably sensitive to PARP inhibitor (Figure S2B and C). Consistent with a lack of sensitivity to IR seen in the RA2630 cells, expression of HA-RAD51 T131P mutant protein in BJ cells did not result in increased IR sensitivity (Figure 2B) despite an increased sensitivity to MMC (Figure 1G).
Figure 2.
Assessment of IR survival and homology directed repair (HDR) of cells expressing RAD51 T131P. (A) IR sensitivity of cells pretreated with NU7026 or DMSO. (B) IR sensitivity of BJ fibroblasts expressing RAD51 wild type (WT) or T131P. (C-E) GFP HDR assay in indicated cells with a stably integrated DR-GFP reporter. BJ-DR, and RA2630-DR (C), RA2630-DR cells depleted of RAD51 (D), and BJ-DR cells stably expressing HA-FLAG tagged WT or T131P RAD51 (E) were assessed. Error bars: s.d. (n=3). (F) Blinded quantification of cells with more than five RAD51 foci after IR. Error bars: s.d. (n=2). (G) RAD51 and γH2AX expression levels after IR. (H) IR-induced foci formation of HA-FLAG tagged RAD51 proteins. Dashed lines mark outline of nuclei. Also see Figure S2.
Since RA2630 cells are not sensitive to IR, we hypothesize that HR is functional in cells expressing T131P RAD51. To determine HR proficiency, we introduced the DR-GFP substrate (Pierce et al., 1999) into RA2630 fibroblast and assayed for the ability of the cell to perform accurate HR repair of two directly-repeated dysfunctional GFP genes. RA2630 cells are capable of restoring GFP expression at a level comparable to that seen in BJ cells (Figure 2C). This is in contrast to a complete abrogation of GFP expression upon RAD51 depletion (Figure 2D), and indicates that HR is proficient in RA2630 cells. Consistently, overexpressing HA-RAD51 T131P did not affect HR in BJ cells (Figure 2E). To further confirm the observation of functional HR in the patient cells, we assessed sister chromatid exchanges (SCEs) in RA2630 cells after either MMC treatment or BLM depletion, and we did not see significant changes in SCE formation (Figure S2D-F). Collectively, the results support the notion that the heterozygous T131P mutation in RAD51 does not abrogate HR. Instead, at the levels present in the patient cells, the T131P RAD51 severely affects ICL repair whilst sparing HR function of WT RAD51.
Given the unexpected finding that HR is proficient in RA2630, we characterized whether the expression of RAD51 T131P affects RAD51 foci formation. We observed a reduction in initial RAD51 foci formation in response to IR, both qualitatively and quantitatively, in RA2630 compared to BJ cells (Figure 2F; Figure S2G). However, with increased time, the foci do become more numerous and brighter in RA2630 cells. The decrease in foci formation is not due to a reduced level of RAD51 protein expression as both BJ and RA2630 showed similar expression levels of RAD51 (Figure 1B, 2G). Although we could readily observe the overexpressed HA-RAD51 WT protein forming foci, we could never detect foci formation of overexpressed HA-RAD51 T131P (Figure 2H). Moreover, high expression of HA-RAD51 T131P abrogated the ability of endogenous RAD51 to form IR-induced foci in BJ cells (Figure S2H). These results suggest that the RAD51 T131P mutant protein does not form stable filaments with DNA and interferes with proper RAD51 filament formation by the WT RAD51 proteins. However, the delay in RAD51 foci formation kinetics due to the presence of T131P mutant protein does not translate into an HR defect in repairing DSBs, presumably because even short filaments can support HR.
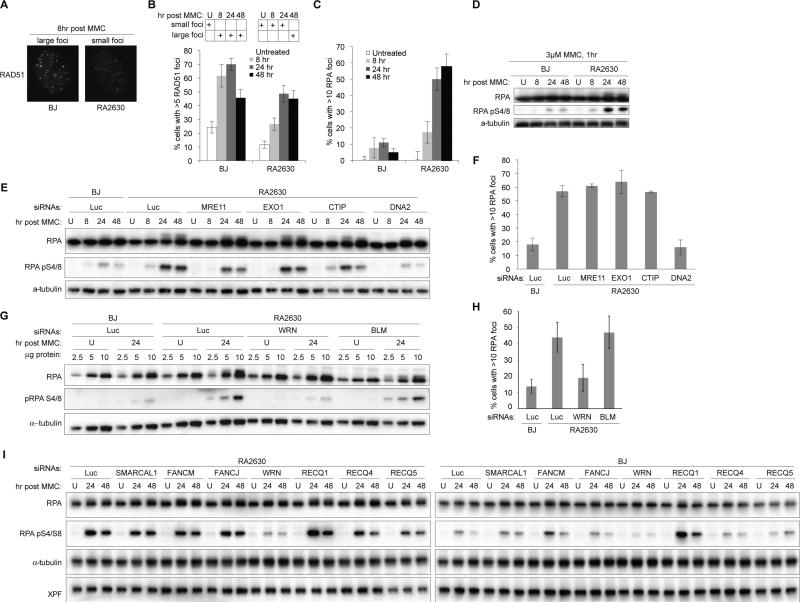
Cells expressing T131P allele show defective ICL repair with aberrant RPA activation
To address the nature of the defect in ICL repair in RA2630 cells, we followed the activation of DNA damage response markers after MMC treatment. As seen for treatment with IR, RAD51 foci were quantitatively and qualitatively diminished in RA2630 as compared to BJ cells (Figure 3A, B). Strikingly, we observed a marked increase in formation of RPA foci and RPA phosphorylation (Figure 3C, D, Figure S3A, B) in RA2630 cells compared to BJ cells, suggesting an accumulation of single stranded (ssDNA) repair intermediates in the presence of RAD51 T131P. To determine the genetic requirement for the hyperactivation of RPA, we depleted nuclease factors implicated in DNA end resection, which generates ssDNA repair intermediates. Depletion of CTIP, MRE11 and EXO1 did not diminish RPA hyperactivation in RA2630 cells (Figure 3E and F; Figure S4A-B and F). This is not due to secondary stimulation of RPA phosphorylation and accumulation at sites of DNA damage associated with their absence, since we did not observe increased RPA phosphorylation after MMC treatment when these nucleases were depleted in wild type BJ cells (Figure S4C).
Figure 3.
DNA2- and WRN-dependent RPA activation in RA2630 cells after MMC treatment. (A) Representative images of RAD51 foci formation in BJ and RA2630 cells eight hours following one-hour 3μM MMC treatment. (B-C) Blinded quantification of cells with more than five RAD51 foci (B) or ten RPA foci (C) following MMC treatment. Error bars: s.e.m. (n=3). (D) Immunoblotting analysis of RPA phosphorylation following MMC treatment. (E-F) RPA phosphorylation (E) and foci formation (F) at indicated hours following MMC treatment in cells transfected with siRNAs targeting different nucleases. (G-H) RPA phosphorylation (G) and foci formation (H) at 24 hours following MMC treatment in cells transfected with siRNAs targeting different nucleases. Error bars: s.e.m. Blinded samples from three independent experiments were counted. (I) RPA phosphorylation at indicated hours following MMC treatment in cells transfected with siRNAs targeting different helicases. Also see Figure S3, S4, and S5.
Because of strong support for the role of MRE11 in the resection of the DNA double strand DNA breaks (Petrini et al., 1995; Xiao and Weaver, 1997; reviewed in Symington, 2014) as well as nascent DNA in cells treated with hydroxyurea (Schlacher et al., 2011; Schlacher et al., 2012; Ying et al., 2012), we have attempted to use mirin, a chemical inhibitor of exonuclease activity associated with MRE11 (Dupre et al., 2008; Garner et al., 2009). Treatment with 50 μM mirin results in a marked decrease in RPA phosphorylation that is not seen at low concentrations of mirin (Figure S5A-E). However, cells treated with 50 μM mirin also exhibit a loss of FANCD2 foci that in our hands remain largely intact in cells depleted of MRE11 (Figure S5D-G), and show changes in the cell cycle profile (Figure S5H). These control experiments suggest that mirin has broader, possibly, non-specific effects on the Fanconi anemia pathway and further studies will be needed to clarify the role of MRE11 in the DNA resection seen when RAD51 T131P is expressed.
Depleting one of the resecting nucleases, DNA2 reduced RPA foci formation and phosphorylation in RA2630 to levels seen in BJ cells (Figure 3E, F; Figure S4F-I), indicating that in the absence of proper RAD51 function in cells expressing RAD51 T131P, DNA2-mediated resection results in increased ssDNA accumulation and hyperactivation of RPA at sites where replication had been blocked by ICLs. DNA2 has been shown to genetically and biochemically interact with BLM helicase (Imamura and Campbell, 2003; Nimonkar et al., 2011) but more recent data show that it also may work together with WRN protein (Sturzenegger et al., 2014). We have depleted BLM and WRN proteins from RA2630 and observed that only WRN depletion led to reduced RPA foci formation and phosphorylation in RA2630 to levels seen in BJ cells (Figure 3G-I and Figure S4 J-N). Depletion of no other candidate helicase tested led to a decrease in RPA phosphorylation (Figure 3I and S4N). Of interest is that depletion of RECQL/RECQ1, already implicated in the maintenance of genome stability, protection of replication forks and a potential breast cancer susceptibility gene (Berti et al., 2013; Cybulski et al., 2015; Sharma et al., 2007; Sun et al., 2015), led to high levels of RPA phosphorylation after MMC treatment even in the wild type BJ cells, suggesting that RECQ1 is necessary for proper cellular response to ICL-inducing agents (Figure 3I).
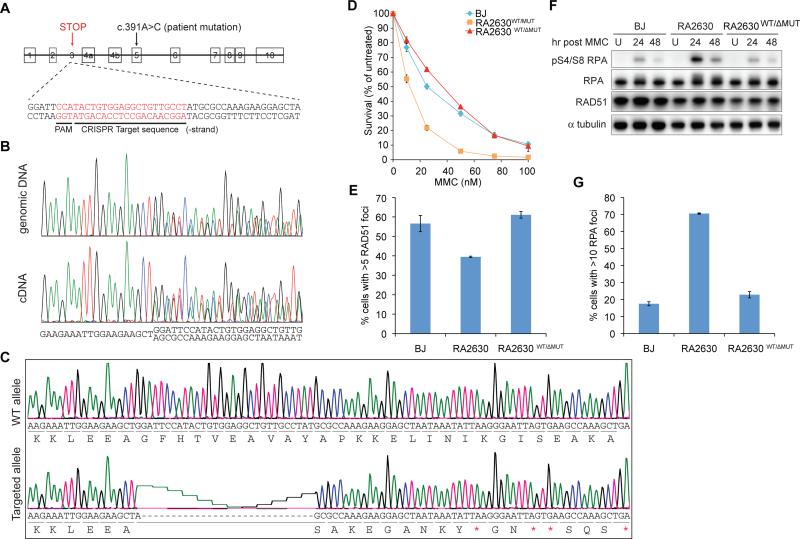
Complementation of patient cellular phenotype by CRISPR-targeted disruption of the RAD51 mutant allele
To demonstrate most directly that the mutation is indeed the disease causing mutation, we reasoned that disruption of the mutant RAD51 allele in the patient fibroblast should revert cellular abnormalities that we have ascribed to the expression of the dominant negative allele. Therefore, we disrupted the RAD51 T131P gene by using clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 nuclease mediated genome editing (reviewed in Hsu et al., 2014) in exon 3 of RAD51 (Figure 4A). This strategy resulted in a cell line with a frameshift mutation in the mutant allele, generating premature stop codons (c.133_162delinsA; p.G45Sfs*10; Figure 4B-C) predicted to make a protein with only the first 44 residues of RAD51. The second allele of RAD51 remained WT. This cell line (indicated as RA2630 WT/ΔMUT) showed rescue of the MMC sensitivity of RA2630 to the BJ wild type level (Figure 4D). Consistently, loss of the mutant allele in this cell line also both restored formation of RAD51 foci and suppressed hyperactivation of RPA to levels seen in BJ wild type cells following MMC treatment (Figure 4E-G). The ability to rescue these phenotypes demonstrates that the c.391A>C mutation is the causative change that results in a defect in ICL repair and is likely responsible for the clinical development of Fanconi anemia-like phenotype in the patient. Similar complementation of patient cellular phenotype was seen in an additional independently derived clone (Figure S3D-F).
Figure 4.
CRISPR/Cas9-directed complementation of MMC sensitivity in RA2630. Mutant allele was targeted to eliminate expression of T131P RAD51. (A) Schematic representation of RAD51 genomic locus and target location and sequence of guide RNA. Exon 4a refers to the exon 4 in NM_133487.3 and NM_001164269.1 Exon 4b refers to the exon 4 sequence in NM_002875.4 and NM_001164270.1. (B) Sequencing traces of genomic DNA and cDNA showing regions of genome editing by CRISPR-Cas9 in targeted cell line (RA2630WT/ΔMUT). (C) Sequencing traces of full length WT and truncated mutant transcripts identified by TOPO cloning of RAD51 cDNA PCR from RA2630WT/ΔMUT. Red asterisks indicate premature stop codons resulting from out of frame deletion. (D-G) Cell survival (D), RAD51 foci formation (E), RPA phosphorylation (F) and foci formation (G) in cells following MMC treatment. Error bars: s.e.m. (n=3). Also see Figure S3D-F.
RAD51 T131P displays unregulated ATPase activity
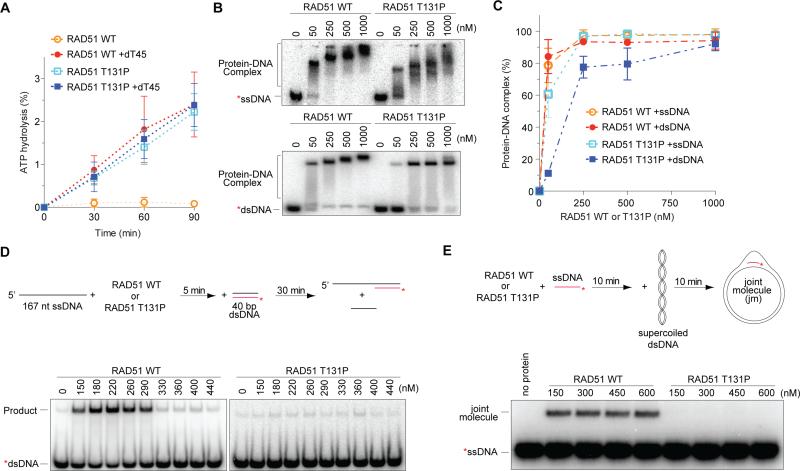
To gain a deeper understanding of how the patient mutation affects the biochemical activities of the RAD51 protein, and also to reconcile the surprising sensitivity to MMC without an apparent HR defect in the patient cells, we purified the mutant RAD51 T131P protein and examined its biochemical functions (Figure S6A). Unlike WT RAD51, which shows little basal ATPase activity that is stimulated by the addition of ssDNA (Figure 5A), the mutant RAD51 exhibits constitutive ATPase activity that is comparable to WT RAD51 but this activity is unexpectedly independent of ssDNA (Figure 5A). The ATPase activity of RAD51 T131P requires Mg2+, and, as expected, is not supported by Ca2+ (Figure S6B). Furthermore, the high basal ATPase activity of the mutant RAD51 is not due to the presence of DNA or RNA contamination in the purified protein (Figure S6C and D).
Figure 5.
In vitro activities of RAD51 WT and T131P proteins. (A) ATPase activity of purified RAD51 WT or T131P with or without ssDNA, dT45, in the presence of MgCl2. (B) EMSA showing titration of RAD51 WT or T131P binding to ssDNA (top) or dsDNA (bottom) at 2 mM ATP with 2 mM CaCl2 and 1 mM MgCl2. (C) Quantification of EMSA in panel B. Error bars: s.d. (n=3). (D) Schematic of DNA strand exchange assay (top) and results with RAD51 WT or T131P (bottom). Quantification of the data is shown in Figure S7A. (E) Joint molecule formation by RAD51 WT or T131P proteins. Reaction schematic of joint molecule formation assay is shown above the gel. Also see Figure S6.
Because ATP hydrolysis results in the dissociation of RecA/RAD51 proteins from DNA (Chi et al., 2006; Menetski and Kowalczykowski, 1985), we next examined whether the mutant protein is capable of binding to DNA. The results from electrophoretic mobility shift assays show that the mutant protein forms less stable complexes with ssDNA than WT protein (Figure 5B, top), and also has a lower affinity for dsDNA (Figure 5B, bottom, and 5C). Even though RAD51 T131P can bind to both ssDNA and dsDNA, the mutant protein completely lacks DNA strand exchange activity (Figure 5D, Figure S7A); the WT protein displays the characteristic optimum that results when protein in excess of the amount needed to bind ssDNA begins to bind to the dsDNA partner (Figure S7A). Furthermore, RAD51 T131P also lacks the capacity to form homologously paired joint molecules (Figure 5E; Figure S7C). These defects arise presumably because RAD51 T131P cannot form stable nucleoprotein filaments. Similar defects in both DNA binding (Figure S6E and F) and DNA strand exchange (Figure S7B) are also seen in reactions using different ATP analogs (AMP-PNP and ATPγS) and cofactors (ATP-Ca2+, ATP-Mg2+, and ADP-Mg2+). Collectively, our biochemical analyses identify RAD51 T131P as a unique mutant with an unregulated ATPase activity that cannot function as a DNA pairing and strand exchange protein.
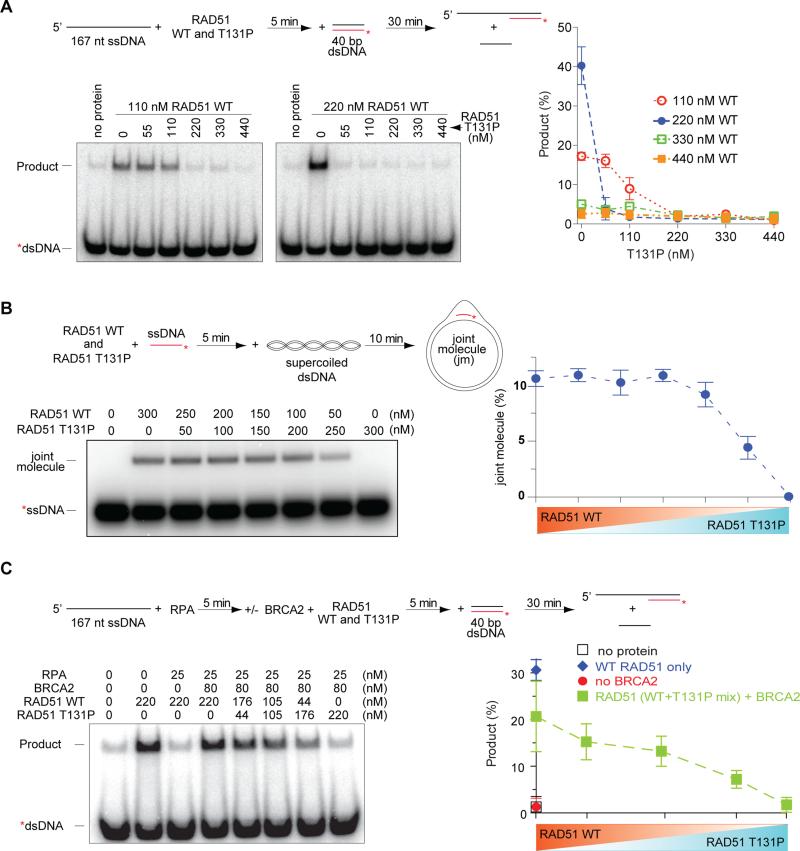
As both WT and T131P RAD51 are present in the patient cells with a heterozygous RAD51 mutation, we reconstituted the physiologically relevant scenario in cells by examining mixtures of both WT and mutant RAD51. In DNA strand exchange reactions, RAD51 T131P demonstrated dominant negative behavior, and reduced WT RAD51 function at multiple concentrations (Figure 6A). At the optimal concentration of WT RAD51, inhibition by RAD51 T131P could occur by a variety of mechanisms, but at a sub-optimal concentration of RAD51 (e.g., 110 nM, Figure S7A), RAD51 T131P may be forming a mixed filament and poisoning WT RAD51 function, just as detailed for RecA protein (Kowalczykowski and Krupp, 1989; Lauder and Kowalczykowski, 1993). Joint molecule formation is similarly affected and this simpler in vitro reaction clearly shows that pairing function is unaffected until the percentage of RAD51 T131P exceed 67% of the mixture (Figure 6B), although inhibition could be alleviated by raising the total concentration of the RAD51 proteins (Figure S7C and D), due to the higher affinity of WT RAD51 outcompeting the mutant (Figure S7C and D). The capacity of RAD51 T131P to disrupt the filaments is demonstrated by decrease of pre-formed functional RAD51WT filaments over time (Figure S7E). The defect in DNA strand exchange was not rescued by BRCA2, which stimulated WT RAD51 function when RPA was present, but did not activate RAD51 T131P; RAD51 T131P nonetheless exhibited a dominant negative effect even in the presence of BRCA2 (Figure 6C). The collective in vitro experiments show that when optimal amounts of WT RAD51 are in excess of RAD51 T131P concentrations, as observed in patient cells (Figure 1C), DNA pairing functions are largely unaffected (Figure 6A-C, Figure S7C and D), consistent with the observation that HR is proficient in the RAD51 patient cells (Figure 2).
Figure 6.
Dominant negative effects of mutant RAD51 T131P on WT RAD51 activities in vitro. (A) DNA strand exchange by WT RAD51 in the presence of increasing amounts of RAD51 T131P. (B) Joint molecule formation by RAD51 WT and T131P pre-mixed at different ratios. (C) DNA strand exchange by RAD51 WT and T131P in the presence of BRCA2. Schematic of DNA strand exchange is shown above the gel and the quantification of DNA strand exchange products is shown to the right of the gel. RAD51 WT and T131P were pre-mixed at different ratios and added to RPA-coated ssDNA together with BRCA2. Error bars: s.d. (n=3). Also see Figure S7.
Discussion
RAD51 is an essential DNA repair factor operating in mammalian cells, and its absence is lethal (Tsuzuki et al., 1996). Haploinsufficiency of RAD51 has been reported in patients who develop mirror movement disorder (Depienne et al., 2012; Gallea et al., 2013), but germline missense pathogenic variants in RAD51 have not been previously documented in humans. Here we identified a previously unidentified heterozygous mutation in RAD51 that results in Fanconi anemia-like phenotype. The resulting mutant protein, unlike other previously studied mutant RAD51 proteins in any system, exhibits constitutively high ATPase activity that prevents the protein from stably associating with DNA and catalyzing the DNA strand exchange activity required for HR. Even though T131P substitution results in a gain of biochemical function (unregulated ATPase activity) for the protein, it results in loss of RAD51 function in vivo, specifically for ICL repair.
At first glance it seems surprising that homologous recombination is proficient in the patient cells, despite T131P RAD51 having a dominant negative effect on the kinetics of WT RAD51 foci formation in vivo and the dominant negative effect of this mutant on DNA strand exchange activity by WT RAD51 in vitro. However, even though patient cells expressing the T131P allele render WT RAD51 filaments less functional, the in vitro experiments reveal that this filament still retains threshold levels of homology pairing and DNA strand invasion needed for sufficient DSB repair, as reflected by the lack of IR sensitivity, normal SCE formation, and normal levels of recombination in a reporter assay. Given the strong intrinsic functional defect observed as a result of the amino acid change on the protein level, homozygous T131P mutation would result in a complete HR defect in cells and consequent lethality. However, the patient cell line with heterozygous mutation creates a unique situation where the expression of T131P RAD51 is sufficient to disrupt function of WT RAD51 in ICL repair whilst sparing HR. This defect points to a role of RAD51 that is independent of its canonical DNA strand exchange activity. This function of RAD51 in ICL repair was not previously possible to delineate in other experimental settings.
Drawing on the earlier finding that RAD51 is present at the ICL early during the repair process (Long et al., 2011), we propose that the T131P mutant interferes with WT RAD51 function at this early step (Figure 7). However, it is possible that RAD51 could have protective functions at other steps during ICL repair. Regardless, expression of the T131P mutant results in lack of proper ICL repair without a defect in HR. This ICL repair specific function of RAD51 is likely also regulated by BRCA2 protein, and identification of specific BRCA2 mutations affecting such function may be possible.
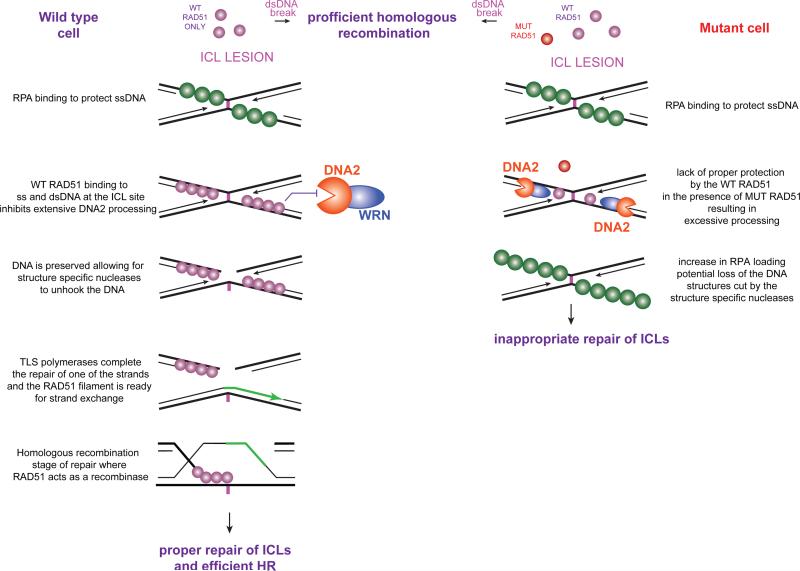
Figure 7.
Proposed model for RAD51 function at the interstrand cross-link (ICL). Although our data are consistent with this model, it is possible that RAD51 has protective functions at other steps during ICL repair. (A) When only wild type RAD51 is present (shown in purple), it is fully capable of displacing RPA (shown in green) in the vicinity of ICL leading to protection of nascent strands from DNA2 and WRN mediated resection/unwinding and setting up proper downstream processing and repair. (B) When T131P RAD51 protein (shown in red) is present in the cell in addition to WT RAD51, less (or none) of the protective RAD51 is loaded at the ICL and DNA2 and WRN activities are not sufficiently inhibited. This situation would result in increased ssDNA and loss of DNA structures necessary for the downstream repair of the ICLs. Of note, the dominant negative effect of T131P RAD51 protein is less deleterious at a double stranded (ds) DNA break, where sufficient RAD51 filament is formed to support HR in vivo. In contrast, the protective function of RAD51 is interfered with in the presence of T131P RAD51 protein.
We propose that the attenuated affinity of the mutant allele for both ssDNA and dsDNA prevents RAD51 from stably binding to DNA at the ICLs in the patient cells. Consequently, in the absence of stable binding by RAD51 in the vicinity of an ICL, the DNA becomes susceptible to the degrading/unwinding activity of DNA2 and WRN. It has been suggested that DNA2 is recruited to ICLs by the ubiquitinated FANCD2/FANCI complex (Karanja et al., 2012; Karanja et al., 2014), and we demonstrated that the activation of this complex is normal in cells expressing RAD51 T131P. It is possible that limited DNA2 and WRN-mediated processing of DNA occurs as part of normal ICL repair; however, excessive DNA2- and WRN-mediated resection or unwinding of DNA as a result of impaired RAD51 function is deleterious for the ICL repair process. This could ultimately change the DNA configuration at the ICL, channeling the repair to an aberrant pathway that would impede the accurate full repair of ICLs (Figure 7). Alternatively, the RAD51 T131P mutant, due to its dominant nature, interferes with yet unknown mechanism of nuclease inhibition by wild type RAD51. Titration of a limiting RAD51-interacting factor necessary for the nuclease inhibition would be one such example.
It has been shown that RAD51 is required for reversed fork formation following MMC treatment (Zellweger et al., 2015), and that both DNA2 and WRN act to degrade reversed replication forks following hydroxyurea treatment (Thangavel et al., 2015). It is therefore possible that such reversed fork structure formed during ICL repair is what RAD51 is protecting from DNA2 and WRN-mediated degradation. It remains to be tested if cells expressing RAD51 T131P accumulate fewer reversed fork structures during ICL repair. Observing the formation of abnormal repair intermediates in cells expressing the unique RAD51 T131P mutation will be critical in deciphering the intricate and possibly multifaceted roles of RAD51 in ensuring replication progression in response to different types of DNA damage. Previous studies have implicated RAD51 in protecting replication forks from being degraded by MRE11 and not DNA2. This mechanism was described for replication forks stalled by hydroxyurea (Schlacher et al., 2011; Schlacher et al., 2012; Ying et al., 2012) or lesions that lead to replication fork uncoupling (Hashimoto et al., 2010), and may be distinct from that operating at the ICLs. Supporting the possibility of a distinct nature of DNA protection mechanism by RAD51 at forks stalled by hydroxyurea versus ICLs, is the finding that cells expressing BRCA2 separation of function S3291A mutant, which are defective in replication fork protection following hydroxyurea treatment but capable of supporting HR, showed very mild sensitivity to MMC (Schlacher et al., 2011; Schlacher et al., 2012), in contrast to the cell line expressing T131P RAD51 mutant protein. Also, it remains to be tested whether increased nascent strand degradation in cells defective in BRCA2 or the FA pathway is also dependent on DNA2 and WRN. It is possible that the MRE11 is only the initiating nuclease, which creates substrate for further DNA2/WRN-dependent processing. Although currently not supported by our data, MRE11 could also be the initiating nuclease upon MMC treatment in the RAD51 T131P-expressing cells.
The phenotype of the subject with the mutant RAD51 T131P protein includes congenital abnormalities that are typical of patients diagnosed with Fanconi anemia. However, the absence of bone marrow failure at the age of 13 years, combined with the intermediate chromosome breakage phenotype in response to ICL-inducing agents, suggests that the subject has a Fanconi anemia-like syndrome akin to patients with mutations in RAD51C/FANCO. It is possible that bone marrow dysfunction will develop in this subject in the future. Regardless, given the remarkable sensitivity of the patient cells to a range of cross-linking agents, we designate an alias of FANCR for the RAD51 gene. This nomenclature would denote the requirement of RAD51 in the common Fanconi anemia ICL repair pathway, with the understanding that there will be clinical phenotypic differences even among different existing FA complementation groups, as is already seen with FA-O and FA-D1, and FA-N complementation groups. Such designation would aid the molecular classification of FA patients not yet assigned a complementation group and also allow for an integrated view of the ICL repair pathway.
Another point of interest is that the subject with the RAD51 T131P mutation lacks the characteristic features of the high burden of childhood AML and embryonal tumors seen in patients with biallelic mutations in BRCA2/FANCD1 or PALB2/FANCN (Howlett et al., 2002; Reid et al., 2007; Xia et al., 2007). We hypothesize that normal levels of HR protects this patient from early tumorigenesis; however, it does not preclude the possibility that the patient will be tumor-prone in the future. Indeed, monoallelic mutations in many of the RAD51-associated factors, including BRCA2, PALB2, RAD51C, and RAD51D are associated with predisposition to cancers in adulthood (reviewed in Filippini and Vega, 2013; Moynahan and Jasin, 2010; Pennington and Swisher, 2012). We hypothesize that RAD51's ability to protect DNA from inappropriate degradation in proliferating cells when endogenous damage is encountered could constitute one of the mechanisms through which chromosomal aberrations and mutations that ultimately drive tumorigenesis are prevented. The endogenous damage may not be limited to interstrand crosslink damage since T131P RAD51 mutation also rendered cells sensitive to PARP inhibitor (Fig S2B). The lack of an HR defect in the patient cells indicates that PARP inhibitor sensitivity is not solely due to HR defect as already suggested (Ying et al., 2012) and this finding has implications for understanding of the synthetically lethal relationship between cancer mutations and PARP inhibition for therapeutic treatment. The unique mutation of RAD51 that we identified, defining a separation of function from its predominant role in HR, will allow more in-depth mechanistic insight on how defective DNA repair results in chromosomal aberrations that contribute to tumorigenesis.
Experimental Procedures
Study subjects and cell lines
DNA samples and cell lines were derived from subjects enrolled in the International Fanconi anemia Registry (IFAR) after obtaining informed consent. The Institutional Review Board of The Rockefeller University, New York, NY, USA, approved these studies.
Cell culture, transfection and viral transduction
Subject's fibroblasts were transformed and/or immortalized by standard procedures. Viruses for generating RAD51 overexpressing fibroblasts and RA2630-Cas9 cells were packaged in HEK293T cells according to the manufacturer's protocol (Mirus). For siRNA transfection, fibroblasts were subjected to reverse transfection followed by forward transfection the following day using RNAiMax (Life Technologies). Sequences of siRNAs, RT-PCR primers, and mutagenesis primers used in this study are in Table S4.
Cellular assays for DNA repair and damage response
Cell survival, cell cycle and chromosomal breakage analyses following DNA damaging agent treatment were performed as described previously (Kim et al., 2013). DNA damage response and foci formation of DNA repair proteins were determined by standard immunoblotting and immunofluorescence procedures as described previously (Wang et al., 2011). For DR-GFP analysis of HR efficiency (Pierce et al., 1999), clonal fibroblasts containing single copy DR-GFP substrate were transduced with adenovirus expressing I-SceI endonuclease and analyzed for percentage GFP positive cells by flow cytometry as previously described (Smogorzewska et al., 2007). SCE levels were determined by procedures as described previously (Garner et al., 2013).
Protein purification and biochemical assays
Recombinant wildtype and T131P mutant RAD51 were purified according to protocol previously described (Hilario et al., 2009). ATPase activity of purified RAD51 was measured by the release of 32Pi from [γ-32P] ATP. The ability of RAD51 proteins to bind 32P labeled oligonucleotide substrates in vitro was determined by standard electrophoretic mobility shift assay. For DNA strand exchange assay, RAD51 proteins were incubated with ssDNA substrate and homologous dsDNA for 30 min at 37°C before deproteinized and analyzed by PAGE. For joint molecule formation assay, RAD51 proteins were first incubated with 32P-labeled JM1 ssDNA for 10 min at 37°C followed by the addition of pSE1 supercoil DNA. Reactions were deproteinized and analyzed by electrophoresis using 1% agarose.
Supplementary Material
Highlights.
Dominant mutation in RAD51 is identified in Fanconi anemia-like patient
RAD51 T131P expressing cells are ICL repair defective but HR-proficient
RAD51 T131P has unregulated ATPase activity poisoning wild type RAD51
Defective RAD51 function results in DNA2/WRN-dependent hyperactivation of RPA
Acknowledgements
We wish to thank the patient and her family, without whom this study would not be possible. We thank Steve West and John Petrini for providing us with anti-RAD51 and anti-MRE11 antibodies, respectively, Nikola Pavletich for RAD51 cDNA construct and Detlev Schindler for sharing the RAD50-mutant cell line. We are grateful to all members of the Smogorzewska laboratory for helpful discussion and critical comments on the manuscript. This work was supported by the Starr Center Consortium grant (AS and GS), Rita Allen Foundation and Burroughs Wellcome Foundation (AS), an Irma T. Hirschl Research Award (AS), Doris Duke Clinical Scientist Development Award (AS) and National Institutes of Health GM62653 (SCK), grant # 8 UL1 TR000043 from the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program. The Fanconi Anemia Registry is partially supported by the R01HL120922 (AS). ATW was partially funded by Congressionally Directed Medical Research Programs, Bone Marrow Failure Research Program Postdoctoral Fellowship Training Award (Grant log #BM120004)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: ATW, TK, BC, FPL, SCK and AS conceived the ideas and designed experiments for this study. ATW, TK, BC, FPL, AH performed the experiments. HM conducted the mass spectrometry analysis. AA and BKC performed bioinformatic analysis of the exomes. SG and CS led the sequencing team at the Broad Institute. AS is the PI and ES a study coordinator of the International Fanconi Anemia Registry (IFAR). ADA directed clinical testing in the proband at the time of IFAR enrollment, and excluded known complementation groups in the patient. ADA, together with JW enrolled the patient, and HZ and JW provided clinical information and clinical care. ATW, SCK, and AS wrote the manuscript with essential input from other authors.
Accession Numbers
cDNA coordinates were mapped against RefSeq: NM_002875.4
Supplemental information
Supplemental information includes Supplemental Experimental Procedures, seven figures, and four tables.
References
- Akkari YMN, Bateman RL, Reifsteck CA, Olson SB, Grompe M. DNA replication is required to elicit cellular responses to psoralen-induced DNA interstrand cross-links. Molecular and Cellular Biology. 2000;20:8283–8289. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter BP, Rosenberg PS, Brody LC. Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J Med Genet. 2007;44:1–9. doi: 10.1136/jmg.2006.043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach AD. Fanconi anemia and its diagnosis. Mutat Res. 2009;668:4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti M, Ray Chaudhuri A, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, Odreman F, Glatter T, Graziano S, Mendoza-Maldonado R, et al. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nature structural & molecular biology. 2013;20:347–354. doi: 10.1038/nsmb.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogliolo M, Schuster B, Stoepker C, Derkunt B, Su Y, Raams A, Trujillo JP, Minguillon J, Ramirez MJ, Pujol R, et al. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am J Hum Genet. 2013;92:800–806. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst) 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Clauson C, Scharer OD, Niedernhofer L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb Perspect Med. 2013;3:a012732. doi: 10.1101/cshperspect.a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski C, Carrot-Zhang J, Kluzniak W, Rivera B, Kashyap A, Wokolorczyk D, Giroux S, Nadaf J, Hamel N, Zhang S, et al. Germline RECQL mutations are associated with breast cancer susceptibility. Nat Genet. 2015 doi: 10.1038/ng.3284. [DOI] [PubMed] [Google Scholar]
- Depienne C, Bouteiller D, Meneret A, Billot S, Groppa S, Klebe S, Charbonnier-Beaupel F, Corvol JC, Saraiva JP, Brueggemann N, et al. RAD51 haploinsufficiency causes congenital mirror movements in humans. Am J Hum Genet. 2012;90:301–307. doi: 10.1016/j.ajhg.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML, Kopelovich L, Jasin M, Baer R, Paull TT, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nature chemical biology. 2008;4:119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini SE, Vega A. Breast cancer genes: beyond BRCA1 and BRCA2. Front Biosci (Landmark Ed) 2013;18:1358–1372. doi: 10.2741/4185. [DOI] [PubMed] [Google Scholar]
- Gallea C, Popa T, Hubsch C, Valabregue R, Brochard V, Kundu P, Schmitt B, Bardinet E, Bertasi E, Flamand-Roze C, et al. RAD51 deficiency disrupts the corticospinal lateralization of motor control. Brain. 2013;136:3333–3346. doi: 10.1093/brain/awt258. [DOI] [PubMed] [Google Scholar]
- Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489:571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Garner E, Kim Y, Lach FP, Kottemann MC, Smogorzewska A. Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced Holliday junctions. Cell Rep. 2013;5:207–215. doi: 10.1016/j.celrep.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner KM, Pletnev AA, Eastman A. Corrected structure of mirin, a small-molecule inhibitor of the Mre11-Rad50-Nbs1 complex. Nature chemical biology. 2009;5:129–130. doi: 10.1038/nchembio0309-129. author reply 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nature structural & molecular biology. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc Natl Acad Sci U S A. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hira A, Yoshida K, Sato K, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Shimamoto A, Tahara H, et al. Mutations in the Gene Encoding the E2 Conjugating Enzyme UBE2T Cause Fanconi Anemia. Am J Hum Genet. 2015;96:1001–1007. doi: 10.1016/j.ajhg.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura O, Campbell JL. The human Bloom syndrome gene suppresses the DNA replication and repair defects of yeast dna2 mutants. Proc Natl Acad Sci U S A. 2003;100:8193–8198. doi: 10.1073/pnas.1431624100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanja KK, Cox SW, Duxin JP, Stewart SA, Campbell JL. DNA2 and EXO1 in replication-coupled, homology-directed repair and in the interplay between HDR and the FA/BRCA network. Cell Cycle. 2012;11:3983–3996. doi: 10.4161/cc.22215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanja KK, Lee EH, Hendrickson EA, Campbell JL. Preventing over-resection by DNA2 helicase/nuclease suppresses repair defects in Fanconi anemia cells. Cell Cycle. 2014;13 doi: 10.4161/cc.28476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiyama K, Nakazawa Y, Pilz DT, Guo C, Shimada M, Sasaki K, Fawcett H, Wing JF, Lewin SO, Carr L, et al. Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. Am J Hum Genet. 2013;92:807–819. doi: 10.1016/j.ajhg.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Spitz GS, Veturi U, Lach FP, Auerbach AD, Smogorzewska A. Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4. Blood. 2013;121:54–63. doi: 10.1182/blood-2012-07-441212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein Douwel D, Boonen RA, Long DT, Szypowska AA, Raschle M, Walter JC, Knipscheer P. XPF-ERCC1 Acts in Unhooking DNA Interstrand Crosslinks in Cooperation with FANCD2 and FANCP/SLX4. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC, Krupp RA. Biochemical events essential to the recombination activity of Escherichia coli RecA protein. II. Co-dominant effects of RecA142 protein on wild-type RecA protein function. Journal of molecular biology. 1989;207:735–747. doi: 10.1016/0022-2836(89)90240-4. [DOI] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- Lauder SD, Kowalczykowski SC. Negative co-dominant inhibition of recA protein function. Biochemical properties of the recA1, recA13 and recA56 proteins and the effect of recA56 protein on the activities of the wild-type recA protein function in vitro. Journal of molecular biology. 1993;234:72–86. doi: 10.1006/jmbi.1993.1564. [DOI] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nature structural & molecular biology. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DT, Raschle M, Joukov V, Walter JC. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333:84–87. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetski JP, Kowalczykowski SC. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. Journal of molecular biology. 1985;181:281–295. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington KP, Swisher EM. Hereditary ovarian cancer: beyond the usual suspects. Gynecol Oncol. 2012;124:347–353. doi: 10.1016/j.ygyno.2011.12.415. [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini JH, Walsh ME, DiMare C, Chen XN, Korenberg JR, Weaver DT. Isolation and characterization of the human MRE11 homologue. Genomics. 1995;29:80–86. doi: 10.1006/geno.1995.1217. [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Räschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- Rickman KA, Lach FP, Abhyankar A, Donovan FX, Sanborn EM, Kennedy JA, Sougnez C, Gabriel SB, Elemento O, Chandrasekharappa SC, et al. Deficiency of UBE2T, the E2 Ubiquitin Ligase Necessary for FANCD2 and FANCI Ubiquitination, Causes FA-T Subtype of Fanconi Anemia. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Sawyer SL, Tian L, Kahkonen M, Schwartzentruber J, Kircher M, University of Washington Centre for Mendelian, G. Consortium FC, Majewski J, Dyment DA, Innes AM, et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer discovery. 2015;5:135–142. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengerova B, Wang AT, McHugh PJ. Orchestrating the nucleases involved in DNA interstrand cross-link (ICL) repair. Cell Cycle. 2011;10:3999–4008. doi: 10.4161/cc.10.23.18385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Stumpo DJ, Balajee AS, Bock CB, Lansdorp PM, Brosh RM, Jr., Blackshear PJ. RECQL, a member of the RecQ family of DNA helicases, suppresses chromosomal instability. Mol Cell Biol. 2007;27:1784–1794. doi: 10.1128/MCB.01620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturzenegger A, Burdova K, Kanagaraj R, Levikova M, Pinto C, Cejka P, Janscak P. DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA-end resection in human cells. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.M114.578823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. The Journal of biological chemistry. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- Sun J, Wang Y, Xia Y, Xu Y, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, et al. Mutations in RECQL Gene Are Associated with Predisposition to Breast Cancer. PLoS genetics. 2015;11:e1005228. doi: 10.1371/journal.pgen.1005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- Symington LS. End resection at double-strand breaks: mechanism and regulation. Cold Spring Harbor perspectives in biology. 2014;6 doi: 10.1101/cshperspect.a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Fukushima T, Morrison C, Albala JS, Swagemakers SM, Kanaar R, Thompson LH, Takeda S. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol Cell Biol. 2000;20:6476–6482. doi: 10.1128/mcb.20.17.6476-6482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavel S, Berti M, Levikova M, Pinto C, Gomathinayagam S, Vujanovic M, Zellweger R, Moore H, Lee EH, Hendrickson EA, et al. DNA2 drives processing and restart of reversed replication forks in human cells. The Journal of cell biology. 2015;208:545–562. doi: 10.1083/jcb.201406100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci U S A. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, Neveling K, Endt D, Kesterton I, Autore F, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42:406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- Waltes R, Kalb R, Gatei M, Kijas AW, Stumm M, Sobeck A, Wieland B, Varon R, Lerenthal Y, Lavin MF, et al. Human RAD50 deficiency in a Nijmegen breakage syndrome-like disorder. Am J Hum Genet. 2009;84:605–616. doi: 10.1016/j.ajhg.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AT, Sengerova B, Cattell E, Inagawa T, Hartley JM, Kiakos K, Burgess-Brown NA, Swift LP, Enzlin JH, Schofield CJ, et al. Human SNM1A and XPF-ERCC1 collaborate to initiate DNA interstrand cross-link repair. Genes & Development. 2011;25:1859–1870. doi: 10.1101/gad.15699211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AT, Smogorzewska A. SnapShot: Fanconi anemia and associated proteins. Cell. 2015;160:354–354. e351. doi: 10.1016/j.cell.2014.12.031. [DOI] [PubMed] [Google Scholar]
- Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Weaver DT. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic acids research. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S, Hamdy FC, Helleday T. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer research. 2012;72:2814–2821. doi: 10.1158/0008-5472.CAN-11-3417. [DOI] [PubMed] [Google Scholar]
- Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, Herrador R, Vindigni A, Lopes M. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. The Journal of cell biology. 2015;208:563–579. doi: 10.1083/jcb.201406099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Walter JC. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair (Amst) 2014 doi: 10.1016/j.dnarep.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Otto EA, Cluckey A, Airik R, Hurd TW, Chaki M, Diaz K, Lach FP, Bennett GR, Gee HY, et al. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet. 2012;44:910–915. doi: 10.1038/ng.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.