Abstract
CRISPR-Cas9 genome editing technology holds great promise for discovering therapeutic targets in cancer and other diseases. Current screening strategies target CRISPR-induced mutations to the 5’ exons of candidate genes1–5, but this approach often produces in-frame variants that retain functionality, which can obscure even strong genetic dependencies. Here we overcome this limitation by targeting CRISPR mutagenesis to exons encoding functional protein domains. This generates a higher proportion of null mutations and substantially increases the potency of negative selection. We show that the magnitude of negative selection reports the functional importance of individual protein domains of interest. A screen of 192 chromatin regulatory domains in murine acute myeloid leukemia cells identifies six known drug targets and 19 additional dependencies. A broader application of this approach may allow comprehensive identification of protein domains that sustain cancer cells and are suitable for drug targeting.
The RNA-guided endonuclease Cas9, a component of the type II CRISPR (clustered regularly interspaced short palindromic repeats) system of bacterial host defense, is a powerful tool for genome editing6. Ectopic expression of Cas9 and a single guide RNA (sgRNA) is sufficient to direct the formation of a DNA double-strand break (DSB) at a specific region of interest7–9. In the absence of a homology-directed repair DNA template, these DSBs are repaired in an error-prone manner via the non-homologous end joining pathway to generate an assortment of short deletion and insertion mutations (indels) in the vicinity of the sgRNA recognition site7, 8. This approach has been widely used to generate gene-specific knockouts in a variety of biological systems6. Recent studies have demonstrated the use of CRISPR mutagenesis for genetic screens in mammalian cell culture, which have relied on sgRNA libraries that target constitutive 5’ coding exons to achieve gene inactivation1–4. The capabilities of CRISPR-based genetic screens are particularly evident in the setting of positive selection, such as identifying mutations that confer drug resistance1–4. In negative selection screens, it has been shown that sgRNA hits are statistically enriched for essential gene classes (ribosomal, RNA processing, and DNA replication factors); however the overall accuracy of CRISPR for annotating genetic dependencies is currently unclear1, 2.
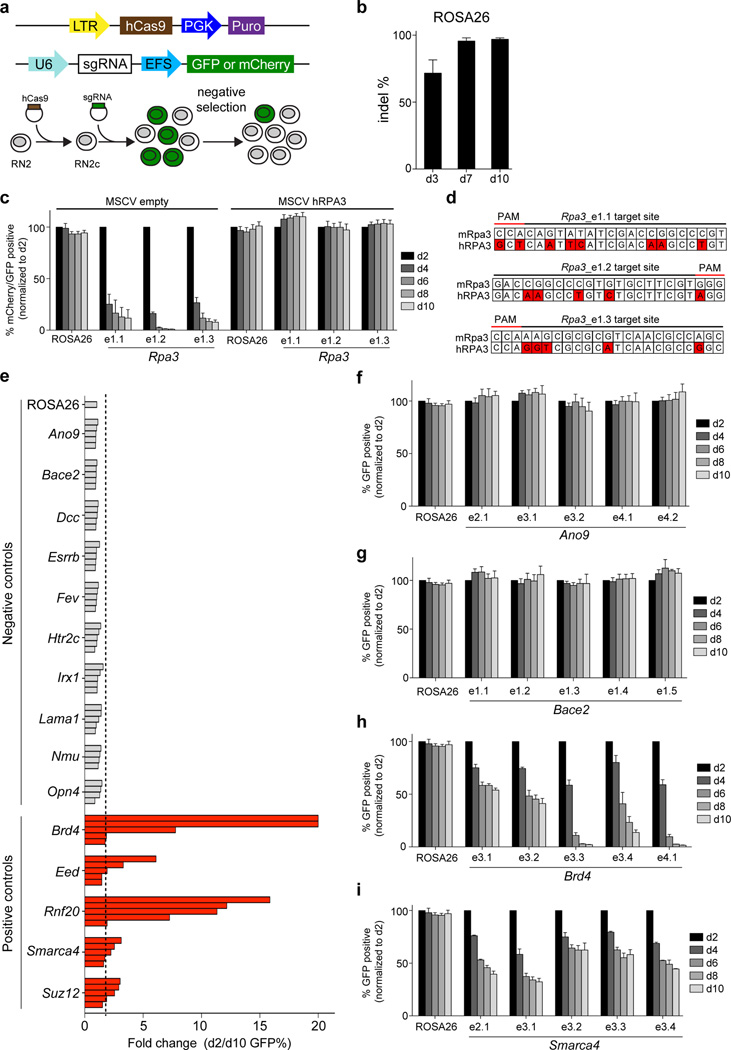
Here we investigated the performance of CRISPR indel mutagenesis to identify essential genes in cancer cells. We employed a murine MLL-AF9/NrasG12D acute myeloid leukemia cell line (RN2), which has been used extensively to identify genetic dependencies and therapeutic targets by RNA interference (RNAi)10, 11. We derived a clonal Cas9+ line (RN2c), which is diploid and remains genomically stable during passaging (Fig. 1a and data not shown). Lentiviral transduction of RN2c cells with a vector expressing GFP and an sgRNA targeting the ROSA26 locus resulted in a high efficiency of indel mutagenesis near the predicted cut site, reaching >95% editing efficiency by day 7 post-infection (Fig. 1b). Next, we designed three sgRNAs targeting the first exon of Rpa3, which encodes a 17 kD protein required for DNA replication12. Unlike the effects of targeting ROSA26, we found that cells expressing Rpa3 sgRNAs were rapidly outcompeted by non-transduced cells over 8 days in culture, as shown by flow cytometry-based tracking of GFP expression (Fig. 1c). These effects were rescued by the presence of a human RPA3 cDNA that contains several mismatches with mouse Rpa3 sgRNAs, indicating that negative selection induced by CRISPR can be attributed to mutational effects at a single essential gene (Fig. 1c, d).
Figure 1. Negative selection CRISPR experiments in murine MLL-AF9/NrasG12D acute myeloid leukemia cells.
(a) Experimental strategy. (top) Vectors used to derive clonal MLL-AF9/NrasG12D leukemia RN2c cells that express a human codon-optimized Cas9 (hCas9) and vectors used for sgRNA transduction. GFP or mCherry reporters were used where indicated to track sgRNA negative selection. LTR: long terminal repeat promoter, PGK: phosphoglycerate kinase 1 promoter, Puro: puromycin resistance gene, U6: a Pol III-driven promoter, sgRNA: chimeric single guide RNA, EFS: EF1α promoter, GFP: green fluorescent protein. (b) Analysis of CRISPR editing efficiency at the ROSA26 locus in RN2c cells. Illumina sequencing was used to quantify PCR-amplified genomic regions corresponding to the ROSA26 sgRNA cut site. (c) Negative selection competition assay that plots the percentage of GFP+/mCherry+ cells over time following transduction of RN2c with the indicated sgRNAs. Experiments were performed in RN2c cells transduced with either an empty murine stem cell virus (MSCV) vector or MSCV expressing human RPA3, which are linked with a GFP reporter. The mCherry/GFP double positive percentage is normalized to the day 2 measurement. e1 labeling of sgRNAs refers to targeting of exon 1. n = 3. (d) Comparison of mouse Rpa3 and human RPA3 sequences at the indicated sgRNA recognition sites. Location of protospacer adjacent motif (PAM) is indicated. Red color indicates mismatches. (e) Summary of negative selection experiments with sgRNAs targeting the indicated genes. Negative selection is plotted as the fold change of GFP positivity (d2/d10) during 8 days in culture. Each bar represents an independent sgRNA targeting a 5’ exon of the indicated gene. The dashed line indicates a two-fold change. The fold change for two Brd4 sgRNAs was >50, but the axis was limited to a 20-fold maximum for visualization purposes. The data shown are the mean value of 3 independent replicates. (f–i) Negative selection timecourse experiments, as described in (c), except a GFP reporter was used exclusively. n=3. All error bars in this figure represent SEM.
To further evaluate the performance of CRISPR mutagenesis as a negative selection screening strategy, we targeted ten additional negative control genes, chosen based on having undetectable expression in RN213. We also targeted five essential genes encoding chromatin regulators (Brd4, Smarca4, Eed, Suz12, and Rnf20) that were previously identified using shRNA-based knockdown10, 13–15. For each gene we designed 4–5 sgRNAs that target constitutive 5’ coding exons, a strategy used in previous CRISPR screens1–4. All 49 sgRNAs targeting non-expressed genes failed to undergo significant negative selection, suggesting a low frequency of false-positive phenotypes conferred by off-target DNA cleavage (Fig. 1e–g). By contrast, a large fraction of the positive control sgRNAs led to depletion of GFP-positivity, with a subset exhibiting robust depletion that exceeded 10-fold changes (Fig. 1e, h, i).
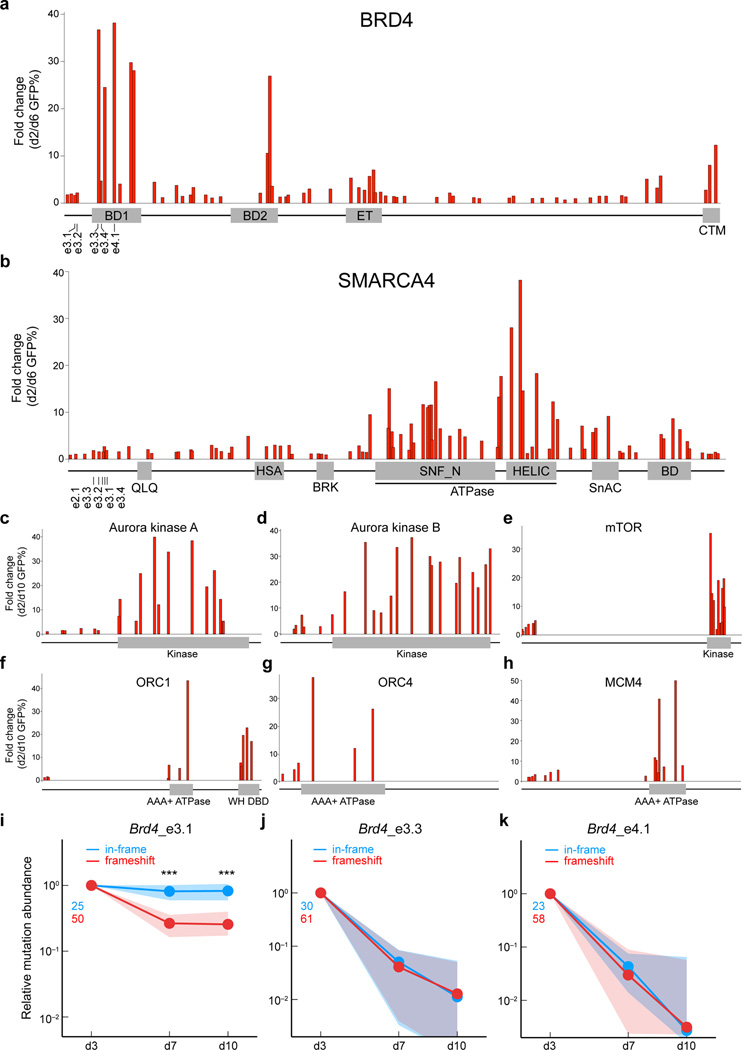
In the experiments described above, we observed dramatic variability in the performance of individual sgRNAs targeting the same gene. For example, two of the Brd4 sgRNAs became depleted >20 fold while two were only depleted ~2-fold over 8 days in culture. Notably, Brd4 sgRNAs causing severe phenotypes targeted sequences that encode bromodomain 1 (BD1), while the sgRNAs causing weaker phenotypes targeted regions that lie outside of BD1 (Fig. 2a). Prior studies have shown that the bromodomains of BRD4 are required for leukemia cell viability, as evidenced by the anti-leukemia activity of small-molecule inhibitors of BRD4 bromodomains10, 16, 17. This prompted us to evaluate whether robust negative selection is generally correlated with the targeting of functionally important protein domains. Using 64 sgRNAs in total, we targeted every exon of Brd4 to evaluate the relative severity of negative selection (Fig. 2a). All of the sgRNAs that achieved >10-fold depletion after 4 days were found to target exons encoding BD1, BD2, or the C-terminal motif (CTM), which are domains of BRD4 involved in transcriptional regulation18. At later timepoints, we also noted that sgRNAs targeting the BRD4 extra-terminal (ET) domain became depleted more than 10-fold (Supplementary Fig. 1). By contrast, many of the sgRNAs targeting regions outside of these domains exhibited minimal phenotypes (Fig. 2a and Supplementary Fig. 1). To further corroborate this relationship, we surveyed all of the exons of Smarca4 and found that sgRNAs targeting the ATPase domain generally produced stronger negative selection phenotypes than sgRNAs targeting other regions (Fig. 2b). This finding is consistent with our prior demonstration that the catalytic function of SMARCA4 is essential in leukemia13. In addition, CRISPR targeting of the catalytic domains of Aurora kinase A and B, the kinase domain of mTOR, the AAA+ ATPase and winged helix DNA binding domains of ORC1, the AAA+ ATPase domain of ORC4, and the AAA+ ATPase domain of MCM4 all led to stronger phenotypes than targeting of 5’ coding exons (Fig. 2c–h).
Figure 2. CRISPR mutagenesis of functional protein domains leads to a higher proportion of null mutations and an enhanced severity of negative selection.
(a) Systematic evaluation of 64 Brd4 sgRNAs in negative selection experiments, targeting each Brd4 exon. The location of each sgRNA relative to the Brd4 protein is indicated along the x-axis. Location of Brd4 sgRNAs used in Figure 1 is indicated. BD1: bromodomain 1, BD2: bromodomain 2, ET: extra-terminal domain, CTM: C-terminal motif. Plotted is the average of three biological replicates. (b) Systematic evaluation of 88 Smarca4 sgRNAs in negative selection experiments, targeting each Smarca4 exon. The relative location of each sgRNA relative to the Smarca4 protein is indicated along the x-axis. Location of Smarca4 sgRNAs used in Figure 1 is indicated. Indicated domains were obtained from the NCBI database. SNF_N and HELIC constitute the ATPase domain. BD: bromodomain. Plotted is the average of three biological replicates. (c–h) Negative selection experiments evaluating sgRNAs targeting 5’ coding exons and domain locations for the indicated proteins. In a-h, the proteins are not drawn to the same scale. WH DBD: winged helix DNA binding domain. Plotted is the average of three biological replicates. (i–k) Deep sequencing analysis of mutation abundance following CRISPR-targeting of different Brd4 regions. This analysis was performed on PCR-amplified genomic regions corresponding to the sgRNA cut site at the indicated timepoints. Indel mutations were categorized into two groups: in-frame (3n) or frameshift (3n+1, 3n+2). Nonsense mutations induced by CRISPR mutagenesis were included in the frameshift category, however such mutations were rare. Green and red numbers indicate the number of distinct in-frame and frameshift mutants that were tracked, respectively. Dots of the same color indicate the median normalized abundance at the indicated time point for all mutations within each group; shaded regions indicate the interquartile range of normalized abundance values. Significant differences between the enrichment values of the in-frame and frameshift mutations were assessed using a Mann-Whitney-Wilcoxon test; ** indicates p < 0.01, and *** indicates p < 0.005. The normalized abundance of each tracked mutation was defined as the ratio of the number of observed mutant sequences divided by the number of wild-type sequences, normalized by the value of this same quantity at day 3.
Using SURVEYOR assays and deep-sequencing analysis, we found that the differences in phenotypic severity between domain-targeting sgRNAs and 5’ exon Brd4 sgRNAs could not be explained by variation in the overall efficiency of mutagenesis (Supplementary Fig. 2). These phenotypic differences were also not attributed to off-target cutting at exons encoding homologous protein domains (Supplementary Fig. 3). Instead, CRISPR-induced mutations within domains became depleted more rapidly from the cell population than mutations introduced outside of the domain (Supplementary Fig. 2). These findings raised the possibility that CRISPR targeting of functionally important domains resulted in a higher proportion of null mutations than targeting outside of critical domains.
We evaluated this hypothesis by deep sequencing of the CRISPR-mutagenized Brd4 exons (PCR-amplified from genomic DNA) during the negative selection time course, which is a means to track how individual mutations impair cellular fitness19. For these experiments, we directly compared BD1 mutations (introduced by sgRNAs e3.3 and e4.1) with mutations introduced outside of BD1 by sgRNA e3.1 (Fig. 2i–k). As expected, roughly two thirds of the mutations generated by the three different sgRNAs resulted in a frameshift, which underwent negative selection when introduced at any of the three Brd4 locations (Fig. 2i–k). By contrast, we found that the in-frame mutations only underwent negative selection when generated within the BD1 region, and not when generated outside of BD1 (Fig. 2i–k). Indeed, the in-frame and frameshift mutations generated within BD1 displayed indistinguishable kinetics of negative selection, suggesting that both mutational classes are genetic nulls (Fig. 2j, k). These findings imply that in-frame mutations occurring within BD1 may compromise the acetyl-lysine recognition function of this domain to inactivate BRD4, whereas in-frame mutations occurring outside of BD1 retain full BRD4 functionality. A similar deep sequencing analysis of Smarca4 also suggests that functional impairment of in-frame mutations contributes to the increased severity of negative selection when targeting the ATPase domain (Supplementary Fig. 4).
In a diploid cell line it is expected that random pairing of in-frame and frameshift CRISPR mutations will generate cell populations of varying genotypes. If in-frame CRISPR mutations are functional and occur at the expected frequency of around 33% (Supplementary Fig. 5), then ~56% of cells in the population will possess at least one functional allele of the essential gene, thereby limiting the overall severity of observed phenotypes (Supplementary Fig. 6). By contrast, if in-frame CRISPR mutations are non-functional, it is anticipated that nearly all cells in the population are capable of experiencing a robust cellular phenotype. Random allele pairing with in-frame variants also explains why frameshift mutations undergo stronger negative selection when occurring within a domain than outside of a domain (see Supplementary Discussion). Finally, we also noted that the deep sequencing-based measurement of allele functionality also provided a means of excluding off-target effects when validating hits obtained from CRISPR-based screens (see Supplementary Discussion).
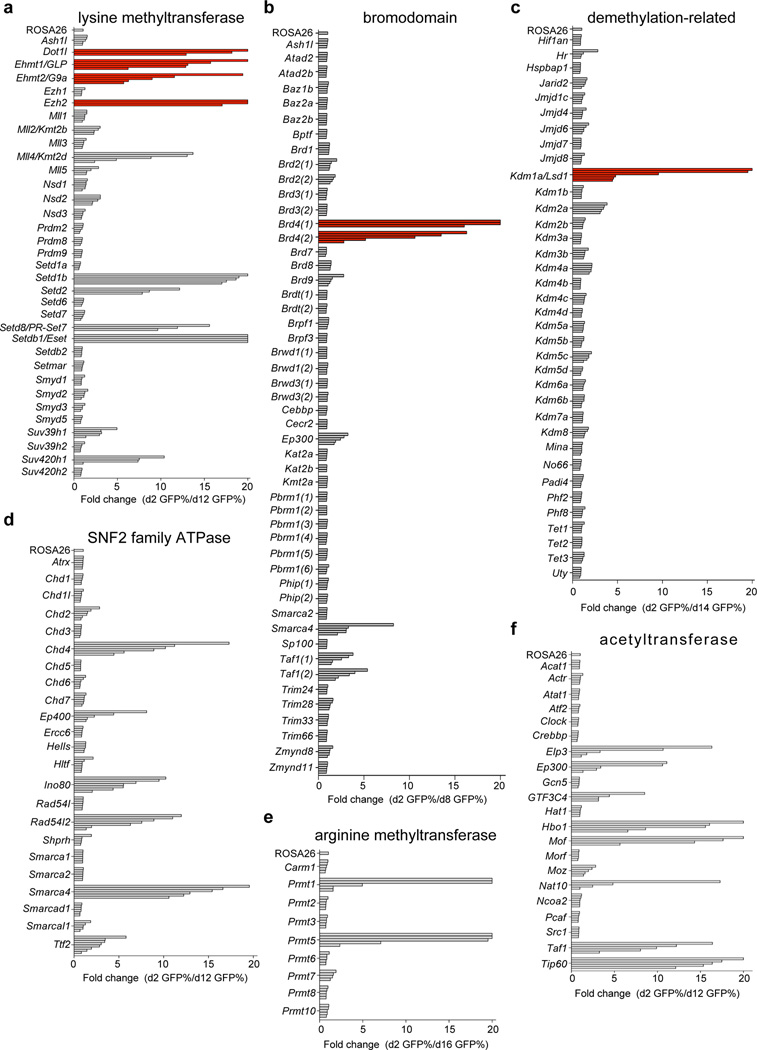
The major implication of the experiments described above is that the severity of negative selection in CRISPR screens reflects, at least in part, the functional importance of the protein region being targeted. Therefore, a CRISPR screening strategy that exclusively targets exons encoding protein domains could be used to nominate individual domains as cancer dependencies and potentially as drug targets. To evaluate this hypothesis, we designed an sgRNA library that targeted 192 domains involved in chromatin regulation. These domains included methyltransferase, demethylase, acetyltransferase, deacetylase, ATPase, and bromodomain regions. The lysine methyltransferase activities of DOT1L, EZH2, EHMT1/2, the bromodomains of BRD4, and the lysine demethylase activity of KDM1A have all been previously validated as therapeutic targets in MLL-AF9 leukemia using small-molecule inhibitors10, 16, 17, 20–24. Hence, we sought to benchmark the capabilities of a domain-focused CRISPR screen in identifying these known drug targets within a systematic survey of chromatin regulator dependencies.
933 sgRNAs targeting these different domains were evaluated individually using negative selection/GFP-depletion assays in RN2c cells. Although CRISPR targeting of many domains failed to cause a phenotype, we identified 25 domains in which targeting with multiple independent sgRNAs led to negative selection effects that exceeded 5-fold. (Fig. 3 and Supplementary Fig. 7). Many of these proteins have-to our knowledge- not been described previously as leukemia dependencies. Notably, sgRNAs targeting Dot1l, Ehmt1, Ehmt2, Ezh2, Brd4, and Kdm1a led to a consistent and pronounced negative selection, with each gene being among the top dependencies identified in the screen (Fig. 3a–c). This suggests that domain-focused CRISPR screens are capable of revealing essential domains that are relevant for pharmacological inhibition. We also noted a high level of internal consistency among independent sgRNAs targeting the same domain, thus providing further confidence to hit identification. To explore the scalability of this approach, we pooled the lysine methyltransferase sgRNA sublibrary and used deep-sequencing to track negative selection in a multiplexed format. The results of the pooled screen were highly consistent with the results obtained by evaluating sgRNAs individually using GFP reporters (Supplementary Fig. 8). These CRISPR screening procedures were also readily implemented in other Cas9+ cell lines to allow identification of cell line-specific dependencies (Supplementary Fig. 9). Collectively, these findings validate domain-focused CRISPR screening as a robust and scalable tool for cancer drug target discovery.
Figure 3. A chromatin regulatory domain-focused CRISPR screen in MLL-AF9 leukemia validates known drug targets and reveals additional dependencies.
(a–f) Summary of negative selection experiments with sgRNAs targeting the indicated domains plotted as fold-change in GFP-positivity. Each bar represents the mean value of three independent biological replicates for an independent sgRNA targeting the indicated domain. Red coloring indicates domains for which prior pharmacological validation of the dependency has been performed. A 20-fold cutoff was applied for visualization purposes. Different timepoints of GFP measurements were chosen based on the severity of the strongest hit in the screen.
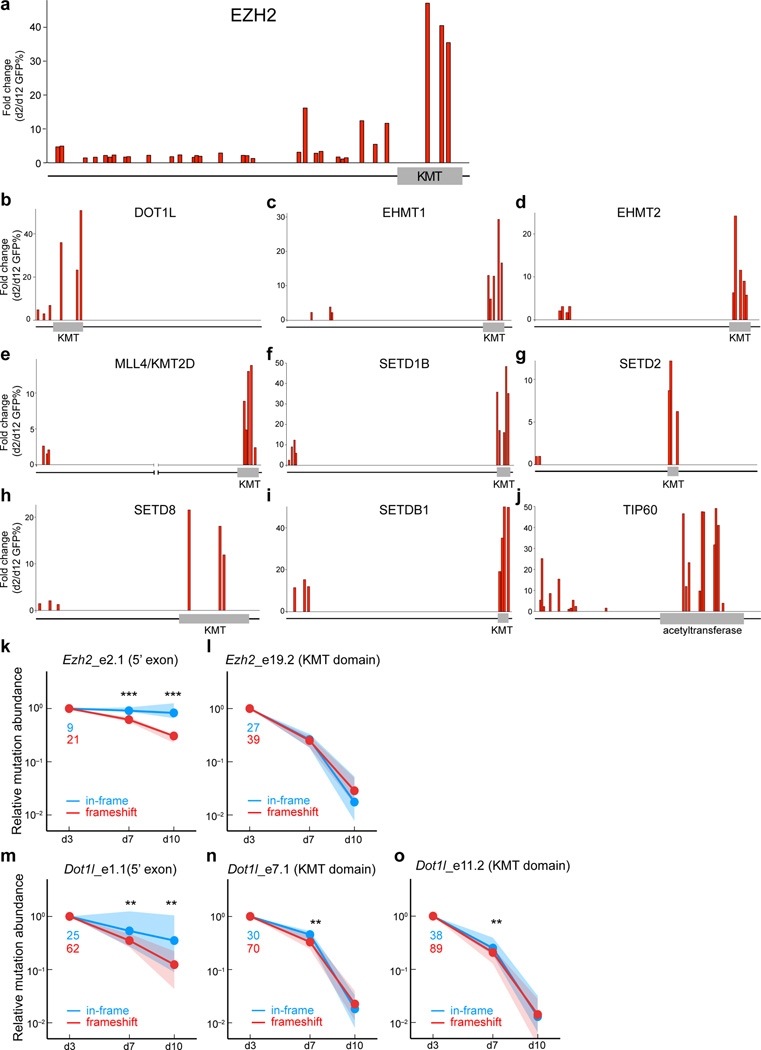
A series of validation experiments performed on hits identified in the chromatin regulator screen further support a domain-focused CRISPR strategy (Fig. 4). For ten of the identified dependencies, we evaluated additional to sgRNAs to show that targeting of enzymatic domains consistently outperformed targeting of 5’ coding exons in negative selection experiments (Fig. 4a–j). Hence, a screening strategy that exclusively targets 5’ coding exons would have failed to identify most of these regulators as cancer cell dependencies. Finally, deep sequencing analyses of Ezh2 and Dot1l mutation abundance was performed at both methyltransferase domain and 5’ exon locations. This analysis confirmed the contribution of functionally-defective in-frame mutations at domain regions as the underlying basis for the increased severity of negative selection (Fig. 4k–o and Supplementary Fig. 10).
Figure 4. CRISPR targeting of enzymatic domains consistently outperforms targeting of 5’ coding exons in negative selection experiments.
(a) Evaluation of 32 Ezh2 sgRNAs in negative selection experiments, targeting each Ezh2 exon. The relative location of each sgRNA relative to the Ezh2 protein is indicated along the x-axis. (b–j) Evaluation of 5’ coding exon and enzymatic domain-focused sgRNAs in negative selection experiments. The relative location of each sgRNA relative to the protein is indicated along the x-axis. Owing to the large size of MLL4/KMT2D, we have cropped out amino acids 2000 to 15,000 for visualization purposes. For a–j, plotted is the average fold change in GFP% of three biological replicates. KMT: lysine methyltransferase domain. The proteins are not drawn to the same scale. (k–o) Deep sequencing analysis of mutation abundance following CRISPR-targeting of different Ezh2 and Dot1l regions. This analysis was performed on PCR-amplified genomic regions corresponding to the sgRNA cut site at the indicated timepoints. Indel mutations were categorized into two groups: in-frame (3n) or frameshift (3n+1, 3n+2). Nonsense mutations induced by CRISPR mutagenesis were also included in the frameshift category, however such mutations were rare. Green and red numbers indicate the number of in-frame and frameshift mutants that were tracked, respectively. Dots of the same color indicate the median normalized abundance at the indicated time point for all mutations within each group; shaded regions indicate the interquartile range of normalized abundance values. Significant differences between the enrichment values of the in-frame and frameshift mutations were assessed using a Mann-Whitney-Wilcoxon test; ** indicates p < 0.01, and *** indicates p < 0.005. The normalized abundance of each tracked mutation was defined as the ratio of the number of observed mutant sequences divided by the number of wild-type sequences, normalized by the value of this same quantity at day 3.
The overall performance of CRISPR for genetic screening is influenced by several experimental parameters influencing the overall efficiency of mutagenesis, including the level of Cas9 expression25 and sgRNA sequence features2, 5. Here we have shown that the performance of CRISPR in generating null mutations is substantially improved when Cas9 cutting is directed to sequences that encode functionally important protein domains. This leads to a potentially useful approach to identify cancer dependencies suitable for pharmacological inhibition: sgRNA libraries should be designed to target exons that encode ‘druggable’ protein domains. In such screens the severity of negative selection phenotypes would directly indicate the functional importance of the domain being targeted. We expect this approach to be critical when targeting genes that encode large multi-domain proteins, but less important for small proteins, like Rpa3. It would also be expected that domain-focused CRISPR screens might be most effective for probing discrete enzymatic active sites, but potentially less amenable for probing extended protein-protein interaction surfaces. Nonetheless, in this study we have targeted a diverse collection of protein domains to highlight the general utility of domain-focused CRISPR screening as a tool for drug target discovery.
Online Methods
Plasmid construction and sgRNA design
The constitutive Cas9 expression construct was derived by subcloning the 5’ 3×FLAG tagged human-codon optimized Cas9 cDNA from Streptococcus pyogenes (addgene: # 49535) into the MSCV-PGK-Puro vector (Clontech: # 634401). The U6-sgRNA-EFS-GFP and the U6-sgRNA-EFS-mCherry vectors were derived from the lentiCRISPR plasmid (addgene: # 49535) by removing the hCas9 cDNA and replacing the Puro cassette with GFP or mCherry. The wild-type RPA3 was PCR cloned directly from human cDNA into the MSCV-IRES-GFP (MigR1) vector. All cloning procedures were performed using the In-Fusion cloning system (Clontech: #638909). sgRNAs were cloned by annealing two DNA oligos and ligating into a BsmB1-digested U6-sgRNA-EFS-GFP/mCherry vectors, as described26. To improve U6 promoter efficiency, we added an extra 5’ G nucleotide to all of the sgRNAs that did not start with a 5’ G.
All sgRNAs in this study were designed using http://crispr.mit.edu/25. The majority of sgRNAs used in this study had a quality score above 70 to minimize off-target effects. In Figure 1, sgRNAs were designed targeting 5’ constitutive coding exons of each target gene. For the chromatin regulatory domain-focused CRISPR screen, sgRNAs were designed to target the catalytic domain or bromodomain of each protein based on the NCBI database annotation.
All sgRNA sequences used in this study are provided in a Supplementary Table 1.
Cell culture, virus production, and sgRNA competition assays to measure negative selection
All of the cell lines used in this study were tested for mycoplasma and were negative. RN2c cells were derived by retroviral transduction of a murine MLL-AF9/NrasG12D acute myeloid leukemia cell line (RN2)11 with MSCV-hCas9-PGK-Puro, followed by puromycin selection and serial dilution to derive single cell-derived clones. Clones were screened by anti-flag Western blotting for high levels of stable Cas9 expression. Multiple independent clones displayed a similar CRISPR editing efficiency as RN2c. RN2c were cultured in RPMI1640 supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin. 38B9 cells were cultured in RPMI1640 supplemented with 10% FBS and 0.055 mM 2-mercaptoethanol. NIH3T3 cells were cultured in DMEM supplemented with 10% fetal calf serum and penicillin/streptomycin. Ecotropic Plat-E cells and HEK293T cells were cultured in DMEM supplemented with 10% FBS and penicillin/streptomycin. Plat-E cells were used for retroviral delivery of hCas9 or RPA3 cDNA expression vectors, following standard procedures27.
sgRNA competition assays were performed using the U6-sgRNA-EFS-GFP or the U6-sgRNA-EFS-mCherry plasmids, where indicated. These plasmids were used to generate lentivirus by transfecting HEK293T cells with sgRNA:pVSVg:psPAX2 plasmids in a 4:2:3 ratio using PEI reagent (Polysciences : #23966). Viral supernatants were collected between the 36 and 72 hour timepoints following transfection. All transfections and viral collections were performed in 96 well plates to allow the evaluation of large numbers of sgRNAs systematically in a one-by-one manner. For sgRNA/GFP competition assays, flow cytometry analysis was performed on 96 well plates of cells using a Guava Easycyte HT instrument (Millipore). Gating was performed on live cells using forward and side scatter, prior to measuring of GFP positivity.
For the Rpa3 sgRNA/cDNA rescue experiment, RN2c were first transduced with empty or RPA3 MigR1, followed by transduction with Rpa3 sgRNAs expressed using the U6-sgRNA-EFS-mCherry vector. Gating was performed on GFP+/mCherry+ cells to evaluate rescue.
SURVEYOR assays
Genomic DNA (gDNA) was isolated at indicated timepoints post-infection using QiAamp DNA mini kit (Qiagen #51304) following the manufacturer’s instructions. To amplify mutagenized DNA for determination of CRISPR editing efficiency, 100 ng of gDNA was PCR-amplified a using primer set specific for the sgRNA-targeted region with 2× Phusion Master Mix (Thermo Scientific #F-548) following the manufacture’s instructions. All of the primers were optimized to ensure their selectivity for a single genomic region. The PCR products were subjected to SURVEYOR assay (Transgenomic #706020) following the manufacturer’s protocol. To calculate the cleavage efficiency, we quantified the intensity of each DNA band using the ImageJ software http://imagej.nih.gov/ij/ resolved by agarose gel electrophoresis, stained with ethidium bromide. SURVEYOR assay-based calculation of indel frequency was performed using a binomial probability distribution of random duplex formation, as described previously26.
All PCR primers for SURVEYOR analysis are provided in Supplementary Table 1.
MiSeq library construction to evaluate mutation abundance
To quantify the abundance of individual indel mutations induced by CRISPR, we prepared gDNA as above and PCR-amplified a 100–200 bp amplicon centered on the sgRNA recognition region. 100 ng gDNA was amplified for 20 cycles with 2× Phusion Master Mix. The PCR product was end repaired with T4 DNA polymerase (NEB), DNA polymerase I (NEB), and T4 polynucleotide kinase (NEB). An A overhang was added to the end-repaired DNA using Klenow DNA Pol Exo- (NEB). The DNA fragment was then ligated with diversity-increased barcoded Illumina adaptors followed by 5 pre-capture PCR cycles. Bar-coded libraries were pooled at equal molar ratio and subjected to massively parallel sequencing using a Mi-Seq instrument (illumina) using paired-end 150 bp sequencing (MiSeq Reagent Kit v2; Illumina MS-102-2002). The deep sequencing data can be obtained through the NCBI Sequence Read Archive (SRA) accession SRP057117.
All primer sequences are listed in Supplementary excel file.
MiSeq analysis of abundance of indel mutations induced using CRISPR
Custom Python scripts were used to reconstruct mutant allele sequences from paired end reads, to analyze these sequences. The scripts for performing these computation and generation of figures are available at https://github.com/jbkinney/14_crispr.
Paired end reads were stitched together to form observed sequences as follows. Customized barcodes were used to separate different samples. Next, the 5’-most 15 bp of each primer sequence was used to identify which genomic region the paired-end read came from. Forward and reverse reads that overlapped by at least 15 bp were then stitched together and classified as “observed sequences.”
Observed sequences were aligned with the corresponding wild-type sequence starting from both ends and moving inward. Forward and reverse breakpoints were called as the positions at which the wild-type sequence and observed sequence became substantially different (defined as 2 consecutive nucleotide mismatches). “Deletions” were defined as the region of wild-type sequence between these two breakpoints. “Insertions” were defined as the region of observed sequence between these breakpoints.
After alignment with the wild-type sequence, each observed sequence was classified as follows. If the sequence exactly matched the endogenous locus, it was classified as “wild-type”. If the sequence contained an insertion and/or deletion mutation that altered the exon reading frame or generated a stop codon, it was classified as a “frameshift” mutant. If the sequence contained an insertion and/or deletion that preserved the reading frame and did not generate a stop codon, it was classified as an “in-frame” mutant. Observed sequences with insertions and/or deletions that disrupted an exon boundary were discarded.
To ensure sufficient counts, only sequences occurring at least 100 times in each d3 sample were tracked. To remove common PCR-induced mutations from the analysis, observed sequences that occurred more than 10 times in the corresponding control sample (ROSA26 sgRNA) were discarded. At each time point t, the enrichment ratio for a sequence s at time point t was defined as
where n(s,t) is the number of observations of sequence s at time point t, wt denotes the wild-type sequence, and d3 is the day 3 time point. Solid colored dots in the figures (Figure 2i–k, Figure 4k–o, and Supplementary Figure 4) show the median enrichment ratio for each class of mutations at each time point. Shaded region indicates the corresponding interquartile range of these enrichment ratios.
The efficiency of CRISPR mutagenesis in each sample was estimated from the following measured quantities:
M: the fraction of observed sequences containing mutations
I: the fraction of cells that were infected and observed to express sgRNA
m: the probability of mutation due to PCR artifacts.
We estimated the efficiency E of CRISPR mutagenesis from these quantities using the relationship
The rationale for this relationship is as follows. If we obtain N observed sequences in a given sample. MN of these observed sequences will have mutations. Of these mutant sequences, NIE will have come from CRISPR-mutagenized alleles inside infected, NI(1 − E)m will come from non-mutated alleles within infected cells and contain PCR-generated mutations, and N(1 − I)m will come from wild-type alleles within non-infected cells and contain PCR-generated mutations. Solving for E we obtain
Due to the uncertainties in M, m, and I, some of these efficiency estimates are greater than 100%. This issue is particularly evident in samples having low values for I (due to strong negative selection). Nevertheless, these estimates all suggest true CRISPR mutagenesis efficiencies that are very close to 100%.
Pooled sgRNA screening and data analysis
The lysine methyltransferase U6-sgRNA-EFS-GFP library was pooled at equimolar ratio and used to generate a lentiviral supernatant as described above. A dilutions series of this virus correlated with GFP positivity in infected cells to derive the viral titer multiplicity of infection (MOI). The total number of RN2c cells was chosen to achieve at least 500-fold representation of each sgRNA in the initially infected cell population. To ensure that a single sgRNA was transduced per cell, the viral volume for infection was chose to achieve an MOI of 0.3–0.4. Genomic DNA was extracted at the indicated time points using QiAamp DNA mini kit (Qiagen #51304), following the manufacturer’s instructions. To maintain >500× sgRNA library representation, 16–20 independent PCR reactions were used to amplify the sgRNA cassette, which were amplified for 20 cycles with 100 ng of starting gDNA using the 2× Phusion Master Mix (Thermo Scientific #F-548). PCR products were pooled and subjected to illumina MiSeq library construction and sequencing, as described above. The sequence data were trimmed to contain only the sgRNA sequence then were mapped to the reference sgRNA library without allowing any mismatches. The read counts were then calculated for each individual sgRNA. To compare the differential representation of individual sgRNAs between day 2 and day 12 timepoints, the read counts for each sgRNA were normalized to the counts of the negative control ROSA26 sgRNA. The deep sequencing data can be obtained through the NCBI Sequence Read Archive (SRA) accession SRP057117.
The PCR primers and conditions are listed in the Supplementary Table 1.
Supplementary Material
Acknowledgements
Ying Jin for assistance with pooled sgRNA screen analysis. C.R.V. is supported by NIH CA174793, Burroughs-Wellcome Fund Career Award for Medical Scientists, Alex’s Lemonade Stand Foundation ‘A’ Award, and NCI Cancer Center Support Grant Development Funds CA45508. J.B.K is supported by the Simons Center for Quantitative Biology at Cold Spring Harbor Laboratory.
Footnotes
Author Contributions
J.S. and C.R.V designed experiments; J.S., E.W., J.P.M. carried out experiments; J.S. analyzed experimental results. J.B.K. analyzed sequencing data and developed analysis tools. Z. W. assisted with Illumina sequencing. J.S., J.B.K, and C.R.V wrote the manuscript.
Competing Financial Interests
No competing interests.
References
- 1.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Y, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 5.Doench JG, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P, et al. RNA-guided human genome engineering via Cas9. Science (New York. N.Y.) 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuber J, et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nature biotechnology. 2011;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McJunkin K, et al. Reversible suppression of an essential gene in adult mice using transgenic RNA interference. Proc Natl Acad Sci U S A. 2011;108:7113–7118. doi: 10.1073/pnas.1104097108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi J, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes & development. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang E, et al. Histone H2B ubiquitin ligase RNF20 is required for MLL-rearranged leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3901–3906. doi: 10.1073/pnas.1301045110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi J, et al. The Polycomb complex PRC2 supports aberrant self-renewal in a mouse model of MLL-AF9;Nras(G12D) acute myeloid leukemia. Oncogene. 2013;32:930–938. doi: 10.1038/onc.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mertz JA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Molecular cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu B, et al. Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood. 2014;125:346–357. doi: 10.1182/blood-2014-06-581082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daigle SR, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris WJ, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer cell. 2012;21:473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 23.Lehnertz B, et al. The methyltransferase G9a regulates HoxA9-dependent transcription in AML. Genes Dev. 2014;28:317–327. doi: 10.1101/gad.236794.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim W, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene therapy. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.