SUMMARY
Precise temporal control of gene expression or deletion is critical for elucidating gene function in biological systems. However, establishment of human pluripotent stem cell (hPSC) lines with inducible gene knockout (iKO) remains challenging. We explored building iKO hPSC lines by combining CRISPR/Cas9-mediated genome editing with the Flp/FRT and Cre/LoxP system. We found that “dual-sgRNA targeting” is essential for biallelic knock-in of FRT sequences to flank the exon. We further developed a strategy to simultaneously insert an activity-controllable recombinase-expressing cassette and remove the drug-resistance gene, thus speeding up the generation of iKO hPSC lines. This two-step strategy was used to establish human embryonic stem cell (hESC) and induced pluripotent stem cell (iPSC) lines with iKO of SOX2, PAX6, OTX2, and AGO2, genes that exhibit diverse structural layout and temporal expression patterns. The availability of iKO hPSC lines will substantially transform the way we examine gene function in human cells.
INTRODUCTION
Human pluripotent stem cells (hPSCs), including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), are useful tools for elucidating regulatory processes during early development and disease pathogenesis under the human genetic background (Takahashi et al., 2007; Thomson et al., 1998; Yu et al., 2007). Genetic modification, including gene knockout (KO), further expands the utility of hPSCs in studying gene function in human embryogenesis or human genetic diseases (Zou et al., 2010). However, constitutive KO of genes that function in hPSC self-renewal or survival may render the cells unable to propagate or survive, thus limiting their utility. Furthermore, phenotypes arising from gene KO in early development may obscure the analysis of gene function at later stages, as many genes exert pleiotropic functions throughout differentiation. Compensatory effects from other genes, unwanted lineage selection, and cellular transformation are also potential concerns of long-term culture of constitutive gene KO hPSCs. Thus, precise temporal control of gene KO in hPSCs is often necessary or highly beneficial for elucidating gene functions and molecular pathways that underlie complex human traits.
In animals, inducible gene KO is typically accomplished by combining homologous recombination-mediated gene targeting with the flippase (Flp)/flippase recognition target (FRT) and Cre/LoxP system (Branda and Dymecki, 2004; Capecchi, 2005). However, generating inducible gene knockout (iKO) hPSCs using the same procedure has proven very difficult and laborious due to their much lower efficiency of homologous recombination (Giudice and Trounson, 2008; Hockemeyer and Jaenisch, 2010; Mali and Cheng, 2012; Zwaka and Thomson, 2003). Additionally, while homozygous targeted mice can be generated through mating, the generation of homozygously targeted hPSCs requires sequential targeting of the two individual alleles (Bu et al., 2010; Song et al., 2010). Furthermore, repeated gene targeting and cellular cloning is not only laborious, but may also be detrimental to the maintenance and differentiation of hPSCs (Baker et al., 2007). Thus, generation of iKO hPSC lines is very challenging and requires new strategies to support the next wave of discovery in human biology.
Several gene targeting strategies based on sequence-specific nucleases (SSNs) have recently been developed to generate double-strand breaks in targeted DNA, including zinc-finger nucleases (ZFNs) (Miller et al., 2007; Porteus and Baltimore, 2003; Urnov et al., 2010; Wood et al., 2010), transcription activator-like effector nucleases (TALENs) (Boch et al., 2009; Christian et al., 2010; Miller et al., 2011; Moscou and Bogdanove, 2009; Wood et al., 2010), and the clustered regularly interspaced short palindromic repeats (CRISPR)/ CRISPR-associated (Cas) system (Cong et al., 2013; Garneau et al., 2010; Gasiunas et al., 2012; Jinek et al., 2012; Jinek et al., 2013; Mali et al., 2013b; Sapranauskas et al., 2011). These targeted double strand breaks are then repaired by either error-prone non-homologous end joining (NHEJ) or high-fidelity homology-directed repair (HDR) (Helleday et al., 2007; Lieber, 2010). Newly-developed site-specific nucleases have facilitated gene targeting in hPSCs including generation of gene KO hPSCs (Ding et al., 2013a; Ding et al., 2013b; Hockemeyer et al., 2009; Hockemeyer et al., 2011; Mali et al., 2013b; Zou et al., 2009). However, an efficient method to generate gene inducible KO hPSC lines has not been shown until recently (Gonzalez et al., 2014). Gonzalez et al created a master hPSC line with doxycycline (DOX)-inducible Cas9 expression cassette inserted in AAVS1 site. After DOX treatment and two-rounds of single guide RNA (sgRNA) transfection at specific times, random indels are introduced into targeted gene sites via NHEJ, and potentially resulted in gene KO. However, the resultant cells are a mixed population of KO and non-KO cells with unpredictable genotypes ascribed to the random indel insertion. So, while the use of this iCRISPR system is a potentially new way to generate a wide array of constitutive KO hPSC lines after subcloning, the inefficiency of KO and randomness of indel insertion make it less practical for inducing gene KO in most of the cells during stem cell differentiation or at a particular stage of the pathological process. Thus, it is highly desirable to establish an efficient inducible gene KO system so that target genes can be deleted in a uniform manner at any given time.
Here, we report an efficient two-step strategy to generate iKO hPSC lines by combining CRISPR/Cas9-mediated genome editing with the Flp/FRT and Cre/LoxP system. We found that “dual-sgRNA targeting” is critical for bi-allelic knock-in of FRT sequences to flank the exon within a gene of interest in one step. In addition, we devised a strategy to simultaneously insert an activity-controllable recombinase-expressing cassette and remove a drug-resistance gene, thus speeding up the generation of iKO hPSC lines. This two-step strategy was reproducible as demonstrated in generation of hESC and iPSC lines with iKO in SOX2, PAX6, OTX2, and AGO2 genes.
RESULTS
Generation of homozygous FRT knock-in hPSC lines by “dual-sgRNA targeting”
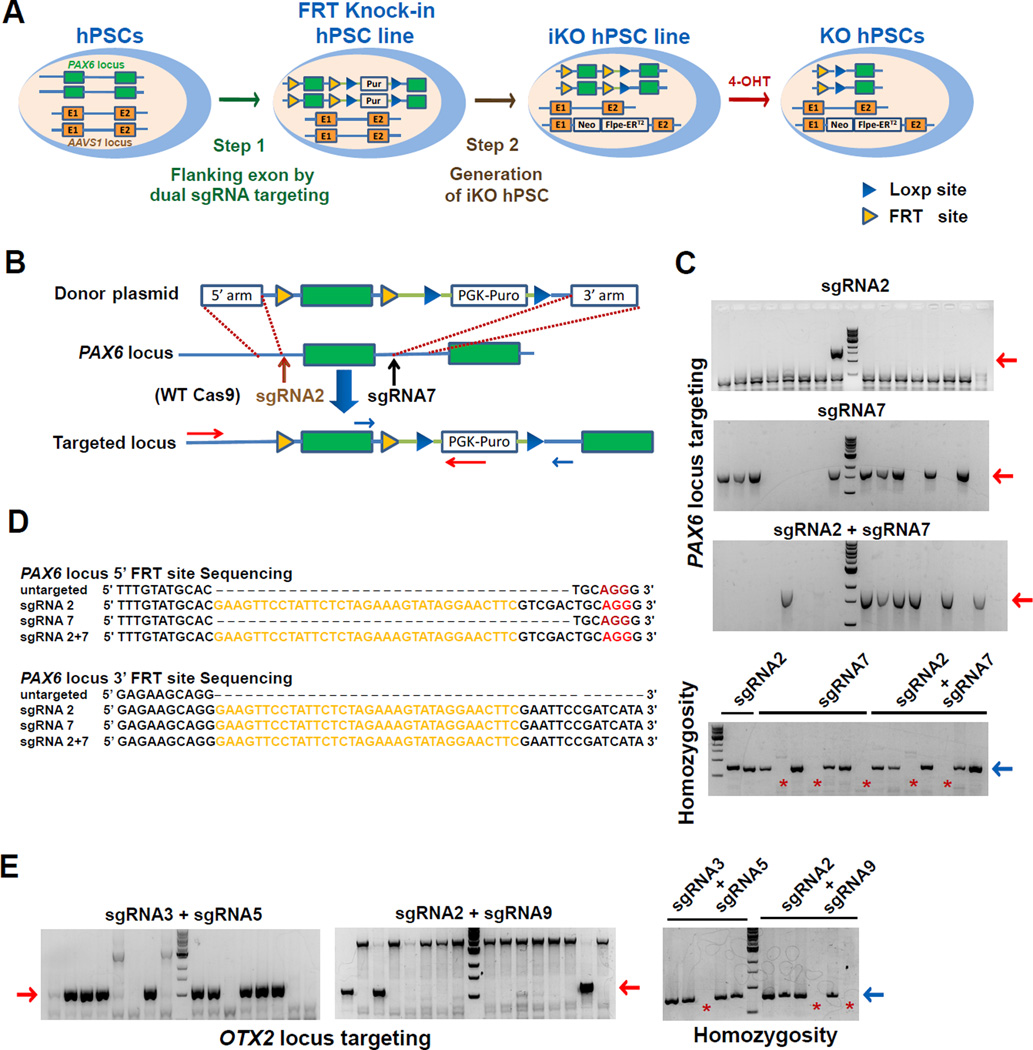
The most critical step in engineering iKO hPSC lines is to ensure predictable gene knockout by knock-in of FRT to flank an exon via CRISPR/Cas9-catalyzed HDR. We developed a one-step genome targeting method using CRISPR/Cas9 to generate homozygous hPSC lines with a FRT-flanked exon within a gene of interest instead of the conventional sequential targeting strategy (Figure 1A). We then devised a unique method for establishing iKO hPSC lines by simultaneous insertion of the activity-controllable enhanced-flippase (Flpe) recombinase expression cassette, named Flpe-ERT2, into the AAVS1 locus and removal of the drug-resistance expression cassette in targeted cells (Figure 1A). We reproduced this strategy using four genes that contain either multiple exons (PAX6, OTX2, and AGO2) or large exons (SOX2) as well as those that are not expressed (PAX6) or expressed at the PSC stage (SOX2, OTX2, and AGO2).
Figure 1. Generation of FRT knock-in hPSC lines using Cas9 nuclease.
(A) Schematic diagram of the two-step strategy for generating iKO hPSC line using PAX6 as an example. The first step is to generate hPSC line with exons in both alleles flanked by FRT sites using our dual-sgRNA targeting strategy. The second step is to remove the drug-resistance cassette and insert Flpe-ERT2 expression cassette into AAVS1 locus to establish iKO hPSC line. Upon treatment with 4-OHT, Flpe-ERT2 will translocate into the nucleus and recombine the FRT-flanked exons, thus resulting in frame-shift of a protein-coding sequence and knockout of the targeted gene. Exons are shown as green or orange boxes, blue triangles as LoxP sites, yellow triangles as FRT sites.
(B) Schematic depiction of the targeting strategy for exon 4 of PAX6 locus. Vertical arrows indicate sgRNA2 and sgRNA7 targeting sites. Red and blue horizontal arrows indicate PCR genotyping primers for assaying PAX6 locus targeting and homozygosity, respectively. Donor plasmids: PGK, phosphoglycerate kinase promoter; Puro, puromycin-resistance gene.
(C) PCR genotyping of hESC clones targeted by sgRNA2 (first panel), sgRNA7 (second panel) or both sgRNA2 and sgRNA7 (third panel). The expected PCR product for correctly targeted PAX6 locus is ~1800bp (red arrows). Correctly targeted clones underwent further homozygosity assay (fourth panel). Clones with the PCR products ~700bp are heterozygous (blue arrow), and those clones without PCR products are homozygous (red asterisk).
(D) Representative sequencing results of targeted heterozygous or homozygous clones in PAX6 locus using sgRNA2 (heterozygous), sgRNA7 (homozygous) or both sgRNA2 and sgRNA7 (homozygous). The PAM sequences and FRT sequences are labeled in red and yellow, respectively.
(E) OTX2 locus targeting. PCR genotyping of hESC clones targeted by both sgRNA3 and sgRNA5 (left panel) or both sgRNA2 and sgRNA9 (middle panel). Expected PCR products for correctly targeted OTX2 locus are ~1200bp (Red arrows). Correctly targeted clones underwent further homozygosity assay (right panel). Those clones with the PCR products of ~750bp are heterozygous (blue arrow), and those clones without these PCR products are homozygous (red asterisks).
See also Figure S1 and S2.
We first screened sgRNAs that effectively cleave DNA for individual genes. For PAX6, a transcription factor expressed in differentiated neural cells, but not in hPSCs (Zhang et al., 2010), we designed seven sgRNAs that target unique sequences in the introns flanking exon 4 (Figure S1A). Genomic deletion assay in 293T cells indicated that sgRNA2 and sgRNA7 exhibited the most effective genome deletion activity as revealed by shortened PCR products (Figure S1B). We then introduced two FRT sequences into the targeting sites by transfecting hESCs with donor plasmids and the Cas9 plasmid, along with either sgRNA2, sgRNA7, or both sgRNA2 and sgRNA7 (Figure 1B). After drug selection, PCR genotyping and sequencing showed that ~50% of the clones were targeted in one (heterozygous) or both (homozygous) alleles in groups using sgRNA7 or both sgRNA2 and sgRNA7 (Figure 1C and Table 1). In the group using sgRNA2 alone, the targeting efficiency was extremely low (2 out of 136 analyzed clones), and there were no homozygous clones (Figure 1C and Table 1). Sequencing results from homozygous clones revealed that the FRT sequence, which was expected to be inserted into the sgRNA7 targeting site and was close to the floxed drug-resistance gene (3' FRT site), was correctly inserted in both alleles of targeted cells using sgRNA7 or both sgRNA2 and sgRNA7 (Figure 1D, Table 1, and Table S1). Surprisingly, the 5' FRT site, which was expected to be inserted into the sgRNA2 targeting site and was far away from the drug-resistance gene expression cassette, was lost in both alleles of all the homozygous clones from the group using only sgRNA7 (Figure 1D, Table 1, and Table S1). In contrast, the 5' FRT site was correctly inserted in both alleles of PAX6 in most of the clones from the group using both sgRNA2 and sgRNA7 (Figure 1D, Table 1, and Table S1). Similar sequencing results were obtained from heterozygous clones (Table S1). These results suggest that using two sgRNAs to target the 5' and 3' FRT insertion sites is necessary for generating correctly targeted homozygous hESC lines. By using two sgRNAs to target the 5' and 3' FRT insertion sites in OTX2 locus, we generated homozygous hESC lines with exon3 of OTX2 flanked by FRT sequences (Figure 1E, Table 1, and Figure S1C–S1E).
Table 1.
Summary of targeting experiments using CRISPR/Cas9
| Targeted clones identified by PCR genotyping | Homozygous clones identified by sequencing | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cell Line |
Targeted gene |
Targeting gRNAs | Cas9 | Clone number |
Hetero zyous |
Homo zygous |
Targeting efficiency (%) |
Homozyous efficiency (%) |
correct / sequenced |
homozygous efficiency (%) |
| H9 | PAX6 | sgRNA2 | WT | 136 | 2 | 0 | 15 | 0 | 0/0 | 0.0 |
| sgRNA7 | WT | 35 | 7 | 10 | 48.6 | 28.6 | 0/10 | 0.0 | ||
| sgRNA2+sgRNA7 | WT | 34 | 10 | 8 | 52.9 | 23.5 | 8/8 | 23.5 | ||
| H9 | SOX2 | sgRNA1A | WT | 66 | 1 | 0 | 1.5 | 0 | 0/0 | 0.0 |
| sgRNA6B | WT | 71 | 37 | 6 | 60.6 | 8.5 | 0/6 | 0.0 | ||
| sgRNA1A+sgRNA6B | WT | 84 | 27 | 11 | 45.2 | 13.1 | 10/11 | 11.9 | ||
| sgRNApair1A+sgRNApair6B | Nickase | 72 | 27 | 13 | 55.6 | 18.1 | 7/12 | 10.5 | ||
| sgRNApair2A+sgRNApair5B | Nickase | 87 | 45 | 14 | 67.8 | 16.1 | 11/14 | 12.6 | ||
| H9 | OTX2 | sgRNA2+sgRNA9 | WT | 110 | 9 | 6 | 13.6 | 5.5 | 4/5 | 4.4 |
| sgRNA3+sgRNA5 | WT | 56 | 31 | 3 | 60.7 | 5.4 | 1/2 | 2.7 | ||
| H9 | AGO2 | sgRNApair8+sgRNApair15 | Nickase | 95 | 34 | 19 | 55.8 | 20 | 14/18 | 15.6 |
| sgRNApair9+sgRNApair14 | Nickase | 81 | 20 | 14 | 42 | 17.3 | 14/14 | 17.3 | ||
| D90A | AGO2 | sgRNApair7+sgRNApair14 | Nickase | 104 | 37 | 16 | 51.0 | 15.4 | 12/15 | 12.3 |
| D90D | SOX2 | sgRNA1A+sgRNA6B | WT | 114 | 41 | 64 | 92.1 | 56.1 | 23/25 | 51.6 |
We then asked if targeting two sites is necessary for genes with different structures and expression patterns. For this, we designed sgRNAs targeting unique sequences flanking the SOX2 gene. In contrast to PAX6, SOX2 is actively expressed in hESCs (Ran et al., 2013; Rizzino, 2013) and the distance between designated FRT insertion sites is approximately 3.5 kilobases. By screening sgRNAs, we identified sgRNA1A and sgRNA6B to be effective in targeting 5' and 3' FRT insertion sites in the SOX2 locus (Figure S2A–S2C). Similar to the results from PAX6 targeting, the correctly targeted homozygous SOX2 clones were obtained only when using both sgRNA1A and sgRNA6B. The use of either sgRNA alone resulted in either low targeting efficiency and no identified homozygous clones (using 5' FRT insertion site targeting sgRNA, sgRNA1A), or loss of the 5' FRT site (using 3' FRT insertion site targeting sgRNA, sgRNA6B) (Figure S2D–S2F, Table 1, and Table S1). Together, our results indicate that utilizing gRNAs targeting both 5’ and 3’ FRT insertion sites is essential to generate correct homozygous hESC lines with FRT-flanked exons. This strategy, named dual-sgRNA targeting, is applicable to genes of varied structure.
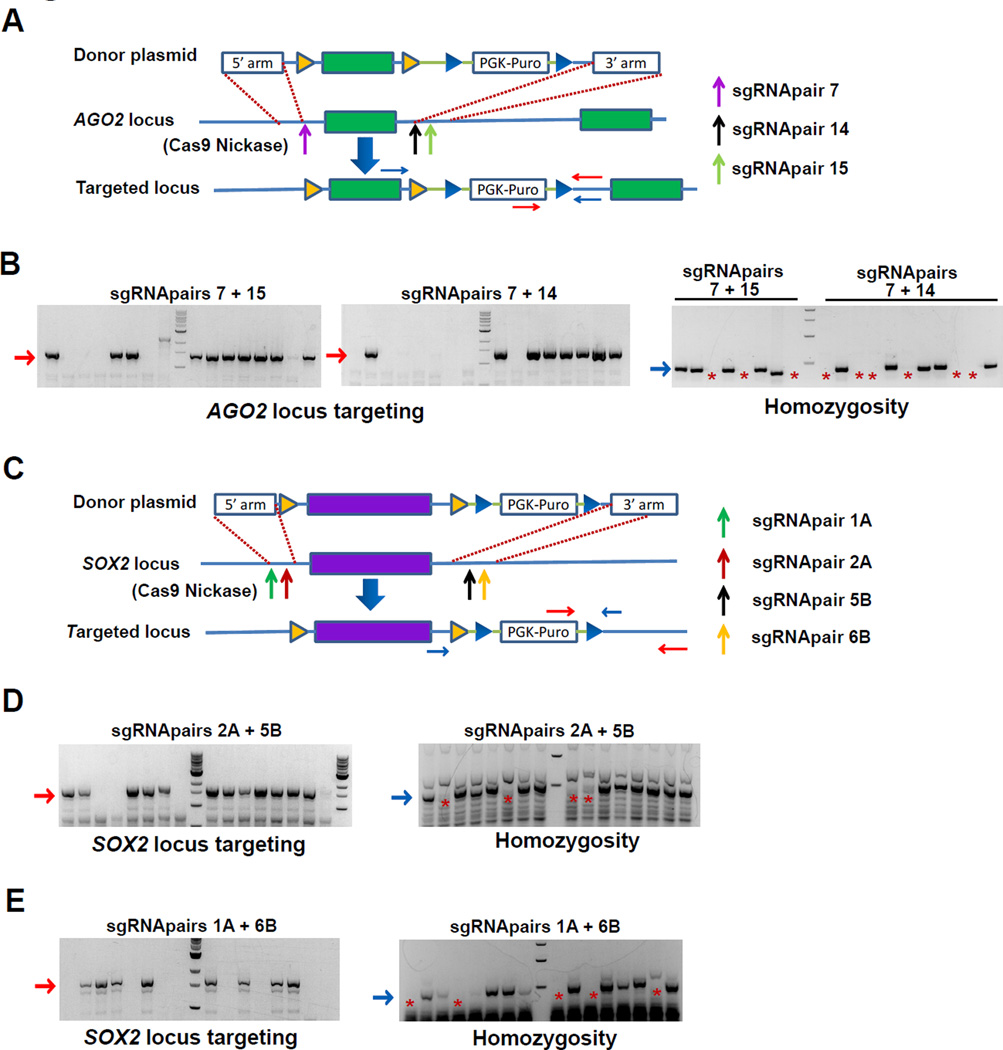
Recently, the paired nickase strategy, which combines the D10A mutant nickase version of Cas9 (Cas9 nickase) with a pair of offset sgRNAs complementary to opposite strands of the target site, has been developed to reduce the off-target effect of Cas9-mediated genome editing since binding of both offset sgRNAs is required for double-strand cleavage (Cho et al., 2014; Mali et al., 2013a; Ran et al., 2013). We thus tested the feasibility of generating homozygous FRT knock-in hESC lines by Cas9 nickase and the dual-sgRNA targeting strategy using AGO2 and SOX2 as examples. sgRNApair 7, sgRNApair 14, and sgRNApair 15 were identified to be effective in targeting the 5' and 3' FRT insertion sites in AGO2 locus (Figure 2A and S3A–S3B). Using Cas9 nickase combined with sgRNApair 7 and sgRNApair 14, or sgRNApair 7 and sgRNApair 15, we generated homozygous hESC lines with exon3 of AGO2 flanked by FRT sequences with an efficiency comparable to wild-type Cas9 (Figure 2A, 2B, and Table 1). Similarly, by using Cas9 nickase and two pairs of offset sgRNAs (sgRNApair 2A and sgRNApair 5B, sgRNApair 1A and sgRNApair 6B) to target the 5' and 3' FRT insertion sites in SOX2 locus, we generated homozygous hESC lines with the SOX2 gene flanked by FRT sequences (Figure 2C–2E, Table 1).
Figure 2. Generation of FRT knock-in hPSC lines using Cas9 nickase.
(A) Schematic depiction of the targeting strategy for exon 3 of AGO2 locus. Vertical arrows indicate targeting sites for sgRNApair 7 (sgRNA7+sgRNA9), sgRNApair 14 (sgRNA14+sgRNA17) or sgRNApair 15 (sgRNA15+sgRNA18). PCR genotyping primers for AGO2 locus targeting test (red arrows) or homozygosity test (blue arrows) are indicated.
(B) PCR genotyping of hESC clones targeted by both sgRNApair 7 and sgRNApair 15 (left panel) or both sgRNApair 7 and sgRNApair 14 (middle panel). Expected PCR products for correctly targeted AGO2 locus are ~1400bp (red arrows). Correctly targeted clones underwent further homozygosity assay (right panel). Those clones with the PCR products of about 420bp are heterozygous (blue arrow), and those without these PCR products are homozygous (red asterisk).
(C) Schematic diagram depicting the targeting strategy for SOX2 locus using Cas9 nickase. The vertical arrows indicate targeting sites by SOX2 sgRNApair 1A (sgRNA1A+sgRNA1B), sgRNApair 2A (sgRNA2A+sgRNA2B), sgRNApair 5B (sgRNA5A+sgRNA5B) or sgRNApair 6B (sgRNA6A+sgRNA6B). PCR genotyping primers for SOX2 locus targeting test (red horizontal arrows) or homozygosity test (blue horizontal arrow) are indicated.
(D–E) SOX2 locus targeting. PCR genotyping of hESC clones targeted using Cas9 nickase combined with sgRNApair 2A and sgRNApair 5B (D), or with sgRNApair 1A and sgRNApair 6B (E). Expected PCR products for correctly targeted SOX2 locus are ~1700bp (red arrow). Those clones with the PCR products of about 550bp are heterozygous (blue arrow), and those clones without these PCR products are homozygous (red asterisks).
See also Figure S3.
Giving the increasing use of iPSCs, we examined the homozygous FRT knock-in efficiency in SOX2 locus or AGO2 locus using two human iPSC lines (Chen et al., 2014). The targeting efficiency and homozygous efficiency in these human iPSCs was comparable to those in hESCs. Both wild-type Cas9 and Cas9 nickase worked well for genome targeting in human iPSCs (Figure S3C–3D and Table 1). Thus, our dual-sgRNA targeting strategy can also be applied to generating homozygous human iPSC lines.
Establishing iKO hPSC lines by simultaneous insertion of an inducible cassette and removal of a drug-resistance expression cassette
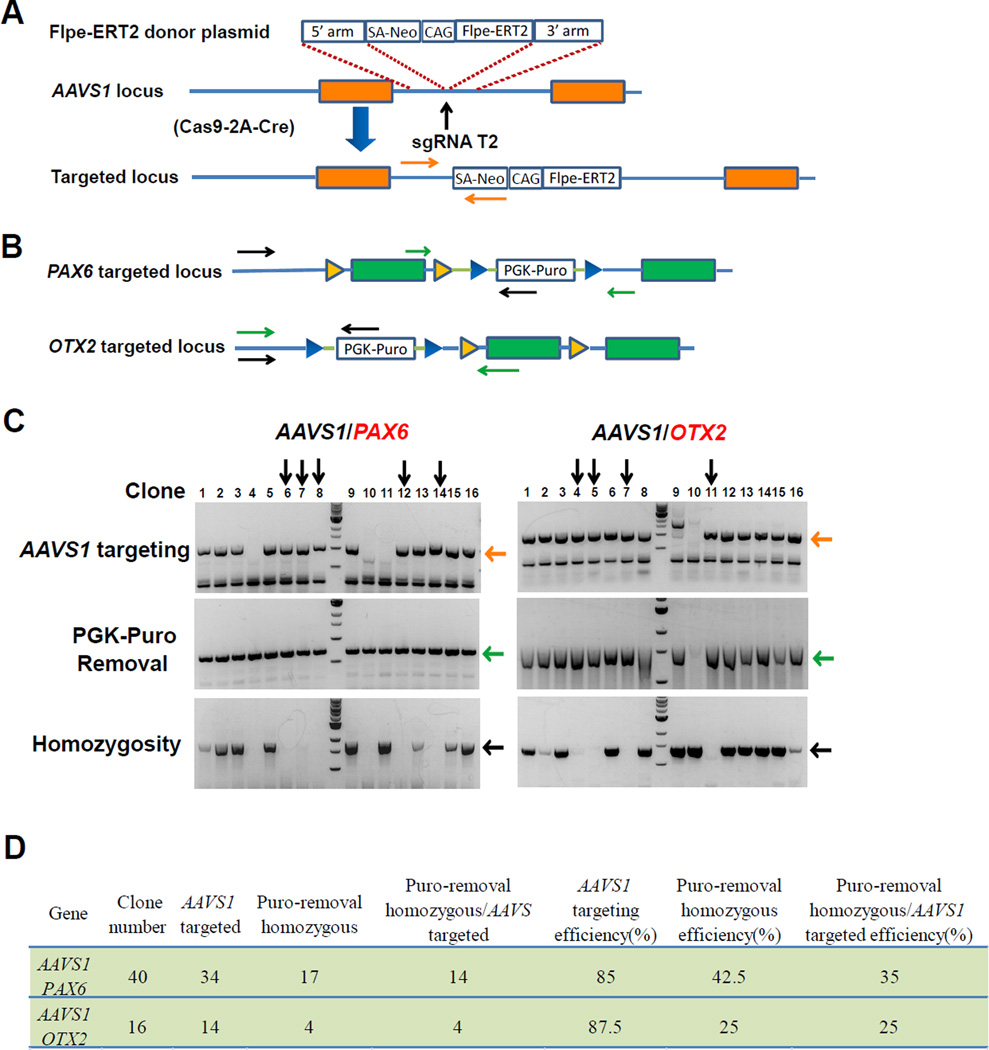
To generate iKO hPSC lines, we need to remove the drug-resistance expression cassette in the FRT-flanked gene locus and insert the control element, Flpe-ERT2, into the AAVS1 locus. This is typically achieved by sequential removal of the drug-resistance cassette through Cre recombinase application, genotyping of resulting clones, expansion of corrected clones, and then a second round of targeting to insert the control element. This laborious process takes approximately six weeks. Considering the high genome targeting efficiency of the CRISPR/Cas9 system, we explored the one-step strategy to simultaneously remove the drug resistance-gene expression cassette and target the Flpe-ERT2 expression cassette into the AAVS1 locus. hESCs with the exon4 of PAX6 flanked by FRT sequences were electroporated with a donor plasmid containing Flpe-ERT2 expression cassette (AAVS1-neo-CAG-FlpeERT2), sgRNA T2 targeting AAVS1 locus, and Cas9-2A-Cre plasmid that enabled co-expression of Cas9 and Cre recombinase in the same cells (Figure 3A). After drug selection, 14 out of 40 clones were identified to be correct with insertion of Flpe-ERT2 expression cassette in the AAVS1 locus and removal of the PGK-puromycin cassette in both alleles of the PAX6 locus (Figure 3B–3D). Using the same strategy, we inserted the CAG-Flpe-ERT2 cassette into the AAVS1 locus and removed the PGK-puromycin cassette in hESCs with the exon3 of OTX2 flanked by FRT sequences at the same time. Four out of sixteen clones were identified as correct (Figure 3B–3D). SOX2 iKO and AGO2 iKO hESC lines as well as SOX2 iKO hiPSC lines were established using a similar strategy with comparable efficiency (Figure S4A–4C). Thus, this one-step approach enables establishment of iKO human ESC or iPSC lines within four weeks.
Figure 3. Generation of iKO hPSC lines.
(A) Schematic depiction of the targeting strategy for AAVS1 locus. Exons are shown as orange boxes. The vertical arrows indicate targeting site by sgRNA T2 in AAVS1 locus. The orange horizontal arrows indicate PCR genotyping primers for AAVS1 locus targeting. Donor plasmids: SA-Neo, splice acceptor sequence followed by a T2A self-cleaving peptide sequence and the neomycin resistance gene; CAG, synthetic CAGGS promoter containing the actin enhancer and the cytomegalovirus early promoter; Flpe, enhanced Flp recombinase; ERT2, mutated ligand-binding domain of estrogen receptor (ER).
(B) Genotyping strategy for Cre recombinase-mediated removal of the resistance gene expression cassette in FRT knock-in PAX6 or OTX2 locus. Exons are shown as green boxes, blue triangles as LoxP sites, yellow triangles as FRT sites. The green and black arrows indicate PCR primers for assaying removal of the Cre recombinase-mediated resistance gene expression cassette and homozygosity, respectively.
(C) PCR genotyping of PAX6- or OTX2-iKO hESCs clones. The expected PCR products for correctly targeted AAVS1 locus are ~1000bp (Orange arrows). The expected PCR products for PGK-Puro removal in flanked PAX6 or OTX2 locus is ~700bp or ~1500bp (Green arrows), respectively. The expected PCR products for homozygosity of PGK-Puro removal (PGK-Puro was removed in both targeted alleles) in FRT knock-in PAX6 or OTX2 locus are ~1800bp or ~1450bp (Black arrows), respectively. Those clones with the positive PCR products in homozygosity test are heterozygous. Vertical arrows indicate clones with correct AAVS1 locus targeting, PGK-Puro removal, and homozygosity of PGK-Puro removal in FRT knock-in PAX6 or OTX2 locus.
(D) Summary of the efficiency of second step targeting in hESC line with the exons of PAX6 or OTX2 flanked by FRT sequences.
See also Figure S4.
Off target effects are a major concern of CRISPR/Cas9 system. By genomic PCR and Sanger sequencing, we sequenced 6–7 potential off-targets for every sgRNA target site in the above iKO hESC lines according to Online tools (http://crispr.mit.edu/). In the total 114 potential off-target sites, we did not detect indel formation in these sites (Table S2), suggesting high specificity of CRISPR/Cas9-based genome targeting in generating gene iKO hPSC lines.
Expression of target genes is depleted upon induction
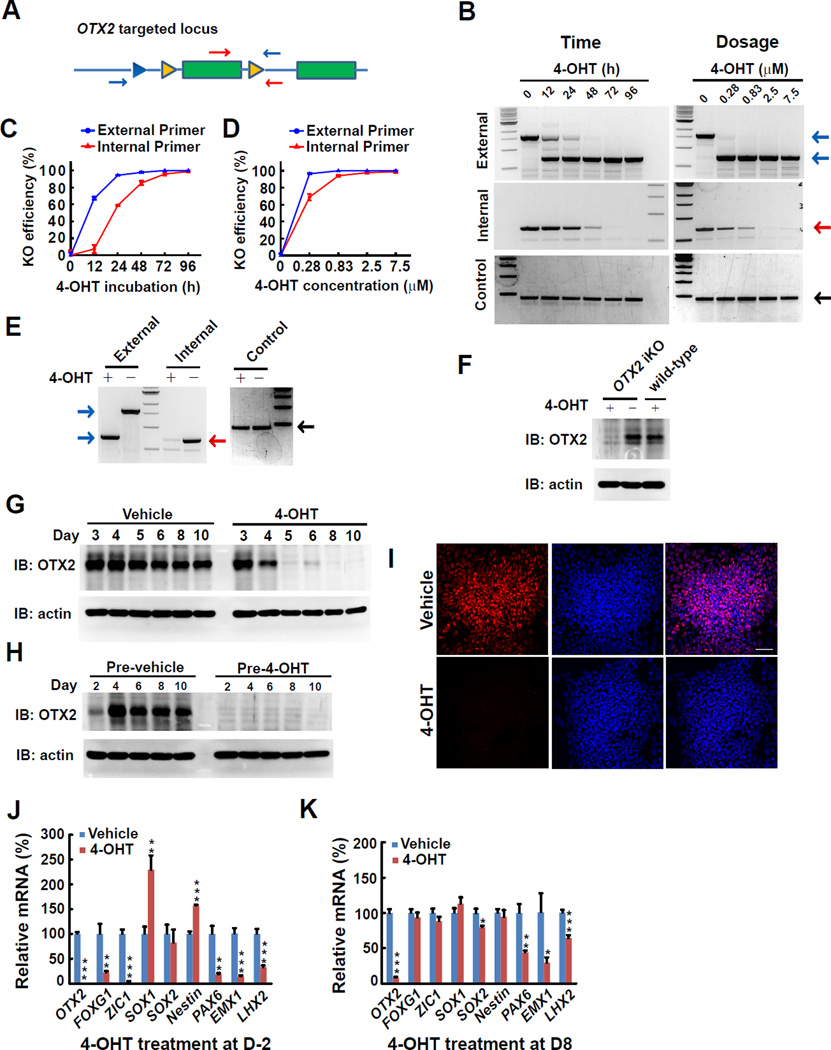
To validate inducible gene KO in our established hPSCs, we treated the cells with 4-hydroxytamoxifen (4-OHT) and assayed iKO efficiency at genomic level. We used external primer pairs to detect both the un-recombined FRT-flanked exons and recombined FRT-flanked exons after 4-OHT treatment. Additionally, we used an internal primer pair as a more sensitive measure to detect the un-recombined FRT-flanked exons (Figure 4A). As shown in figure 4B–D, 4-OHT treatment depleted the FRT-flanked exon in OTX2 locus in a time- and dose-dependent manner. Most of the FRT-flanked exons were depleted within 48 hours of 4-OHT treatment (85.2% for internal primer pair, 97.8% for external primer pair), and almost complete depletion of flanked exons could be obtained after 96 hours of 4-OHT treatment at a concentration of 2.5µM (Figure 4B, left panel, and Figure 4C). Treatment with 4-OHT at a concentration as low as 0.83µM effectively depleted FRT-flanked exons (94.1% for internal primer pair, 100% for external primer pair) (Figure 4B, right panel, and Figure 4D). Depletion of FRT-flanked genome was also achieved in differentiated neural cells from OTX2 iKO hESCs after 4-OHT treatment (Figure 4E). These results suggest that 4-OHT treatment can efficiently deplete FRT-flanked genome at the PSC stage or in differentiated cells. At the protein level, the expression of OTX2 was similar between OTX2 iKO hPSCs and WT hPSCs without 4-OHT treatment, demonstrating that our genome editing process does not affect the normal expression of the target gene (Figure 4F). OTX2 was significantly diminished within 2–3 days of 4-OHT treatment at the PSC stage or during neural differentiation (Figure4F and 4G). Transient pretreatment of OTX2 iKO cells with 4-OHT permanently eliminated expression of OTX2 protein during neural differentiation as revealed by immunoblotting (Figure 4H) and immunofluorescence (Figure 4I). Similarly, PAX6 iKO cells exhibit similar expression of PAX6 protein when compared with WT cells during neural differentiation in the absence of 4-OHT. 4-OHT treatment effectively depleted FRT-flanked exon of PAX6 and PAX6 protein expression in PAX6 iKO cells (Figure S5A–5E and Table S3).
Figure 4. Inducible depletion of OTX2 and functional consequence.
(A) Schematic depiction of PCR primer sets for genotyping. Blue arrows: external primer pair, Red arrows: internal primer pair.
(B–D) Depletion of FRT-flanked exons of OTX2 in a time-(at 2.5µM, left panel) and dose-(at 72 hours, right panel) dependent manner upon 4-OHT treatment. The expected sizes of PCR products using external primer pair for un-recombined FRT-flanked exons (blue arrow, upper) or recombined FRT-flanked exons (blue arrow, lower) is ~1600bp or ~650bp, respectively. The red arrow indicates the PCR products (~650bp) using internal primer pair. The black arrow indicates PCR products (~420bp) amplified from AGO2 locus which did not undergo recombination (control). The KO efficiency was plotted in (C) and (D) by calculating the density of PCR products from 3–4 independent experiments. Data are represented as mean+/− SEM.
(E) Depletion of FRT-flanked exons of OTX2 upon 72 hours of treatment with 2.5µM 4-OHT in OTX2 iKO neuroepithelial cells that underwent neural differentiation for 8 days. The arrows were indicated as in (B)
(F) Western blotting shows depletion of OTX2 protein upon treatment with 1.25µM 4-OHT for 72 hours in OTX2 iKO and parental human ESCs.
(G) Western blotting shows OTX2 protein expression along neural differentiation upon treatment with 1.25µM 4-OHT at day 3.
(H) Western blotting shows permanent depletion of OTX2 expression upon 4 days of treatment with 1.25µM 4-OHT commenced 2 days before neural differentiation. (I) Immunostaining shows depletion of OTX2 protein expression at day 8 following 4 days of treatment with 1.25µM 4-OHT commenced 2 days before neural differentiation. Scale bar, 20µM.
(J–K) RT-qPCR shows genes expression after 4 days of treatment with 1.25µM 4-OHT 2 days before neural differentiation (cells were collected at day 8, J) or 8 days after neural differentiation (cells were collected at day 16, K). Student's t test. Data are represented as mean+/− SEM. *p<0.05; **p<0.01; ***p<0.001.
See also Figure S5, Table S3 and Table S5.
Functional consequence of gene KO during hPSC differentiation
Inducible gene KO hPSCs offer a tool to look into the roles of genes in human cells at different developmental stages. As a proof-of-principle, we examined the roles of OTX2 during early neural development. Animal studies indicate that OTX2 is essential in defining the identities of brain regions, especially the forebrain and midbrain, during development (Acampora et al., 1995; Matsuo et al., 1995). However, its role in human neural development are less well known. We found that OTX2 was expressed at the PSC stage but its expression increased substantially during neural differentiation and plateaued at day 2 (Figure S5F). This result suggests that OTX2 may play a role in early neuroepithelial development as well as neural patterning. As shown in Figure 4J, depletion of OTX2 before neural induction resulted in a severe reduction in the expression of neuroepithelial markers such as ZIC1 and PAX6. The expression of pan-forebrain marker (FOXG1) and dorsal forebrain markers (EMX1, LHX2) were also dramatically decreased. This result suggests that OTX2 is required for inducing anterior neuroectoderm and/or conferring the forebrain character to hPSC-derived neuroectoderm progenitors at an early stage.
We then asked what effect OTX2 KO will have after forebrain neuroepithelial cells are differentiated. Treatment of differentiating hPSCs with 4-OHT at day 8, a stage when the expression of most of forebrain neuroectoderm genes has reached plateau (Figure S5F), did not affect the expression of forebrain transcription factors, FOXG1 and ZIC1, nor hindbrain transcription factors, EN1, HOXB2, HOXA2, and HOXA3 (Figure S5G and S5H), but significantly reduced the expression of dorsal forebrain genes such as PAX6, EMX1 and LHX2 at day 16 (Figure 4K). This result suggests that OTX2 is not required for the maintenance of forebrain identity of the neuroepithelia, but is critically involved in the dorsal-ventral patterning of forebrain neuroepithelia after forebrain neuroepithelial cells are differentiated. Taken together, these results illustrate that our iKO hPSC lines enable dissection of the temporally regulated functions of genes under the human genetic background.
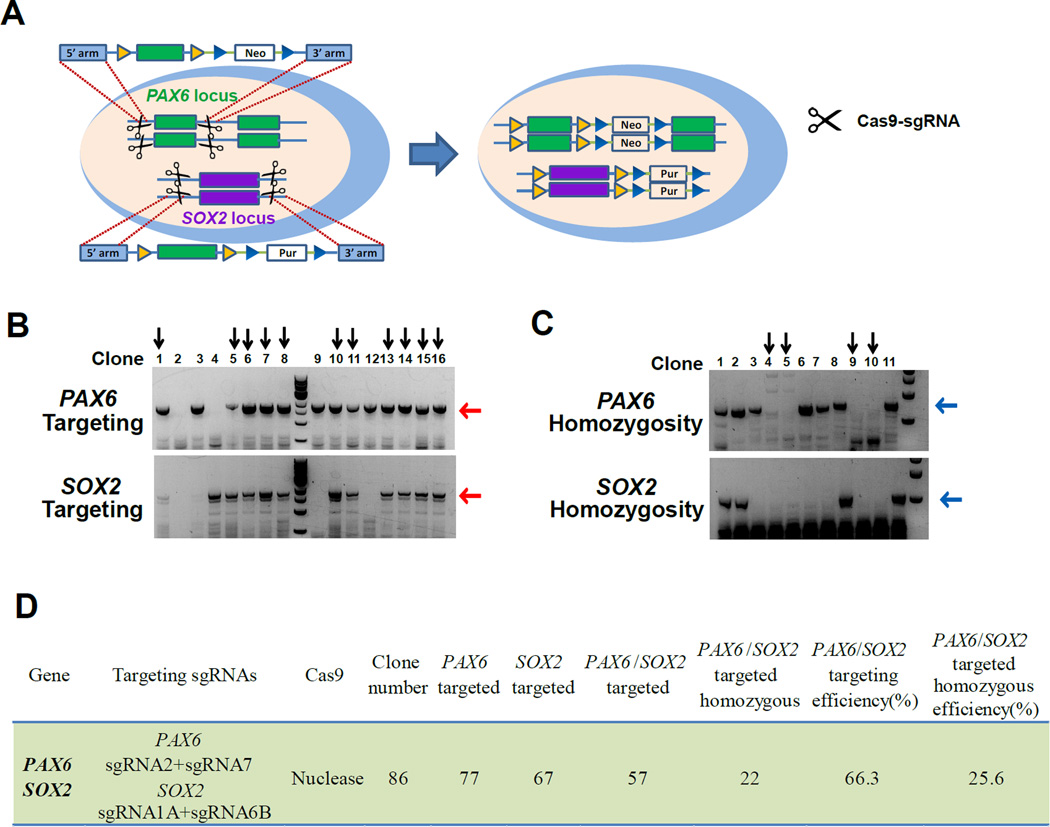
Generation of multiple-gene iKO hPSC line
Many gene products often interact with and/or compensate each other, and it is sometimes necessary to conditionally delete more than one gene in order to elucidate their functions. To determine if our strategy is applicable for generating hPSC lines with iKO of multiple genes, we tested the possibility of generating homozygous hPSC lines with multiple genes flanked by applying our dual-sgRNA targeting strategy. hESCs were electroporated by plasmid encoding Cas9, the two donor plasmids for PAX6 or SOX2 with distinct drug-resistance gene (neomycin for PAX6 and puromycin for SOX2), and two pairs of sgRNAs for PAX6 or SOX2 (Figure 5A). After selection with both neomycin and puromycin for two weeks, we found that 57 out of 86 clones were targeted in both PAX6 and SOX2 locus, and among them, 22 clones were homozygous (Figure 5B–4D). We randomly selected three homozygous clones for sequencing. Two out of three clones were correctly targeted with intact 5' FRT sites and 3' FRT sites in all four alleles in PAX6 and SOX2 loci. We further established the PAX6/SOX2 iKO hPSC line by simultaneous removal of the drug-resistance expression cassette in the FRT-flanked gene locus (puromycin in SOX2 locus and neomycin in PAX6 locus) and insertion of Flpe-ERT2 into the AAVS1 locus by electroporation with the AAVS1-blasticidin-CAG-FlpeERT2 donor plasmid, sgRNA T2, Cas9-2A-Cre and pCAG-Cre plasmids and subsequent blasticidin selection. 1 out of 24 clones (efficiency of 4.2%) was identified as a correct clone. 4-OHT treatment depleted the FRT-flanked exons in both PAX6 locus and SOX2 locus, demonstrating feasibility of multiple-gene inducible KO in human cells using our iKO system (Figure S6).
Figure 5. Flanking exons of multiple genes in one step.
(A) Schematic overview depicting the strategy for simultaneously targeting both PAX6 locus and SOX2 locus. Donor plasmids: Pur, PGK-driven puromycin resistance gene; Neo, PGK-driven neomycin resistance gene.
(B) PCR genotyping of hPSC clones targeted using SOX2 donor plasmid (Puro), PAX6 donor plasmid (Neo), WT Cas9, SOX2 targeting sgRNAs (sgRNA1A and sgRNA6B), and PAX6 targeting sgRNA (sgRNA2 and sgRNA7). Expected PCR products for correctly targeted SOX2 locus or PAX6 locus are ~1700bp or ~1800bp (red arrows) respectively. The vertical arrows indicate clones with both SOX2 locus and PAX6 locus targeted.
(C) Correctly targeted clones in both SOX2 locus and PAX6 locus underwent further homozygosity testing in both SOX2 locus and PAX6 locus. Those clones without the PCR products of ~700bp at PAX6 locus (blue arrow, upper) and without the PCR products of ~550bp at SOX2 locus (blue arrow, lower) are homozygous clones targeted in both SOX2 locus and PAX6 locus (Vertical arrows).
(D) Summary of the targeting efficiency and homozygous efficiency in both SOX2 locus and PAX6 locus.
See also Figure S6.
DISCUSSION
We have established an efficient and effective strategy to generate iKO hPSC lines. It takes advantages of the high bi-allelic targeting efficiency of the CRISPR/Cas9 system for knock-in of FRT sites, but more importantly relies on the dual-sgRNA-mediated gene targeting method we have developed. This strategy enables predictable knockout of genes with different structural organization and/or expression patterns, including those that are silent (PAX6) or actively expressed (SOX2, OTX2, AGO2) in the PSC stage, or those bearing a larger exon (SOX2). Furthermore, we have developed a one-step method to simultaneously remove the drug resistance gene and insert the Flpe-ERT2 cassette at the AAVS1 site, which overcomes the laborious sequential targeting/cloning required by traditional methods. Together, our two-step strategy enables production of iKO human PSC lines, including human ESC and iPSC lines, in as short as 12 weeks. This strategy can also be used to target multiple genes at once without extending the time frame, thus enhancing the utility of this method for the study of multifaceted processes. The hPSC lines established in this way are easy to use and the target genes can be deleted in a uniform manner at any given time by simply applying 4-OHT.
In this study, it is interesting to find that two individual sgRNAs (or sgRNA pairs for nickase) targeting separate FRT insertion sites are necessary to generate correct homozygous FRT knock-in hPSCs. Using single sgRNA targeting the 5' FRT insertion site (5' sgRNA) results in a very low targeting efficiency with no homozygous clones. Using single sgRNA targeting the 3' FRT insertion site, we obtained homozygously targeted clones but the 5' FRT sequence was lost. Only by using both 5' sgRNA and 3' sgRNA (dual sgRNAs) can we obtain correct homozygous FRT-knock-in hPSC clones with FRT sequences inserted in both 5' and 3' FRT insertion sites (Figure 1, Table 1 and Table S1). The reason behind the inefficient targeting by a single sgRNA is not clear. One potential explanation is the incomplete HDR using the donor plasmid as the template. After the Cas9/sgRNA mediated double-strand break at the designated 3' FRT insertion site, the cells may use the donor plasmid as a template to repair the double-strand break, resulting in correct insertion of the FRT sequence and drug resistance gene in the 3' FRT insertion site. At the 5' FRT insertion site, however, cells tend to use cognate genomic DNA, instead of the donor plasmid, as a template for repair, thus resulting in the loss of the FRT sequence. Regardless of potential mechanisms, our findings demonstrate that the dual-sgRNA targeting strategy is critical for generating correct homozygous FRT knock-in hPSCs. Technically, our optimized transfection, drug selection, and culture condition for selected individual cells, as detailed in material and method, should make it feasible for other labs to generate FRT knock-in hPSC lines.
Off target effects are a major concern of CRISPR/Cas9 system. Our sequencing analysis in the total of 114 potential off-target sites showed no indel formation in any of these sites (Table S2), suggesting relative specific genome targeting of the CRISPR/Cas9 system in hPSCs. This is consistent with recent studies using whole-genome sequencing analysis (Smith et al., 2014; Veres et al., 2014) or capture sequencing (Mandal et al., 2014). Thus, the CRISPR/Cas9 system is effective and specific in generating gene iKO hPSC lines.
Generation of inducible KO hPSCs has been reported by Gonzalez et al using their iCRISPR system (Gonzalez et al., 2014). It will first create a common inducible line by integrating the Tet inducible system in the AAVS1 site and then introduce sgRNA to target a specific DNA site to induce KO. While the intention is ideal and it potentially allows generation of versatile new constitutive KO lines, its dependence on Cas9/sgRNA-mediated DNA double strand break and subsequent random indel formation makes it nearly impossible to generate clonal inducible KO lines. In order to induce KO in differentiated cells, cells need to be pretreated with doxycycline (to induce Cas9 expression), and then transfected with sgRNA (for targeting a gene to induce gene KO) (Gonzalez et al., 2014). Transfection without cloning will unlikely achieve a uniform population, even with two rounds of transfection. That will result in a mixed cell population consisting of those with and those without gene targeting. Even among the sgRNA-targeted cells, cells will exhibit distinct genotypes from each other after random indel formation in one or both alleles of a target gene. Only those with the out-of-frame indel formation in both alleles will exhibit gene KO. Therefore, the resultant cells will always be a mixture and the proportion of KO cells depends on the rate of out-of-frame indel formation. Indeed, the average KO (out-of-frame) efficiency by iCRISPR is 64% from 5 individual genes (Table S3). The inefficient inducible KO also applies to the modified iCRISPR method which simultaneously knocks in sgRNA expression cassette together with Tet-on Cas9 to bypass the sgRNA transfection step (Table S3).
In contrast, our iKO system is based on Flpe-recombinase-mediated high efficient excision of the DNA segment between FRT sites, resulting in predictable exon loss and frame-shift, which leads to gene KO in nearly all the cells. While it takes longer time to generate a line (10–12 weeks), gene KO can be induced at any time by simply applying 4-OHT without cell manipulations (Table S3). This is particularly important for dissecting gene function not only in the stem cell stage but more importantly during stem cell differentiation and/or pathological processes. For example, we found that OTX2, a homeodomain protein that is critical for defining the fore-midbrain boundary based on animal studies (Acampora et al., 1995; Matsuo et al., 1995), may also be essential for generation of human forebrain progenitors at an early stage and for maintaining the dorsal forebrain identity of presumptive forebrain precursors at a later stage. The same strategy may be used to delete a gene in functional cells that are differentiated from patient iPSCs, thus dissecting its role in a pathological process. It may also enable controlling the fate of transplanted human cells in vivo. For example, after transplantation, treatment with 4-OHT can deplete a cell cycle gene to prevent proliferation of transplanted cells, thus avoiding potential tumorigenesis. The easiness in use and precise time resolution for gene deletion of the iKO hPSCs established with our new method will enable discovery of novel roles of known genes in human biology and pathology.
EXPERIMENTAL PROCEDURES
Construction of donor plasmids, sgRNAs, and Cas9 plasmids
Human codon-optimized Streptococcus pyogenes wild-type Cas9 (Cas9-2A-GFP) and Cas9 nickase (Cas9D10A-2A-GFP) were obtained from Addgene (plasmid #44719 and plasmid #44720) (Ding et al., 2013b). Cas9-2A-Cre was constructed by replacing GFP in the Cas9-2A-GFP with the Cre cDNA amplified from pCAG-Cre (Addgene plasmid #13775) (Matsuda and Cepko, 2007). sgRNA T2 was obtained from Addgene (plasmid #41818) (Mali et al., 2013b). To facilitate the sgRNA construction, we generated Cas9 sgRNA vector. A previously described chimeric guide RNA expression cassette was ordered as gBlocks and cloned into the gRNA cloning vector (Addgene plasmid #41824) (Mali et al., 2013b). The new sgRNA cloning vector (Cas9 sgRNA vector) includes two BbsI restriction sites for rapid cloning of sgRNA (Table S4). Briefly, Cas9 sgRNA vector was digested with BbsI and gel purified. A pair of oligos including targeting sequences was annealed and cloned into the BbsI-digested Cas9 sgRNA vector. The guide sequence of individual sgRNA could be found in Table S2. PL552 donor plasmid vector containing a floxed PGK-puromycin expression cassette was constructed by replacing the neomycin gene in the PL452 (Frederick National Lab) with puromycin gene. To generate the donor plasmid for FRT-knock-in, DNA fragments of about 1–1.25 kb in length were PCR-amplified from the genomic DNA beyond the designated 5' and 3' FRT insertion sites of a targeted gene. The DNA fragment between the designated 5' and 3' FRT insertion sites (containing exon) was also amplified from genomic DNA. FRT sequences were included in the PCR primers. These three fragments were then cloned into the multiple cloning sites of plasmid PL552. For simultaneously targeting PAX6 and SOX2, the PL452 PAX6 donor plasmid was generated by inserting the DNA fragments into the PL452 vector containing a floxed PGK-neomycin expression cassette. To generate AAVS1-neo-CAG-FlpeERT2 donor plasmid, we replaced the puromycin resistance gene in the AAVS1-pur-CAG-hrGFP plasmid (Addgene plasmid #52344) (Qian et al., 2014) by neomycin resistance gene to obtain the AAVS1-neo-CAG-hrGFP. We next amplified mammalian codon optimized Flpe recombinase cDNA and ERT2 cDNA by PCR from pDIRE (Addgene plasmid #26745) (Osterwalder et al., 2010) and pCAG-FlpeERT2 (Addgene plasmid #14756) (Matsuda and Cepko, 2007), respectively. The mammalian codon-optimized Flpe fused with ERT2 was inserted into the AAVS1-neo-CAG-hrGFP to replace GFP to get the AAVS1-neo-CAG-FlpeERT2 donor plasmid. AAVS1-blasticidin-CAG-FlpeERT2 donor plasmid was constructed by replacing the neomycin resistance gene in the AAVS1-neo-CAG-FlpeERT2 donor plasmid with blasticidin resistance gene.
Cell culture
Human ESCs (line WA09 (WiCell), passages 20–40), D90A, and D90D (Chen et al., 2014)) were maintained on a feeder layer of irradiated embryonic mouse fibroblasts (MEF) in hPSC medium consisting of DMEM/F12 (Life Technologies), 1× Non-Essential Amino Acids (Life Technologies), 0.5× GlutaMAX (Life Technologies), 0.1 mM β-mercaptoethanol (Sigma). 4 ng/ml FGF-2 (R&D Systems) were added when feed cells, as previously described (Zhang et al., 2001). D90A hiPSC line was generated from fibroblasts of an ALS patient with SOD-D90A mutation by Sendai virus. D90D hiPSC line was a genetically corrected line from D90A hiPSCs (Chen et al., 2014).
Electroporation
Electroporation was performed using the Gene Pulser Xcell System (Bio-Rad) at 250 V, 500µF in a 0.4 cm cuvettes (Phenix Research Products). Details are given in Supplemental Experimental Procedures.
Genomic deletion assay
Pairs of sgRNA plasmids with Cas9 plasmids were co-transfected into HEK293T cells via calcium phosphate transfection. 72 hours later, 1×106 cells were collected and genomic DNA was extracted. To detect the effective genome deletion caused by a pair of sgRNAs, genomic PCR was performed with a pair of primers flanking the deletion.
Off-target analysis of established hPSC lines
The potential off target sites were selected according to online tools provided by Feng Zhang's lab (http://crispr.mit.edu/) (Hsu et al., 2013). These sites were amplified by genomic PCR and then undergone Sanger sequencing. 6–7 potential off targets were sequenced for every sgRNA target site. The information about the selected off-target sites was listed in Table S2.
Western Blotting, Immunocytochemistry and Quantitative RT-PCR
These procedures were performed using standard methods. Details are given in Supplemental Experimental Procedures. Primers used are listed in Table S5.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Bhattacharyya for helpful comments on the manuscript, R. Bradley for helpful suggestions. This study was supported in part by the NIH-NINDS (NS045926, NS076352, NS086604), NIH-NIMH (MH099587, MH100031), the Bleser Family Foundation, the Busta Foundation, and the NICHD (P30 HD03352),
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Y.C. conceived the study. Y.C. and J.C. and M.X. performed most of the experiments. Y.D. A.P., Y.T. and C.H. did the off-target sequencing and prepared the reagents. Y.C. and J.C. collected and analyzed data. Y.C., J.C., M.X., Z.D., A.P., and S.Z. wrote the manuscript. S.Z supervised the project.
Supplemental Information for this article includes six figures, five tables and Supplemental Experimental Procedures can be found with this article online.
REFERENCES
- Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brulet P. Forebrain and midbrain regions are deleted in Otx2−/− mutants due to a defective anterior neuroectoderm specification during gastrulation. Development. 1995;121:3279–3290. doi: 10.1242/dev.121.10.3279. [DOI] [PubMed] [Google Scholar]
- Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, Heath PR, Holden H, Andrews PW. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- Bu L, Gao X, Jiang X, Chien KR, Wang Z. Targeted conditional gene knockout in human embryonic stem cells. Cell Res. 2010;20:379–382. doi: 10.1038/cr.2010.23. [DOI] [PubMed] [Google Scholar]
- Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- Chen H, Qian K, Du Z, Cao J, Petersen A, Liu H, Blackbourn LWt, Huang CL, Errigo A, Yin Y, et al. Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell. 2014;14:796–809. doi: 10.1016/j.stem.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, Kuperwasser N, Motola DL, Meissner TB, Hendriks WT, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013a;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Regan SN, Xia Y, Oostrom LA, Cowan CA, Musunuru K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013b;12:393–394. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice A, Trounson A. Genetic modification of human embryonic stem cells for derivation of target cells. Cell Stem Cell. 2008;2:422–433. doi: 10.1016/j.stem.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Gonzalez F, Zhu Z, Shi ZD, Lelli K, Verma N, Li QV, Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Jaenisch R. Gene targeting in human pluripotent cells. Cold Spring Harb Symp Quant Biol. 2010;75:201–209. doi: 10.1101/sqb.2010.75.021. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013a;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Cheng L. Concise review: Human cell engineering: cellular reprogramming and genome editing. Stem Cells. 2012;30:75–81. doi: 10.1002/stem.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013b;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal PK, Ferreira LM, Collins R, Meissner TB, Boutwell CL, Friesen M, Vrbanac V, Garrison BS, Stortchevoi A, Bryder D, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell. 2014;15:643–652. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci U S A. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 1995;9:2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Osterwalder M, Galli A, Rosen B, Skarnes WC, Zeller R, Lopez-Rios J. Dual RMCE for efficient re-engineering of mouse mutant alleles. Nat Methods. 2010;7:893–895. doi: 10.1038/nmeth.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- Qian K, Huang CT, Chen H, Blackbourn LWt, Chen Y, Cao J, Yao L, Sauvey C, Du Z, Zhang SC. A simple and efficient system for regulating gene expression in human pluripotent stem cells and derivatives. Stem Cells. 2014;32:1230–1238. doi: 10.1002/stem.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzino A. Concise review: The Sox2-Oct4 connection: critical players in a much larger interdependent network integrated at multiple levels. Stem Cells. 2013;31:1033–1039. doi: 10.1002/stem.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Gore A, Yan W, Abalde-Atristain L, Li Z, He C, Wang Y, Brodsky RA, Zhang K, Cheng L, et al. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014;15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Chung SK, Xu Y. Modeling disease in human ESCs using an efficient BAC-based homologous recombination system. Cell Stem Cell. 2010;6:80–89. doi: 10.1016/j.stem.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, Erdin S, Cowan CA, Talkowski ME, Musunuru K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2010;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nature biotechnology. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- Zhang X, Huang CT, Chen J, Pankratz MT, Xi J, Li J, Yang Y, Lavaute TM, Li XJ, Ayala M, et al. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7:90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Cochran R, Cheng L. Double knockouts in human embryonic stem cells. Cell Res. 2010;20:250–252. doi: 10.1038/cr.2010.29. [DOI] [PubMed] [Google Scholar]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.