Abstract
Natural antisense transcripts (NATs) are a class of long noncoding RNAs (lncRNAs) that are complementary to other protein-coding genes. Although thousands of NATs are encoded by mammalian genomes, their functions in innate immunity are unknown. Here, we identify and characterize a novel NAT, AS-IL1α that is partially complementary to IL-1α. Similar to IL-1α, AS-IL1α is expressed at low levels in resting macrophages and is induced following infection with Listeria monocytogenes or stimulation with TLR ligands (Pam3CSK4, LPS, PolyI:C). Inducible expression of IL-1α mRNA and protein were significantly reduced in macrophages expressing shRNA that target AS-IL1α. AS-IL1α was located in the nucleus and did not alter the stability of IL-1α mRNA. Instead, AS-IL1α was required for the recruitment of RNA Polymerase II (RNAPII) to the IL-1α promoter. In summary, our studies identify AS-IL1α as important regulator of IL-1α transcription during the innate immune response.
Introduction
Recent advances in large-scale RNA-sequencing have led to the identification of novel RNA species encoded in the genome, many of which are long noncoding RNAs (lncRNAs) (1, 2). lncRNAs are non-coding RNAs that are arbitrarily defined as having 200 or more nucleotides to discriminate them from small non-coding RNAs. These RNAs are further categorized based on their genomic location relative to neighboring protein-coding genes. Several comprehensive studies have demonstrated that lncRNAs are important regulators of cell differentiation and developmental pathways (3, 4). A growing literature also supports their importance in immunity (5).
Natural antisense transcripts (NATs) are a class of lncRNAs defined as being complementary to one or more protein-coding genes (6). Approximately 50-70% of lncRNAs are classified as NATs thus far, making them a substantial proportion of the noncoding genome (2, 6). Although the functions of some lncRNAs are well defined, relatively few NATs are functionally characterized. NATs can both activate and repress the expression of complementary coding genes (7, 8). Similar to the broader class of lncRNAs, NATs probably function in a cell-type specific manner. For example, in Th2 cells, lincR-Ccr2-5’AS regulates expression of Ccr2, Ccr3, and Ccr5 and trafficking of Th2 cells to the lung (9). However, the molecular functions of NATs in innate immunity are currently unknown.
Macrophages are the first line of defense against microbial pathogens. Following infection, host cells produce pro-inflammatory cytokines. In particular, IL-1α and IL-1β are rapidly induced and amplify inflammation via IL-1 receptors expressed on neighboring cells (10). As these cytokines can cause significant tissue damage their production must be tightly regulated. Caspase-1 dependent inflammasomes convert IL-1β into the mature biologically active cytokine. In contrast, IL-1α is active in its full-length form upon release from damaged or dying cells (11).
Here, we describe the first example of a functional innate immune NAT, AS-IL1α, a novel NAT encoded within the IL-1α locus. Characterization of this NAT shows that AS-IL1α functions as an additional layer of regulation important in controlling IL-1α transcription.
Materials and methods
In vivo infections
C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME) were maintained in specific pathogen-free conditions and used in accordance with the Institutional Animal Care and Use Committee. Listeria monocytogenes (clinical isolate 10403s) was from V. Boyartchuk (NTNU, Trondheim, Norway) and infected as described (12) for 24 hours before harvesting the spleen. PBS was used as a control.
RNA-sequencing
Single cell suspensions from spleens were used to make total RNA. 4 µg of total RNA was used to generate libraries for RNA-sequencing (Illumina unstranded RNA kits). Samples were sized, quantified and validated on a Bioanalyzer. Libraries were sequenced on a HiSeq 2000 (Illumina, San Diego, CA) as paired-end 100 reads. Sequence reads were aligned to the mouse genome (NCBI m37/mm9) using TopHat. Gene-level read counts were calculated using HTSeq and the Ensembl64 transcript annotation file. The DESeq package was used to normalize gene counts, calculate fold-change values, and p-values. The Circos program was used to visualize genome-wide gene expression changes. As previously described (3), all protein coding genes annotated with particular GO IDs were classified as immune genes. All RNA-seq data are available from ArrayExpress (E-MTAB-3315).
Macrophage stimulations
Primary bone marrow derived macrophages (BMDM) were generated and infected with Listeria or stimulated with lipopolysaccharide (LPS) (100 ng/mL), Pam3CSK4 (100 nM), Poly(I:C) (25 µg/mL) and ISD (3 µM) oligonucleotides (13) as described in (12). After 2 hours incubation, total RNA was harvested from all conditions (RNeasy, Qiagen).
Polysome profiling
shRNA-mediated silencing
Lentiviral particles were generated using pLKO.1 TRC as described (14). Knockdown efficiency was assessed by one-step qRT-PCR (BioRad). Sequences/primers are listed in Supplemental Table 1.
Western blot
Immunoblotting was performed using mouse IL-1α/IL-1F1 antibody (R&D Systems AB-400NA) and anti-goat IgG) (BioRad 172-1011).
Nanostring Analysis
nCounter CodeSets were constructed for detecting selected mouse-specific immune genes and levels of RNA measured using the nCounter Digital Analyzer as described (3, 15).
Cell fractionation
Cytosolic and nuclear fractions were prepared as described (16). The cell lysis buffer contained 0.15% NP-40; the sucrose cushion did not contain detergent. After fractionation, cytoplasmic and nucleoplasmic RNA was purified using Qiagen RNeasy columns. GAPDH and 7SK were used for cytoplasmic and nuclear controls respectively. Primers are listed in Supplemental Table 1.
Chromatin Immunoprecipitation (ChIP)
Cells were stimulated as described and fixed in 1% formaldehyde, lysed, and sonicated. 5 µg of chromatin was immunoprecipitated (IP) with Dynabeads Protein G (10009D, Novex/Life Technologies) and antibodies to RNAPII (Active Motif 102660), H3K9-Ac (Abcam AB4441), or IgG isotype (Abcam AB37415). Purified chromatin DNA was used for qPCR amplification at the IL-1α, IP-10, or Gapdh promoter.
Statistical Analysis
Statistical analysis was calculated using a Student’s t test using GraphPad Prism Software.for bivariate studies and one-way ANOVA for multivariate studies. p value ≤ of 0.05 was considered statistically significant.
Results
Identification of AS-IL1α, a natural antisense non-coding RNA in the IL-1α locus
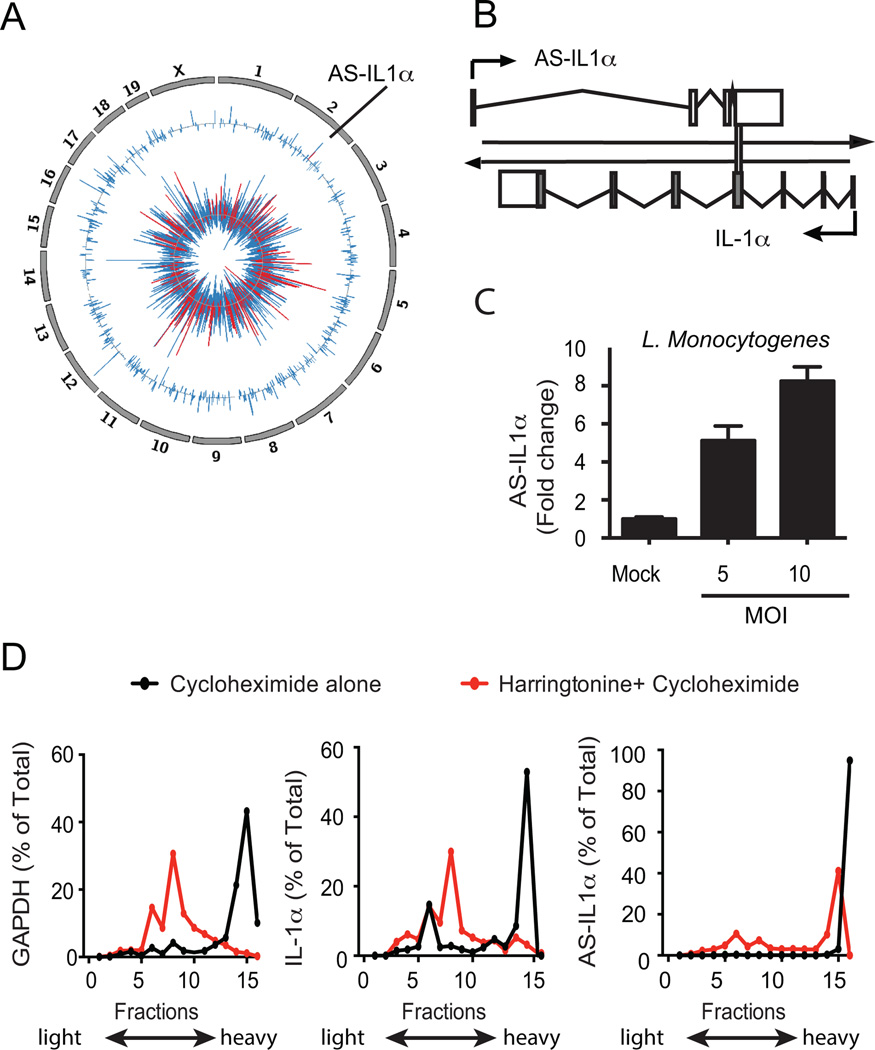
To assess lncRNA expression during an active infection, we infected C57Bl/6 mice with Listeria monocytogenes and prepared libraries for RNA-sequencing from splenocytes isolated from infected and non-infected mice (24 hrs). Consistent with previous reports (17), Listeriainduced expression of a wide range of protein-coding immune genes (Figure 1a, inner track). We also detected many lncRNAs that were differentially expressed following Listeria infection (Figure 1a, outer track).
Figure 1.
Identification of AS-IL1α. (A) Circos plot showing differentially expressed genes in splenocytes from L. Monocytogenes infected mice. Chromosomes are indicated on the outer tracks. The middle track shows log2 fold-change values for all lncRNAs and AS-IL1α (colored red). The inner track shows log2 fold-change values for protein-coding genes, with immune genes colored in red. (B) AS-IL1α schematic in murine macrophages. AS-IL1α chromosomal localization overlaps with IL-1α protein coding gene on chromosome 2 indicated by vertical bar. (C) Macrophages infected with L. Monocytogenes for 3 hours at MOI 5 and MOI 10. Data were normalized with GAPDH, fold change calculated and are representative of 2 independent experiments (D) Polysome profiling of GAPDH, IL-1α and AS-IL1α transcripts. Immortalized macrophages were stimulated with LPS and either treated with cycloheximide alone, or harringtonine prior to cycloheximide treatment to determine translational potential of AS-IL1α.
Strikingly, one of these lncRNAs (hereafter referred to as AS-IL1α) was induced 14-fold and was encoded by the opposite strand of the IL-1α locus on chromosome 2. We used PCR to confirm the orientation and gene structure of AS-IL1α (Figure 1b, KR095173 http://www.ncbi.nlm.nih.gov/genbank). Similar to the in vivo infection, AS-IL1α was induced in macrophages infected with Listeria (Figure 1c). The Coding Potential Calculator (http://cpc.cbi.pku.edu.cn/) classified AS-Il1α as noncoding (score=-0.87), which was also supported by polysome profiling. Cells were treated with cycloheximide, which traps ribosomes along their RNA strands and these RNA were detected in the heavier fractions of a sucrose density gradient. In parallel, cells were pretreated with harringtonine prior to cycloheximide, which inhibits translation and polysome formation by causing ribosomes to accumulate at their initiation sites. As expected, harringtonine prevented polysome formation in both GAPDH and IL-1 α mRNA (Figure 1d), and both mRNAs shifted to lighter fractions in the sucrose gradient. In contrast, AS-IL1α was largely unaffected by harringtonine treatment, indicating that it is unlikely to associate with ribosomes and encode a protein product.
AS-IL1α is inducible in macrophages exposed to TLR ligands via NF-κB
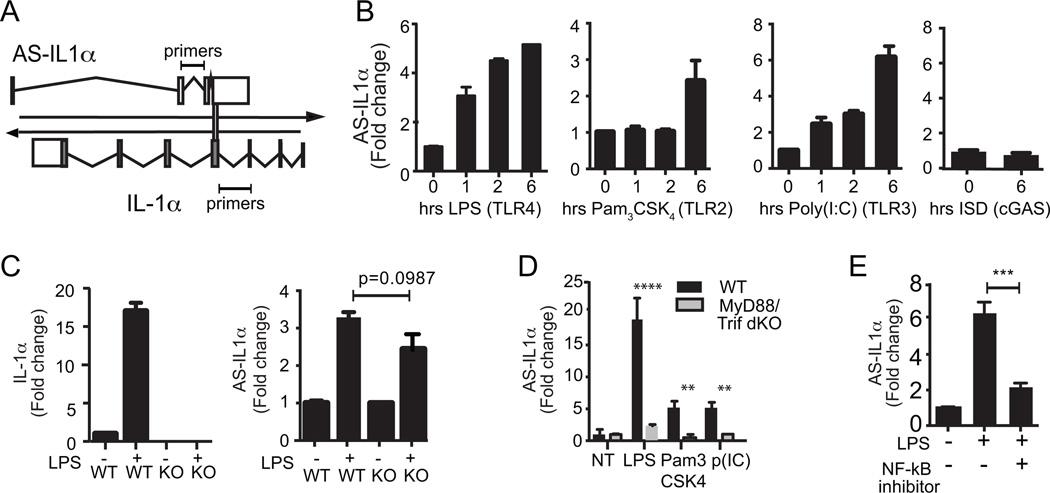
We next examined the expression of AS-IL1α in bone marrow derived macrophages (BMDMs) stimulated with ligands for TLRs and other sensors. Figure 2a depicts the location of our qRT-PCR primers, which do not overlap with IL-1α itself. AS-IL1α was induced by LPS (TLR4), Pam3CSK4 (TLR1/2) and PolyI:C (TLR3) but not Interferon stimulatory DNA (ISD), which activates cGAS (Figure 2b). We also measured expression of AS-IL1α in macrophages from IL-1α-deficient mice. In these cells, LPS induced expression of AS-IL1α but not IL-1α, indicating that transcription of AS-IL1α is regulated independently of IL-1α (Figure 2c). We next compared LPS-inducible levels of AS-IL1α in WT and MyD88/TRIF DKO cells and cells treated with Bay11-7085, an inhibitor of NF-κB, In the absence of MyD88/TRIF or following inhibition of NF-κB, LPS failed to induce AS-IL1α (Figure 2d and 2e).
Figure 2.
Inducibility of AS-IL1α by TLR ligands. (A) A schematic of AS-IL1α and IL-1α and qRT-PCR primers for experiments B-E (not scaled). (B) LPS, Pam3CSK4, Poly(I:C) or ISD were stimulated on iBMDMs for 0, 1, 2, or 6 hours and AS-IL1α expression was measured by qRT-PCR (C) Wildtype and IL-1α knockout macrophages were stimulated with LPS for 6 hours. AS-IL1α expression was induced even in the absence of IL-1α. (D) WT and MyD88/Trif double knockout macrophages were not-treated (NT), LPS, Pam3CSK4 or Poly(I:C) treated for 6 hours. AS-IL1α expression was measured by qRT-PCR. (**** p<0.0001, ** p<0.01 by one way ANOVA). (E) An NF-κB inhibitor, Bay11-7082, was treated on cells 1 hour prior to LPS stimulation (6 hours), and AS-IL1α expression was measured by qRT-PCR. (*** p<0.001 by two-tailed T test). Data were normalized to GAPDH, fold change calculated and are means + S.D. representative of 3 replicates.
Knocking down AS-IL1α inhibits inducible IL-1α expression
We next generated macrophage cell lines in which AS-IL1α expression was specifically silenced by shRNA. We made two independent shRNAs that targeted AS-IL1α exons that did not overlap with those of IL-1α(Figure 3a). We confirmed that AS-IL1α was significantly silenced in LPS-stimulated cell lines that expressed these shRNA (Figure 3b). In shRNA-GFP lines, LPS strongly induced IL-1α mRNA levels whereas LPS only induced low levels of IL-1α mRNA in cells expressing shRNA-AS-IL1α (Figure 3c). LPS also failed to induce the high levels of IL-1α protein normally detected by immunoblotting (Figure 3d). These results indicate that AS-IL1α is required for the inducible expression of IL-1α. We subsequently used RNA profiling (Nanostring) to simultaneously analyze mRNA expression levels of a broader selection of immune genes following LPS stimulation (6 hours) in control and AS-IL1α silenced macrophages (Figure 3e). IL-1α was the most significantly altered gene in the shRNA lines. To a lesser extent, IL-1β levels were also decreased.
Figure 3.
AS-IL1α regulates IL-1α expression. (A) shRNA 1 and shRNA 2 target AS-IL1α in red regions (not to scale) that do not overlap with IL-1α. (B) AS-IL1α RNA levels (qRT-PCR) were reduced in both shRNA 1 and shRNA 2 cell lines. shRNA GFP is a control that targets the GFP gene. (C) IL-1α mRNA levels (qRT-PCR) were reduced in shRNA 1 and shRNA 2 cell lines. (D) IL-1α p33 protein expression (Western blot) was decreased in shRNA 1 and shRNA 2 cell lines stimulated with LPS. β-actin was used as a loading control. (E) Nanostring analysis was performed on the cell lines to determine if knocking down AS-IL1α affects the expression of other immune genes. Immune genes with a 15-fold change in expression following LPS stimulation are shown. For B and C, P-values were <0.0001**** (one way ANOVA) and data were calculated as fold-change relative to GAPDH and are representative of 3 biological replicates.
AS-IL1α is localized to the nucleus and enhances IL-1α expression at the transcriptional level
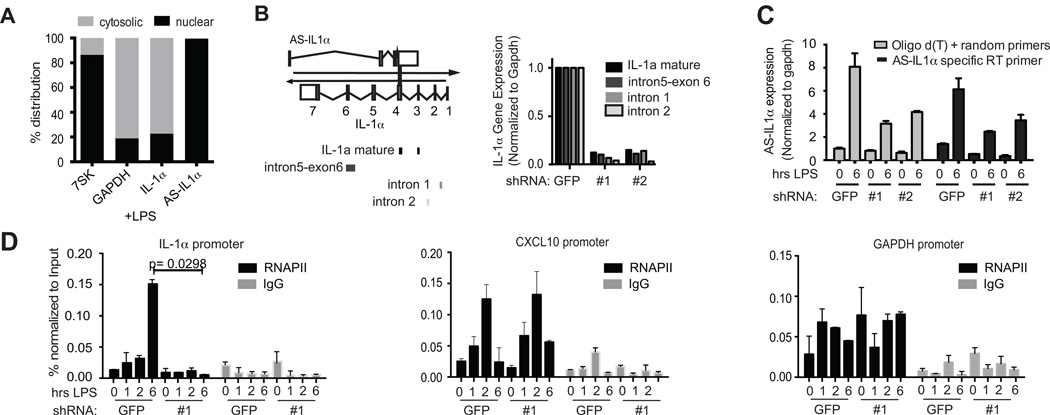
To understand how AS-IL1α might regulate IL-1α, we first examined its localization by performing subcellular fractionation of nuclear and cytosolic compartments and analyzing RNA levels by qRT-PCR. We also measured levels of GAPDH, IL-1α and the nuclear RNA, 7SK. As expected, the mature IL-1α and GAPDH transcripts were localized to the cytosol, while 7SK RNA was confined to the nucleus. AS-IL1α was primarily nuclear (Figure 4a).
Figure 4.
AS-IL1α regulates IL-1α transcription. (A) Localization of AS-IL1α in LPS-induced iBMDMs. Cytosolic and nuclear fractions were separated via a sucrose gradient and qRT-PCR was performed on fractionated RNA. Gapdh (cytosolic), IL-1α (cytosolic) and 7SK (nuclear) were also measured as controls. Data is represented as % cytosolic or % nuclear fractions over cytosolic+nuclear (100%) total RNA. (B) qRT-PCR was performed on shRNA-GFP and two AS-IL1α KD cell lines. Schematic shows regions to be amplified by primers in mature IL-1α mRNA (exon-exon junction), and pre-spliced IL-1α mRNA. RNA levels shown as a percentage of expression in shRNA-GFP (representative of 2 experiments). (C) A comparison of poly-d(T) and random hexamer priming for Reverse Transcription to an AS-IL1α specific primer to demonstrate specificity of amplified region prior to qRT-PCR. (D) Chromatin immunoprecipitation (ChIP) on the cell lines were performed. Antibodies against RNAPII or IgG isotype control were used. Regions of the IL-1α gene, near the transcription start site (+22) were measured for RNAPII binding. RNAPII recruitment did not decrease at the CXCL10 (IP10) or Gapdh promoter. Data were normalized with Input chromatin before immunoprecipitation. (p<0.05 by two-tailed T test). Same comparisons were made for CXCL10 and GAPDH. Bars without * are not significant.
Since AS-IL1α was localized to the nucleus, we hypothesized that AS-IL1α regulated the transcription of IL-1α. We used qRT-PCR primers that targeted intron-containing sequences of the IL-1α pre-mRNA as well as primers targeting spliced IL-1α transcripts. Consistent with an effect on transcription, both spliced and unspliced pre-mRNA IL-1α levels were decreased in the three AS-IL1α shRNA lines (Figure 4b). We also made reverse transcription primers that targeted AS-IL1α only and compared it to our normal method which uses poly-d(T) and random hexamer primers in order to confirm specific amplification of our qRT-PCR amplicon from the AS-IL1α cDNA. The trends were comparable indicating the abrogated AS-IL1α expression levels were specific to AS-IL1α and not the result of promiscuous priming (Figure 4c).
We also performed chromatin immunoprecipitation (ChIP-PCR) assays to measure RNAPII occupancy at the region surrounding the transcription start site (TSS) of the IL-1α locus (+22nt). As expected, LPS treatment in the shRNA-GFP control cells resulted in robust RNAPII binding at this region. This was greatly reduced in macrophages expressing AS-IL1α shRNA (Figure 4d). We observed similar results when we measured the epigenetic mark H3K9-acetylation, an indicator of active transcription at this same locus (Supplementary Figure 1). The effect of AS-IL1α shRNA was specific to the IL-1α locus, as recruitment of RNAPII to Cxcl10 or GAPDH were unaffected in the knockdown line. These results indicate that AS-IL1α is required to facilitate RNA polymerase II recruitment to the IL-1α locus during LPS-induced transcription.
Discussion
lncRNAs are emerging as important regulators of gene expression in diverse biological contexts including immunity. LPS has been shown to induce widespread changes in lncRNA expression in immune cells (18). lncRNAs have also been shown to promote or repress inflammatory gene expression in immune cells (19). The identification of AS-IL1α as a natural antisense lncRNA that enhances IL-1α gene transcription adds to our understanding of cytokine-mediated inflammation. Indeed, disruption of AS-IL1α function could limit IL-1α transcription and potentially alleviate the damaging effects of excessive IL-1α levels during infection and inflammatory disease.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (A1095213, a T32 Training Grant) to J.C and (AI067497) to K.A.F. as well as by grants from the American Heart Association (to M.A), the German Research Fund, DFG (to R.E.) and the Arthritis National Research Foundation (to S.C.).
References
- 1.Consortium, R. G. E. R. G. and G. S. G. (Genome N. P. C. G. and the F. Antisense Transcription in the Mammalian Transcriptome. Science (80-.) 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 2.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, O’Neill LAJ, Moore MJ, Caffrey DR, Fitzgerald KA. A long noncoding RNA mediates both activation and repression of immune response genes. Science (80.) 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinn JL, Chang HY. Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpenter S, Fitzgerald KA. Transcription of inflammatory genes: long noncoding RNA and beyond. J. Interferon Cytokine Res. 2015;35:79–88. doi: 10.1089/jir.2014.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Werner A, Carlile M, Swan D. What do natural antisense transcripts regulate? RNA Biol. 2009;6:43–48. doi: 10.4161/rna.6.1.7568. [DOI] [PubMed] [Google Scholar]
- 7.Matsui K, Nishizawa M, Ozaki T, Kimura T, Hashimoto I, Yamada M, Kaibori M, Kamiyama Y, Ito S, Okumura T. Natural antisense transcript stabilizes inducible nitric oxide synthase messenger RNA in rat hepatocytes. Hepatology. 2008;47:686–697. doi: 10.1002/hep.22036. [DOI] [PubMed] [Google Scholar]
- 8.Vigetti D, Deleonibus S, Moretto P, Bowen T, Fischer JW, Grandoch M, Oberhuber A, Love DC, Hanover JA, Cinquetti R, Karousou E, Viola M, D’Angelo ML, Hascall VC, De Luca G, Passi A. Natural antisense transcript for hyaluronan synthase 2 (HAS2-AS1) induces transcription of HAS2 via protein O-GlcNAcylation. J. Biol. Chem. 2014 doi: 10.1074/jbc.M114.597401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, Zhu J, Zhao K. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat. Immunol. 2013;14:1190–1198. doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C-J, Kono H, Golenbock D, Reed G, Akira S, Rock KL. Identification of a key pathway required for the sterile inflammatory response triggered by dying cells. Nat. Med. 2007;13:851–856. doi: 10.1038/nm1603. [DOI] [PubMed] [Google Scholar]
- 11.Kim B, Lee Y, Kim E, Kwak A, Ryoo S, Bae SH, Azam T, Kim S, Dinarello CA. The interleukin-1α precursor is biologically active and is likely a key alarmin in the IL-1 family of cytokines. Front. Immunol. 2013;4 doi: 10.3389/fimmu.2013.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Severa M, Islam SA, Waggoner SN, Jiang Z, Kim ND, Ryan G, Kurt-Jones E, Charo I, Caffrey DR, Boyartchuk VL, Luster AD, Fitzgerald KA. The transcriptional repressor BLIMP1 curbs host defenses by suppressing expression of the chemokine CCL8. J. Immunol. 2014;192:2291–2304. doi: 10.4049/jimmunol.1301799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixit E, Boulant S, Zhang Y, Lee ASY, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, Nibert ML, Superti-Furga G, Kagan JC. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smale DBAP-JA-JTIBMLGNDBS, Pandya-Jones A, Tong A-J, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. Transcript Dynamics of Proinflammatory Genes Revealed by Sequence Analysis of Subcellular RNA Fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leber JH, Crimmins GT, Raghavan S, Meyer-Morse NP, Cox JS, Portnoy DA. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 2008;4:e6. doi: 10.1371/journal.ppat.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atianand MK, Fitzgerald KA. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol. Med. 2014;20:623–631. doi: 10.1016/j.molmed.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.