SUMMARY
Many human cancers share similar metabolic alterations, including the Warburg effect. However, it remains unclear whether oncogene-specific metabolic alterations are required for tumor development. Here we demonstrate a “synthetic lethal” interaction between oncogenic BRAF V600E and a ketogenic enzyme 3-hydroxy-3-methylglutaryl-CoA lyase (HMGCL). HMGCL expression is upregulated in BRAF V600E-expressing human primary melanoma and hairy cell leukemia cells. Suppression of HMGCL specifically attenuates proliferation and tumor growth potential of human melanoma cells expressing BRAF V600E. Mechanistically, active BRAF upregulates HMGCL through an octamer transcription factor Oct-1, leading to increased intracellular levels of HMGCL product, acetoacetate, which selectively enhances binding of BRAF V600E but not BRAF wild type to MEK1 in V600E-positive cancer cells to promote activation of MEK-ERK signaling. These findings reveal a mutation-specific mechanism by which oncogenic BRAF V600E “rewires” metabolic and cell signaling networks and signals through the Oct-1-HMGCL-acetoacetate axis to selectively promote BRAF V600E-dependent tumor development.
INTRODUCTION
The importance of metabolic alterations in cancer has been increasingly recognized over the past decade. Identification of metabolic vulnerability of human cancers has informed development of therapeutic strategies to treat cancer. However, although increasing evidence emerges and suggests that different human cancers may share common metabolic properties such as the Warburg effect, it remains unclear whether distinct oncogenic backgrounds in different cancer types require different metabolic properties for tumor development.
Melanoma is one of the most common human cancers, which, according to American Cancer Society, accounts for >76,600 cases in US in 2013 with ~9,000 death each year. More than 50% of melanomas express BRAF V600E mutant, which represents a therapeutic target due to its pathogenic role. However, despite the success of BRAF mutant and MEK inhibitors in clinical trials for BRAF V600E positive melanoma patients, clinical resistance invariably develops (Bollag et al., 2012; Gibney et al., 2013; Johnson and Sosman, 2013). Thus, identification of alternative “targets” in BRAF V600E positive melanomas may inform effective long-term treatment strategies.
Herein we approached this question by identifying “metabolic vulnerabilities” specifically required by oncogenic BRAF V600E mutant, but not other oncogenes such as NRas Q61R/K in human melanomas. We found that HMG-CoA lyase (HMGCL), a key enzyme in ketogenesis producing ketone bodies, was selectively essential in melanoma cells expressing BRAF V600E, but not in control cells containing NRas mutants or WT BRAF and NRas. Ketogenesis mainly occurs in the mitochondria of liver cells, which normally produces ketone bodies as a result of fatty acid breakdown to generate energy when glucose levels in the blood are low (Balasse and Fery, 1989; McPherson and McEneny, 2012). β-oxidation breaks down fatty acids to form acetyl-CoA, which, under normal conditions, is further oxidized in the TCA cycle. However, if TCA cycle activity is low, or the acetyl-CoA generation rate of β-oxidation exceeds the capacity of the TCA cycle, ketogenesis will be activated to convert acetyl-CoA to ketone bodies via HMG-CoA. HMGCL converts HMG-CoA to acetyl-coA and a ketone body, acetoacetate (AA), which can be further converted to two other ketone bodies, including D-β-hydroxybutyrate (3-HB) and acetone. Ketone bodies can be transported from liver to other tissues, where AA and 3-HB but not acetone will be further oxidized via the TCA cycle to produce acetyl-CoA for energy production. Organs including heart and brain can use AA and 3HB for energy. AA, if not used for energy, will be decarboxylated to acetone that is removed as waste (Cotter et al., 2013; Morris, 2005). However, although the ketogenic diet (high-fat, adequate-protein and low-carbohydrate) has been evaluated for cancer prevention and treatment purposes with the hope of attenuating tumor development by limiting carbohydrate supply, it is unknown whether and how ketogenesis and/or ketone bodies may contribute to cancer metabolism and tumor growth.
Here we report that active BRAF upregulates HMGCL via an octamer transcription factor Oct-1. Consistently, BRAF V600E expression results in increased HMGCL gene expression in cancer cells. HMGCL, however, selectively promotes BRAF V600E dependent phosphorylation and activation of MEK1 by controlling intracellular levels of its product AA, which specifically promotes BRAF V600E (but not BRAF WT) binding to MEK1 and subsequent MEK1 phosphorylation in cancer cells.
RESULTS
HMGCL is a “synthetic lethal” partner of BRAF V600E in human melanoma cells
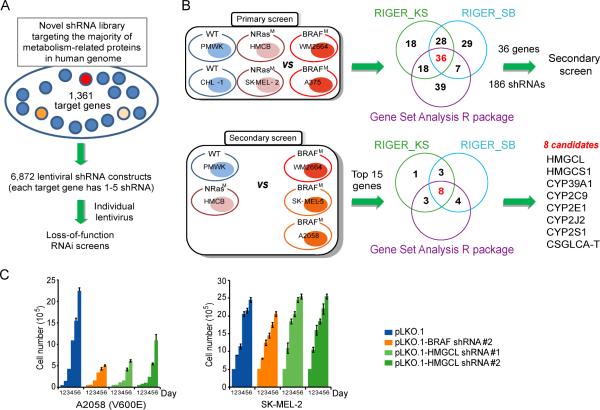
To identify “metabolic vulnerabilities” specific to oncogenic BRAF V600E mutant, but not other oncogenes such as NRas Q61R/K in human melanomas, we designed and constructed a shRNA library that targets 1,361 out of 1,417 genes encoding known metabolism-related enzymes and protein factors in the human genome (search at http://www.phosphosite.org/psrSearchAction.do by selecting “containing ‘metabolism’” in “Protein type” section), which are available in the whole genome shRNA library that we purchased from OpenBioSystems. The library contains 6,872 lentiviral-based shRNA constructs, where each gene is individually targeted by 1-5 different shRNA constructs that target different regions of the target gene (Figure 1A and Table S1). We performed a systematic RNAi screen in two BRAF V600D/E mutant expressing melanoma cells lines including WM2664 (V600D) and A375 (V600E), as well as control BRAF WT-expressing melanoma cells including PMWK, CHL-1, HMCB (NRas Q61K) and SK-MEL-2 (NRas Q61R) (Figure 1B). We then used 2 RIGER methods (RIGER_SB and RIGER_KS) and overlapped these against another method, Gene Set Analysis R package to analyze the normalized B-scores for each cell line (Barbie et al., 2009; Gould et al., 2006; Malo et al., 2006; Sims et al., 2011). The top-ranked 100 genes identified by each method were overlapped and 36 genes were enriched as top candidate synthetic lethal partners of BRAF V600E (Figure 1B and Tables S3-S6). In a secondary screen, we validated the 36 candidates (186 shRNAs) using additional BRAF V600E expressing SK-MEL-5 and A2058 melanoma cells, compared to control BRAF WT-expressing PMWK and HMCB cells (Figure 1B). Results analyzed by RIGER_SB, RIGER_KS and Gene Set Analysis R package, and 8 genes were enriched using the top 15 genes identified in the primary screen (Figure 1B and Tables S7).
Figure 1. “Metabolism targeted” RNAi screens identify HMGCL as a synthetic lethal partner of BRAF V600E.
(A) Construction of a shRNA library systematically targeting human genes related to metabolism.
(B) Primary and secondary screening strategy. Supervised analysis of viability data (B-score) identified candidate genes that, when knocked down by shRNAs, distinguish BRAF V600E human melanoma cells (BRAFM) from mutant NRas cells (NRasM) and cells expressing wild-type BRAF and NRas (WT). Overlapped results of indicated statistical methods identified top 8 candidate genes.
(C) Effect of BRAF or HMGCL knockdown on melanoma cell proliferation rates assessed by daily cell counting. Data are mean±s.d.; n=3 each; p values were obtained by a two-tailed Student's t test.
Among these candidates, we validated the two key ketogenic enzymes, HMG-CoA lyase (HMGCL; Figures 1C and S1A-S1B) and HMG-CoA synthase 1 (HMGCS1; Figure S1C) as synthetic lethal partners of BRAF V600E, using an alternative cell number-based cell proliferation rate assay. Suppression of HMGCL and HMGCS1 resulted in more attenuated cell proliferation rates in BRAF V600E-expressing melanoma cells compared to cells expressing BRAF WT, suggesting selective importance of ketogenesis in BRAF V600E induced melanoma transformation.
Expression of BRAF V600E upregulates HMGCL in cells
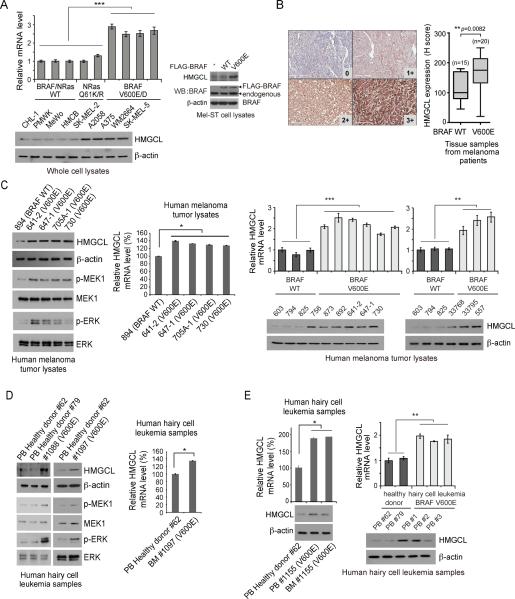
Ketogenesis mainly occurs in the mitochondria of liver cells, which normally converts acetyl-CoA to ketone bodies via HMG-CoA as a result of fatty acid breakdown to generate energy when glucose levels in the blood are low. HMGCL converts HMG-CoA to acetyl-coA and a ketone body, acetoacetate (AA), which can be further converted to two other ketone bodies, including D-β-hydroxybutyrate (3-HB) and acetone (Balasse and Fery, 1989; McPherson and McEneny, 2012). We found that HMGCL is upregulated in a group of BRAF V600E-expressing human melanoma cell lines compared to cells expressing BRAF WT (Figure 2A left), and in the immortalized melanocyte Mel-ST cells expressing BRAF V600E but not BRAF WT (Figure 2A right).
Figure 2. Expression of BRAF V600E upregulates HMGCL in cells.
(A) Left: RT-PCR and Western blot results show increased HMGCL expression in human melanoma cells expressing BRAF V600E/D compared to cell expressing BRAF WT. Right: Western blot results of HMGCL expression in Mel-ST cells with FLAG-BRAF WT or V600E. Data are mean±s.d. n=3 each; p values were obtained by a two-tailed Student's t test.
(B) HMGCL immunohistochemistry. Left: Positive staining of HMGCL was determined by histochemical score (H score = 3 × percentage of strong staining + 2 × percentage of moderate staining + 1 × % of weak staining + 0 × % of no staining; score range 0~300). Representative IHC staining images for 0 (WT; no staining), 1+ (WT; weak staining), 2+ (V600E; moderate staining), and 3+ (V600E; strong staining) scores of human melanoma tissue samples are shown (20×). Right: H scores are presented by Box and Whisker plots. Medians, interquartile, maximum, and minimum are shown.
(C-E) Western blot and RT-PCR results show increased HMGCL expression with increased MEK1 and ERK phosphorylation in human primary melanoma (C) and hairy cell leukemia tissue samples (D-E). PB: peripheral blood; BM: bone marrow. Data are mean±s.d.; n=3 each; p values were obtained by a two-tailed Student's t test.
Also see Figure S2.
Consistent with these findings, HMGCL expression assessed by immunohistochemistry (IHC) staining is significantly upregulated in primary tumor samples of BRAF V600E-positive melanoma patients compared to control tumor tissue samples from BRAF WT-expressing melanoma patients (Figure 2B). In addition, immunoblotting results confirmed upregulated HMGCL protein and mRNA levels in primary human tumor tissue samples from BRAF V600E-positive melanoma patients, which also demonstrated enhanced phosphorylation and activation of MEK1 and ERK, compared to primary melanoma sample from one representative patient with BRAF WT (Figure 2C).
BRAF V600E mutation has been identified in other human malignancies such as colorectal cancer, multiple myeloma (Benlloch et al., 2006; Chapman et al., 2011) and hairy cell leukemia (HCL). HCL is a chronic B cell lymphoproliferative disease and nearly 100% of classic HCL patients harbor somatic BRAF V600E mutation (Arcaini et al., 2012; Golomb et al., 1982; Schnittger et al., 2012; Tiacci et al., 2011). Consistently, protein and mRNA levels of HMGCL and phosphorylation of MEK1 and ERK were upregulated in primary human leukemia cells from HCL patients compared to control peripheral blood samples from healthy donors (Figure 2D-2E).
Furthermore, HMGCL protein levels were increased in tumor and bone marrow samples of BRAF V600E conditional knock-in mouse models of melanoma (Dankort et al., 2009) or HCL (Chung, 2014) compared to control skin or bone marrow samples from normal mice, respectively (Figure S2A). These results together suggest that HMGCL is upregulated in BRAF V600E-positive human malignancies.
HMGCL is required for BRAF V600E-induced transformation
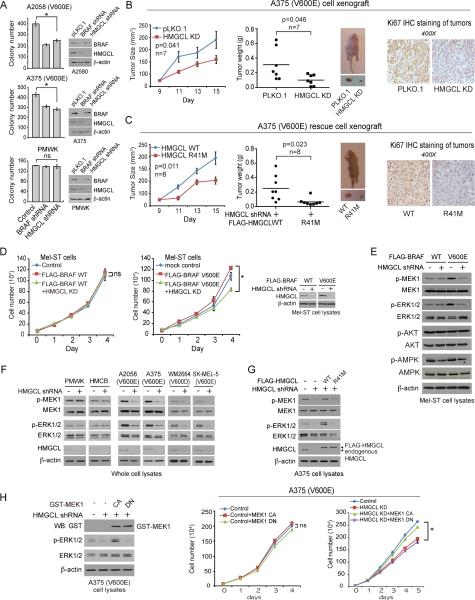
We next examined whether HMGCL is required for BRAF V600E-induced transformation. We found that stable knockdown (KD) of HMGCL or BRAF resulted in attenuated colony formation potential of BRAF V600E expressing A2058 and A375 cells but not control PMWK cells (Figure 3A). Moreover, in a xenograft nude mouse model, tumors derived from HMGCL KD A375 cells demonstrated decreased growth rate and masses with decreased cell proliferation rate assessed by reduced immunohistochemistry (IHC) staining of Ki-67, compared to those derived from control A375 cells (Figure 3B). Similar results were obtained using A2058 cells, whereas tumors derived from BRAF WT-expressing PMWK and PMWK HMGCL KD cells, or HMCB and HMCB HMGCL KD cells, in xenograft mice were indistinguishable (Figure S2B-S2D).
Figure 3. HMGCL promotes MEK-ERK activation and is specifically required for cell proliferation and tumor growth potential of BRAF V600E-expressing melanoma cells.
(A) Anchorage-independent growth of melanoma cells with or without stable knockdown of BRAF or HMGCL. Duplicate experiment; Data are mean ± SEM. p values were obtained by a two-tailed Student's t test.
(B-C) Left two panels: Tumor growth and size of xenograft mice injected with parental or HMGCL knockdown (KD) BRAF V600E-positive A375 cells (B), or HMGCL KD cells with rescue expression of FLAG-HMGCL wild type (WT) or enzyme deficient R41M mutant (C). Middle panels show the dissected tumors in representative mice. Right panels show representative images of IHC staining of Ki-67 of tumors (brown color). Data are mean ± SEM. p values were obtained by a paired two-tailed Student's t test.
(D) Effect of HMGCL knockdown on cell proliferation rates of Mel-ST cells expressing BRAF WT or V600E by daily cell counting. Right: Immunoblotting of HMGCL upon shRNA-mediated knockdown. FLAG-BRAF WT and V600E expression in Mel-ST cells is shown in Figure 2A right. Data are mean±s.d.; n=3 each; p values were obtained by a two-tailed Student's t test.
(E) Immunoblotting of phosphorylation levels of MEK1, ERK1/2, AKT, and AMPK in Mel-ST cells expressing BRAF WT or V600E upon HMGCL knockdown.
(F) Effect of HMGCL knockdown on phosphorylation levels of MEK1 and ERK1/2 in melanoma cell lines.
(G) Effect of rescue expression of HMGCL WT or R41M on phosphorylation levels of MEK1 and ERK1/2 in melanoma cell lines with HMGCL knockdown.
(H) Left: Immunoblotting of expression of GST-tagged MEK1 constitutively active (CA) or dominant-negative (DN) forms, as well as phosphorylation levels of ERK1/2 in BRAF V600E-expressing A375 cells. Right panels: Effect of expressing MEK1 CA or DN forms on cell proliferation of parental or HMGCL KD A375 cells. Data are mean±s.d.; n=3 each; p values were obtained by a two-tailed Student's t test.
Also see Figures S2-S4.
We next generated “rescue” A375 cell lines with stable knockdown of endogenous HMGCL and rescue expression of wild-type (WT) or an enzyme-dead R41M mutant (Fu et al., 2010) of a shRNA-resistant, FLAG-tagged human HMGCL form. Rescue expression of FLAG-HMGCL WT, but not R41M mutant, significantly reversed the reduced cell proliferation upon HMGCL knockdown in A375 cells but not control PMWK cells (Figures S3A-S3B). Moreover, rescue A375 cells expressing HMGCL WT but not R41M demonstrated restored potential of tumor formation and growth (Figure 3C). These data together suggest that the synthetic lethal interaction between BRAF V600E and HMGCL requires HMGCL activity.
HMGCL selectively enhances activation of MEK-ERK in BRAF V600Eexpressing cells
We next sought to explore the underlying molecular mechanism. Although HMGCL knockdown did not selectively confer any metabolic alteration to BRAF V600E-expressing melanoma cells (Figure S3C-S3H), silencing HMGCL resulted in significantly decreased cell proliferation of Mel-ST cells expressing BRAF V600E but not BRAF WT (Figure 3D) with decreased phosphorylation of MEK1 and ERK1/2 but not AKT and AMPK (Figure 3E). These results suggest not only that BRAF V600E conferred HMGCL-reliance to Mel-ST cells but also that HMGCL is specifically involved in BRAF V600E-dependent activation of MEK-ERK signaling.
Furthermore, we found that HMGCL knockdown selectively attenuated phosphorylation of MEK1 and ERK1/2 only in BRAF V600E-expressing melanoma cells (Figure 3F), while rescue expression of HMGCL WT, but not R41M mutant, reversed the decreased phosphorylation of MEK1 and ERK1/2 in A375 HMGCL KD cells (Figure 3G). Consistently, expressing a constitutively active (CA) S222D (Mansour et al., 1994) form of MEK1, but not a catalytically inactive, dominant negative (DN) K97M mutant of MEK1 (Mansour et al., 1994) reversed the reduced cell proliferation in A375 HMGCL KD cells, but did not affect control A375 cells (Figure 3H). Similar results were obtained using A2058 cells (Figure S3I).
HMGCL signals through its product acetoacetate to promote BRAF V600E activated MEK-ERK signaling cascade
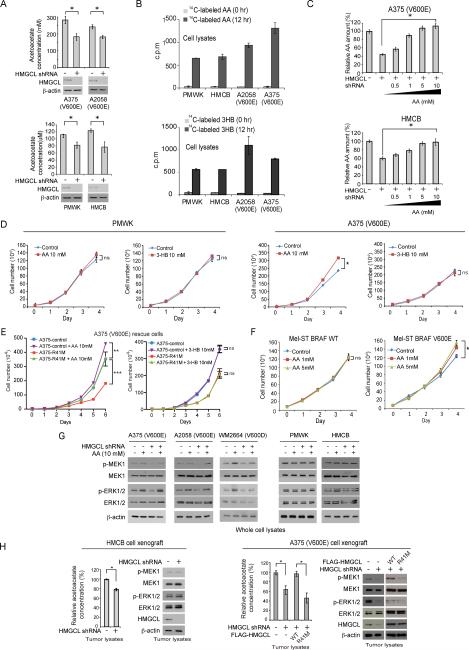
HMGCL knockdown resulted in decreased intracellular concentration of its product, acetoacetate (AA) in melanoma cells (Figure 4A). Note that the intracellular levels of AA are lower in melanoma cells expressing BRAF WT compared to BRAF V600E-expressing cells (Figure 4A), which correlates with the differential expression levels of HMGCL in these cells (Figure 2A). We next tested whether acetoacetate mediates the synthetic lethal importance of HMGCL only in BRAF V600E expressing melanoma cells. Consistent with literature, both AA and its subsequent ketone body 3-HB can readily enter cells ((Patel et al., 1981); Figures 4B and S4A), and we also found that addition of AA up to 10 mM in the culture media eventually significantly reversed the decreased intracellular AA levels in HMGCL knockdown cells (Figures 4C and S4B).
Figure 4. HMGCL's product acetoacetate specifically promotes MEK-ERK activation in BRAF V600E-expressing cells.
(A) Intracellular concentration of acetoacetate in melanoma cells with HMGCL knockdown.
(B) Cell permeability of acetoacetate (AA; upper) or D-β-hydroxybutyrate (3-HB; lower) was examined by increased scintillation counting of 14C in the cell lysates using human melanoma cells cultured in the presence of 14C-labeled acetoacetate or 3HB for 12 hours.
(C) Effect of adding increasing concentrations of AA on A375 (upper) or HMCB (lower) cells in culture media on reduced intracellular levels of acetoacetate upon HMGCL knockdown.
(D-F) Effect of adding AA or 3-HB in culture media on cell proliferation rates of melanoma cell lines (D), HMGCL R41M rescue A375 cells (E) and Mel-ST cells expressing BRAF WT or V600E (F). Cell proliferation rates were determined by daily cell counting.
(G) Effect of adding AA in culture media on phosphorylation levels of MEK1 and ERK1/2 in melanoma cell lines and cells with HMGCL knockdown.
(H) Intracellular AA levels (left panels) and immunoblotting results detecting MEK1, ERK1/2 and HMGCL protein levels as well as phosphorylation levels of MEK1 and ERK1/2 (right panels) using tumor lysates are shown. The tumors were from xenograft nude mice presented in Figures 3B-3C and S2D. Data with error bars in Figure 4 are represented as mean±s.d.; n=3 each; p values were obtained by a two-tailed Student's t test.
Also see Figures S4-S5.
Interestingly, addition of AA but not 3-HB in the culture media selectively promoted cell proliferation of BRAF V600E-expressing A375 and A2058 cells but not control PMWK and HMCB cells (Figures 4D and S4C-S4D), and significantly reversed the reduced cell proliferation of A375 and A2058 rescue cells with HMGCL knockdown (Figure S4D) or cells expressing HMGCL enzyme-dead mutant R41M (Figures 4E and S4E). In addition, AA treatment only promoted proliferation of Mel-ST cells expressing BRAF V600E (Figure 4F).
AA treatment reversed the decreased phosphorylation of MEK1 and ERK1/2 due to HMGCL knockdown only in BRAF V600E-expressing cells (Figure 4G), whereas 3-HB treatment had no effect on MEK1 and ERK phosphorylation (Figure S5A). Moreover, despite the fact that acetoacetate levels were commonly decreased in tumors derived from HMGCL KD melanoma cells (Figures 4H and S5B; left panels), we found decreased phosphorylation levels of MEK1 and ERK1/2 only in tumors derived from A2058 and A375 HMGCL KD cells and A375 rescue cells expressing R41M mutant, but not in tumors derived from control PMWK and HMCB HMGCL KD cells (Figures 4H and S5B; right panels).
To examine the clinical impact of our findings, we measured the intracellular levels of ketone bodies in human primary tissues from hairy cell leukemia patients. We observed increased intracellular levels of ketone bodies including AA and 3-HB in primary human leukemia cells from HCL patients compared to control peripheral blood samples from a healthy donor (Figure S5C; left two panels), which is consistent with the fact that HCL cells are BRAF V600E positive with upregulated HMGCL. In addition, increased AA levels were also detected in tumor and bone marrow samples from BRAF V600E conditional knock-in mouse models of melanoma or HCL compared to control skin or bone marrow samples from normal mice, respectively (Figure S5C; right two panels). These findings are consistent with the results showing increased HMGCL expression in primary leukemia cells from HCL patients (Figure 2D-2E) and tumors expressing BRAF V600E (Figure S2A).
Acetoacetate selectively enhances BRAF V600E-MEK1 binding
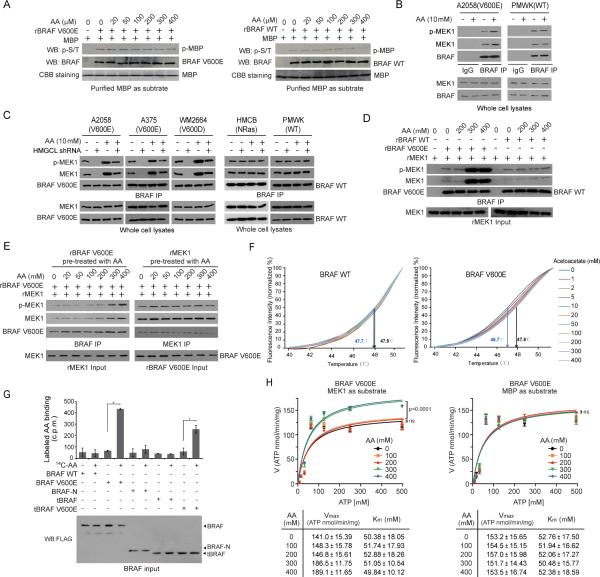
We next tested the hypothesis that acetoacetate might affect BRAF V600E kinase activity. We thus performed cell-free, in vitro kinase assays using purified recombinant BRAF V600E or BRAF WT incubated with purified myelin basic protein (MBP) as a non-specific substrate or purified MEK1, in the presence of increasing amounts of AA. We found that not BRAF WT but BRAF V600E dependent phosphorylation of MEK1 was increased in the presence of AA (Figure S5D). Although both BRAF V600E or BRAF WT phosphorylates MBP but such phosphorylation was not affected in the presence of increasing concentrations of acetoacetate or 3-HB (Figure 5A and S5E-S5F), suggesting that acetoacetate may specifically affect BRAF V600E dependent phosphorylation of MEK1.
Figure 5. Acetoacetate selectively enhances BRAF V600E-MEK1 binding.
(A) Effect of acetoacetate on phosphorylation of MBP in an in vitro kinase assay using purified recombinant BRAF V600E or BRAF WT incubated with purified MBP as a substrate.
(B-C) Effect of adding cell-permeable acetoacetate (AA) in culture media on BRAF-MEK1 binding and MEK1 phosphorylation in melanoma cells (B) and cells with HMGCL knockdown (C). IP: immunoprecipitates.
(D) Effect of acetoacetate on BRAF-MEK1 binding and MEK1 phosphorylation in cell-free, in vitro assays using purified recombinant BRAF (rBRAF) and MEK1 (rMEK1).
(E) Effect of pre-treatment of rBRAF V600E (left) or rMEK1 (right) with increasing concentrations of acetoacetate on BRAF-MEK1 binding and MEK1 phosphorylation in cell-free, in vitro assays.
(F) Thermal melt shift assay was performed to examine the protein (BRAF WT or V600E) and ligand (acetoacetate) interaction. Change of melting temperature (Tm) in a dose-dependent manner at concentrations from 0 μM to 400 μM demonstrates that acetoacetate may directly bind to BRAF V600E but not BRAF WT protein. Arrows in each panel indicate melting temperatures at 0 μM (left) and 300 μM (right), since 300 μM represents the physiological acetoacetate level in BRAF V600E-expressing human melanoma cells.
(G) Radiometric metabolite-protein interaction analysis using 14C-labeled acetoacetate incubated with purified BRAF variants. Data are mean±s.d.; n=3 each; p values were obtained by a two-tailed Student's t test.
(H) Vmax and Km of BRAF V600E were measured using purified BRAF V600E protein (100 ng) incubated with increasing concentrations of ATP in the presence and absence of increasing concentration of acetoacetate, using excessive amount of purified MEK1 (left) or MBP (right) as substrates. Data are mean±s.d.; n=3 each; p values were obtained by a two-tailed Student's t test. Also see Figures S5-S6.
Further mechanistic studies revealed that AA treatment resulted in increased MEK1 binding to BRAF V600E as well as increased phosphorylation of V600E-bound MEK1 in A2058, A375 and WM2664 cells, but not to BRAF WT in control PMWK and HMCB cells (Figures 5B and S6A) or CRAF in A2058 cells (Figure S6B). In contrast, 3-HB treatment did not affect BRAF-MEK1 binding or MEK1 phosphorylation (Figure S6C). Moreover, knockdown of HMGCL resulted in decreased V600E/D-MEK1 binding and MEK1 phosphorylation only in A2058, A375 and WM2664 (V600D) cells, which were reversed by treatment with AA (Figure 5C) but not 3HB (Figure S6D).
To determine whether AA directly or indirectly affects the BRAF V600E-MEK1 complex, we performed cell-free, in vitro binding and kinase assays using purified recombinant BRAF V600E or BRAF WT incubated with recombinant purified MEK1 as substrate, in the presence of increasing amounts of AA. We found that AA treatment promoted MEK1 binding to BRAF V600E and phosphorylation of V600E-bound MEK1 in a dose-dependent manner, whereas such phenotypes were absent in MEK1 incubated with BRAF WT (Figure 5D). Notably, increasing AA concentrations from 200μM to 300μM caused a significant increase in BRAF V600E-bound and phosphorylated MEK1 (Figure 5D), which were physiologically consistent with the AA levels determined in HMGCL KD A375 and A2058 cells compared to control cells (Figure 4A).
Moreover, we found that purified BRAF V600E pre-treated with increasing concentrations of AA (Figure 5E; left) but not 3-HB (Figure S6E; left) showed increased binding ability to MEK1, whereas neither AA (Figure 5E; right) nor 3-HB (Figure S6E; right) pre-treated MEK1 demonstrated increased binding ability to BRAF V600E. These results are consistent with our observation that AA might directly bind to BRAF V600E but not BRAF WT in a thermal melt shift assay using purified BRAF WT or V600E incubated with increasing concentrations of acetoacetate (Figure 5F). Furthermore, we performed a radiometric metabolite-protein interaction analysis using 14C-labeled acetoacetate incubated with purified BRAF variants. Labeled acetoacetate specifically binds to BRAF V600E and a V600E mutant of an active, truncated C-terminal domain of BRAF (tBRAF, 416-766 aa) (Brummer et al., 2006), but not to control proteins including BRAF WT, tBRAF WT or a N-terminal domain of BRAF (BRAF-N, 1-415 aa), which further suggest the direct association between acetoacetate and V600E mutant of BRAF (Figure 5G). Consistently, treatment with 300 or 400 μM of acetoacetate resulted in increased Vmax and slightly decreased Km of BRAF V600E using MEK1 as a substrate, whereas treatment with lower concentrations (0, 100, 200 μM) of acetoacetate did not affect Vmax or Km of V600E (Figure 5H; left panels). In contrast, treatment with acetoacetate did not affect Vmax or Km of BRAF V600E using MBP as a substrate (Figure 5H; right panels). Together, these data suggest an important role of AA in BRAF V600E-dependent transformation.
Active BRAF signals through Oct-1 to upregulate HMGCL and its product acetoacetate, which, however, selectively enhances BRAF V600E-dependent activation of MEK1
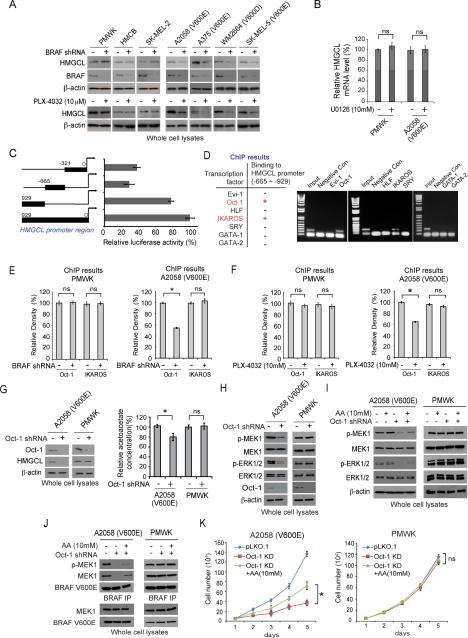
We found that stable knockdown of BRAF V600E but not BRAF WT in melanoma cells resulted in decreased HMGCL protein (Figure 6A; upper) and mRNA (Figure S7A; upper) levels. Similar results were obtained using treatment with BRAF mutant small molecule inhibitor PLX-4032 (Figures 6A and S7A; lower panels), whereas treatment with MEK1 inhibitors U0126 (Figure 6B), selumatinib and trametinib (Figure S7B) did not affect HMGCL mRNA levels in PMWK, A375, and A2058 cells. These results suggest that active BRAF V600E but not its downstream MEK1-ERK signaling is involved in HMGCL upregulation.
Figure 6. BRAF V600E upregulates HMGCL expression through an octamer transcription factor Oct-1.
(A) Western blot to detect HMGCL protein levels in human melanoma cells with stable knockdown of BRAF WT or V600E/D mutant (upper) or treatment with BRAF V600E inhibitor PLX-4032 (lower).
(B) RT-PCR to examine the effect of treatment with MEK1 inhibitor, U0126 on HMGCL mRNA levels in PMWK (BRAF WT) and A2058 (V600E) melanoma cells.
(C) Luciferase reporter assay revealed a functional HMGCL promoter region (−929~−665).
(D) ChIP results detecting binding ability of a group of transcription factors to the functional region of HMGCL promoter. Positive binding of Oct-1 and IKAROS is indicated with “+” in red color.
(E-F) ChIP results detecting binding ability of Oct-1 or IKAROS to HMGCL promoter region in melanoma cells with BRAF knockdown (E) or treatment with BRAF V600E inhibitor PLX-4032.
(G-H) Effect of Oct-1 knockdown on HMGCL expression and intracellular AA levels (G) and phosphorylation levels of MEK1 and ERK1/2 (H).
(I-K) Effect of adding AA in culture on phosphorylation levels of MEK1 and ERK1/2 (I), phosphorylation of MEK1 and BRAF-MEK1 association (J), and cell proliferation (K) in melanoma cells with Oct-1 knockdown. Data with error bars in Figure 6 are represented as mean±s.d.; n=3 each; p values were obtained by a two-tailed Student's t test.
Also see Figure S7.
To decipher how BRAF V600E upregulates HMGCL in cells independent of MEK1, using a series of luciferase reporter constructs, we identified −929~−665 as the functional region of HMGCL promoter (Figure 6C). Sequence analysis by TFSEARCH (http://mbs.cbrc.jp/research/db/TFSEARCH.html) revealed 7 potential transcription factors that may bind directly to −929~−665 and modulate HMGCL promoter activity. Among these factors, ChIP assay suggested that both Oct-1 and IKAROS could bind to HMGCL promoter (Figure 6D). Further experiments revealed that knockdown of BRAF V600E in A2058 cells resulted in decreased binding of Oct-1 but not IKAROS to HMGCL −929~−665 promoter region, whereas knockdown of BRAF WT in PMWK cells did not affect either Oct-1 or IKAROS binding to this region (Figure 6E). Similar results were obtained using A375 and HMCB cells with BRAF knockdown (Figure S7C), or these cells treated with PLX-4032 (Figures 6F and S7D).
Consistent with these findings, we found that Oct-1 knockdown resulted in decreased HMGCL expression and intracellular AA levels in BRAF V600E-expressing A2058 and A375 cells but not in control PMWK and HMCB cells expressing BRAF WT (Figures 6G and S7E). Moreover, Oct-1 knockdown resulted in decreased phosphorylation of MEK1 and ERK1/2 in A2058 and A375 cells but not in control PMWK and HMCB cells (Figures 6H and S7F), which was reversed by adding AA in culture media (Figures 6I and S7G). In addition, treatment with AA also reversed reduced protein amount and phosphorylation of MEK1 bound to BRAF V600E in A2058 and A375 cells with Oct-1 knockdown, but not in control PMWK and HMCB cells (Figures 6J and S7H). Consistently, AA treatment also partially reversed decreased cell proliferation in A2058 and A375 cells with Oct-1 knockdown, but not in control PMWK and HMCB cells (Figures 6K and S7I). This suggests that Oct-1 plays an essential role in BRAF V600E-dependent transformation, and HMGCL is one of the important downstream effectors of Oct-1.
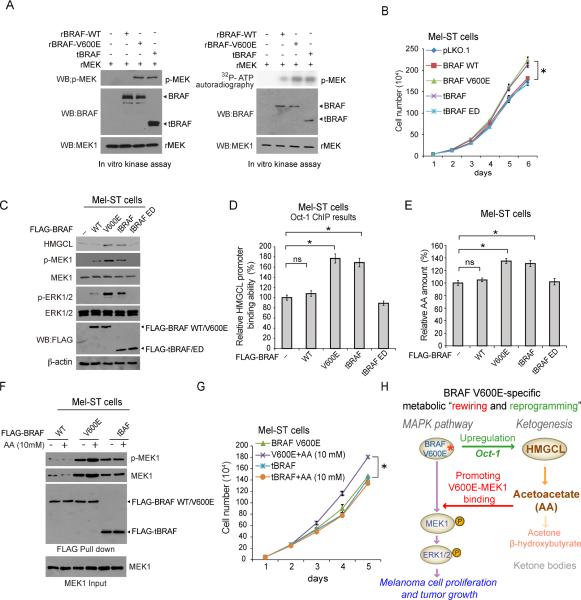
In order to determine whether active BRAF activates the Oct-1-HMGCL-acetoacetate axis in cells, we included an active, N-terminally truncated BRAF (tBRAF, 416-766 aa). We found that both BRAF V600E and tBRAF demonstrated comparably increased kinase activity in an in vitro kinase assay compared to BRAF WT (Figure 7A). In addition, stable expression of BRAF V600E and tBRAF in Mel-ST cells resulted in comparably enhanced cell proliferation (Figure 7B), increased HMGCL expression (Figure 7C) along with increased Oct-1 binding to HMGCL promoter region (Figure 7D) and elevated intracellular AA levels (Figure 7E), compared to cells expressing control BRAF WT or an enzyme dead (ED; K482M (Sievert et al., 2013)) form of tBRAF.
Figure 7. Active BRAF signals through Oct-1 to upregulate HMGCL and its product acetoacetate, which, however, selectively enhances BRAF V600E-dependent activation of MEK1.
(A) Results of an in vitro kinase assay using purified recombinant BRAF (rBRAF) WT, V600E or a truncated form of BRAF (tBRAF) that is constitutively active incubated with recombinant MEK1 (rMEK1) as exogenous substrate.
(B-E) Effects of stable expression of BRAF variants in Mel-ST cells on cell proliferation (B), HMGCL expression and phosphorylation of MEK1 and ERK1/2 (C), Oct-1 binding ability of promoter region of HMGCL (D), and intracellular acetoacetate levels (E). ED: enzyme dead. Data are mean ± s.d; n=3 each; p values were obtained by a two-tailed Student's t test.
(F-G) Effects of adding AA in culture media on protein amount and phosphorylation of MEK1 bound to BRAF (F) and cell proliferation (G) in Mel-ST cells expressing BRAF WT, or constitutively active V600E or tBRAF. Data are mean ± s.d; n=3 each; p values were obtained by a two-tailed Student's t test.
(H) Proposed working model: constitutively active, oncogenic BRAF V600E activates Oct-1 to upregulate HMGCL (reprogramming), leading to increased levels of acetoacetate that specifically binds to BRAF V600E and promotes BRAF V600E-MEK1 binding (rewiring).
However, only expression of BRAF V600E and tBRAF but not the enzyme dead mutant of tBRAF resulted in increased phosphorylation of MEK1 and ERK1/2 (Figure 7C). Moreover, treatment with AA selectively promotes BRAF V600E but not tBRAF to bind and phosphorylate MEK1 (Figure 7F), leading to increased cell proliferation in Mel-ST cells (Figure 7G). These results together suggest that active BRAF commonly activates Oct-1, leading to upregulated HMGCL and consequently intracellular AA levels, whereas AA selectively promotes BRAF V600E-dependent activation of MEK-ERK signaling cascade.
DISCUSSION
The current understanding of metabolic alterations in human cancers is befuddling. We propose to distinguish metabolic “rewiring” and “reprogramming” in cancer cells, wherein metabolic reprogramming describes “software” changes induced by growth factors in normal proliferative cells that are “hijacked” by oncogenic signals in cancer cells, while metabolic rewiring represents “hardware” changes that are “forged” due to “neo-function” of oncogenic mutants, but not found in normal cells. Our findings suggest a “mutation-specific” function in which BRAF V600E upregulates HMGCL, leading to increased intracellular levels of acetoacetate that specifically promote BRAF V600E binding to MEK1 and subsequent MEK1 phosphorylation (Figure 7H). These results also support the emerging “metabolic rewiring” concept describing metabolic alterations in cancer cells that are required for a group of oncogenes but not found in normal cells. In our case, we demonstrated that oncogenic BRAF V600E links ketogenesis to BRAF-MEK-ERK signaling cascade, representing a “wiring” between metabolic and cell signaling pathways. This in part shares conceptual similarity with the mutations in isocitrate dehydrogenase (IDH) 1 and 2 identified in glioma and acute myeloid leukemia, which enable the enzymes to produce oncometabolite 2-hydroxyglutamate (Dang et al., 2009; Mardis et al., 2009; Parsons et al., 2008; Yan et al., 2009).
In addition, our findings provide evidence that supports a concept suggesting that a group of oncogenes may require different metabolic alterations for tumor growth. We found that active BRAF commonly activates Oct-1 to promote HMGCL gene expression, which in turn leads to increased intracellular levels of acetoacetate in cells. However, it appears that only BRAF proteins with substitution of V600 or mutations within the V600 flanking region may benefit from the elevated levels of acetoacetate, which selectively promotes MEK1 binding. Thus, although acetoacetate levels are commonly elevated upon activation of BRAF, only cancer cells expressing BRAF V600E/D mutant can benefit from increased intracellular acetoacetate, where increased acetoacetate specifically promotes V600E-MEK1 signaling, providing an “evolutionary advantage” that may explain why V600E is the predominant mutation of BRAF identified in human malignancies. Further studies are warranted to determine whether other cancer-associated active BRAF mutants may similarly upregulate HMGCL expression and acetoacetate levels but cannot respond to acetoacetate in terms of MEK1 binding and phosphorylation.
In addition, future structural studies are also warranted to explore the molecular basis underlying how acetoacetate specifically promotes BRAF V600E-MEK1 binding, and elucidate the molecular mechanism by which active BRAF mutants, which, however, may not be necessarily limited to BRAF V600E, upregulate HMGCL gene expression in cancer cells. It will also be interesting to explore the molecular mechanism by which active BRAF activates Oct-1 in a MEK1-independent manner, which may involve direct or indirect phosphorylation of Oct-1 since BRAF kinase activity is important to promote Oct-1 binding to HMGCL promoter. In addition, there may be additional Oct-1 transcription targets besides HMGCL that are important to mediate BRAF V600E-dependent transformation in cancer cells, which may explain why AA treatment only partially reversed the decreased cell proliferation in BRAF V600E positive melanoma cells with Oct-1 knockdown (Figures 6K and S7I).
Our findings also link the ketogenic pathway to oncogenic BRAF V600E-MEK-ERK signaling cascade, suggesting a signaling function of acetoacetate that is independent of its role in cell metabolism. These findings add to emerging evidence that supports a concept suggesting that metabolites could function as signaling molecules to allow crosstalk between metabolic pathways and cell signaling networks. For example, we previously reported that glycolytic intermediate 3-phosphoglycerate is a competitive inhibitor of 6-phosphoglyconate dehydrogenase (6PGD) in the oxidative pentose phosphate pathway (Hitosugi et al., 2012), while others have found that AMP is an allosteric activator for AMPK. On the other hand, there has been accumulating evidence showing that post-translational modifications including tyrosine phosphorylation (Fan et al., 2011; Hitosugi et al., 2011; Hitosugi et al., 2009; Hitosugi et al., 2013) and lysine acetylation (Choudhary et al., 2009; Fan et al., 2014; Kim et al., 2006; Wang et al., 2010; Zhao et al., 2010) of metabolic enzymes are common and important to link cell signaling pathways to metabolic pathways in cancer cells. These findings together represent a realm of crosstalk with “back and forth” signal flows between metabolic and cell signaling networks that “acutely” regulate cell metabolism and proliferation, which, unfortunately, are “hijacked” by cancer cells.
Lastly, our results suggest that HMGCL inhibitor or non-metabolizable acetoacetate derivatives that can compete with acetoacetate for BRAF V600E binding may represent alternative therapies to treat oncogenic BRAF V600E driven cancers. However, organs including heart and brain can use ketone bodies including AA and 3-HB for energy (Cotter et al., 2013; Morris, 2005). This warrants further detailed toxicity and pharmacokinetics studies to evaluate the proposed anti-HMGCL or anti-acetoacetate therapies in cancer treatment.
EXPERIMENTAL PROCEDURES
RNAi screens and data analysis
The identities of known metabolism-related enzymes and protein factors in the human genome were provided by the Phosphosite Plus website of Cell Signaling Technology (http://www.phosphosite.org/psrSearchAction.do). We constructed a “metabolism-targeted” shRNA library that targets 1,361 out of 1,417 genes encoding known enzymes and protein factors in the human genome, which are available in the whole genome shRNA library that we purchased from OpenBioSystems. The library contains 6,872 lentiviral-based shRNA constructs, where each gene is individually targeted by 1-5 different shRNA constructs that target different regions of the target gene (Table S1). In brief, RNAi screens were performed in 96-well format such that each well contained a single shRNA species, and each transcript was covered, on average, by 1~5 different shRNAs. Assay conditions (cell number per well, viral dose, puromycin concentration) were optimized for each cell line prior to screening. Cells were seeded, incubated for 24 hours, infected with lentivirus, and incubated for 5 days. All lentiviral infections were performed with 2 replicates selected with puromycin during the final 5 days of incubation and the other 2 replicates left untreated. Cell viability and proliferation were measured 5 days after lentiviral infection using a CyQUANT Direct Cell Proliferation Assay kit (Invitrogen). For both primary and secondary screens, we used 2 RIGER methods (RIGER_KS and RIGER_SB) and another method, Gene Set Analysis R package (Barbie et al., 2009; Gould et al., 2006; Malo et al., 2006; Sims et al., 2011) to analyze the normalized B-scores for each cell line.
Xenograft studies and primary tissue samples from patients with melanoma or hairy cell leukemia and healthy donors
Approval of use of mice and designed experiments was given by the Institutional Animal Care and Use Committee of Emory University. Approval of use of human specimens was given by the Institutional Review Board of Emory University School of Medicine. All clinical samples were obtained with informed consent with approval by the Emory University Institutional Review Board. Clinical information for the patients was obtained from the pathologic files at Emory University Hospital under the guidelines and with approval from the Institutional Review Board of Emory University School of Medicine and according to the Health Insurance Portability and Accountability Act. Detailed experimental procedures using mice and human primary tissue samples are described in Supplemental Extended Experimental Procedures.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Cancer Tissue and Pathology Core of Emory University for providing primary cancer patient tissue samples. This work was supported in part by NIH grants CA140515, CA183594, CA174786 (J.C.), CA175316 (S.K.), GM071440 (C.H.), AR47901 (J.L.A.) and the Pharmacological Sciences Training Grant T32 GM008602 (S.E.), DoD grant W81XWH-12-1-0217 (J.C.), National Natural Science Funds of China No.20902013 (L.Z.), the Jamie Rabinowitch Davis Foundation (J.L.A.), the Charles Harris Run For Leukemia, Inc. (H.J.K.) and the Hematology Tissue Bank of the Emory University School of Medicine and the Georgia Cancer Coalition (H.J.K.). H.J.K., F.R.K., S.K. and J.C. are Georgia Cancer Coalition Distinguished Cancer Scholars. S.K. and J.C. are American Cancer Society Basic Research Scholars. J.C. is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
H.K., J.F. and R.L. contributed equally to this work. J.L.A., C.C., D.B., H.M.M., E.K., O.A.-W., T.M., B.P.P., K.F., R.R.K., D.H.L., G.S., M.R., S.L., H.J.K. and F.R.K. provided critical reagents. Q.J. and C.H. performed biochemical analysis of purified proteins incubated with acetoacetate and analyzed the data. T.J.B. performed structural analysis. B.H.L. performed the histopathological analyses. S.F., C. Scholl, P.T., and D.A.B. contributed to experimental design of screens and statistical analyses. H.K., J.F., R.L., S.E., L.Z., L.J., C. Shan, and J.-H.S. performed all other experiments. H.K., J.F., R.L., S.K. and J.C. designed the study. S.K. and J.C. are senior authors, and jointly managed the project and wrote the paper.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, 7 figures and 7 tables.
REFERENCES
- Arcaini L, Zibellini S, Boveri E, Riboni R, Rattotti S, Varettoni M, Guerrera ML, Lucioni M, Tenore A, Merli M, et al. The BRAF V600E mutation in hairy cell leukemia and other mature B-cell neoplasms. Blood. 2012;119:188–191. doi: 10.1182/blood-2011-08-368209. [DOI] [PubMed] [Google Scholar]
- Balasse EO, Fery F. Ketone body production and disposal: effects of fasting, diabetes, and exercise. Diabetes Metab Rev. 1989;5:247–270. doi: 10.1002/dmr.5610050304. [DOI] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlloch S, Paya A, Alenda C, Bessa X, Andreu M, Jover R, Castells A, Llor X, Aranda FI, Massuti B. Detection of BRAF V600E mutation in colorectal cancer: comparison of automatic sequencing and real-time chemistry methodology. J Mol Diagn. 2006;8:540–543. doi: 10.2353/jmoldx.2006.060070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag G, Tsai J, Zhang J, Zhang C, Ibrahim P, Nolop K, Hirth P. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nature reviews. Drug discovery. 2012;11:873–886. doi: 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- Brummer T, Martin P, Herzog S, Misawa Y, Daly RJ, Reth M. Functional analysis of the regulatory requirements of B-Raf and the B-Raf(V600E) oncoprotein. Oncogene. 2006;25:6262–6276. doi: 10.1038/sj.onc.1209640. [DOI] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Chung S, Kim E, Park JH, Chung YR, Lito P, Feldstein J, Hu W, Beguilin W, Monette S, Duy C, Rampal R, Telis L, Patel M, Kim MK, Melnick AM, Rosen N, Tallman MS, Park CY, Abdel-Wahab O. Hematopoietic Stem Cell Origin of BRAFV600E Mutations in Hairy Cell Leukemia. Science Translational Medicine. 2014;6:238ra71. doi: 10.1126/scitranslmed.3008004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter DG, Schugar RC, Crawford PA. Ketone body metabolism and cardiovascular disease. Am J Physiol Heart Circ Physiol. 2013;304:H1060–1076. doi: 10.1152/ajpheart.00646.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr., You MJ, DePinho RA, McMahon M, Bosenberg M. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nature genetics. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Hitosugi T, Chung TW, Xie J, Ge Q, Gu TL, Polakiewicz RD, Chen GZ, Boggon TJ, Lonial S, et al. Tyrosine phosphorylation of lactate dehydrogenase A is important for NADH/NAD(+) redox homeostasis in cancer cells. Mol Cell Biol. 2011;31:4938–4950. doi: 10.1128/MCB.06120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Shan C, Kang HB, Elf S, Xie J, Tucker M, Gu TL, Aguiar M, Lonning S, Chen H, et al. Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Molecular cell. 2014;53:534–548. doi: 10.1016/j.molcel.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Runquist JA, Montgomery C, Miziorko HM, Kim JJ. Functional insights into human HMG-CoA lyase from structures of Acyl-CoA-containing ternary complexes. J Biol Chem. 2010;285:26341–26349. doi: 10.1074/jbc.M110.139931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibney GT, Messina JL, Fedorenko IV, Sondak VK, Smalley KS. Paradoxical oncogenesis--the long-term effects of BRAF inhibition in melanoma. Nat Rev Clin Oncol. 2013;10:390–399. doi: 10.1038/nrclinonc.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb HM, Davis S, Wilson C, Vardiman J. Surface immunoglobulins on hairy cells of 55 patients with hairy cell leukemia. American journal of hematology. 1982;12:397–401. doi: 10.1002/ajh.2830120411. [DOI] [PubMed] [Google Scholar]
- Gould J, Getz G, Monti S, Reich M, Mesirov JP. Comparative gene marker selection suite. Bioinformatics. 2006;22:1924–1925. doi: 10.1093/bioinformatics/btl196. [DOI] [PubMed] [Google Scholar]
- Hitosugi T, Fan J, Chung TW, Lythgoe K, Wang X, Xie J, Ge Q, Gu TL, Polakiewicz RD, Roesel JL, et al. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Molecular cell. 2011;44:864–877. doi: 10.1016/j.molcel.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Science signaling. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Zhou L, Elf S, Fan J, Kang HB, Seo JH, Shan C, Dai Q, Zhang L, Xie J, et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell. 2012;22:585–600. doi: 10.1016/j.ccr.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Zhou L, Fan J, Elf S, Zhang L, Xie J, Wang Y, Gu TL, Aleckovic M, LeRoy G, et al. Tyr26 phosphorylation of PGAM1 provides a metabolic advantage to tumours by stabilizing the active conformation. Nat Commun. 2013;4:1790. doi: 10.1038/ncomms2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DB, Sosman JA. Update on the targeted therapy of melanoma. Current treatment options in oncology. 2013;14:280–292. doi: 10.1007/s11864-013-0226-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Molecular cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nature biotechnology. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- Mansour SJ, Matten WT, Hermann AS, Candia JM, Rong S, Fukasawa K, Vande Woude GF, Ahn NG. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. The New England journal of medicine. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson PA, McEneny J. The biochemistry of ketogenesis and its role in weight management, neurological disease and oxidative stress. J Physiol Biochem. 2012;68:141–151. doi: 10.1007/s13105-011-0112-4. [DOI] [PubMed] [Google Scholar]
- Morris AA. Cerebral ketone body metabolism. J Inherit Metab Dis. 2005;28:109–121. doi: 10.1007/s10545-005-5518-0. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MS, Russell JJ, Gershman H. Ketone-body metabolism in glioma and neuroblastoma cells. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:7214–7218. doi: 10.1073/pnas.78.11.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger S, Bacher U, Haferlach T, Wendland N, Ulke M, Dicker F, Grossmann V, Haferlach C, Kern W. Development and validation of a real-time quantification assay to detect and monitor BRAFV600E mutations in hairy cell leukemia. Blood. 2012;119:3151–3154. doi: 10.1182/blood-2011-10-383323. [DOI] [PubMed] [Google Scholar]
- Sievert AJ, Lang SS, Boucher KL, Madsen PJ, Slaunwhite E, Choudhari N, Kellet M, Storm PB, Resnick AC. Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5957–5962. doi: 10.1073/pnas.1219232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims D, Mendes-Pereira AM, Frankum J, Burgess D, Cerone MA, Lombardelli C, Mitsopoulos C, Hakas J, Murugaesu N, Isacke CM, et al. High-throughput RNA interference screening using pooled shRNA libraries and next generation sequencing. Genome Biol. 2011;12:R104. doi: 10.1186/gb-2011-12-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiacci E, Trifonov V, Schiavoni G, Holmes A, Kern W, Martelli MP, Pucciarini A, Bigerna B, Pacini R, Wells VA, et al. BRAF mutations in hairy-cell leukemia. The New England journal of medicine. 2011;364:2305–2315. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. The New England journal of medicine. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.