Summary
Pervasive transcription of the human genome generates RNAs whose mode of formation and functions are largely uncharacterized. Here, we combine RNA-Seq with detailed mechanistic studies to describe a transcript type derived from protein-coding genes. The resulting RNAs, which we call DoGs for downstream of gene containing transcripts, possess long non-coding regions (often >45 kb) and remain chromatin bound. DoGs are inducible by osmotic stress through an IP3 receptor signaling-dependent pathway, indicating active regulation. DoG levels are increased by decreased termination of the upstream transcript, a previously undescribed mechanism for rapid transcript induction. Relative depletion of polyA signals in DoG regions correlates with increased levels of DoGs after osmotic stress. We detect DoG transcription in several human cell lines and provide evidence for thousands of DoGs genome-wide.
Introduction
Most of the human genome is transcribed; but many of the resulting transcripts, and the factors regulating their transcription, remain uncharacterized. A significant proportion of this pervasive transcription generates long non-coding RNAs (lncRNAs), non-coding RNAs (ncRNAs) of >200 nt (Hangauer et al., 2013; Kapranov et al., 2010; Ulitsky and Bartel, 2013). Some lncRNAs – such as large intervening ncRNAs (lincRNAs) – are transcribed from independent genes in a regulated fashion (Guttman et al., 2009). The synthesis of others, such as promoter-associated RNAs (Kapranov et al., 2007; Seila et al., 2008) and enhancer RNAs (De Santa et al., 2010; Kim et al., 2010), is associated with transcription of other genes. Additional transcript types coincide with the 3′ ends of protein-coding genes, including termini-associated, short RNAs (Kapranov et al., 2007) and 3′ untranslated regions (UTRs) that are expressed independently of their upstream mRNAs (Mercer et al., 2011). However, the observed pervasive transcription (Hangauer et al., 2013; Kapranov et al., 2007) cannot be fully explained by these transcript types, hinting at other possible contributors to genomic output.
Here, we set out to study putative lncRNAs reported to correlate with bad prognosis in neuroblastoma, a childhood tumor of the sympathetic nervous system (Maris, 2010; Mestdagh et al., 2010). We found that the lncRNA uc.145 is highly induced by osmotic stress and is part of a >50 kb transcript generated by readthrough transcription from the upstream protein-coding gene, CXXC4. This insight led us to discover a new class of long, chromatin-associated transcripts, highly inducible by osmotic stress and generated by readthrough transcription. Throughout this study, we refer to the transcribed regions downstream of genes as “DoG regions” at the genomic level and the resulting transcripts as “DoGs” for DoG-containing transcripts at the RNA level. A DoG is composed of a 5′ segment corresponding to the upstream gene, referred to as DoG-associated, and a 3′ segment specified by the DoG region.
Results
The uc.145-containing transcript is highly enriched after KCl treatment
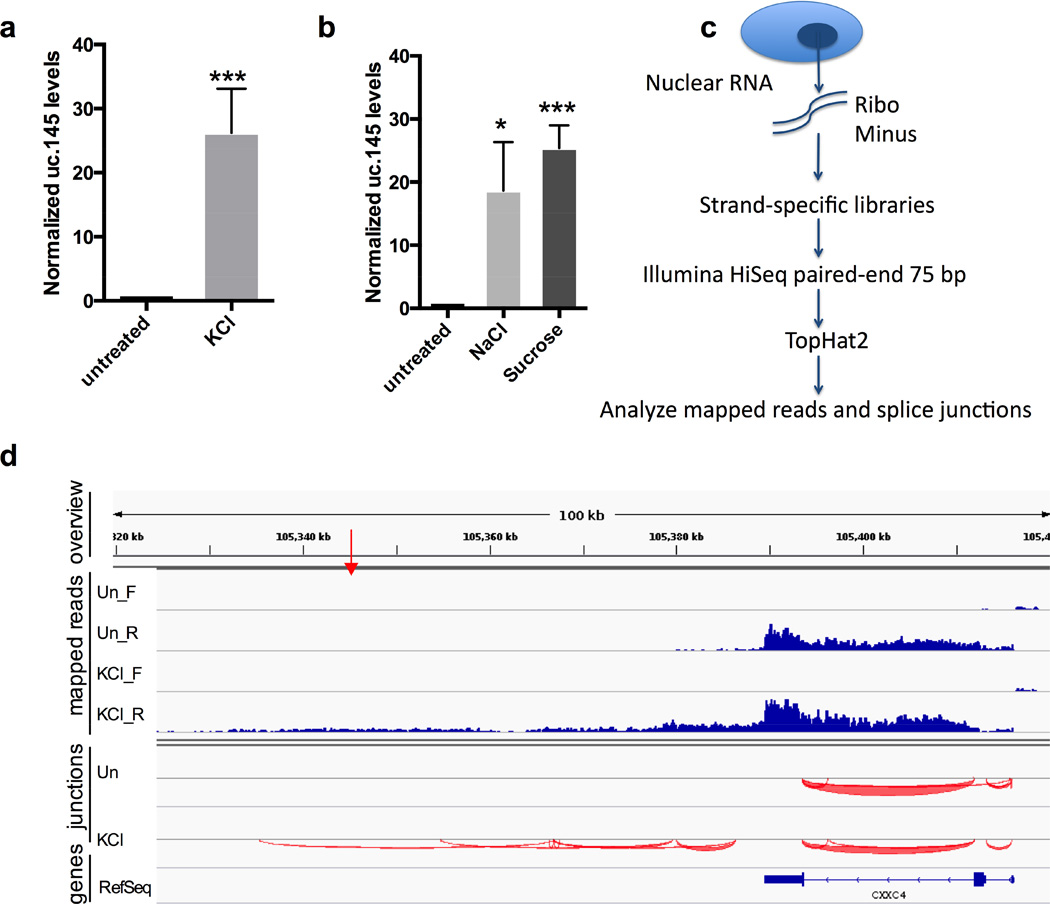
We asked whether the level of any putative lncRNA associated with bad prognosis in neuroblastoma (Mestdagh et al., 2010) were upregulated by KCl, which broadly activates transcription in neuronal cells (Kim et al., 2010). Interestingly, qRT-PCR analysis of neuroblastoma SK-N-BE(2)C cells exposed to 80 mM KCl for 1 h revealed a ~30-fold induction of the putative lncRNA uc.145, while expression levels of other lncRNAs examined were unaffected (Figure 1a and data not shown). This induction of uc.145 by KCl was validated in 3 out of 4 other cell lines tested (Figure S1b). Therefore, we focused on uc.145.
Figure 1. The uc.145 transcript is contained within a DoG induced by osmotic stress.
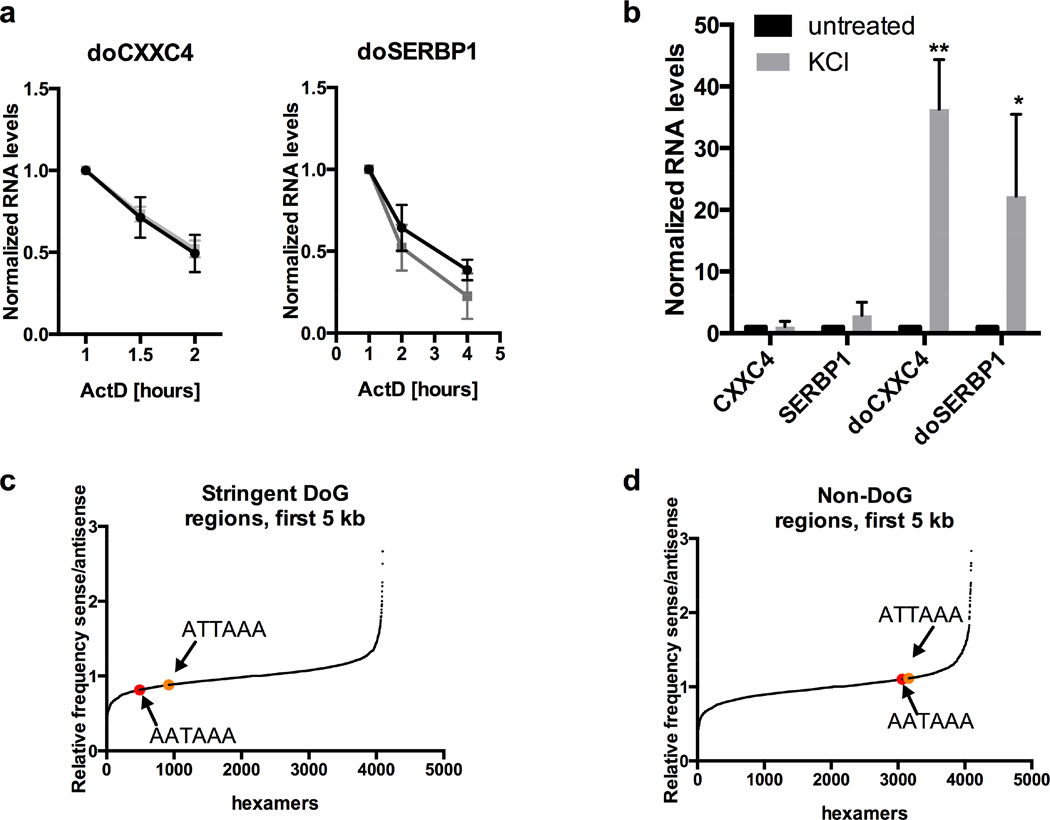
a) qRT-PCR quantitation of KCl-mediated induction (80 mM, 1h) of uc.145 in SK-N-BE(2)C cells, n=4. Figures of pooled data show mean and SD. p-values of 0.05-0.01 are denoted by *, 0.01-0.001 by **, and ≤0.001 by ***. uc.145 levels are normalized to average of GAPDH and GUSB mRNAs in a, and to GAPDH mRNA in b. b) uc.145 is induced by treatment with osmotic stress agents NaCl (80 mM, 1 h) or sucrose (200 mM, 1 h), n=3. A higher dose of sucrose is required because it produces 1 solute while KCl and NaCl produce 2. c) Outline of RNA-Seq experiment. d) Mapped reads and splice junctions surrounding uc.145 from a representative replicate of the RNA-Seq experiment. Top panel (“overview”) shows genomic coordinates and the red arrow indicates the location of the 248 nt uc.145. Second panel (“mapped reads”) shows reads mapping to the forward (_F) and reverse (_R) strands separately. “Un” stands for untreated. Third panel shows splice junctions (“junctions”) from the reverse strand depicted below the indicator line. The bottom panel shows RefSeq genes. See also Figure S1.
uc.145 is upregulated in response to osmotic stress
KCl increases transcription in neuronal cells either by depolarizing cell membranes, thus inducing neuronal cell signaling (Kim et al., 2010), or by inducing osmotic stress (Oh et al., 1995). Because uc.145 levels increase in some non-neuronal cell lines (Figure S1b), the induction mechanism cannot rely entirely on neuronal cell signaling. We treated SK-N-BE(2)C cells with 80 mM NaCl or 200 mM sucrose, inducers of osmotic stress (Oh et al., 1995) that do not cause neuronal membrane depolarization. We observed an upregulation of uc.145 similar to that by KCl, indicating that uc.145 levels increase as a result of osmotic stress (Figure 1b).
RNA-Seq identifies a large readthrough transcript overlapping uc.145
Because the intergenic uc.145 does not overlap any annotated human transcripts, we performed RNA-Seq on nuclear ribo-minus treated RNA from SK-N-BE(2)C cells incubated with or without KCl in 3 biological replicates (Figure 1c). We mapped reads to the human genome using TopHat2 (Kim et al., 2013) and inspected the resulting reads (converted to BigWig format) and the predicted splice junctions in the IGV browser (Robinson et al., 2011; Thorvaldsdottir et al., 2013) (Figures 1d and S1c).
Indeed, RNA-Seq confirmed induced uc.145 expression after KCl treatment. Surprisingly, we observed continuous reads and splice junctions mapping to the upstream CXXC4 gene and ~60 kb further downstream (Figure 1d). Thus uc.145 (red arrow in Figure 1d) appears to be included in a transcript generated by readthrough from the upstream CXXC4 gene. As defined in the introduction, we use the terminology DoG regions for transcribed regions downstream of genes and DoGs for readthrough transcripts; the prefix “do” before the gene name indicates the portion of the DoG that is downstream of the coding gene.
Both we (Figure S1d) and others (Calin et al., 2007) have detected transcripts overlapping uc.145 from both strands downstream of CXXC4 using arrays and qRT-PCR. However, the DoG transcript coming from the same strand as CXXC4 is expressed at higher levels, shows greater variation in expression levels between tissues (Calin et al., 2007) and is specifically induced by KCl (Figure S1d).
Global analysis reveals KCl-inducible transcription downstream of genes
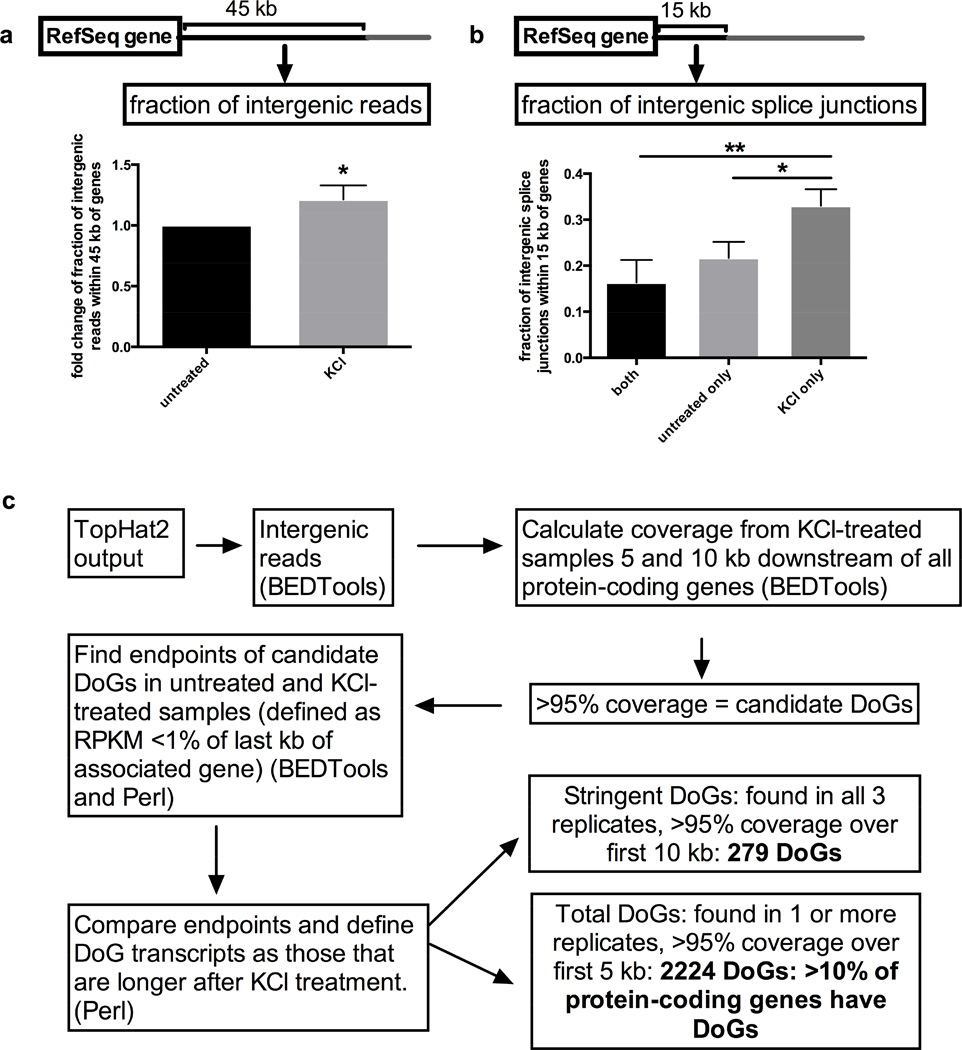
Preliminary inspection of our mapped sequencing data suggested the existence of many KCl-inducible DoGs. To investigate a possible global effect of KCl treatment on transcription directly downstream of genes, we first asked whether KCl treatment increases intergenic RNA levels within 45 kb downstream of all RefSeq genes. Analysis of read coverage in treated versus untreated samples revealed a modest but significant increase in such reads (Figure 2a). Second, we analyzed the percentage of intergenic splice junctions found only after KCl treatment, only in untreated samples or in both treated and untreated samples. For this analysis, we chose a distance of 15 kb downstream of genes because splice junctions are more common within this range and have a greater number of supporting reads. Interestingly, we found a significant increase in the fraction of such intergenic splice junctions present only in KCl-treated samples (Figure 2b). These data indicate that KCl induces the production of spliced transcripts downstream of genes.
Figure 2. Genome-wide KCl-mediated upregulation of DoGs.
a) Increase in intergenic reads within 45 kb downstream of RefSeq genes in KCl-treated relative to untreated samples, n=3; figures of pooled data show mean and SD, p-values are denoted as in Figure 1. b) Fraction of intergenic splice junctions present within 15 kb downstream of RefSeq genes either in both KCl-treated and untreated, in untreated only, or in KCl-treated only samples, with a statistically significant upregulation for splice junctions found in KCl-treated only samples, n=3. c) Outline of the analysis used to identify DoGs. All annotated DoGs are listed in Table S1. See also Figure S2.
A DoG-finding pipeline identifies more than 2000 DoGs genome-wide
Next, we asked whether the KCl-inducible transcription downstream of other genes shares characteristics with doCXXC4; i.e. apparent continuity with the transcripts from the upstream associated genes and increased transcript length after KCl treatment. We created a bioinformatic pipeline that identifies DoG regions and analyzed the 20,314 human protein-coding genes for DoG transcription, defining a DoG as an intergenic transcript that a) is continuous with the upstream associated gene for at least 5 or 10 kb and b) is longer after KCl treatment, based on defining putative DoG endpoints as the location where the reads per kilobase per million (RPKM, calculated iteratively over each kb of the DoG) mapped reads dropped below 1% of that of the last 1 kb of the associated gene transcript. Note that our definition of endpoints depends on the expression level of the associated gene and therefore provides relative estimates rather than absolute endpoints. In the RNA-Seq data, DoGs appear to lack defined endpoints. Instead, transcription decreases over the length of the DoG region, such that DoG length depends at least in part on the expression level of DoG and associated gene transcript. We chose relative estimates of DoG endpoints because they are not influenced by KCl-mediated changes in expression of the associated genes and thus allow comparisons between samples.
The DoG-finding pipeline yielded 279 DoGs identified in all three biological replicates with continuous transcription at least 10 kb downstream of the associated genes (referred to as “stringent DoGs”) and 2224 DoGs identified in at least 1 biological replicate with continuous transcription at least 5 kb downstream of the associated genes (referred to as “total DoGs”) (Figure 2c, Extended Experimental Procedures). All annotated DoGs with coordinates, estimated lengths, and absolute and relative RPKM for the first 5 kb are listed in Table S1. KCl-induced DoG transcripts are therefore generated downstream of more than 10% of human protein-coding genes.
To determine the fraction of intergenic transcription in SK-N-BE(2)C cells attributable to DoGs, we calculated the percentage of intergenic reads and TopHat2 predicted splice junctions that overlap our total DoG annotation. This percentage is 21% in untreated and 31% in KCl-treated cells for reads, and 14% and 18%, respectively, for splice junctions (Figure S2a–b). Next, we determined the percentage of KCl-upregulated intergenic reads attributable to DoGs. We first used the TopHat2 output to create lists of genomic regions and splice junctions ranked in descending order with respect to number of supporting reads. We then selected genomic regions or splice junctions among the top 5% of the ranked list after KCl treatment, but absent from the top 30% in untreated samples, and calculated the percentage that overlapped with our annotation of total DoGs (Figure S2c). We found 68% of upregulated intergenic transcription defined by mapped reads and 41% defined by splice junctions attributable to DoGs, demonstrating that DoGs are a significant source of intergenic transcription in general and of osmotic stress-inducible transcription in particular.
Validation of KCl-mediated induction of specific DoGs
To characterize DoGs further, six DoG candidates in addition to doCXXC4 were selected from the total DoG annotation (see Extended Experimental Procedures and Table S1). Mapped reads and splice junctions for two of these (doRAB11A and doSERBP1) are shown in Figure S3.
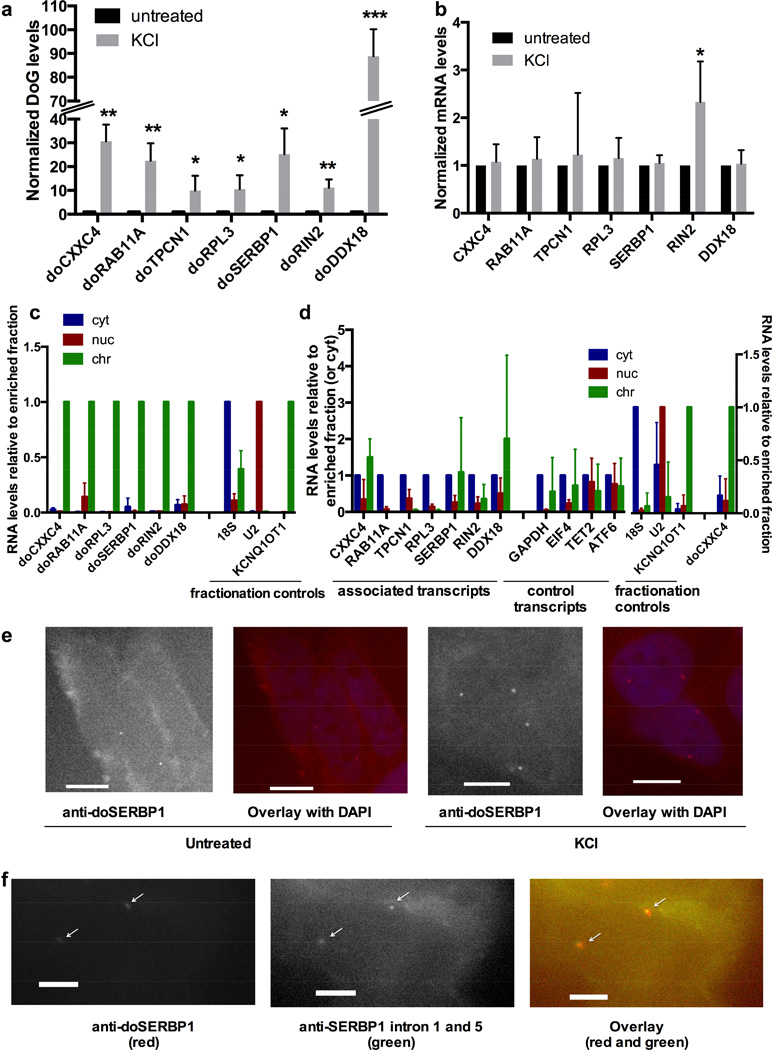
qRT-PCR on total RNA from SK-N-BE(2)C cells confirmed that – like doCXXC4 – the six additional DoG candidates are induced between 10- and 90-fold by KCl treatment (80 mM, 1h) (Figure 3a). Next, we asked whether this upregulation is specific for DoGs or is observed also for their associated mRNAs. Six associated mRNAs were unaffected by KCl as determined by qRT-PCR while one, RIN2, was induced 2-fold (Figure 3b); its corresponding DoG, doRIN2, was induced 10-fold (Figure 3a). We conclude that DoG induction after KCl treatment is not mirrored by a similar increase in associated mRNA levels.
Figure 3. Characterization of DoGs.
a) Validation of DoG induction by KCl (80 mM, 1 h), n=3, except for doTPCN1, where n=4. doCXXC4 from Figure 1a is included for comparison. Figures of pooled data show mean and SD, p-values are denoted as in Figure 1. RNA levels are normalized to GAPDH mRNA in a and b. b) mRNA transcribed from DoG-associated genes are not induced by KCl with the exception of RIN2, which is induced 2-fold, while doRIN2 is induced 10-fold (Figure 3a), n=3, except for RIN2, where n=4. c) qRT-PCR quantitation of DoGs in RNA purified from cytoplasmic, soluble nuclear, or the chromatin fraction. KCNQ1OT1 lncRNA, U2 snRNA and 18S rRNA served as markers for the chromatin (chr), nuclear (nuc), and cytoplasmic (cyt) fraction, respectively, n=3. The enriched fraction is set to 1. All DoGs are significantly enriched in chromatin. d) As in c for DoG-associated mRNAs and control mRNAs, none of which are enriched in chromatin. Due to lack of material, 5 biological replicates were used to reach an n=3 for each target. Fractionation controls were significantly enriched in their respective fractions in both triplicate combinations used, but for simplicity, controls are shown as the mean and SD of all 5 replicates. doCXXC4 is an additional control to verify DoG localization to chromatin. doCXXC4, fractionation controls, RAB11A, RIN2, RPL3, and TPCN1 are significantly enriched in the fraction set to 1 compared to either of the other fractions. Other targets that showed no statistically significant enrichment in any fraction are shown normalized to the cytoplasmic fraction. e) Representative images of FISH using Stellaris probes (Biosearch) for doSERBP1 in untreated and KCl-treated SK-N-BE(2)C cells, confirming KCl-mediated induction and nuclear localization of doSERBP1. Images are at 40x magnification and scale bars indicate 10 µM. f) Representative images of co-staining with FISH Stellaris probes (Biosearch) for intron 1 and 5 of SERBP1 in KCl-treated cells demonstrating that doSERBP1 remains at the site of transcription. Images are at 100x magnification and scale bars indicate 5 µM. See Extended Experimental Procedures for details on FISH probes. See also Figures S3 and S4.
DoGs, but not associated mRNAs, are strongly enriched in the chromatin fraction
We fractionated cells into cytoplasm, soluble nuclear extract and the chromatin fraction (including chromatin and other insoluble nuclear structures). Analysis of purified RNA from each fraction by qRT-PCR revealed that all DoGs investigated are strongly enriched in chromatin (Figure 3c). Chromatin localization of DoGs is in contrast to their corresponding mRNAs and a number of unrelated mRNAs, which either are enriched in the cytoplasmic fraction or show no particular enrichment (Figure 3d). DoG levels estimated by both qRT-PCR and RNA-Seq are significantly lower than levels of associated mRNA (≤10%, in many cases ≤1%). Hence, the small fraction of associated mRNAs that remain connected to the transcribed DoG region and localize to chromatin is not expected to affect the overall observed location of the associated mRNA (see below).
KCl-mediated induction and chromatin localization were verified for one of the more abundant DoGs, doSERBP1, through RNA FISH in SK-N-BE(2)C cells (Figures 3e and S4a–c). doSERBP1 FISH staining localizes to 1 to 4 distinct nuclear dots per cell after KCl treatment (Figure 4b). Because SK-N-BE(2)C cells are largely diploid, this staining is in agreement with doSERBP1 remaining at its site of synthesis on either one or both alleles in cells before or after DNA replication. This notion was confirmed by colocalization of FISH signals for doSERBP1 with those of 2 introns from the associated gene transcript SERBP1 (Figure 3f). doSERBP1 was also detectable by Northern blot (Figure S4d).
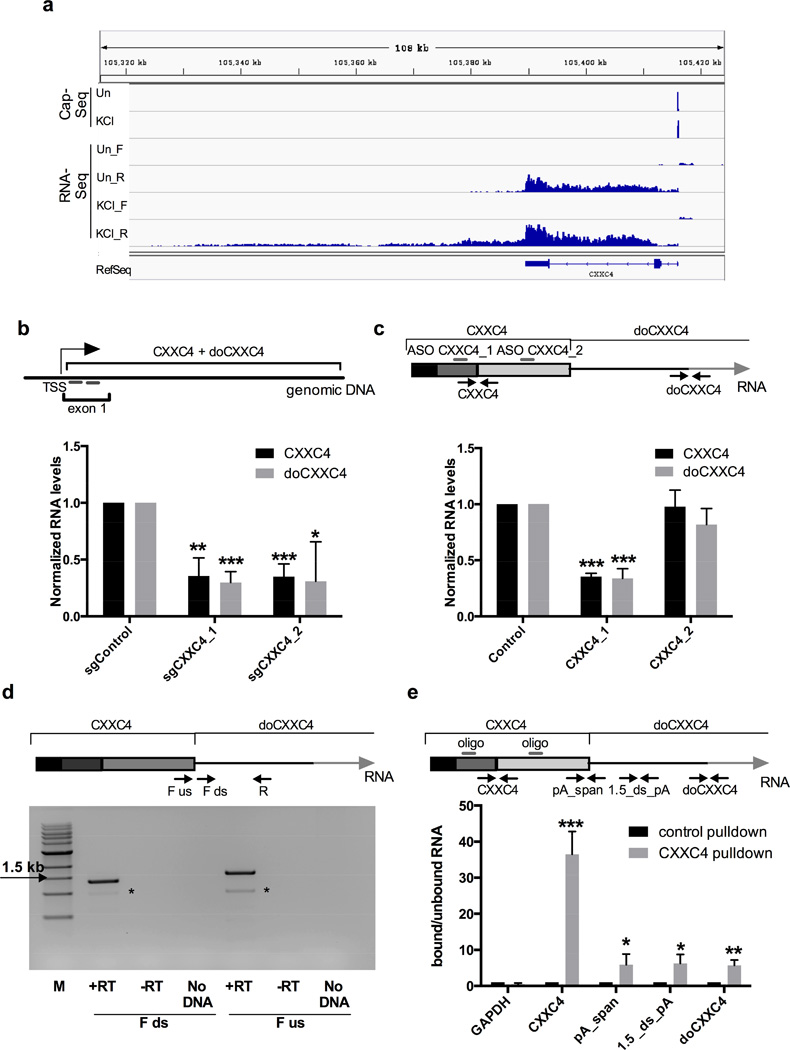
Figure 4. DoGs are part of the same transcript as the associated mRNA.
a) CapSeq identifies the annotated TSS for CXXC4 but no downstream TSSs. Genomic coordinates are shown on top, CapSeq data is shown in the upper panel. The two following panels show mapped reads from Figure 1d and RefSeq genes annotated as in Figure 1d. Un stands for untreated. b) Inhibiting transcription through CXXC4 by targeting dCas9 to its first exon with two sgRNAs in 293T cells reduces the levels of both CXXC4 and doCXXC4, n=3. Figures of pooled data show mean and SD, p-values are denoted as in Figure 1. RNA levels are normalized to the average of GAPDH and GUSB mRNAs in b, c and e. Top panel shows the location of sgRNAs (short grey bars) relative to TSS of CXXC4. Diagram is not to scale. c) An ASO (CXXC4_1) effective in knocking down CXXC4 mRNA also reduces the levels of doCXXC4 in SK-N-BE(2)C cells, while ASO CXXC4_2 designed to target CXXC4 alters neither CXXC4 nor doCXXC4 levels, n=4. Top panel shows CXXC4 and part of doCXXC4 (different exons are depicted in different shades of gray). ASOs are shown as bars and qRT-PCR primers as arrows. Diagram is not to scale. d) Agarose gel showing comparable amounts of products from RT-PCR analysis of nuclear RNA with a reverse primer (R) located ~1.5 kb downstream of the CXXC4 polyA site combined with a forward primer (F) located just downstream (ds) or just upstream (us) of the CXXC4 polyA site as schematized in the top panel. Diagram is not to scale. M indicates 1 kb+ molecular marker (NEB). Asterisks indicate an unspecific band. e) Naked RNA pulldown using a mixture of biotinylated oligonucleotides (shown as grey bars in the top panel) targeting exon 2 and 3 of CXXC4 followed by qRT-PCR. Included primer pairs (shown as arrows) either target CXXC4, or span the CXXC4 polyA site (pA_span, this pair should not generate product if there were cleavage), are located 1.5 kb downstream of the polyA site (1.5_ds_pA), or span the first splice junction in doCXXC4 (doCXXC4). Diagram is not to scale. Figure shows the relative RNA levels in the bound versus unbound fraction after pulldown with the CXXC4 probe normalized to control (EBER2) probe, n=3.
Further characterization of DoGs
To investigate the abundance of DoGs, we spiked in an in vitro transcribed fragment of doCXXC4 into RNA from untreated SK-N-BE(2)C cells, performed qRT-PCR and generated a standard curve. We estimate the copy number of doCXXC4 after KCl treatment to be ~30 per cell (Figure S4e).
To characterize the 3′ ends of DoGs, we asked whether they are polyadenylated. We analyzed the enrichment of doCXXC4 and doSERBP1 in polyA+ and polyA− RNA by qRT-PCR and found approximately equal levels of doCXXC4 and doSERBP1 in the two fractions, while 18S rRNA was clearly enriched in the polyA− fraction and GAPDH mRNA in the polyA+ fraction (Figure S4f). We conclude that each DoG exists in both polyadenylated and non-polyadenylated form. Together with the chromatin localization of DoGs, this result argues that transcribed DoG regions are not simply extremely long 3′-UTR extensions; they would be expected to localize with the upstream mRNA and be polyadenylated. Instead DoGs appear to represent a different transcript type.
Absence of KCl-inducible transcriptional start sites (TSSs) in DoG regions or DoG-associated genes indicates that DoGs are readthrough transcripts
DoGs are generated by at least 5 kb of continuous transcription downstream of associated genes (Figure 2b), suggesting that DoGs are readthrough transcripts. Alternatively, DoGs could be derived from activation of KCl-inducible promoters overlapping, or just downstream of, the associated genes. The latter hypothesis is supported by the reported presence of TSSs in the 3′-UTRs of many genes (Mercer et al., 2011). To distinguish between these possibilities, we performed a CapSeq experiment (Xie et al., 2013) to identify and compare TSSs in untreated and KCl-treated SK-N-BE(2)C cells. We fragmented nuclear RNA and purified capped RNA fragments by pulldown with the cap-binding eIF-4E protein. Purified RNA was subjected to phosphatase treatment and subsequent de-capping, leaving only previously capped RNA fragments accessible for cloning using standard protocols for small RNA library preparations (see Extended Experimental Procedures). RNA libraries were subjected to Illumina HiSeq, and reads were mapped to the human genome by TopHat2 and further processed to reduce background and identify peaks corresponding to TSSs (see Extended Experimental Procedures).
CapSeq data for 3 DoGs are shown in Figures 4a (doCXXC4) and S5a (doSERBP1 and doRAB11A). We readily detected the expected KCl-mediated increase in FOS transcription after KCl treatment (Figure S5b), confirming that our method can indeed detect KCl-mediated activation of TSSs. If DoGs are generated by activation of downstream TSSs, these should overlap or localize immediately downstream of the DoG-associated genes – perhaps with enrichment in 3′-UTRs (Mercer et al., 2011). Therefore, we compared the number of observed peak reads mapping anywhere in both total and stringent DoG-associated genes and 2 kb downstream, or in the 3′-UTRs of DoG-associated genes and 2 kb downstream. We did not observe any increase in TSSs after KCl treatment (Table S2). We repeated the analysis, seeking TSS reads mapping anywhere in total or stringent DoG regions, with the same result (Table S2). The average RPKM for the first 2 kb of the intergenic portion of total and stringent DoGs was 1.8 and 4.1, respectively. As we can identify TSSs for transcripts expressed at levels as low as 0.5 RPKM (Figure S5b), our CapSeq method is likely sensitive enough to detect a large fraction of KCl-inducible TSSs, if they exist. In conclusion, our data provide no evidence to suggest that DoGs are generated from independent, KCl-inducible promoters. These results therefore support the conclusion that DoGs are readthrough transcripts.
DoG production depends on transcription of the associated gene
To further support DoGs being readthrough transcripts, we used a mutant version of the clustered regularly interspaced short palindromic repeats (CRISPR) system that inhibits transcription when targeted to the promoter or first exon of a gene (Chen et al., 2013; Gilbert et al., 2013). We chose 293T cells for this assay because they are readily transfectable with plasmids, while SK-N-BE(2)C cells are not. 293T cell lines stably expressing dCas9 or dCas9-KRAB (the latter fusion protein connects dCas9 to the transcriptional repressor KRAB) were first generated and then transfected with sgRNAs targeting either CXXC4 or SERBP1 (2 sgRNAs for each gene were included in parallel transfections, see Figures 4b and S5c). In both cases, inhibiting transcription through the gene reduced levels of both the associated mRNA and the DoG, analyzed by qRT-PCR (Figures 4b and S5c).
We further knocked down CXXC4 transcript levels using antisense oligonucleotides (ASOs), which induce RNaseH-mediated cleavage of the target RNA and are effective in the nucleus. Of 2 different ASOs designed to target CXXC4, only 1 (CXXC4_1) reduced CXXC4 mRNA levels to 36% (black bars in Figure 4c). Importantly, this ASO also reduced doCXXC4 levels comparably (grey bars in Figure 4c). These data indicate that DoGs are indeed created by readthrough transcription from the upstream gene.
DoGs are continuous with their associated mRNAs
Next, we addressed whether DoGs retain the associated upstream mRNA sequence. We readily detected RT-PCR products spanning the CXXC4 polyA site, arguing that a substantial proportion of doCXXC4 molecules are physically linked to the CXXC4 coding sequence (Figure 4d).
To confirm the continuity of doCXXC4 with the CXXC4 coding sequence, we used streptavidin beads coated with biotinylated oligonucleotide probes complementary to CXXC4 to pull down CXXC4-containing transcripts from denatured RNA. We analyzed doCXXC4 co-selection by qRT-PCR and observed significant enrichment of doCXXC4 in the CXXC4 pulldown compared to the control (Figure 4e). Furthermore, we detected ~4-fold induction in doCXXC4 levels upon knockdown of CPSF73, the catalytic subunit of the cleavage and polyadenylation complex, indicating that inhibiting cleavage and polyadenylation enhances DoG levels (Figure S5d).
KCl enhances DoG transcription, not stability
To rule out the possibility that KCl acts by increasing the stability of DoGs, we analyzed half-lives of doCXXC4 and doSERBP1 by inhibiting transcription with Actinomycin D (ActD). Their half-lives of ~1 h were not significantly altered by KCl treatment (Figure 5a).
Figure 5. DoGs are induced at the level of transcription, not RNA stability.
a) There is no difference in the half-lives of doSERBP1 or doCXXC4 in untreated versus KCl-treated samples. ActD was used to inhibit transcription; 1 h (when ActD is fully effective) was set to 1. doCXXC4 could be monitored for only 1 additional h because of low signal. N=3. RNA levels are normalized to the average of GAPDH and GUSB mRNAs in a and b. b) KCl regulates transcription through DoG regions, but not of associated mRNAs. Newly synthesized RNA was selected after 5-EU incorporation as described in the main text and analyzed by qRT-PCR, demonstrating a 20- to 40-fold increase in newly synthesized doSERBP1 and doCXXC4 from cells treated with KCl compared to untreated cells with no effect on associated mRNAs. Due to lack of material, a total of 4 replicates was used to give n=3 for each target. c) Representation of the frequency on the sense versus antisense strand of all hexamers found in regions 5 kb downstream of 279 stringent DoG-associated genes (Table S1) compared to the counts of all other possible hexamers (4096 in total). PASs AATAAA is shown in red and ATTAAA in orange. d) As in c but for regions 5 kb downstream of 209 non-DoG genes with no sign of DoG transcription. See also Figure S6 and Table S4.
Enhanced KCl-mediated transcription through DoG regions was corroborated by incubating cells with the uridine analogue 5-ethynyl uridine (EU) for 25 min to metabolically label newly-transcribed RNA. EU was then attached to biotin (Click-iT technology, Invitrogen), and labeled RNA was selected on streptavidin beads and analyzed by qRT-PCR. We observed ~30-fold induction of newly-synthesized doCXXC4 and doSERBP1 after KCl treatment (Figure 5b), strengthening the conclusion that KCl regulates the transcription, not the stability, of DoG transcripts.
Finally, we performed Pol II ChIP and qPCR for one of the most abundantly transcribed DoG regions, doSERBP1. We observed an increase in Pol II occupancy in doSERBP1 and at the promoter of the known KCl-inducible gene FOS after KCl treatment, with no change in Pol II occupancy at the GAPDH promoter (Figure S6a–b). Because the levels of the upstream transcripts that produce DoGs are not increased by KCl (Figure 5b), together these data argue that KCl does not cause increased initiation at the promoter of the associated mRNA – such regulation should cause similar increases in both DoG and associated mRNA levels. Instead, KCl most likely regulates Pol II termination to promote increased readthrough.
DoG regions are depleted of polyA signals (PASs)
A gene expressed at a high level is more likely to generate detectable readthrough, and indeed we observe that the genes upstream of our 279 stringent DoGs have a higher average RPKM (of 5.6) calculated over the whole gene, including introns, than a set of “non-DoG” genes: 209 genes chosen based on the absence of any detectable readthrough (Table S3) (RPKM 1.5). However, high expression alone is not sufficient to generate DoGs, as some highly expressed genes including GAPDH (RPKM 89.9) lack DoGs.
Alternatively, continued transcription through DoG regions could be due to low probability of cleavage and polyadenylation. To address this possibility, we first assessed the strength of the annotated PASs of the genes associated with our set of stringent DoGs as well as the PASs of all protein-coding genes and of the 209 non-DoG genes (see Extended Experimental Procedures). We did not observe any difference between the PAS of DoG-associated genes and the average PAS of all protein-coding genes (Table S4). However, the non-DoG genes had somewhat stronger PASs than the average protein-coding genes (Table S4). This observation suggests that under certain conditions, limited readthrough likely occurs downstream of most genes lacking exceptionally strong PASs, and in some cases this readthrough may generate DoGs.
To look for downstream signals that might tip the balance in favor of continued DoG transcription, we next calculated the frequency of AAUAAA (~50% of PASs in mammalian cells (Almada et al., 2013; Beaudoing et al., 2000)) on the sense versus the antisense DNA strand in the 5 kb downstream of the stringent DoG-associated genes, of the non-DoGs, and of all protein-coding genes. Interestingly, we found that these 5 kb of DoG regions have on average a lower frequency of AAUAAAs (ratio of AAUAAA in sense vs. antisense strand of 0.8) compared to the 5 kb downstream of both all protein-coding genes (ratio of 0.9) and non-DoGs (ratio of 1.1) (Table S4) (Figure 5c–d). No difference in the frequency of U1 snRNP binding sites, recently shown to antagonize cleavage and polyadenylation (Berg et al., 2012; Kaida et al., 2010), was observed in either DoG regions or in regions downstream of non-DoG genes (data not shown). We conclude that DoG regions are relatively depleted of PASs, causing termination in these regions to be less efficient. Therefore, any shift that favors transcriptional elongation over termination would have greater impact in DoG regions than regions with a higher frequency of PASs. Interestingly, we found a reduced frequency (ratio of 0.9) of AAUAAAs in the sense versus the antisense strand in the 5kb downstream of all protein-coding genes (Figure S6c), suggesting an evolutionary pressure to allow readthrough transcription downstream of many genes.
Transcription of DoG regions is regulated by IP3 receptor mediated Ca2+ release
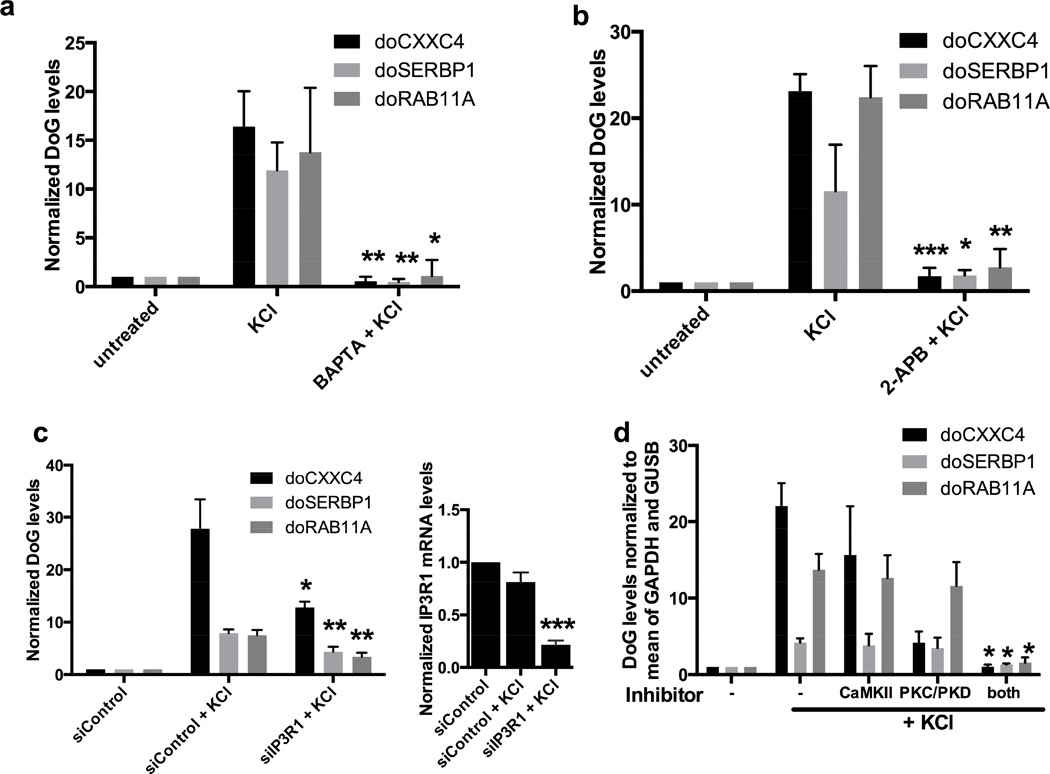
To probe the signaling pathways through which KCl-mediated osmotic stress induces DoG expression, we investigated the importance of intracellular Ca2+, a common second messenger regulating many cellular processes including transcription (Berridge et al., 2000). We pretreated SK-N-BE(2)C cells with either a membrane permeable (BAPTA) or a non-permeable (EGTA) Ca2+ chelator followed by KCl treatment and analyzed the levels of doCXXC4, doSERBP1 and doRAB11A. These DoGs are all inducible by osmotic stress in SK-N-BE(2)C cells (Figures 1a and S7a), as well as in the neuroblastoma cell line SH-SY-5Y (Figures S1b and S7b). Figures 6a and S7c show that KCl-mediated DoG induction is abolished by pretreatment with BAPTA but unaffected by EGTA pretreatment. FOS mRNA, whose induction by KCl is mediated by influx of extracellular Ca2+ (Bading et al., 1993), was reduced by EGTA treatment as expected (Figure S7c). Together, these data suggest that intracellular Ca2+ release is required for DoG induction.
Figure 6. DoG levels are regulated by Ca2+ signalling.
a) qRT-PCR quantitation showing loss of DoG induction if KCl treatment (80 mM, 1 h) follows a 30-min treatment with 50 mM BAPTA, a membrane permeable Ca2+ chelator, n=3; statistical significance indicated compares KCl and BAPTA+KCl. All figures show mean and SD, p-values are denoted as in Figure 1. RNA levels are normalized to GAPDH mRNA in a and to the average of GAPDH and GUSB mRNAs in b-d. b) As in a but KCl treatment followed a 30-min treatment with 100 µM 2-APB, an IP3R inhibitor, n=3. c) qRT-PCR quantitation showing reduced KCl-mediated DoG induction in cells transfected with siRNA targeting IP3R1, compared to KCl treatment of control siRNA-transfected cells, n=3. Significant knockdown of IPR3R1 mRNA is shown by the graph to the right. d) KCl treatment (80 mM, 1 h) followed a 30-min treatment with either 10 µM Gö6976, a PKC/PKD inhibitor; 10µM KN-93, a CaMKII inhibitor; or both. N=3, statistical significance indicated compares KCl alone with KCl + both inhibitors. See also Figure S7.
The main intracellular source of Ca2+ is the endoplasmic reticulum (ER), from which Ca2+ is released in part via IP3 receptor (IP3R) signaling (Berridge, 2009). Therefore, we investigated the involvement of IP3R in DoG induction by pretreating SK-N-BE(2)C cells with the IP3R inhibitor 2-ABP and found that KCl-mediated DoG induction was abolished (Figure 6b). We then individually knocked down all 3 IP3R isoforms (Figure 6c and data not shown); knockdown of IP3R1, but not the other IP3R isoforms, significantly reduced KCl-mediated DoG induction (Figure 6c and data not shown).
A recent report (Sharma et al., 2014) demonstrates a link between Ca2+ signaling and regulation of transcriptional elongation and alternative splicing mediated by activation of Ca2+/calmodulin-dependent protein kinase II (CaMKII) and Protein Kinase C/Protein Kinase D (PKC/D). To investigate if these pathways play a role in DoG induction, we pretreated SK-N-BE(2)C cells with inhibitors for CaMKII and PKC/PKD (KN-93 and Gö6976, respectively); subsequent KCl treatment failed to induce DoG levels (Figure 6d). We conclude that IP3R-mediated Ca2+ release and activation of CaMKII or PKC/PKD are necessary for DoG induction by osmotic stress.
A role for DoGs in reinforcing the nuclear scaffold after stress?
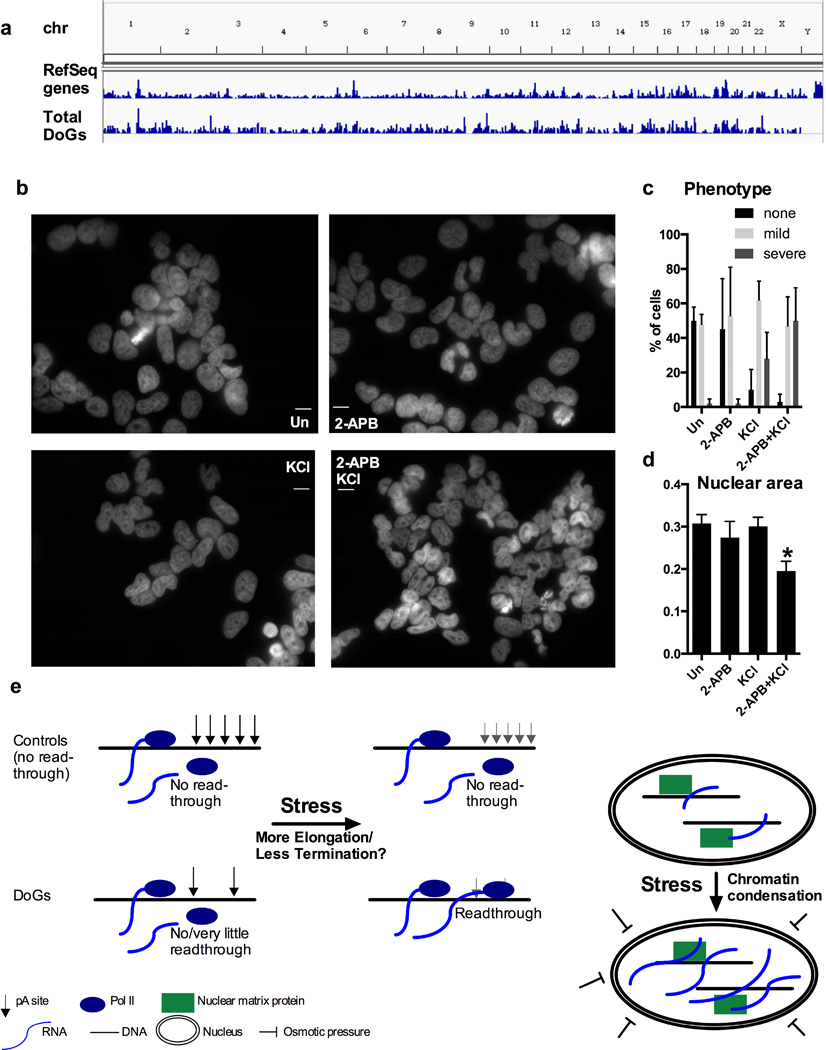
The coordinated stress-mediated induction of thousands of DoGs distributed evenly over euchromatin (Figure 7a) and the retention of DoGs at their sites of transcription suggest that DoGs could participate in a stress response to support functional chromatin organization. Hyper-osmotic stress – the type of stress used in this study – causes nuclear shrinkage and chromatin collapse because it forces water to leave the cell (Finan and Guilak, 2010). We hypothesize that DoGs help to reinforce the nuclear scaffold in the event of stress. This hypothesis is difficult to test because the large number of DoGs presumably act together – thus, inhibiting the expression of one or a few DoGs would likely not have a significant phenotype since neighboring DoGs could compensate for their loss.
Figure 7. Inhibiting stress-induced DoG induction correlates with aggravated nuclear response to osmotic stress.
a) DoG regions are distributed over all euchromatin except for the Y chromosome, demonstrated by a comparison of total DoGs and RefSeq genes. b) DAPI nuclear staining of SK-N-BE(2)C cells either treated with 100 µM 2-APB for 30 min prior to treatment with 80 mM KCl for 1 h, or treated with 2-APB or KCl separately, or left untreated. Images are taken at 63x magnification and scale bars indicate 20 µM. c) Unbiased quantification of the phenotype in b based on 20 cells each from 5 images per treatment (100 cells/treatment). The mild phenotype is significantly more prominent than severe phenotype in all samples except of 2-ABP + KCl, where both phenotypes are detected at similar frequencies. d) Measurement of nuclear area of 20 cells each from 5 images per treatment (100 cells/treatment). Statistical significance indicate a difference between 2-ABP + KCl and all other samples. e) Models for the regulation of DoG induction (left panel) and for DoG function (right panel).
Others have employed general methods, such as transcriptional inhibition and RNase treatment, to investigate the effect of depleting large groups of cellular RNAs (Hall et al., 2014). To test the possible role of DoGs in maintaining nuclear integrity after stress, we chose a similar approach: inhibition of the IP3R, which prevents DoG induction after osmotic stress (Figure 6b). We pretreated SK-N-BE(2)C cells with the IP3R inhibitor 2-APB, followed by KCl treatment, cell fixation and DAPI staining to visualize nuclei. After KCl treatment, we found evidence of chromatin collapse (Finan and Guilak, 2010), demonstrated by less uniform DAPI staining and the appearance of large holes in many nuclei (Hall et al., 2014) (Figure 7b–c). This phenotype was aggravated in cells pretreated with 2-APB (Figure 7b–c), and these nuclei were also smaller (Figure 7d). This more severe phenotype in response to osmotic stress after inhibition of IP3R signaling and DoG induction is consistent with a possible role for DoGs in maintaining nuclear integrity after stress.
Discussion
We describe a transcript class, DoGs, which are generated by readthrough transcription downstream of as many as 10% of protein-coding genes in response to osmotic stress. The lack of increase in the levels of the associated upstream mRNAs argues that regulation of transcription through DoG regions after osmotic stress occurs at the level of transcription termination. DoGs localize to chromatin, and we provide evidence for one DoG that it remains at its site of transcription. Perhaps a fraction of DoGs are tethered to chromatin through continued Pol II transcription, which would explain why DoGs as a group are partially non-polyadenylated. We show that osmotic stress enhances DoG levels by inducing IP3R-mediated Ca2+ release and signaling through the PKD/PKC and CaMKII pathways. Finally, we provide evidence suggesting a collective role for DoGs in maintaining nuclear integrity after stress.
DoGs and intergenic transcription
Widespread transcription downstream of genes potentially explains a significant fraction of pervasive transcription. Individual DoGs appear to be expressed at low levels: our estimate of doCXXC4 copy numbers is ~1 copy per cell before and ~30 after KCl treatment (Figure S4c). Nonetheless, total DoGs account for 15–30% of intergenic transcripts in both treated and untreated samples. This prevalence, in combination with the length of each individual DoG, represents a substantial contribution to overall transcriptional output, suggesting that many annotated lncRNAs may be DoGs. Indeed, transcripts overlapping all of our seven well-characterized DoGs appear in a study identifying lncRNAs (Hangauer et al., 2013); two of these transcripts (overlapping doCXXC4 and doRIN2) were annotated as lincRNAs, and the remainder as 3′-UTRs. Such classifications, although reasonable when based on sequencing data alone, demonstrate the importance of mechanistic insights for proper annotation.
DoGs are clearly different from lincRNAs in that they are not independent transcripts. Additionally, although mRNA 3′-UTR extensions of up to 20 kb have been described in neuronal cells (Miura et al., 2013), our observations that DoGs are strongly enriched in chromatin, while their associated mRNAs are not, and that they exist in both polyadenylated and non-polyadenylated forms, strongly suggest that DoGs are a distinct transcript type. The chromatin localization of DoGs argues that their 5′-coding sequences may be irrelevant to their nuclear function(s) (see below).
DoG regulation through termination of the upstream transcript
Osmotic stress ultimately reduces transcription termination of the upstream transcript, allowing extension through the DoG region. To the best of our knowledge, this mode of transcriptional regulation has not been described previously, but appears to be a mechanism with exceptional capacity for rapidly upregulating transcript levels. Such a mechanism circumvents the need for new transcription initiation, involving recruitment of transcription factors and assembly of active transcription complexes. Instead, Pol II that is already engaged in productive transcription continues transcribing through the DoG region.
Mechanisms regulating transcription termination by Pol II are incompletely understood. Pol II pauses close to the PAS, presumably allowing time for termination to happen (Gromak et al., 2006). Cleavage and polyadenylation create an unprotected 5′ end on the downstream nascent RNA, which then succumbs to Xrn-2 mediated degradation, ultimately dislodging Pol II from the DNA template (West et al., 2004). The upstream transcript is protected from premature cleavage and polyadenylation at cryptic polyA sites by binding of the U1 snRNP (Berg et al., 2012; Kaida et al., 2010). Knockdown of CPSF73, the catalytic subunit of the cleavage and polyadenylation complex, led to a 4-fold induction of DoG levels (Figure S6b), suggesting its role in suppressing transcription through DoG regions under normal conditions.
Interestingly, a recent report connects Ca2+ signaling through activation of PKD and CaMKII in enhancing Pol II elongation rates and the regulation of alternative splicing (Sharma et al., 2014). PKC/PKD and CaMKII signaling are also required for DoG induction (Figure 6d), suggesting a possible connection between PKD- and CaMKII-mediated increases in Pol II elongation rate (Sharma et al., 2014) and DoG transcription. Alternatively, PKC/PKD and CaMKII may regulate one or several factors involved in cleavage and polyadenylation to reduce their efficiency, thus decreasing transcriptional termination. We find that DoG regions are relatively depleted in PASs within the first 5 kb (Figure 5c). While many yet-to-be identified factors likely affect DoG transcription, this relative PAS depletion suggests that favoring elongation over termination promotes transcription of DoG regions (Figure 7e, left panel). Additionally, we find that genes generating DoGs are often expressed at relatively high levels, which is consistent with the putative role of DoGs in a stress response to maintain euchromatin. If DoG expression keeps euchromatin open, it makes sense that DoGs are generated wherever genes are being actively transcribed at high levels; these would be the locations where DoGs are most needed.
Potential functions of DoGs
The facts that transcription through DoG regions is regulated and that regions downstream of genes genome-wide show depletion of the PAS AATAAA suggest that DoGs are functional. Most mammalian tissues are protected from osmotic stress by the tight control of osmotic balance provided by the kidneys. However, in the event of kidney malfunction, severe dehydration or local injury, osmotic pressure can change drastically. Neuronal cells are highly sensitive to such changes (Finan and Guilak, 2010; Lin et al., 2005), correlating with our observations of pronounced DoG induction in cells of neuronal origin.
Signaling pathways induced by osmotic stress are not fully understood, but include ion channels, Ca2+ signaling and direct mechanical effects on the nucleus (Finan and Guilak, 2010; Guilak et al., 2002; Martins et al., 2012). Hyper-osmotic conditions cause water to leave the nucleus, leading to nuclear shrinkage and chromatin condensation. Osmotic stress therefore affects processes as diverse as DNA repair, replication and transcription, as these events are dependent on intact genomic structure (Finan and Guilak, 2010; Martins et al., 2012).
Recently, it was shown that a large group of heterogeneous transcripts consisting of repetitive element-containing RNAs tethered to the nuclear scaffold surrounding chromosomes help maintain euchromatin (Hall et al., 2014). Possibly, stress-inducible DoGs reinforce this chromosome-associated nuclear scaffold in situations where mechanical pressure forces chromatin to condense. Our observation that doSERBP1 remains at its site of transcription (Figure 3e) agrees with the idea of DoGs acting to stabilize their genomic regions of origin after stress. Indeed, we show that in SK-N-BE(2)C cells exposed to osmotic stress in the absence of DoG induction (orchestrated by IP3R inhibition), the osmotic stress-associated phenotype of nuclear shrinkage and chromatin collapse is aggravated. In fact, we do not observe nuclear shrinkage in cells treated with KCl alone – likely due to the mild dose and short time used – but only when KCl treatment follows IP3R inhibition and prevention of DoG induction. These data suggest a possible function for DoGs in maintaining nuclear integrity after stress (Figure 7e, right panel).
The possibility that DoGs are induced as part of a nuclear stress response suggests that other stresses may also induce DoG levels. Heat shock is known to alter the transcriptional landscape by reducing splicing (Shalgi et al., 2014) and is associated with a weakening of the nuclear scaffold due to the heat labile nature of nuclear matrix proteins (Roti Roti et al., 1997). These results prompted us to investigate its influence on DoG expression. Indeed, we observed a 10- to 30-fold induction of three DoGs examined after exposing SK-N-BE(2)C cells to 44°C for 2 or 4 h, compared to 37°C (Figure S7d). We conclude that increased DoG expression in response to stress is not limited to osmotic stress but may be a more general mechanism activated by nuclear scaffold stress.
In conclusion, we describe a class of long, chromatin-bound transcripts, which are generated by readthrough from upstream genes and are highly inducible by osmotic stress. DoGs contribute significantly to observed intergenic transcription and may play a role in protecting chromatin from osmotic stress.
Experimental Procedures
Statistical analysis
Evaluation of significance was performed using the Student’s t-test.
Accession number
The SRA Accession number for sequencing data is SRP058633.
RNA-Seq and Cap-Seq
Nuclear, ribo-minus treated RNA from untreated or KCl-treated (80 mM,1h) SK-N-BE(2)C cells were subjected to Illumina HiSeq 2000 according to the manufacturer’s protocols. For Cap-Seq, we used a modified version of the previous protocol (Xie et al., 2013). See Extended Experimental Procedures for further details.
FISH
We performed Stellaris FISH according to the manufacturer’s protocols with custom-made probes. See Extended Experimental Procedures for further details.
Supplementary Material
Highlights.
DoGs are a new transcript type generated by readthrough transcription.
DoGs are chromatin-bound transcripts highly inducible by osmotic stress.
DoGs are induced downstream of more than 10% of human protein-coding genes.
DoG induction is mediated through IP3 receptor signaling.
Acknowledgments
We thank Luke Gilbert and Jonathan Weissman for the kind gift of dCas9 system plasmids, Karen Adelman and Telmo Henriques for input on the CapSeq protocol, and Reut Shalgi for advice in designing the DoG-finding pipeline. We are grateful to Mingyi Xie, Nara Lee, Genaro Pimienta and Eric Guo for plasmids, reagents and protocols, Johanna Withers and Jessica Brown for critical discussion and Angela Miccinello for editorial assistance. This work was supported by grant GM026154 from the National Institutes of Health. A.V. is supported by the Wenner-Gren Foundations, the Swedish Society for Medical Research and the Sweden-America Foundation. J.A.S is an Investigator at the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–363. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bading H, Ginty DD, Greenberg ME. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science. 1993;260:181–186. doi: 10.1126/science.8097060. [DOI] [PubMed] [Google Scholar]
- Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ. Inositol trisphosphate and calcium signalling mechanisms. Biochim Biophys Acta. 2009;1793:933–940. doi: 10.1016/j.bbamcr.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan JD, Guilak F. The effects of osmotic stress on the structure and function of the cell nucleus. J Cell Biochem. 2010;109:460–467. doi: 10.1002/jcb.22437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak N, West S, Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilak F, Erickson GR, Ting-Beall HP. The effects of osmotic stress on the viscoelastic and physical properties of articular chondrocytes. Biophys J. 2002;82:720–727. doi: 10.1016/S0006-3495(02)75434-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Carone DM, Gomez AV, Kolpa HJ, Byron M, Mehta N, Fackelmayer FO, Lawrence JB. Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell. 2014;156:907–919. doi: 10.1016/j.cell.2014.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kapranov P, St Laurent G, Raz T, Ozsolak F, Reynolds CP, Sorensen PH, Reaman G, Milos P, Arceci RJ, Thompson JF, et al. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ un-annotated RNA. BMC Biol. 2010;8:149. doi: 10.1186/1741-7007-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Liu SJ, Lim IT. Disorders of water imbalance. Emerg Med Clin North Am. 2005;23:749–770. doi: 10.1016/j.emc.2005.03.001. ix. [DOI] [PubMed] [Google Scholar]
- Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins RP, Finan JD, Guilak F, Lee DA. Mechanical regulation of nuclear structure and function. Annu Rev Biomed Eng. 2012;14:431–455. doi: 10.1146/annurev-bioeng-071910-124638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Wilhelm D, Dinger ME, Solda G, Korbie DJ, Glazov EA, Truong V, Schwenke M, Simons C, Matthaei KI, et al. Expression of distinct RNAs from 3’ untranslated regions. Nucleic Acids Res. 2011;39:2393–2403. doi: 10.1093/nar/gkq1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, Fredlund E, Pattyn F, Rihani A, Van Maerken T, Vermeulen J, Kumps C, Menten B, De Preter K, Schramm A, et al. An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours. Oncogene. 2010;29:3583–3592. doi: 10.1038/onc.2010.106. [DOI] [PubMed] [Google Scholar]
- Miura P, Shenker S, Andreu-Agullo C, Westholm JO, Lai EC. Widespread and extensive lengthening of 3’ UTRs in the mammalian brain. Genome Res. 2013;23:812–825. doi: 10.1101/gr.146886.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SK, Chua FK, Choo AB. Intracellular responses of productive hybridomas subjected to high osmotic pressure. Biotechnol Bioeng. 1995;46:525–535. doi: 10.1002/bit.260460605. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roti Roti JL, Wright WD, VanderWaal R. The nuclear matrix: a target for heat shock effects and a determinant for stress response. Crit Rev Eukaryot Gene Expr. 1997;7:343–360. doi: 10.1615/critreveukargeneexpr.v7.i4.30. [DOI] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R, Hurt JA, Lindquist S, Burge CB. Widespread inhibition of posttranscriptional splicing shapes the cellular transcriptome following heat shock. Cell Rep. 2014;7:1362–1370. doi: 10.1016/j.celrep.2014.04.044. [DOI] [PubMed] [Google Scholar]
- Sharma A, Nguyen H, Geng C, Hinman MN, Luo G, Lou H. Calcium-mediated histone modifications regulate alternative splicing in cardiomyocytes. Proc Natl Acad Sci U S A. 2014;111:E4920–E4928. doi: 10.1073/pnas.1408964111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Gromak N, Proudfoot NJ. Human 5’ → 3’ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- Xie M, Li M, Vilborg A, Lee N, Shu MD, Yartseva V, Sestan N, Steitz JA. Mammalian 5’-capped microRNA precursors that generate a single microRNA. Cell. 2013;155:1568–1580. doi: 10.1016/j.cell.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.