Abstract
Background
The establishment of new cell lines is of vital importance to the research of cancer.
Methods
Six new head and neck squamous cell carcinoma cell lines were established using a novel fluorescence-activated cell sorting method in order to overcome the barrier of fibroblast overgrowth and the susceptibility of primary tumors to fail in vitro.
Results
Antibodies chosen for specific targeting of epithelial cells and fibroblasts successfully separated cells for line establishment in six out of twelve attempts, providing an alternative method of establishing head and neck squamous cell carcinoma (HNSCC) cell lines. Each attempt at cell line establishment resulted in an epithelial carcinoma population, which was genotyped and catalogued as a unique cell line, and a corresponding fibroblast population.
Conclusions
The selection of antibody markers could be optimized to aid in the establishment of any cancer cell line derived from any tumor tissue; this method is not limited to head and neck cancer.
Keywords: Cell Line, Flow Cytometry, Fibroblasts, Squamous Cell, Carcinoma
Introduction
Cancer cell lines provide an invaluable research tool for the study of this diverse and deadly disease. Cell culture techniques were developed in the early twentieth century involving animal cells(1–3), with immortalized mouse cells being established in 1943(4). The first human continuous cancer line, HeLa cells, was cultured in 1951 at John Hopkins Hospital in Baltimore, Maryland(5,6). Under laboratory conditions that are quite different than those of the modern era, the establishment of this cervical cancer cell line allowed institutions from all over the world to study the disease in the laboratory without limitations due to specimen availability. After the initial breakthrough of HeLa establishment, human cells were cultured with greater frequency and efficacy(7–9). Culturing techniques have improved as years have passed, including the introduction of antibiotics, sterile conditions and laminar flow hoods, as well as the optimization of tissue culture medium formulas(10–13). As a result of improved cell line establishment methods and culture conditions, cell lines of a variety of cancers have been made available for research, with the most recent cancer cell line encyclopedia containing information on 947 different cell lines from 36 tumor types(14).
The availability of a library of cancer cell lines is especially important in the study of head and neck cancer, which includes a diverse group of biologically similar cancers from multiple sites. 90% of head and neck cancers are squamous cell carcinomas, mostly occurring in the oral cavity, larynx and pharynx, with roughly 40,000 new diagnoses each year in the United States(15–16). Worldwide collections of head and neck cell lines are now being assembled as valuable repositories to reflect the different varieties of the disease(17–20). Recent interest in the role of human papilloma virus (HPV) in the pathogenesis of head and neck cancer has driven research to compare HPV-positive and HPV-negative tumor types and has increased the need for newly established HPV-positive cell lines.(21–22).
Current methods for establishing cell lines from primary tissue of the head and neck include tumor explant in tissue culture or mechanical or enzymatic digestion of the tissue and then in vitro growth of attached epithelial cells from single-cell suspensions or partially digested tumor tissue(17,23). A key concern that arises in these methods is fibroblast overgrowth of the culture. Fibroblasts accompany the primary tumor tissue and usually divide faster than the epithelial population of cancer cells, while also competing for media nutrients and area for expansion. Fibroblasts are typically removed from culture through a series of partial trypsinizations, as they will detach from culture flasks or plates before the epithelial population, but other methods of fibroblast elimination have been described(24,25). Partial trypsinizations are performed multiple times until the fibroblasts are eliminated or become senescent, after an average of 50 population doublings known as the Hayflick limit(26). This method can take several months before fibroblast growth is arrested, and also risks loss of cancer cells during each successive trypsinization. From 1978 to 1994 our laboratory established over 112 unique UM-SCC cell lines from 95 different patients including 17 cell lines from eight patients who provided more than one tumor from either different sites or from different times in the course of their disease. Nearly all of these were established using the partial tripsinization method(27–29). The overall success rate during this time varied from 30–35% of attempts. The average time to successful passage of the tumor cells from mixed epithelial and fibroblast cultures ranged from 195 days for recurrent and metastatic tumors to more than 250 days for previously untreated primary tumors. Using similar methodology, from 1992 until 1997, the University of Pittsburgh established 52 new head and neck cell lines out of 199 tumor samples utilizing the partial trypsinization culture method, successfully establishing cell lines in 26% of the attempts(19). 101 of the tumor samples were deemed unsuccessful due to fibroblast overgrowth or growth of only fibroblasts when culture was attempted. The remaining attempts failed due to contamination of the culture flasks. While good, these rates of success fall below a level that would allow study of most patients’ tumors in a timely manner for identifying targets for therapy. Thus, improvements in cultivation techniques would be of great value.
Herein, we describe a novel method of cell line establishment that removes the need for a series of partial trypsinizations to eliminate fibroblast overgrowth. Careful selection of antibodies specific to surface markers exclusive to either squamous cell carcinomas or to fibroblasts allows for complete separation of the two populations by fluorescence-activated cell sorting (FACS). We chose epithelial cell adhesion molecule (EpCAM) and fibroblast surface protein (FSP). EpCAM is a pan-epithelial carcinoma-associated antigen that is expressed in a majority of carcinomas(30–32). FSP is a surface protein on human fibroblasts that is absent on human epithelial cells(33–35). This technique allows for isolation of a carcinoma population within several hours of tissue acquisition, as well as conserving the fibroblasts as a separate population if desired. There exists a potential benefit in being able to expand the cancer cell population without the time and effort involved in having to first eliminate the fibroblasts. This antibody-based sorting method may help to improve upon the frequency that lines are established, as many potential cell lines were lost due to fibroblast overgrowth in culture using traditional partial trypsinization methods.
Materials and Methods
Patients
Patients were recruited from the Head and Neck Oncology Division of the Department of Otolaryngology at the University of Michigan and asked to sign an Institutional Review Board approved informed consent to study their tissue, including permission to establish a permanent cell line.
Preparation and Digestion of Tissue
Primary tumor tissue was transported from the operating room to the lab and was washed extensively in Hank’s balanced salt solution containing penicillin, streptomycin, and amphotericin B. The tissue was then minced by scalpel blade and digested in Dulbecco’s Modified Eagle Medium (DMEM)/F12 (Gibco, Grand Island, New York, USA) with 1× collagenase/hyaluronidase (Stem Cell Technologies, Vancouver, Canada). After two hours of digestion at 37°C, the mixture was strained through a 70 um sieve and the cells were counted before being prepared for flow cytometry. When tumor was not collected on the same day as the cell sort, digested cells were placed in a culture flask and remaining tumor pieces were placed in a separate flask for a further digestion on the day of the sort. All primary samples were sorted by flow cytometry within 72 hours of collection.
Flow Cytometry
Epithelial cell adhesion molecule (EpCAM) and fibroblast surface protein (FSP) expression were detected using primary antibodies (Neomarkers Marseille, France, CAT#MS-181-P; Abcam Cambridge, Massachusetts, USA, CAT#ab11333) and fluorophore-conjugated secondary antibodies (BD Pharmingen; eBioscience). Cells were suspended in Hank’s Balanced Salt Solution (HBSS; Gibco) with 2% Heat Inactivated Calf Serum (HICS) added to a concentration of 1 million cells per mL. Five uL of primary antibody was added per mL of cell suspension, and left to incubate on ice for 20 minutes. Cells were then pelleted down and re-suspended in HBSS to a concentration of 1 million cells per mL. Five uL of secondary antibody was added per mL of cell suspension for 20 minutes on ice. For EpCAM expression, cell-sorting gates were established using an unstained control population in the allophycocyanin (APC) channel with excitation and emission wavelengths of approximately 650 nm/ 660 nm. For FSP expression, cell-sorting gates were established using an unstained control population in the Phycoerythrin (PE) channel with excitation and emission wavelengths of approximately 565 nm/ 578 nm.
Cell Line Establishment
Cells collected from flow cytometry were cultured in DMEM containing 20% fetal bovine serum (Gibco) containing penicillin, streptomycin, non-essential amino acids, and amphotericin B. Once growth without contamination was confirmed, cells were grown in 10% FBS media without amphotericin B. Fibroblasts were frozen down before five passages, and cell lines were frozen down as often as possible until establishment was confirmed at twenty passages as described previously(27). Lines were then further passaged until at least fifty passages and 100 doublings were achieved.
Genomic DNA Purification for Genotyping
Cells were harvested and washed in phosphate buffered saline (PBS), then frozen at −80°C. The thawed cell pellets were re-suspended in 600 uL of Promega Nuclei Lysis Solution (Promega) for one hour at 55°C, then allowed to cool to room temperature. 200 uL of Promega Protein Precipitation Solution (Promega, San Luis Obispo, California, USA) was added to each sample on ice for five minutes before being centrifuged at 13,000 revolutions per minute (RPM) for two minutes. Supernatant was transferred to a tube containing 600 uL of ispropanol and centrifuged at 13,000 RPM for one minute. Supernatant was aspirated and the DNA pellet washed in 200 uL of 70% ethanol and re-suspended in 50 uL of nuclease-free water.
Analysis of Genetic Loci
DNA samples were diluted to 0.10 ng/L and were analyzed at the University of Michigan DNA Sequencing Core using the Profiler Plus PCR Amplification Kit (Invitrogen, Carlsbad, California, USA) in accord with the manufacturer’s protocol. The 10 loci D3S1358, D5S818, D7S820, D8S1179, D13S317, D18S51, D21S11, FGA, vWA, and AMEL were analyzed and compared with ladder control samples as described previously(18).
Luciferase Transduction
UM-SCC-105, UM-SCC-106, and UM-SCC-110 were transduced with human immunodeficiency virus (HIV) with a luciferase reporter, a lentiviral vector containing a pLentiloxbackbone and a cytomegalovirus promoter. Polybrene was added to increase efficiency of the transduction. Successful gene delivery was confirmed via green fluorescent protein (GFP) visualization in a side-by-side transduction of the HIV-GFP vector under identical conditions.
Flank Injections
100,000 cells of luciferase-transduced cells were suspended in a mixture of 100 uL of DMEM and 100 uL of Matrigel extracellular matrix (BD Biosciences, San Jose, California, USA). The resulting 200 uL volumes were injected subcutaneously into the left flanks of NOD/SCID mice (Jackson Laboratories, Bar Harbor, Maine, USA) to examine xenograft potential. 100,000 cells of luciferase-transduced cells were mixed with 100,000 tumor-associated fibroblasts (TAFs) derived from the same tumor and injected subcutaneously into the right flanks of the same mouse. Tumor growth was allowed to persist for 12 weeks until harvested for sectioning and digestion. The xenografts were confirmed to match the initial cell lines.
Bioluminescence Imaging
All animals injected with luciferase-transduced cells were imaged with the Xenogen IVIS-200 imaging system. Treated mice were given intraperitoneal injections of 100 uLluciferin at a concentration of 40 mg/mL and allowed to sit for 10 minutes before being anesthetized with isofluorane and imaged.
Results
Primary Tissue
Previous attempts at cell line establishment using traditional partial trypsinization methods for fibroblast removal resulted in limited success (Figure 1). Six cell lines were established out of twelve attempts from digested tumor tissue using flow cytometry. The site and staging of each case at the time the tissue was obtained is noted (Table 1). Five of the established lines were derived from T4 tumors, and one line was established from a T3 tumor.
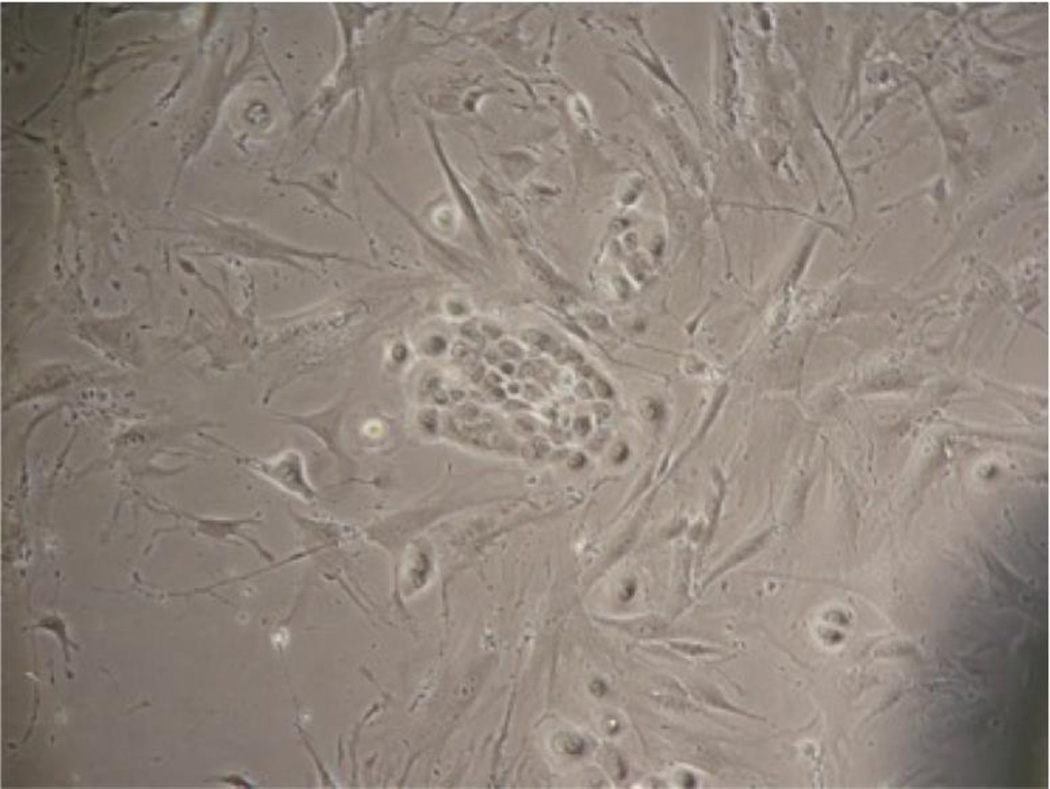
Figure 1.
An early attempt at cell line establishment from a primary tumor, using traditional partial-trypsinization methods. An island of epithelial cells is seen surrounded by fibroblast overgrowth.
Table 1.
Six cell lines were established from primary tumors out of twelve attempts.
| Sample | Site | Classification | Sex | Age | Cell Line |
|---|---|---|---|---|---|
| HN162 | Lateral tongue | T4N2bM0 | Male | 57 yo | |
| HN165 | Larynx | T4N0M0 | Male | 51 yo | UM-SCC-105 |
| HN166 | Oral tongue | T4N2bM0 | Male | 36 yo | UM-SCC-106 |
| HN171 | Floor of mouth | T4N0M0 | Male | 60 yo | |
| HN172 | Ventral tongue | T4N2cM0 | Female | 47 yo | |
| HN173 | Lateral tongue | T4N0M0 | Female | 30 yo | UM-SCC-108 |
| HN174 | Lateral tongue | T4N2bM0 | Male | 25 yo | |
| HN175 | Floor of mouth | T4N1M0 | Female | 69 yo | |
| HN176 | Base of tongue | T4N1M0 | Male | 49 yo | UM-SCC-109 |
| HN177 | Anterior tongue | T3N0M0 | Male | 39 yo | UM-SCC-110 |
| HN178 | Floor of mouth | T3N0M0 | Male | 51 yo | |
| HN181 | Lateral tongue | T4N2cM0 | Male | 60 yo | UM-SCC-111 |
Flow Cytometry
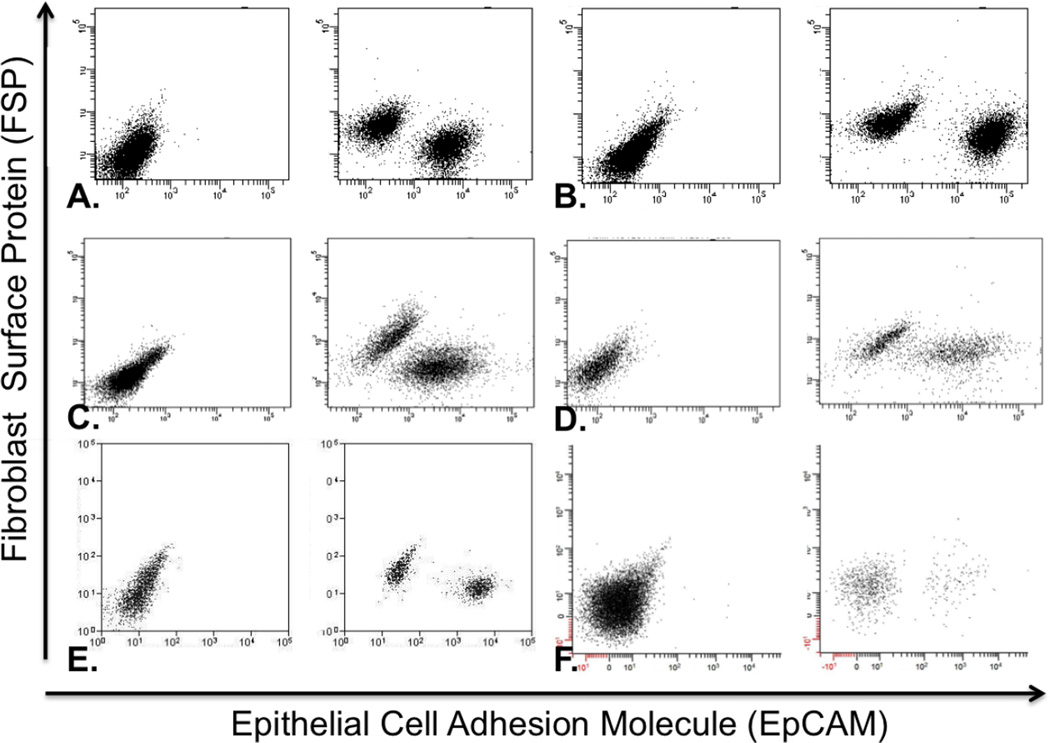
Cells were collected within 72 hours of surgery as epithelial populations (EpCAM+ / FSP−) and fibroblast populations (EpCAM− / FSP+). Gates were created in the flow cytometry software from cell suspensions that lacked antibody staining (Figure 2). Cells stained for markers that fell into these gated regions were collected and grown in culture.
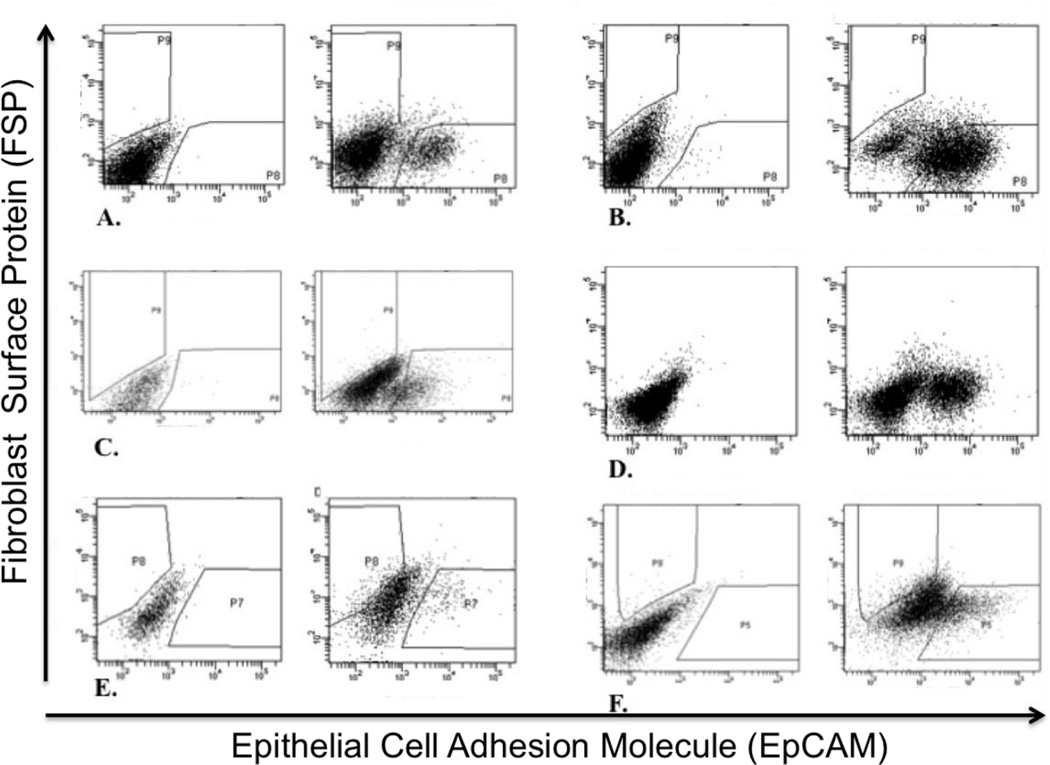
Figure 2.
FACS plots of each primary tumor that became a successful cancer cell line with the unstained plot on the left and the stained plot on the right. Cells expressing the EpCAM marker shift to the right along the X-axis, whereas cells expressing the FSP marker shift up along the Y-axis. Debris and other cell types do not stain for either antibody. Primary tumors shown are A) HN165; B) HN166; C) HN173; D) HN176 E) HN177; F) HN181.
Cell Culture
Sorted populations were cultured separately as EpCAM+ and FSP+ populations. The cells cultured as EpCAM+ showed epithelial phenotypes after 24 hours of culture (Figure 3). The cells cultured as FSP+ showed fibroblast phenotypes after 24 hours of culture (Figure 4). After twenty passages, the EpCAM+ populations were genotyped as unique new cell lines. Immortalization has been confirmed past fifty passages and at least 100 population doublings for each cell line. The FSP+ populations were frozen at early passages as tumor-associated fibroblasts (TAFs) paired with each new cell line, as these cells are not expected to be immortalized.
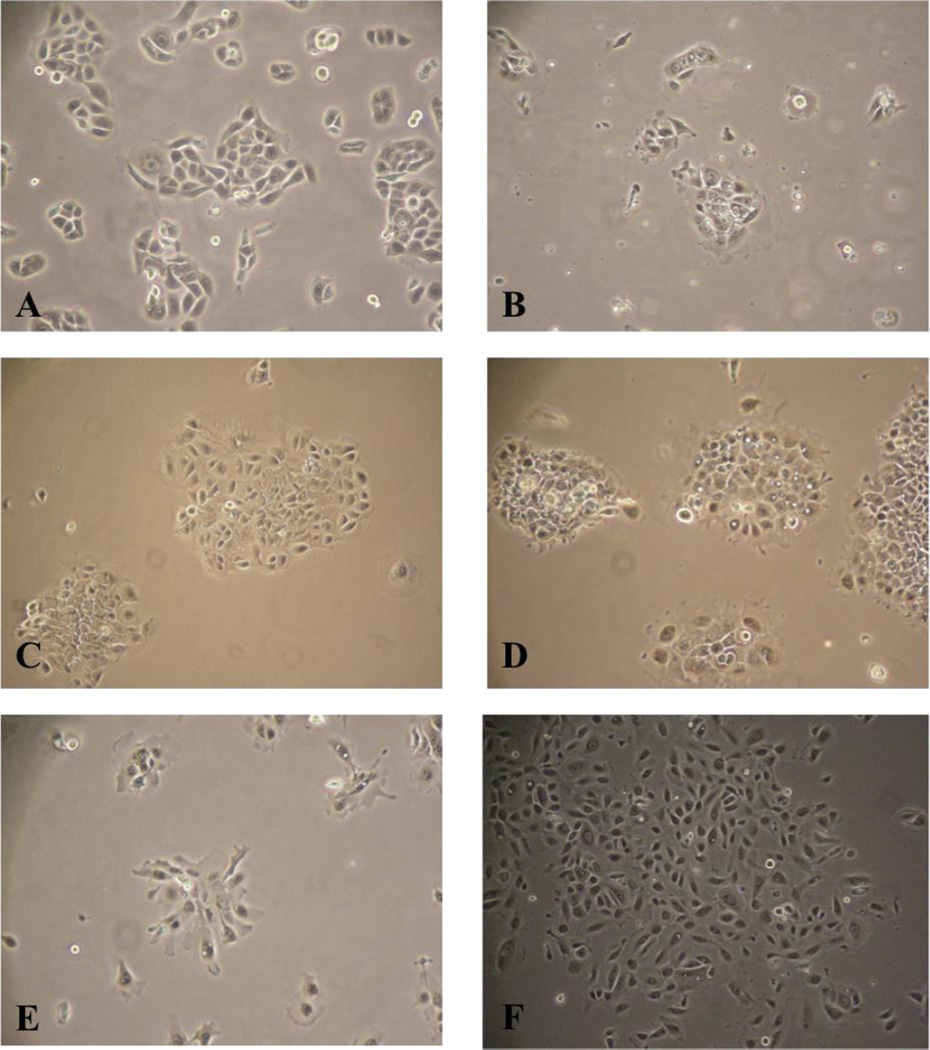
Figure 3.
Cells from the EpCAM+ / FSP− population were collected and grown in culture. These cells were established as new carcinoma lines A) UM-SCC-105; B) UM-SCC-106; C) UM-SCC-108; D) UM-SCC-109; E) UM-SCC-110; F) UM-SCC-111.
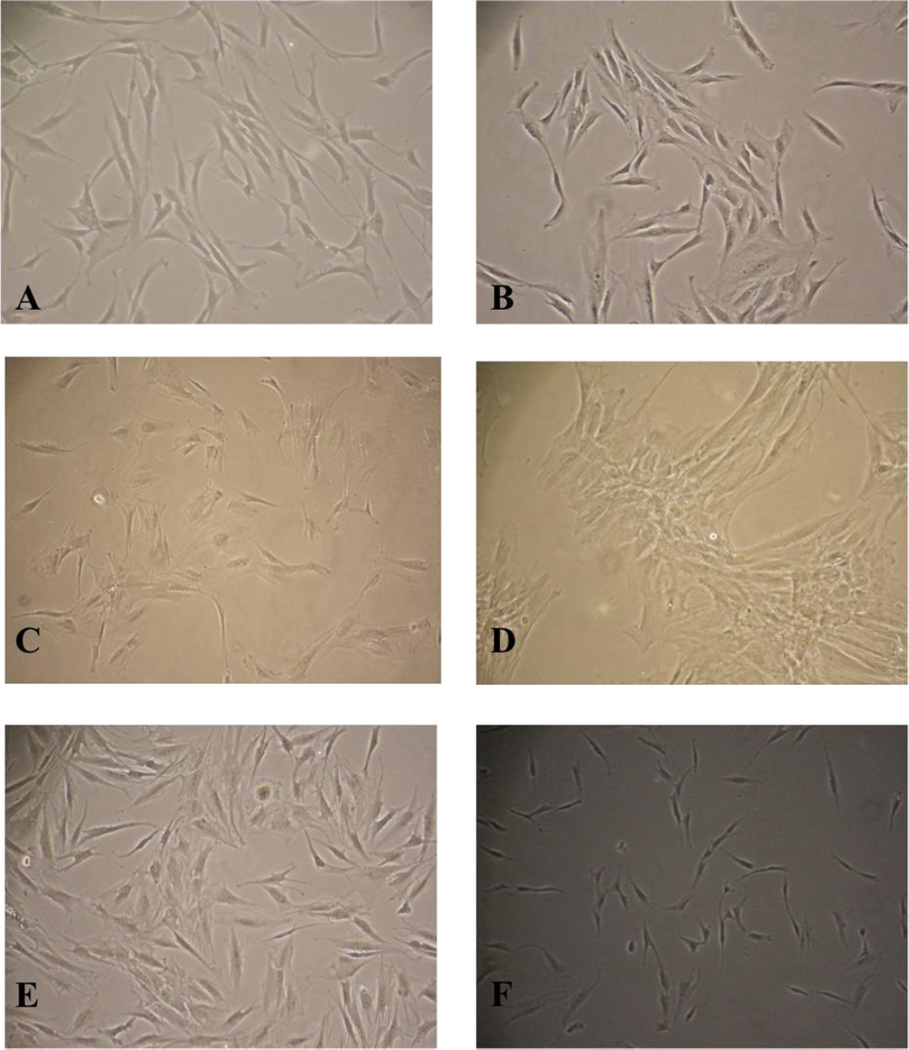
Figure 4.
Cells from the EpCAM− / FSP+ population were collected and grown in culture. These cells were stored as tumor-associated fibroblasts (TAFs) in conjunction with the established cell lines A) UM-SCC-105; B) UM-SCC-106; C) UM-SCC-108; D) UM-SCC-109; E) UM-SCC-110; F) UM-SCC-111.
Population Resorting
To confirm the validity of this new approach to cell line establishment, 100,000 cells from each line were mixed with an equal number of TAFs derived from the same primary tumor. Every mixed population was easily separated into two distinct populations, each composed of roughly 50% of the sample (Figure 5).
Figure 5.
FACS plots of each established cell line when mixed with an equal number of TAFs and separated. The unstained plot is shown on the left and the stained plot is shown on the right. Cells expressing the EpCAM marker shift to the right along the X-axis, whereas cells expressing the FSP marker shift up along the Y-axis. Cell lines with their corresponding TAFs are A) UM-SCC-105; B) UM-SCC-106; C) UM-SCC-108; D) UM-SCC-109; E) UM-SCC-110; F) UM-SCC-111.
Genotyping
Each pure population of EpCAM+ cells was genotyped as a unique cell line. Each paired population of FSP+ cells was genotyped to demonstrate the cell line is consistent with the genotype of the donor (Table 2). Interestingly there is frequent loss of heterozygosity in the tumor cells, for example loss of the allele 16 in UM-SCC-106 at the D35.1358 locus.
Table 2.
Each EpCAM+ cell population was genotyped as a unique cell line. Each FSP+ cell population was genotyped consistently with its paired cell line as tumor-associated fibroblasts (TAFs).
| Cell Line | AMEL | D3S1358 | vWA | FGA | D8S1179 | D21S11 | D18S51 | D5S818 | D13S317 | D7S820 | Results |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UM-SCC-105 | X,Y | 15, 18 | 15, 17 | 18, 21 | 12, 15 | 29, 30 | 12, 16 | 11 | 11, 12 | 9, 11 | Unique Cell Line |
| UM-SCC-105 Fibroblasts | X, Y | 15, 18 | 15, 17 | 18, 21 | 12, 15 | 29, 30 | 12, 16 | 11 | 11, 12 | 9, 11 | Consistent with UM-SCC-105 |
| UM-SCC-106 | X | 14 | 17 | 20, 21 | 14 | 30, 31 | 12 | 11 | 10, 11 | 9, 12 | Unique Cell Line |
| UM-SCC-106 Fibroblasts | X, Y | 14, 16 | 17 | 20, 21 | 12, 14 | 30, 31 | 12, 14 | 11, 13 | 10, 11 | 9, 12 | Consistent with UM-SCC-106 |
| UM-SCC-108 | X | 17 | 14, 18 | 21 | 12, 13 | 29, 30.2 | 15 | 11 | 9, 11 | 8, 12 | Unique Cell Line |
| UM-SCC-108 Fibroblasts | X | 17 | 14, 18 | 21, 24 | 12, 13 | 29, 30.2 | 15 | 11 | 9, 11 | 8, 12 | Consistent with UM-SCC-108 |
| UM-SCC-109 | X | 16 | 14, 17 | 22 | 8, 13 | 30, 32.2 | 12 | 12, 13 | 11 | 8, 11 | Unique Cell Line |
| UM-SCC-109 Fibroblasts | X | 14, 16 | 14, 17 | 21, 22 | 8, 13 | 30, 32.2 | 12, 15 | 12, 13 | 11, 13 | 8, 11 | Consistent with UM-SCC-109 |
| UM-SCC-110 | X, Y | 15 | 17, 20 | 21 | 8, 9 | 29 | 15 | 9 | 13 | 9, 11 | Unique Cell Line |
| UM-SCC-110 Fibroblasts | X, Y | 15, 18 | 17, 19 | 21, 23 | 8, 9 | 29, 31.2 | 14, 15 | 9, 11 | 11, 13 | 9, 11 | Consistent with UM-SCC-110 |
| UM-SCC-111 | X | 17 | 17, 18 | 20, 24 | 11, 14 | 31 | 13, 19 | 11 | 8, 14 | 11 | Unique Cell Line |
| UM-SCC-111 Fibroblasts | X, Y | 16, 17 | 17, 18 | 20, 24 | 11, 14 | 30, 31 | 13, 19 | 9, 11 | 8, 13 | 11 | Consistent with UM-SCC-111 |
Xenografts
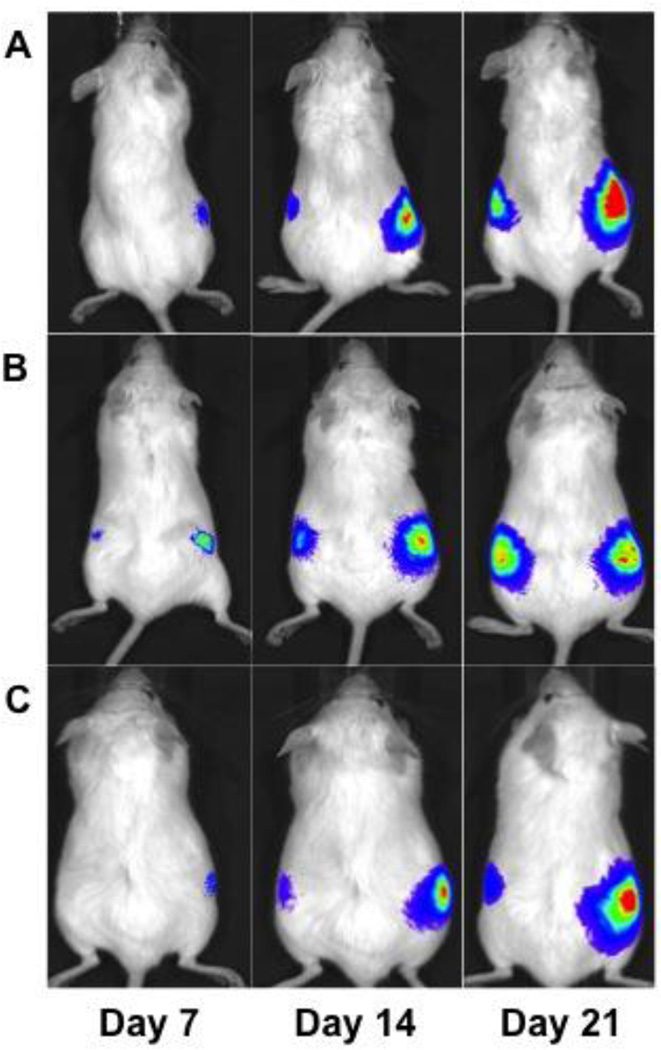
Three cell lines (UM-SCC-105, UM-SCC-106, UM-SCC-110) were transduced with luciferase and injected into the flanks of NOD/SCID mice to test for tumorigenicity. 100,000 cells were injected into the left flanks, while another 100,000 cells mixed with an equal number of TAFs were injected into the right flank in the same volume (Figure 6). The fibroblasts were not transduced, so measured bioluminescence was solely from the cancer cells. All injections were able to form flank tumors. The injections with fibroblasts initiated tumor growth earlier and promoted tumor growth when compared to the injections without fibroblasts, as measured by the bioluminescence of the xenografts.
Figure 6.
Flank xenografts of the newly established cell lines after transduction with luciferase. 100,000 transduced cells were injected into the left flank, while 100,000 transduced cells along with 100,000 non-transduced tumor-associated fibroblasts were injected into the right flank of NOD/SCID mice. Tumors initiated earlier when co-injected with fibroblasts and grew larger than tumors that did not have fibroblasts co-injected. Cell lines used are A) UM-SCC-105; B) UM-SCC-106; C) UM-SCC-110.
Discussion
Initial attempts to separate the fibroblasts by flow cytometry utilized only the FSP antibody, which did not provide staining that was strong enough to properly separate two different cell populations. After addition of the EpCAM antibody, the two cell populations were easily separated along two axes on a dot plot. The fibroblasts shifted upwards along the Y-axis, while the epithelial cells shifted to the right along the X-axis. Each cell line was unique in its staining pattern. Most cell lines had a strong separation of EpCAM+ cells from the remaining population, but UM-SCC-110 and UM-SCC-111 displayed an EpCAM+ population that only slightly disassociated itself from the other cell types. This suggests that certain carcinomas may express EpCAM at higher levels, and may even express EpCAM as a gradient in a heterogeneous population. The literature suggests that cell lines which express high levels of EpCAM may be more aggressive and capable of metastasis(30).
Of particular note is the successful establishment of UM-SCC-110. The primary tumor tissue for this cell line was grown in the lab for 72 hours before sorting was attempted, and each culture flask appeared to be completely overgrown with fibroblasts. No cells with an epithelial phenotype could be seen under the microscope, but FACS analysis did detect a very small EpCAM+ population that was sorted and cultured. The successful establishment of this cell line would most likely not have been possible using traditional partial trypsinization methods.
Separation of the two cell populations was immediate and thorough. Within 24 hours, the sorted cells were successfully growing in culture and displayed either an epithelial phenotype or a fibroblast phenotype. Once enough cells were grown in culture, 100,000 EpCAM+ and 100,000 FSP+ cells from the previous sort were mixed together before being re-stained and re-analyzed by flow cytometry. In each of the six cell lines, the two cell types were easily re-separated. The staining patterns differed from the original primary tumor tissue, and we hypothesize that the large population of cells that do not stain for either FSP or EpCAM in the primary tissue are cells that are neither mesenchymal nor epithelial, and constitute other cell types such as blood cells and lymphocytes. These cells were not collected, but blood draws were conducted for each patient in order to acquire lymphocytes.
There exists much potential from the immediate isolation of a carcinoma population of cells from primary tumor tissue. These cells can be immediately expanded for future applications, rather than having to wait for a pure population devoid of fibroblasts. Our own research involving cells acquired from primary tumors was limited to less than 1.0 × 106 viable tumor cells from each sample, or about the same number of cells in a confluent T-25 culture flask(36–38). Immediate expansion of sorted cancer cells provides us with the opportunity to produce a much larger number of cells within a matter of weeks. Our future research will rely on this method of cell line establishment in order to grow and harvest large numbers of cancer stem cells within a short period of time for specific applications. This includes the priming of dendritic cells with cancer stem cell lysate in a time frame that allows appropriate treatment for the patient from which the cell line was established, which has been shown to confer anti-tumor immunity in an animal model(39,40).
Another benefit from this method of cell line establishment is the acquisition of a pure fibroblast population at the time that the primary tissue is available. As previously mentioned, fibroblasts are not immortal and only go through a limited number of cell divisions before entering senescence. This novel method allows for immediate isolation of a population of tumor-associated fibroblasts (TAFs), stromal cells that help contribute to tumor growth(41,42). Research into the effects of TAFs on the cancer environment has been gaining momentum in the past few years, and a consistent collection of cancer cell lines paired with TAFs from the same source is a useful resource on which to draw(43). Our initial experiments in a mouse model suggest that the TAFs not only help to initiate tumor growth when grown in conjunction with their patient-specific cancer cell lines, but also help to fuel growth over time. This is an interesting area to explore in the future, as more information about the tumor niche and crosstalk with other cell types is learned.
Genotyping results from these six new head and neck cell lines were also made more quickly and more efficiently through this cell line establishment method. Carcinoma cells and fibroblasts from each patient were genotyped as pure populations. Each cell line showed significant locus deviation from the fibroblast population, with the exception of UM-SCC-105, which matched the fibroblast companion on every locus. UM-SCC-105 was the only cell line out of the six that tested positive for HPV-18. This cell line was established from a never smoker, consistent with fewer chromosome aberrations in HPV induced tumors than in tumors derived from patients with tobacco smoke carcinogenesis. These results infer that UM-SCC-105 is a viral-driven cancer that does not rely on cell-cycle mutation but rather E6 and E7 protein inhibition of cell-cycle checkpoints(21). UM-SCC-110 indicated genetic instability leading to DNA polymerase slippage from 19 to 20 at the D3S1358 locus, as well as frequent loss of heterozygosity at six of the loci. UM-SCC-111 also indicated genetic instability due to DNA polymerase slippage or absence of mismatch repair from 13 to 14 at the D13S317 and the loss of heterozygosity at three of the loci.
Conclusions
Fluorescence-activated cell sorting is a useful tool for the continued accumulation of new cancer cell lines, and provides an alternative antibody-based method to the traditional partial trypsinization technique used previously. In a group of twelve primary tumors of the head and neck, six were successfully established as cancer cell lines. Identifying and isolating the cancer cells from the accompanying fibroblasts allows for immediate growth of pure cell populations. This method significantly decreases the length of time required for cell line establishment as well as providing an isolated population of tumor-associated fibroblasts unique to the established cell line.
Acknowledgements
The work was supported by NIH NIDCR R01 DE019126, NIH NIDCD P30 DC05188 core support grant, NCI P50 CA97248 (Head and Neck SPORE), NIH NCI P30 CA46592 (U of M Comprehensive Cancer Center Core Grant), and a gift from Patricia Korican of Korican Real Estate. Thank you to the University of Michigan Comprehensive Cancer Center Flow Cytometry Core.
References
- 1.Carrel A. On the permanent life of tissues outside the organism. J. Exp. Med. 1912;15:516–528. doi: 10.1084/jem.15.5.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrison RG. Observations on the living developing nerve fiber. Proc. Soc. Exp. Biol. Med. 1907;4:140–143. [Google Scholar]
- 3.Burrows MT. The cultivation of tissues of the chick embryo outside the body. J. Am. Med. Assoc. 1910;55:2057–2058. [Google Scholar]
- 4.Earle WR. Production of malignancy in vitro. IV. The mouse fibroblast cultures and changes seen in living cells. J. Natl Cancer Inst. 1943;4:165–212. [Google Scholar]
- 5.Gey GO, Coffman WD, Kubicek MT. Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Cancer Res. 1952;12:264–265. [Google Scholar]
- 6.Masters JR. HeLa cells 50 years on: the good, the bad and the ugly. Nature Reviews Cancer. 2002;2:315–319. doi: 10.1038/nrc775. [DOI] [PubMed] [Google Scholar]
- 7.Chang RS. Continuous subcultivation of epithelial-like cells from normal human tissues. Proc. Soc. Exp. Biol. Med. 1954;87:440–443. doi: 10.3181/00379727-87-21406. [DOI] [PubMed] [Google Scholar]
- 8.Fjelde A. Human tumor cells in tissue culture. Cancer. 1955;8:845–851. doi: 10.1002/1097-0142(1955)8:4<845::aid-cncr2820080434>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 9.Henle G, Deinhardt FJ. The establishment of strains of human cells in tissue culture. J. Immunol. 1957;79:54–59. [PubMed] [Google Scholar]
- 10.Barnes D, Sato G. Serum-free cell culture: a unifying approach. Cell. 1980;22:649–655. doi: 10.1016/0092-8674(80)90540-1. [DOI] [PubMed] [Google Scholar]
- 11.Eagle H. Nutrition Needs of Mammalian Cells in Tissue Culture. Science. 1955;122:501–504. doi: 10.1126/science.122.3168.501. [DOI] [PubMed] [Google Scholar]
- 12.Ham R. Clonal growth of mammalian cells in a chemically defined, synthetic medium. Proc. Natl. Acad. Sci. 1965;53(2):288–293. doi: 10.1073/pnas.53.2.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi I, Sato G. Replacement of serum by hormones permits the growth in a defined medium. Nature. 1976;259:132–134. doi: 10.1038/259132a0. [DOI] [PubMed] [Google Scholar]
- 14.Barretina J, Caponigro G, Stransky N, et al. The cancer cell line encyclopedia enables predictive modeling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forastiere AA, Ang KK, Brizel D, et al. Head and neck cancers. J. Natl. Compr. Canc. Netw. 2008 Aug;6(7):646–695. doi: 10.6004/jnccn.2008.0051. [DOI] [PubMed] [Google Scholar]
- 16.Shah JP, Lydiatt W. Treatment of cancer of the head and neck. CA Cancer J Clin. 1995;45(6):352–368. doi: 10.3322/canjclin.45.6.352. [DOI] [PubMed] [Google Scholar]
- 17.Lin CJ, Grandis ER, Carey TE, et al. Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck. 2007 Feb;29(2):163–188. doi: 10.1002/hed.20478. [DOI] [PubMed] [Google Scholar]
- 18.Brenner JC, Graham MP, Kumar B, et al. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck. 2010;32(4):417–426. doi: 10.1002/hed.21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White JS, Weissfeld JL, Ragin CC, et al. The Influence of Clinical and Demographic Risk Factors on the Establishment of Head and Neck Squamous Cell Carcinoma Cell Lines. Oral Oncol. 2007;43(7):701–712. doi: 10.1016/j.oraloncology.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao M, Sano D, Pickering CR, et al. Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites. Clin Cancer Res. 2011;17:7248–7264. doi: 10.1158/1078-0432.CCR-11-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goon PK, Stanley MA, Ebmeyer J, et al. HPV & head and neck cancer: a descriptive update. Head & Neck Oncology. 2009;1:36. doi: 10.1186/1758-3284-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang AL, Hauff SJ, Owen JH, et al. UM-SCC-104: a new human papillomavirus-16-positive cancer stem cell-containing head and neck squamous cell carcinoma cell line. Head Neck. 2012 Oct;34(10):1480–1491. doi: 10.1002/hed.21962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liebertz DJ, Lechner MG, Masood R, et al. Establishment and characterization of a novel head and neck squamous cell carcinoma cell line USC-HN1. Head Neck Oncol. 2010 Feb;22:2–5. doi: 10.1186/1758-3284-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linge C, Green MR, Brooks RF. A method for removal of fibroblasts from human tissue culture systems. Exp. Cell Res. 1989 Dec;185(2):519–528. doi: 10.1016/0014-4827(89)90320-0. [DOI] [PubMed] [Google Scholar]
- 25.Boateng SY, Hartman TJ, Ahluwalia N, Vidula H, Desai TA, Russell B. Inhibition of fibroblast proliferation in cardiac myocyte cultures by surface microtopography. Am J Physiol Cell Physiol. 2003;285(1):171–182. doi: 10.1152/ajpcell.00013.2003. [DOI] [PubMed] [Google Scholar]
- 26.Hayflick L. The cell biology of aging. Clin. Geriatr. Med. 1985;1(1):15–27. [PubMed] [Google Scholar]
- 27.Krause CJ, Carey TE, Ott RW, Hurbis C, McClatchey KD, Regezi JA. Human squamous cell carcinoma: Establishment and characterization of new permanent cell lines. Arch Otolaryngol. 1981;107:703–710. doi: 10.1001/archotol.1981.00790470051012. [DOI] [PubMed] [Google Scholar]
- 28.Carey TE. Establishment of Epidermoid Carcinoma Cell Lines. In: Wittes RE, editor. Head and Neck Cancer. London UK: John Wiley & Sons, Ltd.; 1985. pp. 287–314. [Google Scholar]
- 29.Lansford C, Grenman R, Bier H, et al. Head and Neck Cancers. In: Masters JRW, Palsson B, editors. Human Cell Culture Vol 2, Cancer Cell Lines Part 2. Dordrecht (Holland): Kluwer Academic Press; 1999. pp. 185–255. [Google Scholar]
- 30.Shiah SG, Tai KY, Wu CW. Epigenetic Regulation of EpCAM in tumor invasion and metastasis. Journal of Cancer Molecules. 2008;3(6):165–168. [Google Scholar]
- 31.Schartinger VH, Schmutzhard J, Wurm M, et al. The expression of EGFR, HER2 and EpCAM in head and neck squamous cell carcinomas. MEMO. 2009;2(1):45–50. [Google Scholar]
- 32.Van der Gun BTF, Melchers LJ, Ruiters MHJ, de Leij LF, McLaughlin PM, Rots MG. EpCAM in carcinogenesis: the good, the bad or the ugly. Carcinogenesis. 2010;31(11):1913–1921. doi: 10.1093/carcin/bgq187. [DOI] [PubMed] [Google Scholar]
- 33.Singer KH, Scearce RM, Tuck DT, Whichard LP, Denning SM, Haynes BF. Removal of fibroblasts from human epithelial cell cultures with use of a complement fixing monoclonal antibody reactive with human fibroblasts and monocytes/macrophages. J Invest Dermatol. 1989;92:166–170. doi: 10.1111/1523-1747.ep12276685. [DOI] [PubMed] [Google Scholar]
- 34.Ronnov-Jessen L, Celis JE, Van Deurs B, Petersen OW. A fibroblast-associated antigen: characterization in fibroblasts and immunoreactivity in smooth muscle differentiated stromal cells. J Histochem Cytochem. 1992;40:475–486. doi: 10.1177/40.4.1552184. [DOI] [PubMed] [Google Scholar]
- 35.Esterre P, Melin M, Serrar M, Grimaud JA. New specific markers of human and mouse fibroblasts. Cell Mol Biol. 1992;38:297–301. [PubMed] [Google Scholar]
- 36.Krishnamurthy S, Dong Z, Vodopyanov D, et al. Endothelial cell-initiated signaling promotes the survival and self-renewal of cancer stem cells. Cancer Res. 2010 Dec;70(23):9969–9978. doi: 10.1158/0008-5472.CAN-10-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tabor MH, Clay MR, Owen JH, et al. Head and neck cancer stem cells: the side population. The Laryngoscope. 2011;121(3):527–533. doi: 10.1002/lary.21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clay MR, Tabor M, Owen JH, et al. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck. 2010 Sep;32(9):1195–1201. doi: 10.1002/hed.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pellegatta S, Finocchiaro G. Dendritic cell vaccines for cancer stem cells. Methoids in Molecular Biology. 2009;568:233–247. doi: 10.1007/978-1-59745-280-9_15. [DOI] [PubMed] [Google Scholar]
- 40.Ning N, Pan Q, Zheng F, et al. Cancer stem cell vaccination confers significant antitumor immunity. Cancer Res. 2012 Apr;72:1853–1864. doi: 10.1158/0008-5472.CAN-11-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall B, Dembinski J, Sasser AK, Studeny M, Andreeff M, Marini F. Mesenchymal stem cells in cancer: tumor-associated fibroblasts and cell-based delivery vehicles. International Journal of Hematology. 2007 Jul;86(1):8–16. doi: 10.1532/IJH97.06230. [DOI] [PubMed] [Google Scholar]
- 42.Ostman A, Augsten M. Cancer-associated fibroblasts and tumor growth- bystanders turning into key players. Current Opinion in Genetics & Development. 2009 Feb;19(1):67–73. doi: 10.1016/j.gde.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 43.Rosenthal E, McCrory A, Talbert M, Young G, Murphy-Ullrich J, Gladson C. Elevated expression of TGF-Beta1 in head and neck cancer-associated fibroblasts. Molecular Carcinogenesis. 2004 Jun;40(2):116–121. doi: 10.1002/mc.20024. [DOI] [PubMed] [Google Scholar]