To the Editor
Inhaled glucocorticoids are a primary therapy for controlling persistent asthma symptoms.1 However, approximately 30% of patients continue to experience poor symptom control.2 Criteria to identify patients who might respond more favorably to a specific inhaled glucocorticoid could be beneficial.3
We previously identified a genetic variant in the cytochrome P450 3A4 enzyme (CYP3A4*22) that was associated with improved asthma control among children treated with inhaled fluticasone.4 Patients with the CYP3A4*22 allele have been reported to feature 1.6-6.3 fold lower CYP3A4 mRNA expression and enzyme activity in the liver.5 Fluticasone is efficiently metabolized by CYP3A4 and is a potent mechanism-based inhibitor of CYP3A5 and to a lesser extent CYP3A4.4 It was hypothesized that decreased levels of CYP3A4 combined with inhibition of CYP3A5 by fluticasone may extend the half-life of fluticasone within lung cells and in the systemic circulation, thereby increasing its therapeutic effectiveness. We speculated that similar processes may occur for other inhaled glucocorticoids.4
Beclomethasone is a commonly prescribed inhaled glucocorticoid that features potent anti-inflammatory effects and effectively reduces bronchial hyperresponsiveness.6 In this study, a convenience sample of beclomethasone-treated children 2-17 years of age with a physician-confirmed diagnosis of asthma were recruited at Primary Children's Hospital (Salt Lake City, UT). Details of the study design and genotyping have been described previously.4 Briefly, saliva samples were collected and tested for nine single nucleotide polymorphisms (SNPs) in CYP3A4, CYP3A5, and CYP3A7. Demographic and clinical data were obtained through patient and/or parent survey and medical record abstraction. Asthma control was assessed using a questionnaire modified from the National Heart Lung and Blood Institute.4, 7 Asthma control scores were analyzed as a numeric variable that ranged from 0 (well controlled) to 15 (poorly controlled). Logistic regression models were developed in R 3.1.1 (R Foundation for Statistical Computing, Vienna, Austria) to assess the effect of CYP3A polymorphisms upon asthma control with inhaled beclomethasone. The Bonferroni correction was applied to adjust for three pairwise group comparisons (3 SNPs) for each CYP3A enzyme. The daily dose of beclomethasone dipropionate (mcg/day) was evaluated as a covariate in the regression analyses; however, the dose did not significantly alter the beta-coefficient in any of the models tested (P > 0.5) and was not retained in the final analyses.
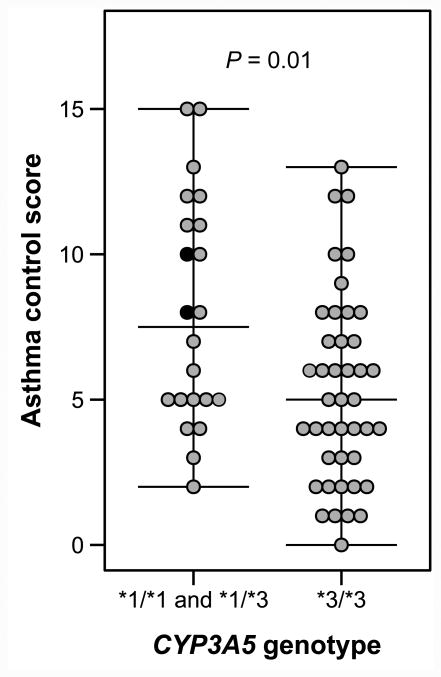
A total of 64 beclomethasone-treated asthmatic children were recruited. The median age was 8 (interquartile range [IQR]: 5.5-11) years and 42 (66%) were male. The median asthma control score was 5.5 (IQR: 4-8). Saliva was collected from all participants and CYP3A genotyping results are presented in the Table. Allelic variants in CYP3A4 and CYP3A7 were not significantly associated with asthma control scores, following adjustment for multiple comparisons. Two SNPs in CYP3A5 were associated with improved asthma control. The CYP3A5*3/*3 genotype was associated with a 2.7 (95% confidence interval: 0.9-4.6) point improvement in the asthma control score when compared to patients with the CYP3A5*1/*1 or CYP3A5*1/*3 genotypes (Figure). As previously reported, the CYP3A5*1D SNP (rs15524) was found in the same patients carrying the CYP3A5*3 SNP (rs776746), presumably explaining the concurrent association with asthma control.8, 9 The CYP3A5*3 SNP was found to be in complete linkage disequilibrium with the CYP3A5*1D SNP (Hedrick's multiallelic D' = 1.0).10
Table.
Association of CYP3A genetic polymorphisms and asthma control scores among 64 children receiving daily inhaled beclomethasone dipropionate.
| Polymorphism | Reference SNP (rs #) | Association with Asthma Control Scores (P-value) ⊗ | Reference Allele Frequency | Variant Allele Frequency |
|---|---|---|---|---|
| CYP3A4 | ||||
| CYP3A4*22 | rs35599367 | N/A | 1.00 | 0.00 |
| CYP3A4 int 7 | rs2246709 | 0.39 | 0.60 | 0.40 |
| CYP3A4 int 7 | rs4646437 | 0.10 | 0.70 | 0.30 |
| CYP3A5 | ||||
| CYP3A5*3 | rs776746 | 0.01 | 0.66 | 0.34 |
| CYP3A5*6 | rs10264272 | 0.57 | 0.98 | 0.02 |
| CYP3A5*1D | rs15524 | 0.01 | 0.66 | 0.34 |
| CYP3A7 | ||||
| CYP3A7 | rs2687133 | 0.13 | 0.84 | 0.16 |
| CYP3A7*2 | rs2257401 | 0.14 | 0.81 | 0.19 |
| CYP3A7 6 nt 5′ of ex 14 | rs2740565 | 0.07 | 0.77 | 0.23 |
Bonferroni adjustment for multiple comparisons.
No patients identified with the variant allele (N/A).
All SNPs were in Hardy-Weinberg Equilibrium.
Figure.
Asthma control scores among 64 children receiving inhaled beclomethasone, stratified by CYP3A5 genotype. Two patients featured the CYP3A5*1/*1 genotype and are represented as solid black circles. Asthma control scores are scaled from 0 (well controlled) to 15 (poorly controlled).
Using recombinant CYP3A enzymes, we have previously shown that beclomethasone is efficiently inactivated by CYP3A5.6 Several CYP3A5 genetic polymorphisms have been shown to alter mRNA expression and enzyme function – the most common is CYP3A5*3, which codes for an inactive form of CYP3A5.8 A majority of Caucasians are homozygous for CYP3A5*3 and therefore do not express active CYP3A5, whereas other racial and ethnic groups commonly express the CYP3A5*1 allele, which codes for an active form of CYP3A5.9 However, it is important to note that in this study, 88% of individuals with the CYP3A5*1/*1 or *1/*3 genotype self-reported their race as White, demonstrating the importance of genetic testing when variations in CYP3A genotypes are considered as a potential basis for personalizing therapy.
CYP3A5*1D is found in the 3′-UTR of the CYP3A5 gene, although its effect on CYP3A5 function has not been established. It is possible that this SNP may influence the efficacy of beclomethasone treatment; however, previous studies have found that the CYP3A5*1D allele was only identified in patients who had the CYP3A5*3 allele.8, 9 In this study, all patients that were homozygous for the CYP3A5*1D SNP (rs15524) were also homozygous for the CYP3A5*3 SNP (rs776746). Therefore, the relative contributions of these SNPs to the clinical effects we observed could not be differentiated, although it is well known that CYP3A5*3 causes an alternative splicing event that results in non-functional CYP3A5 protein. Additional studies are needed to clarify the role of CYP3A5*1D and its effect(s), if any, on the expression and function of CYP3A5. Based on the current state of the field, the current data support the hypothesis that the relationship between improved asthma control scores among beclomethasone-treated children with the CYP3A5*1D/*1D genotype is ultimately due to the inactivation of CYP3A5 caused by the CYP3A5*3 SNP.
The physiological basis for the association between improved asthma control with beclomethasone and CYP3A5*3 is not completely understood. This inactivating SNP abolishes CYP3A5 activity both in the lung and the liver.8 Reduced pulmonary and hepatic enzyme activity is likely to prolong the presence of active beclomethasone within the airway, thereby increasing the duration of its anti-inflammatory effects. Additionally, further investigation is warranted to determine whether diminished CYP3A5 activity may be associated with higher systemic concentrations of beclomethasone, which has the potential to increase the risk of adverse effects, including suppression of the hypothalamic-pituitary-adrenal axis.
Interpretation of our findings should be considered in light of several limitations. First, the precision of our effect estimates is limited by our sample size (n = 64). Second, it was not possible to directly measure CYP3A5 expression or tissue-specific activity; however, these studies are ongoing. Lastly, we did not obtain pulmonary function tests because standard spirometry measurements require a degree of patient cooperation that is difficult to achieve in the youngest of children.
The clinical relevance of this observed association requires further mechanistic explanation and additional study with larger sample sizes. Nevertheless, these data support an association between improved asthma control with inhaled beclomethasone and the loss of function CYP3A5*3 allele and are consistent with our earlier work in which asthma control was improved among children treated with fluticasone who had a genotype consistent with reduced CYP3A4 activity.4 When genetic testing is clinically available, these findings may be useful in selecting an appropriate therapeutic agent for patients who do not achieve optimal control with their currently prescribed inhaled glucocorticoid.
Acknowledgments
This project was supported by Grant Number R01HD060559 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health & Human Development or the National Institutes of Health. Additional support was provided by the Primary Children's Medical Center Research Foundation and the Center for Clinical and Translational Sciences (CTSA 5UL1RR025764-02). CS is supported by the American Foundation for Pharmaceutical Education's Clinical Pharmaceutical Sciences Fellowship. We thank Bradley W. Thomas and Amber Bagherian for their technical support.
Funding: This project was supported by Grant Number R01 HD060559 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. CS is supported by the American Foundation for Pharmaceutical Education's Clinical Pharmaceutical Sciences Fellowship.
Abbreviations
- SNPs
single nucleotide polymorphisms
- CYP
cytochrome P450
- CI
confidence interval
- NIH
National Institutes of Health
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tantisira KG, Lasky-Su J, Harada M, Murphy A, Litonjua AA, Himes BE, et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med. 2011;365:1173–83. doi: 10.1056/NEJMoa0911353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szefler SJ, Phillips BR, Martinez FD, Chinchilli VM, Lemanske RF, Strunk RC, et al. Characterization of within-subject responses to fluticasone and montelukast in childhood asthma. J Allergy Clin Immunol. 2005;115:233–42. doi: 10.1016/j.jaci.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 3.Drazen JM. A step toward personalized asthma treatment. N Engl J Med. 2011;365:1245–6. doi: 10.1056/NEJMe1102469. [DOI] [PubMed] [Google Scholar]
- 4.Stockmann C, Fassl B, Gaedigk R, Nkoy F, Uchida DA, Monson S, et al. Fluticasone propionate pharmacogenetics: CYP3A4*22 polymorphism and pediatric asthma control. J Pediatr. 2013;162:1222–7. 7 e1–2. doi: 10.1016/j.jpeds.2012.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J. 2010 doi: 10.1038/tpj.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts JK, Moore CD, Ward RM, Yost GS, Reilly CA. Metabolism of beclomethasone dipropionate by cytochrome P450 3A enzymes. J Pharmacol Exp Ther. 2013;345:308–16. doi: 10.1124/jpet.112.202556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Expert Panel Report 3 (EPR-3): Guidelines for the Diagnosis and Management of Asthma-Summary Report 2007. J Allergy Clin Immunol. 2007;120:S94–138. doi: 10.1016/j.jaci.2007.09.043. [DOI] [PubMed] [Google Scholar]
- 8.Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–91. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 9.Quaranta S, Chevalier D, Allorge D, Lo-Guidice JM, Migot-Nabias F, Kenani A, et al. Ethnic differences in the distribution of CYP3A5 gene polymorphisms. Xenobiotica. 2006;36:1191–200. doi: 10.1080/00498250600944300. [DOI] [PubMed] [Google Scholar]
- 10.Hedrick PW. Gametic disequilibrium measures: proceed with caution. Genetics. 1987;117:331–41. doi: 10.1093/genetics/117.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]