Abstract
The mucin 1 (MUC1) oncoprotein has been linked to the inflammatory response by promoting cytokine-mediated activation of the NF-κB pathway. The TGF-β-activated kinase 1 (TAK1) is an essential effector of proinflammatory NF-κB signaling that also regulates cancer cell survival. The present studies demonstrate that the MUC1-C transmembrane subunit induces TAK1 expression in colon cancer cells. MUC1 also induces TAK1 in a MUC1+/−/IL-10−/− mouse model of colitis and colon tumorigenesis. We show that MUC1-C promotes NF-κB-mediated activation of TAK1 transcription and, in a positive regulatory loop, MUC1-C contributes to TAK1-induced NF-κB signaling. In this way, MUC1-C binds directly to TAK1 and confers the association of TAK1 with TRAF6, which is necessary for TAK1-mediated activation of NF-κB. Targeting MUC1-C thus suppresses the TAK1→NF-κB pathway, downregulates BCL-XL, and in turn sensitizes colon cancer cells to MEK inhibition. Analysis of colon cancer databases further indicates that MUC1, TAK1 and TRAF6 are upregulated in tumors associated with decreased survival and that MUC1-C-induced gene expression patterns predict poor outcomes in patients. These results support a model in which MUC1-C-induced TAK1→NF-κB signaling contributes to intestinal inflammation and colon cancer progression.
Keywords: MUC1-C, TAK1, TRAF6, NF-κB, colitis, colon cancer
Introduction
Mucin 1 (MUC1) is a heterodimeric transmembrane protein that is activated in cytokine-mediated inflammatory responses (1). MUC1 is aberrantly overexpressed in human colon cancers and is associated with invasion, metastases and a poor prognosis (2–8). However, there is limited information regarding a functional role for MUC1 in colon cancer cells.
MUC1 consists of an extracellular N-terminal subunit (MUC1-N) with variable numbers of glycosylated tandem repeats and a C-terminal transmembrane subunit (MUC1-C) (1). MUC1-N is shed from the epithelial cell surface, where it contributes to the intestinal mucosal barrier. In turn, MUC1-C signals growth and survival responses to the interior of cells constituting the epithelial layer (1). The available evidence indicates that colon and other cancer cells have exploited MUC1-C-induced survival signals by aberrantly expressing this subunit. For example, MUC1-C contributes to activation of the WNT/β-catenin pathway, which is frequently activated in colon cancer (9), by binding directly to and stabilizing β-catenin (10). The MUC1-C cytoplasmic domain also interacts directly with TCF7L2/TCF4 and thereby promotes β-catenin/TCF4-mediated transcription of WNT target genes, such as cyclin D1 (11).
Other studies have linked MUC1-C to the constitutive activation of NF-κB in human carcinomas (12). In this context and like activated NF-κB, MUC1-C contributes to transformation and blocks apoptosis by a mechanism that involves in part upregulation of BCL-XL expression (13; 14). These MUC1-C-induced responses are conferred by interaction of the MUC1-C cytoplasmic domain with the high-molecular weight IκB (IKK) complex (15). In turn, MUC1-C promotes IKKβ activation, resulting in phosphorylation and degradation of IκBα (15). Other work has demonstrated that MUC1-C interacts directly with NF-κB p65 and contributes to activation of NF-κB target genes, such as BCL-XL (12). The interaction between MUC1-C and NF-κB has also been linked to the induction of ZEB1, a transcriptional repressor that drives EMT and cancer progression (16).
The transforming growth factor β-activated kinase 1 (TAK1) is a proinflammatory effector that contributes to activation of the IKK complex and thereby the NF-κB pathway (17). TAK1 is a key regulator of the innate immune response and inflammation (18; 17). TAK1 has also been linked to colon cancer cell survival and the control of cell death (19–22). However, little is know about the control of TAK1 levels in inflammation and cancer. The present studies demonstrate that MUC1-C induces TAK1 expression in colon cancer cells. We show that (i) MUC1-C induces TAK1 expression by promoting NF-κB-mediated activation of the TAK1 promoter, and (ii) MUC1-C binds directly to TAK1 and confers the formation of a TAK1 complex with TRAF6, which in turn activates TAK1→NF-κB signaling. In concert with these results, targeting MUC1-C with silencing or with an inhibitor suppresses the TAK1→NF-κB pathway. These in vitro studies were performed on colon cancer cells that harbor KRAS mutations; however, the focus of the present work is on MUC1-C-induced activation of TAK1 and not on a role for MUC1-C in the context of mutant KRAS. Our studies extend to a MUC1 transgenic model of inflammatory bowel disease and colon tumorigenesis and provide further support for MUC1-C-mediated induction of TAK1→NF-κB signaling. Additionally, analysis of gene array databases demonstrates that MUC1-C, TAK1 and TRAF6 associated expression patterns predict poor outcomes in colon cancer patients.
Results
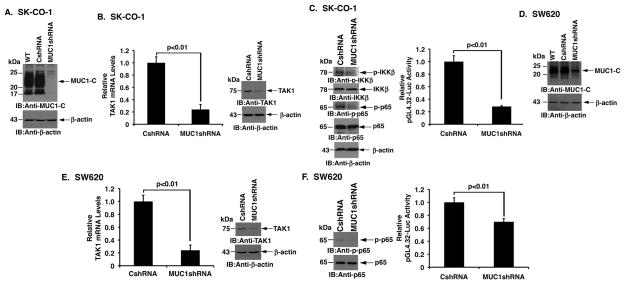
Silencing MUC1-C decreases TAK1 signaling in colon cancer cells
SK-CO-1 colon cancer cells are dependent on TAK1 for survival (21). MUC1-C was therefore stably silenced in SK-CO-1 cells to determine whether MUC1-C affects TAK1 signaling (Fig. 1A). Notably, silencing MUC1-C in SK-CO-1/MUC1shRNA cells was associated with marked downregulation of TAK1 mRNA levels as compared to that in control SK-CO-1/CshRNA cells (Fig. 1B, left). Silencing MUC1-C was also associated with decreases in TAK1 protein (Fig. 1B, right). In addition, MUC1-C was necessary for activation of phospho-IKKβ and phospho-NF-κB p65 (Fig. 1C, left and Supplemental Fig. S1A, left and right). Intriguingly, treatment of SK-CO-1 cells with the NF-κB inhibitor BAY11-7085 (23) suppressed TAK1 mRNA levels (Supplemental Fig. S1B), indicating that MUC1-C may activate a TAK1-NF-κB auto-inductive loop. In concert with these results, silencing MUC1-C decreased activation of a NF-κB p65-driven pGL4.32 promoter-Luc reporter (Fig. 1C, right). To extend this analysis, MUC1-C was downregulated in SW620 colon cancer cells (Fig. 1D). As found with SK-CO-1 cells, MUC1-C suppression in SW620 cells was associated with decreases in TAK1 expression (Fig. 1E, left and right). We also found that silencing of MUC1-C in SW620 cells results in suppression of NF-κB signaling (Fig. 1F, left and right). These results indicate that MUC1-C contributes to activation of the TAK1→NF-κB pathway in colon cancer cells.
Figure 1. Silencing MUC1-C in colon cancer cells suppresses the TAK1→NF-κB pathway.
A. SK-CO-1 cells were infected with lentiviruses to stably express a control CshRNA or a MUC1shRNA. Lysates from the wild-type (WT) and transduced cells were immunoblotted with the indicated antibodies. B. TAK1 mRNA levels in the indicated SK-CO-1 cells were determined by qRT-PCR (left). The results are expressed as relative TAK1 mRNA levels (mean±SD of three determinations) as compared to that obtained for the SK-CO-1/CshRNA cells (left). Lysates were immunoblotted with the indicated antibodies (right). C. Lysates from SK-CO-1/CshRNA and SK-CO-1/MUC1shRNA cells were immunoblotted with the indicated antibodies (left). The indicated cells were transfected with pGL4.32-Luc for 48 h and then assayed for luciferase activity. The results are expressed as the relative luciferase activity (mean±SD of three determinations) compared to that obtained for the SK-CO-1/CshRNA cells (right). D. SW620 cells were infected with lentiviruses to stably express a control CshRNA or a MUC1shRNA. Lysates from the wild-type (WT) and transduced cells were immunoblotted with the indicated antibodies. E. TAK1 mRNA levels in the indicated SW620 cells were determined by qRT-PCR (left). The results are expressed as relative TAK1 mRNA levels (mean±SD of three determinations) as compared to that obtained for the SW620/CshRNA cells (left). Lysates were immunoblotted with the indicated antibodies (right). F. Lysates from the SW620/CshRNA and SW620/MUC1shRNA cells were immunoblotted with the indicated antibodies (left). The indicated cells were transfected with pGL4.32-Luc for 48 h and then assayed for luciferase activity. The results are expressed as the relative luciferase activity (mean±SD of three determinations) compared to that obtained for the SW620/CshRNA cells (right).
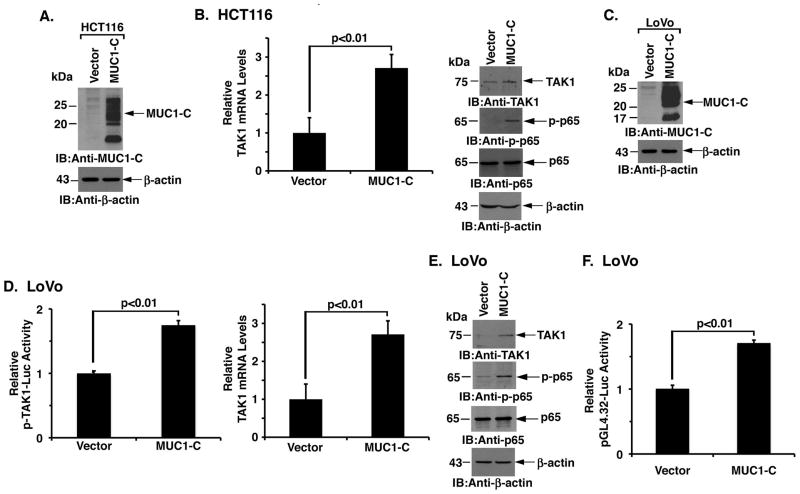
MUC1-C induces TAK1 expression
HCT116 colon cancer cells have low to undetectable levels of MUC1-C as compared to that in SK-CO-1 and SW620 cells (Supplemental Fig. S2A). Accordingly, we stably overexpressed MUC1-C in these cells (Fig. 2A) to further define the interaction between MUC1-C and TAK1. In concert with the finding that silencing MUC1-C downregulates TAK1 expression, we found that TAK1 mRNA levels are significantly increased in HCT116/MUC1-C, as compared to HCT116/vector, cells (Fig. 2B, left). Expression of MUC1-C was also associated with increases in TAK1 protein (Fig. 2B, right and Supplemental Fig. S2B, left) and phospho-NF-κB p65 (Fig. 2B, right and Supplemental Fig. S2B, right). Intriguingly, stable silencing of NF-κB p65 in HCT116/MUC1-C cells blocked MUC1-C-induced TAK1 expression (Supplemental Fig. S2C), providing further support for a potential auto-inductive loop involving MUC1-C, TAK1 and NF-κB. Similarly, in LoVo colon cancer cells transduced to stably overexpress MUC1-C (Supplemental Fig. S2A and Fig. 2C), we found increased activation of the TAK1 promoter (Fig. 2D, left) and upregulation of TAK1 mRNA levels (Fig. 2D, right). Expression of MUC1-C was also associated with increases in TAK1 protein and in phospho-NF-κB p65 (Fig. 2E). Consistent with activation of NF-κB p65, expression of MUC1-C in the LoVo cell model resulted in activation of the pGL4.32 promoter-Luc reporter (Fig. 2F). These findings demonstrate that MUC1-C is sufficient for induction of TAK1 and the NF-κB pathway.
Figure 2. MUC1-C is sufficient for activation of the TAK1→NF-κB pathway.
A. Lysates from HCT116 colon cancer cells transduced to stably express an empty vector or MUC1-C were immunoblotted with the indicated antibodies. B. TAK1 mRNA levels in the indicated HCT116 cells were determined by qRT-PCR (left). The results are expressed as relative TAK1 mRNA levels (mean±SD of three determinations) as compared to that obtained for the HCT116/vector cells (left). Lysates were immunoblotted with the indicated antibodies (right). C. Lysates from LoVo colon cancer cells transduced to stably express an empty vector or MUC1-C were immunoblotted with the indicated antibodies. D. The indicated LoVo cells were transfected with a TAK1 promoter-luciferase reporter (pTAK1-Luc) for 48 h and then assayed for luciferase activity. The results are expressed as the relative luciferase activity (mean±SD of three determinations) compared to that obtained for the LoVo/vector cells (left). qRT-PCR analysis of TAK1 mRNA in the indicated LoVo cells is expressed as relative levels (mean±SD of three determinations) as compared to that obtained for the LoVo/vector cells (right). E. Lysates from the LoVo/vector and LoVo/MUC1-C cells were immunoblotted with the indicated antibodies. F. The indicated LoVo cells were transfected with pGL4.32-Luc for 48 h and then assayed for luciferase activity. The results are expressed as the relative luciferase activity (mean±SD of three determinations) compared to that obtained for the LoVo cells.
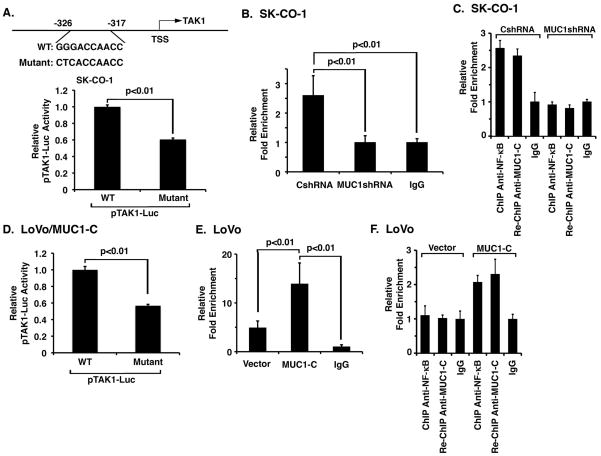
MUC1-C promotes NF-κB p65 occupancy of the TAK1 promoter
Based on the above observations, we searched the human TAK1 (MAP3K7) promoter region using the annotated sequence available at the Ensembl genomic database (ENST00000369329; NCBI Reference Sequence: NG_011966.2) and found a potential NF-κB binding sequence at position -326 to -317 upstream to the transcription start site (Fig. 3A, upper panel). To determine whether this putative NF-κB site contributes to activation of the TAK1 promoter, we compared activation of a wild-type (GGGACCAACC) and mutant (CTCACCAACC) TAK1 promoter-luciferase reporter (pTAK1-Luc) in SK-CO-1 cells. Mutation of the putative NF-κB site was associated with a significant decrease in TAK1 promoter activation (Fig. 3A, lower panel). Based on these results, ChIP studies were performed which demonstrated that NF-κB p65 occupies the TAK1 promoter (Fig. 3B). We also found that silencing MUC1-C in SK-CO-1 cells decreases NF-κB p65 occupancy (Fig. 3B). Re-ChIP analysis further demonstrated that NF-κB p65 occupies the TAK1 promoter with MUC1-C (Fig. 3C). In concert with these findings, activation of the pTAK1-Luc reporter in LoVo/MUC1-C cells was significantly attenuated by mutation of the NF-κB site (Fig. 3D). Moreover, NF-κB p65 occupancy on the TAK1 promoter was increased in LoVo/MUC1-C, as compared to LoVo/vector, cells (Fig. 3E). Re-ChIP studies using the LoVo cells further confirmed that NF-κB occupies the TAK1 promoter with MUC1-C (Fig. 3F). These results collectively provided support for a model in which MUC1-C also promotes NF-κB-mediated activation of TAK1 expression.
Figure 3. MUC1-C promotes NF-κB-mediated activation of TAK1 expression.
A. Schematic representation of the TAK1 promoter with positioning of the NF-κB binding site (upper panel). SK-CO-1 cells were transfected with wild-type or mutant pTAK1-Luc for 48 h and then assayed for luciferase activity. The results are expressed as the relative luciferase activity (mean±SD of three determinations) compared to that obtained with wild-type pTAK1-Luc (lower panel). B. Soluble chromatin from SK-CO-1/CshRNA and SK-CO-1/MUC1shRNA cells was precipitated with anti-NF-κB p65 and, as a control, IgG. The final DNA samples were amplified by qPCR with pairs of primers for the NF-κB binding region (NBR; -326 to -317). The results (mean±SD of three determinations) are expressed as the relative fold enrichment compared to that obtained with the IgG control. C. Soluble chromatin from the indicated SK-CO-1 cells was precipitated with anti-NF-κB p65. The precipitates were released, reimmunoprecipitated with anti-MUC1-C and then analyzed for TAK1 promoter sequences. The results (mean±SD of three determinations) are expressed as the relative fold enrichment compared to that obtained with the IgG control. D. LoVo/MUC1-C cells were transfected with wild-type or mutant pTAK1-Luc for 48 h and then assayed for luciferase activity. The results are expressed as the relative luciferase activity (mean±SD of three determinations) compared to that obtained with wild-type pTAK1-Luc. E. Soluble chromatin from LoVo/vector and LoVo/MUC1-C cells was precipitated with anti-NF-κB p65 and, as a control, IgG. The final DNA samples were amplified by qPCR with pairs of primers for the NF-κB binding region (NBR). The results (mean±SD of three determinations) are expressed as the relative fold enrichment compared to that obtained with the IgG control. F. Soluble chromatin from the indicated LoVo cells was precipitated with anti-NF-κB p65. The precipitates were released, reimmunoprecipitated with anti-MUC1-C and then analyzed for TAK1 promoter sequences. The results (mean±SD of three determinations) are expressed as the relative fold enrichment compared to that obtained with the IgG control.
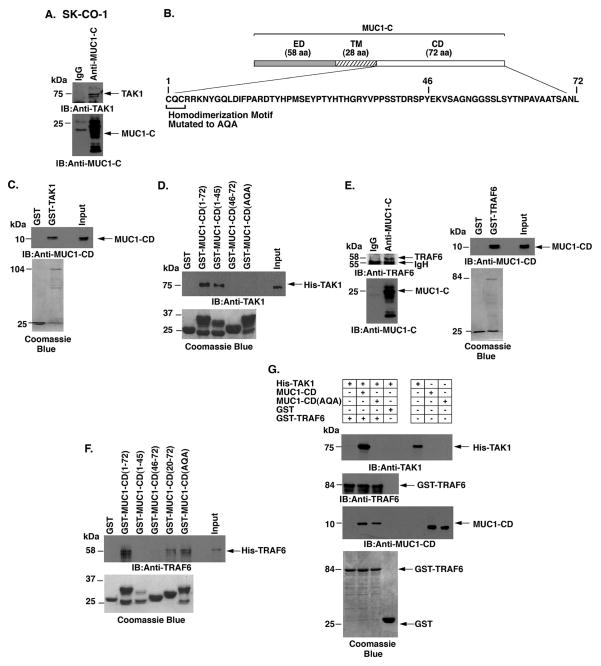
MUC1-C forms a complex with TAK1 and TRAF6
TAK1 phosphorylates IKKβ on Ser-181 and thereby activates the NF-κB pathway (24; 25). The demonstration that MUC1-C associates with IKKβ (15) prompted studies to determine whether MUC1-C also forms a complex with TAK1. Indeed, coimmunoprecipitation (co-IP) experiments using lysates from SK-CO-1 cells showed that MUC1-C associates with TAK1 (Fig. 4A). In concert with these co-IP results and the previous demonstration that MUC1-C interacts with the IKK complex (15), immunodepletion of MUC1-C from SK-CO-1 cell lysates was associated with decreases in TAK1, p-IKKβ and IKKβ (Supplemental Fig. S3A). As additional controls, MUC1-C/TAK1 complexes were also detectable when co-IP studies were performed on HCT116/MUC1-C, but not HCT116/vector, cell lysates (Supplemental Fig. S3B). These results collectively supported the association of MUC1-C and TAK1 in cells. In vitro binding studies further demonstrated that the MUC1-C cytoplasmic domain (MUC1-CD; Fig. 4B) interacts directly with GST-TAK1 (Fig. 4C). To define the region of MUC1-CD that confers the interaction, we incubated GST-MUC1-CD deletion mutants with His-TAK1 (Fig. 4D). The results showed that MUC1-CD(1-45), but not MUC1-CD(46-72), binds to TAK1 (Fig. 4D). We also found that mutation of the MUC1-CD CQC motif to AQA abrogated binding to TAK1 (Fig. 4D), indicating that the Cys residues are of importance for the interaction.
Figure 4. MUC1-C cytoplasmic domain binds directly to TAK1.
A. Lysates from SK-CO-1 cells were precipitated with anti-MUC1-C or a control IgG. The precipitates were immunoblotted with the indicated antibodies. B. Schematic representation of MUC1-C (ED, extracellular domain; TM, transmembrane domain) and the amino acid (aa) sequence of the cytoplasmic domain (CD). Highlighted is the CQC motif that is necessary for MUC1-C homodimerization and has been mutated to AQA in MUC1-CD. C. GST or GST-TAK1 was incubated with purified MUC1-CD. The adsorbates were immunoblotted with anti-MUC1-CD. Input of the GST proteins was assessed by Coomassie blue staining. D. GST, GST-MUC1-CD (full-length; 1-72) or the indicated GST-MUC1-CD mutants were incubated with His-TAK1. The adsorbates were immunoblotted with anti-TAK1. Input of the GST proteins was assessed by Coomassie blue staining. E. Lysates from SK-CO-1 cells were precipitated with anti-MUC1-C or a control IgG. The precipitates were immunoblotted with the indicated antibodies (left). GST or GST-TRAF6 was incubated with purified MUC1-CD. The adsorbates were immunoblotted with anti-MUC1-CD (right). Input of the GST proteins was assessed by Coomassie blue staining (right). F. GST, GST-MUC1-CD(1-72) or the indicated GST-MUC1-CD mutants were incubated with His-TRAF6. The adsorbates were immunoblotted with anti-TRAF6. Input of the GST proteins was assessed by Coomassie blue staining. G. His-TAK1 was incubated with GST or GST-TRAF6 in the presence of MUC1-CD or MUC1-CD(AQA). Adsorbates and input proteins were immunoblotted with the indicated antibodies. Input of the GST-TRAF6 and GST proteins was assessed by Coomassie blue staining.
TNF receptor-associated factor 6 (TRAF6) is an E3 ubiquitin ligase that functions upstream to TAK1 and NF-κB activation (26). Based on the known interaction between TAK1 and TRAF6, we asked if, like TAK1, MUC1-C also associates with TRAF6. Indeed, coimmunoprecipitation studies demonstrated that MUC1-C forms a complex with TRAF6 (Fig. 4E, left). In addition, we found that the MUC1-C cytoplasmic domain (MUC1-CD) binds directly to GST-TRAF6 (Fig. 4E, right). However, in contrast to TAK1, the interaction with TRAF6 was conferred by MUC1-CD(20-72) and not MUC1-CD(1-45) (Fig. 4F). The lack of detectable binding of MUC1-CD(46-72) to TRAF6 (Fig. 4F) further supported involvement of a region spanning aa 45-46 and indicated that distinct regions of MUC1-CD bind to TAK1 and TRAF6. Moreover, and in contrast to TAK1, binding to TRAF6 was not affected by mutating MUC1-CD at CQC to AQA (Fig. 4F). The observation that MUC1-CD interacts with both TAK1 and TRAF6 invoked the possibility that MUC1-CD might facilitate the formation of TAK1/TRAF6 complexes. Significantly, there was little if any association of His-TAK1 and GST-TRAF6 in the absence of MUC1-CD (Fig. 4G). By contrast, binding of TAK1 and TRAF6 was clearly observed in the presence of MUC1-CD and this interaction was abrogated by mutation of the CQC motif to AQA (Fig. 4G).
TRAF6 bridges the RAS and NF-κB pathways in lung cancer cells (27); however, little is known about involvement of TRAF6 in colon cancer cells. Of interest in this regard, silencing MUC1-C in SK-CO-1 and SW620 cells was associated with downregulation of TRAF6 mRNA and protein (Supplemental Figs. S4A and S4B, left and right). Consistent with these results, expression of MUC1-C in HCT116 cells resulted in upregulation of TRAF6 mRNA and protein levels (Supplemental Fig. S4C, left and right). Moreover, we found that MUC1-C induces TRAF6 expression in LoVo cells (Supplemental Fig. S4D, left and right), indicating that MUC1-C is sufficient to drive the upregulation of both TAK1 and TRAF6.
Targeting the MUC1-C CQC motif blocks TAK1→NF-κB signaling
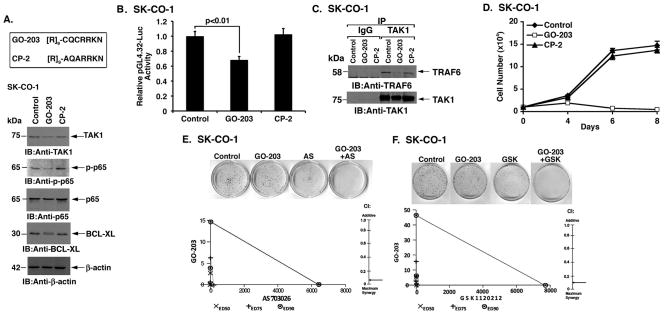
The findings that MUC1-C induces TAK1 expression and binds directly to TAK1 prompted studies with the cell-penetrating peptide GO-203 that targets the MUC1-C CQC motif and blocks MUC1-C function (28) (Fig. 5A, upper panel). As a control, we used another cell-penetrating peptide, designated CP-2, in which the critical Cys residues for binding to endogenous MUC1-C were mutated to Ala (Fig. 5A, upper panel). Treatment of SK-CO-1 cells with GO-203, but not CP-2, was associated with suppression of TAK1 expression and decreases in NF-κB p65 phosphorylation (Fig. 5A, lower panels). We also found that treatment with GO-203 downregulates expression of the NF-κB target gene, BCL-XL (Fig. 5A, lower panels). These effects of GO-203 were conferred, at least in part, by inhibition of NF-κB-mediated transactivation as determined by studies with the pGL4.32 promoter-Luc reporter (Fig. 5B). GO-203 also blocked the interaction between TAK1 and TRAF6 in SK-CO-1 cells (Fig. 5C). Consistent with the demonstration that TAK1 is necessary for survival of colon cancer cells (21), we found that GO-203 inhibited SK-CO-1 cell growth (Fig. 5D). By contrast, GO-203 had no apparent effect on growth of HCT116 cells (Supplemental Fig. S5). TAK1 contributes to the activation of both (i) NF-κB and (ii) β-catenin/TCF4 signaling in colon cancer cells (21). Accordingly, we asked if targeting MUC1-C and thereby downregulating TAK1 expression also affects the β-catenin/TCF4 pathway. Indeed, silencing MUC1-C in SK-CO-1 and SW620 cells was associated with decreases in activation of the β-catenin/TCF4-driven TOP-Flash promoter-reporter (Supplemental Figs. S6A and S6B). Silencing MUC1-C was also associated with decreases in AXIN2, the product of a β-catenin/TCF4-dependent target gene (Supplemental Figs. S6C and S6D), indicating that MUC1-C contributes to activation of the TAK1→NF-κB and β-catenin/TCF4 pathways.
Figure 5. Targeting MUC1-C with GO-203 suppresses TAK1→NF-κB signaling.
A. D-amino acid sequences of the GO-203 and CP-2 peptides (upper panel). SK-CO-1 cells were left untreated or treated with 2.5 μM GO-203 or CP-2 each day for 48 h. Lysates were immunoblotted with the indicated antibodies (lower panels). B. SK-CO-1 cells were transfected with pGL4.32-Luc, treated with 2.5 μM GO-203 or CP-2 each day for 48 h and then assayed for luciferase activity. The results are expressed as the relative luciferase activity (mean±SD of three determinations) compared to that obtained with control untreated cells. C. Lysates from SK-CO-1 cells left untreated or treated with 2.5 μM GO-203 or CP-2 each day for 24 h were precipitated with anti-TAK1. The precipitates were immunoblotted with the indicated antibodies. D. SK-CO-1 cells were left untreated (solid diamonds) and treated with 2.5 μM GO-203 (open squares) or CP-2 (solid triangles) each day for the indicated days. Viable cell number (mean±SD of three replicates) was determined by trypan blue staining. E. SK-CO-1 cells were seeded at 100 cells/well in 6-well plates and left untreated (Control) or treated each day with 0.61 μM GO-203 alone, 1.17 μM AS703026 alone on day 0, or the GO-203/AS703026 combination for 3 d. Colonies were stained with crystal violet on day 14 after treatment (upper panel). SK-CO-1 cells were treated with (i) fixed IC50 ratios of GO-203 alone on days 0, 1, 2 and 3, (ii) fixed IC50 ratios of AS703026 alone on day 0, and (iii) the GO-203/AS703026 combination for 4 d. The multiple effect isobologram analyses on day 4 are shown for the ED50 (X), ED75 (+) and ED90 (o) values (lower left panel). The combination index (CI) is indicated with the arrow (lower right panel). F. SK-CO-1 cells were seeded at 100 cells/well in 6-well plates and left untreated (Control) or treated each day with 1.22 μM GO-203 alone, 9.63 nM GSK1120212 alone on day 0, or the GO-203/GSK1120212 combination for 3 d. Colonies were stained with crystal violet on day 14 after treatment (upper panel). SK-CO-1 cells were treated with (i) fixed IC50 ratios of GO-203 alone on days 0, 1, 2 and 3, (ii) fixed IC50 ratios of GSK1120212 alone on day 0, and (iii) the GO-203/GSK1120212 combination for 4 d. The multiple effect isobologram analyses on day 4 are shown for the ED50 (X), ED75 (+) and ED90 (o) values (lower left panel). The combination index (CI) is indicated with the arrow (lower right panel). CI values are included in Supplemental Table S2.
Nonetheless, the novelty of a MUC1-C-dependent TAK1 and NF-κB auto-inductive loop that confers upregulation of BCL-XL prompted our focus on that pathway. Previous studies had demonstrated that MEK inhibitors are highly effective against KRAS mutant cancers when combined with agents that target BCL-XL (29). Therefore, based on the finding that GO-203 decreases BCL-XL, we treated SK-CO-1 cells with GO-203 in combination with the MEK inhibitor AS703026. Inhibitory effects of the GO-203+AS703026 combination on SK-CO-1 colony formation were more pronounced than with either agent alone (Fig. 5E, upper panel). Moreover, isobologram analysis of the combination showed marked synergy with a combination index of less than 0.1 (Fig. 5E, lower panels; Supplemental Table S2). Similar results were obtained when GO-203 was combined with the MEK inhibitor GSK1120212 (Fig. 5F, upper and lower panels; Supplemental Table S2). These findings with SK-CO-1 cells were confirmed when SW620 cells were treated with GO-203 (Supplemental Fig. S7A) and the combination of GO-203 and MEK inhibitors (Supplemental Figs. S7B and S7C; Supplemental Table S2), indicating that GO-203 is synergistic with different MEK inhibitors.
MUC1-C regulates TAK1 and NF-κB in a MUC1 transgenic model of colitis and tumorigenesis
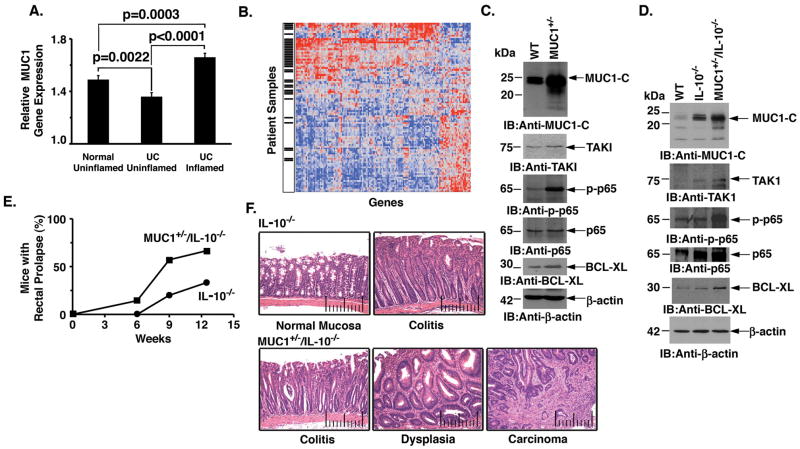
Intestinal inflammation increases the risk of developing colon cancer, conceivably through recurrent cycles of damage and repair of the mucosa (30). Aberrant expression of MUC1 protein has been linked to inflammatory bowel disease (31–33); however, to our knowledge, there is no available analysis of MUC1 and its relationship with NF-κB signaling in gene array databases from patients with colitis. MUC1 gene expression was therefore assessed in tissue samples from inflamed colonic mucosa derived from patients with ulcerative colitis as compared to corresponding uninflamed colonic mucosa from either ulcerative colitis patients or normal healthy controls. The data demonstrate significantly elevated levels of MUC1 gene expression in inflamed colonic mucosa as compared to that in corresponding uninflamed mucosa or that from healthy controls (Fig. 6A), supporting the potential involvement of MUC1 in colitis. Analysis of 438 NF-κB-regulated genes further showed that 34% are differentially expressed in colitis mucosa samples versus uninflamed controls. Of these differentially expressed genes, the majority (82%) were overexpressed in colitis samples. In addition, upregulation of MUC1 was associated with overexpression of NF-κB target genes, including TAK1 and TRAF6 (Fig. 6B).
Figure 6. Regulation of TAK1 in a MUC1 transgenic model of colitis and colon tumorigenesis.
A. Tissue specimens from healthy colonic mucosa (normal uninflamed; n=63), uninflammed ulcerative colitis (UC) mucosa (UC uninflammed; n=62) and inflamed UC mucosa (n=61) were assessed for MUC1 gene expression. Differences in gene expression between groups was determined using a 2-tailed Student’s t-test. The results are presented as mean±SEM. B. Hierarchical clustering of 150 differentially expressed (p<0.01) NF-κB-regulated genes in inflamed UC mucosa samples and normal controls. Red indicates high expression, while blue denotes low expression. Black hash marks denote UC samples. There is a significant association between inflamed histology and overexpression of NF-κB-regulated genes (p<0.0001, Fisher’s exact test). In addition, MUC1 expression was significantly higher in samples over-expressing NF-κB pathway genes (p<0.0001, 2-tailed t-test). C. Lysates from colon tissue obtained from wild-type (WT) and MUC1+/− mice were immunoblotted with the indicated antibodies. D. Lysates from colon tissue from WT, IL-10−/−, and MUC1+/−/IL-10−/− mice were immunoblotted with the indicated antibodies. E. The percentage of MUC1+/−/IL-10−/− (n=7) and IL-10−/− (n=6) mice with rectal prolapse is expressed at the indicated ages. F. Images of colon tissue from IL-10−/− mice (upper panels). The normal mucosa has short straight crypts with very few inflammatory cells between crypts (upper left panel). With the development of colitis, the mucosa is thickened and the crypts are elongated with infiltrates of lymphocytes between the crypts (upper right panel). Images of colon tissue from MUC1+/−/IL-10−/− mice (lower panels). Colitis is seen with a thickened mucosa and elongated crypts. Adjacent crypts are separated by infiltrates of inflammatory cells and the presence of crypt abscess (lower left panel). With progression to dysplasia, the crypts are enlarged, twisted and branched with inflammatory cells between the crypts (lower middle panel). The crypts at the top of the image are twisted and tortuous and at the bottom extend into an adenocarcinoma (lower right panel).
To extend this analysis to a mouse model, we first studied mice transgenic for human MUC1 (MUC1+/−) that express MUC1 under control of the endogenous human MUC1 promoter and exhibit a pattern of expression similar to that found in humans (34). Immunoblot analysis of colon tissue from MUC1+/− mice demonstrated increased MUC1-C levels as compared to that in wild-type mice (Fig. 6C and Supplemental Fig. S8A). In addition and in concert with the results obtained with the SK-CO-1 and SW620 cells, upregulation of MUC1-C in the MUC1+/− mouse colon was associated with increases in TAK1 and phospho-NF-κB p65 (Fig. 6C and Supplemental Fig. S8A). BCL-XL levels were also upregulated in colon tissue from the MUC1+/− mouse (Fig. 6C and Supplemental Fig. S8A). The MUC1+/− mice did not develop colitis as evidenced by a normal colonic mucosa (data not shown). By contrast, IL-10−/− mice develop colitis with epithelial hyperplasia and inflammatory infiltrates due to aberrant regulation of the innate immune response to intestinal flora (35). To assess the effects of MUC1-C on TAK1 signaling in a model of colitis and tumorigenesis (36), we crossed MUC1+/− mice with IL-10−/− mice and then the F1 MUC1+/−/IL-10+/− mice were bred with IL-10−/− mice. Analysis of colon tissue from IL-10−/− mice demonstrated marked upregulation of endogenous mouse Muc1-C expression, but at levels somewhat lower than that found in the MUC1+/−/IL-10−/− mouse colon (Fig. 6D and Supplemental Fig. S8B). We also found increases in TAK1, phospho-NF-κB p65 and BCL-XL that were more pronounced in the MUC1+/−/IL-10−/− mouse colon (Fig. 6D and Supplemental Fig. S8B). The presence of colitis as assessed by the development of rectal prolapse supported increased inflammation of the bowel in MUC1+/−/IL-10−/−, as compared to IL-10−/−, mice (Fig. 6E). Moreover, and consistent with previous studies (36), the MUC1+/−/IL-10−/− mice exhibited more severe colitis with dysplasia and progression to carcinomas (Fig. 6F).
MUC1-C-induced gene expression in human colon cancers
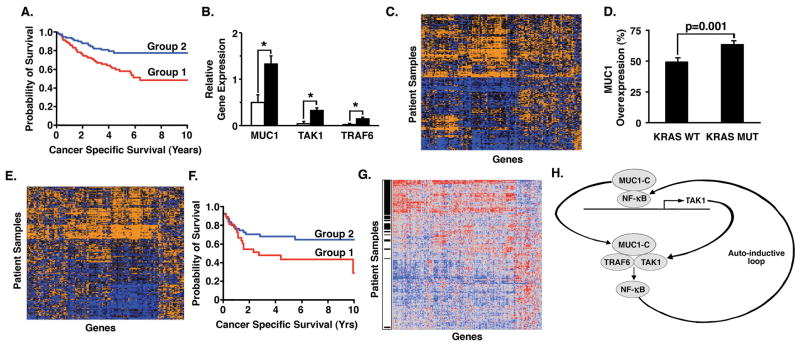
Our experimental results obtained from cell based and genetically engineered mouse models lend support to a functional association of MUC1-C with colon cancer. However, there is no evidence that MUC1-C is linked to outcomes of patients with this disease. MUC1-C interacts with certain transcription factors, such as NF-κB, and contributes to the activation of downstream target genes (15; 12; 37; 10; 11). We therefore investigated a model of MUC1-C-induced genes (38) and, of these, 261 were present on the Affymetrix Human Genome U133 Plus 2.0 GeneChip. Analysis of the expression of these 261 genes across a clinical dataset of 232 colon cancers defined two groups of patients with differing patterns of gene expression. Groups 1 and 2 included 114 and 118 patients, respectively. Of these, 226 had survival data. Using Kaplan-Meier survival analysis, we found a significant (p=0.0020, log-rank test) difference in 10-year cancer-specific survival between groups (group 1: 48% vs. group 2: 77%) (Fig. 7A). The group 10-year survival was 60.7%. In addition, tumors belonging to the poor prognosis cohort significantly overexpressed MUC1, TAK1 and TRAF6 as compared to those in the good prognosis cohort (Fig. 7B). Significance Analysis of Microarrays (SAM) identified 127 of these genes as having differential expression between groups (Fig. 7C and Supplemental Table S3). The majority of these genes were overexpressed in the poor prognosis cohort. Importantly, expression of this reduced set of 127 genes remained highly predictive of outcome. K-means clustering based on expression of the MUC1-C-induced genes was associated with a significantly increased risk for cancer-specific death in univariate (HR=2.26, p=0.0013) and multivariate (HR=1.93, p=0.01) analyses after controlling for clinical stage (Supplemental Table S4).
Figure 7. Activation of a MUC1-C-induced gene set in colon cancers is associated with a decrease in disease-specific survival.
A. Kaplan-Meier survival curves of patient groups defined by K-means clustering of expression of 261 MUC1-C-induced genes. Group 1 is represented by the red curve (n=112), while Group 2 is represented by the blue curve (n=114). B. Relative gene expression of MUC1, TAK1, TRAF6 in group 1 tumors (solid bars) as compared to group 2 tumors (open bars). The results are presented as mean±SEM. Significant differences in gene expression were found between groups as determined using a 2-tailed Student’s t-test (*; p<0.0005). C. Hierarchical clustering of a subset of 127 differentially expressed MUC1-C-induced genes in 232 colon cancers demonstrating contrasting patterns of expression across patient samples. Orange indicates high expression, while blue denotes low expression. D. Differences in the frequency of MUC1 over-expression in KRAS wild-type (WT) tumors (n=328) as compared to KRAS mutant (MUT) tumors (n=217) as determined by Fisher’s exact test (p=0.0011). The results are presented as mean±SEM. E. Hierarchical clustering of a subset of 127 differentially expressed MUC1-C-induced genes in 217 KRAS mutant tumors. F. Kaplan-Meier survival curves of relapse-free survival of KRAS mutant patient groups receiving adjuvant chemotherapy as defined by K-means clustering of 127 MUC1-C-induced genes. Group 1 is denoted by the red curve (n=38), while Group 2 is denoted by the blue curve (n=56). G. Hierarchical clustering of 199 differentially expressed (p<0.005) NF-κB-regulated genes in poor and good prognosis KRAS mutant colon cancers. Red indicates high expression, while blue denotes low expression. Black hash marks denote poor prognosis patients. H. Schema depicting the proposed MUC1-C-mediated auto-inductive loop involving activation of TAK1 and NF-κB. MUC1-C promotes NF-κB-mediated induction of TAK1 transcription by interacting with NF-κB p65 on the TAK1 promoter and increasing NF-κB occupancy. MUC1-C also forms a complex with TAK1 that can facilitate binding of TAK1 and TRAF6, which is necessary for activation of the downstream NF-κB pathway. In a positive regulatory loop, activated NF-κB interacts with MUC1-C and drives TAK1 expression.
Approximately 40% of colon cancers harbor KRAS gene mutations (9). Therefore, in an independent clinical dataset of 545 human colon cancers with a similar distribution of clinical stages, we extended the above results by examining overexpression of MUC1 as a function of KRAS mutation status. The results demonstrate a significantly higher incidence of MUC1 overexpression in the 217 KRAS mutated tumors as compared to the 328 KRAS wild-type tumors (64% vs. 49%, Fisher’s exact test p=0.0011) (Fig. 7D). Hierarchical clustering analysis of KRAS mutated tumors by expression of the 127 differentially expressed MUC1-C-induced genes identified differences in gene expression patterns across patients (Fig. 7E). Using K-means clustering analysis to define patient groups and Kaplan-Meier analysis to compare survival, we confirmed a significant (p=0.05, log-rank test) difference in 10-year relapse-free survival between groups (group 1: 29% vs. group 2: 65%) as is demonstrated in Figure 7F. The group 10-year survival was 50%. Taken together, these results demonstrate that MUC1-C-induced gene expression patterns predict poor outcomes in patients with colon cancer, including those with KRAS mutations. Consistent results were obtained in both colon cancer datasets in that 74–82% of the NF-κB genes were significantly over-expressed in the poor prognostic cohorts defined by MUC1-C-induced gene expression (Supplemental Fig. S9 and Fig. 7G).
Discussion
MUC1 is aberrantly expressed in colon cancers with aggressive features (2–8). However, remarkably little is known about the potential involvement of MUC1 and, specifically, the oncogenic MUC1-C subunit in colon cancer cells. The present studies were performed based in part on the finding that TAK1, a key regulator of innate immunity, is of importance for cancer cell survival (21; 39). In this context, MUC1-C and TAK1 both function as upstream activators of the canonical NF-κB pathway (26; 40; 15), supporting potential cross-talk between these two effectors. Moreover, the demonstration that silencing TAK1 in TNFα-stimulated non-malignant epithelial cells suppresses MUC1-C-mediated activation of IKKβ and NF-κB (15), indicated that MUC1-C and TAK1 are functionally linked in the inflammatory response. Significantly, silencing MUC1-C in colon cancer cells was associated with downregulation of TAK1 expression. We also found that overexpression of MUC1-C increases TAK1 mRNA and protein levels. To our knowledge, there are no available reports that address the regulation of TAK1/MAP3K7 gene transcription. In this regard and interestingly, we identified an NF-κB binding motif in the TAK1 promoter and found that mutation of that site suppresses MUC1-C-induced TAK1 expression, indicating that both MUC1-C and NF-κB contribute to the induction of TAK1 expression. Previous work has shown that MUC1-C interacts with NF-κB p65, occupies the promoters of NF-κB target genes in a complex with p65, and enhances NF-κB-mediated transcriptional activation (12). In concert with the MUC1-C/NF-κB interaction, we found that MUC1-C associates with NF-κB p65 on the TAK1 promoter and increases NF-κB p65 occupancy. TAK1 mediates NF-κB pathway activation in the regulation of innate immunity and proinflammatory responses (26). Our results in colon cancer cells thus further indicate that NF- κB can in turn increase TAK1 expression in a positive regulatory loop (Fig. 7H).
TAK1-mediated activation of NF-κB signaling in the response to cytokine stimulation requires the formation of a complex with TRAF6 (26). TRAF6 is also of functional importance to activation of the NF-κB pathway in lung cancer cells (27). In addition, depletion of TRAF6 in certain lung cancer cells is associated with loss of survival, indicating that TRAF6 promotes oncogenesis (27). Amplification of the TRAF6 locus and overexpression of TRAF6 has been identified in lung cancer cells (27); however, little is known about the regulation of TRAF6 expression in other carcinomas, including colon cancer. In this regard and as found for TAK1, our studies demonstrate that silencing MUC1-C in colon cancer cells is associated with downregulation of TRAF6 expression. We also found that MUC1-C increases TRAF6 mRNA and protein levels. How MUC1-C activates TRAF6 expression will require further study. Nonetheless, the finding that MUC1-C induces expression of both TAK1 and TRAF6 is of potential interest in that TRAF6 forms a complex with TAK1 that is necessary for TAK1-mediated activation of NF-κB (26). The MUC1-C cytoplasmic domain is an intrinsically disordered protein that has structural plasticity for interactions with diverse effectors (41). The present studies demonstrate that the MUC1-C cytoplasmic domain binds directly to TAK1 and that this interaction is dependent on the MUC1-C CQC motif (Fig. 7H). In addition, the finding that an adjacent region of the MUC1-C cytoplasmic domain binds to TRAF6 invoked the possibility that MUC1-C might contribute to the formation of TAK1-TRAF6 complexes. In support of this notion, we found that the MUC1-C cytoplasmic domain confers the association between TAK1 and TRAF6 in vitro (Fig. 7H). Moreover, targeting the MUC1-C CQC motif with GO-203 showed that MUC1-C promotes the TAK1-TRAF6 interaction in cells. TAK1 and TRAF6 therefore contribute to activation of NF-κB signaling (26) and, in a positive feed-forward loop, NF-κB p65 induces TAK1 expression (Fig. 7H). MUC1-C thus contributes to this auto-inductive regulatory loop by driving TAK1 and TRAF6 expression and facilitating TAK1-TRAF6 complexes (Fig. 7H).
MUC1 is an important component of the intestinal mucosal barrier (1). The epithelial lining of the intestinal tract is exposed to luminal contents that include proteases, bile, ingested toxins and the microbiome of bacteria. For protection, the intestinal epithelium is covered by a MUC1 containing mucin layer, which provides a physical barrier that limits damage to the mucosa. Levels of MUC1 in the intestinal mucosa are increased in the innate immune response to infection with pathogenic bacteria (42). Increased MUC1 expression has also been reported in inflammatory bowel disease (31–33). The present studies further demonstrate that upregulation of MUC1 in ulcerative colitis is associated with overexpression of NF-κB target genes. In mice, dextran sulfate sodium (DSS)-induced colitis results in upregulation of MUC1 expression (43). Muc1 knockout mice are also more resistant to DSS-induced colitis, indicating that MUC1 promotes the inflammatory response (44). As additional support for a functional role in the immune response, crossing MUC1 transgenic mice with the IL-10−/− mouse model of colitis confers a significant increase in inflammation (36). In the present work, upregulation of Muc1-C as observed in the IL-10−/− mouse colon was associated with increases in TAK1 and activation of NF-κB signaling. More pronounced activation of the TAK1→NF-κB pathway was also found in the highly inflammed and dysplastic MUC1+/−/IL-10−/− mouse colon. The MUC1+/−/IL-10−/− mouse exhibits a much higher incidence of colon cancers as compared to the IL-10−/− mouse (36). Thus, taken together with the DSS models of colitis, these findings indicate that MUC1-C is associated with intestinal inflammation and the progression of colitis to colon cancer. Persistent IKKβ→NF-κB activation has linked the inflammatory response with tumor development in mouse models of colitis-associated colon cancer (45). In turn, activated NF-κB induces IL-6 and STAT3 signaling that further contribute to colitis and colon cancer (46). One interpretation of our findings is that the MUC1-C-induced TAK1→NF-κB pathway functions in the upregulation of innate immune and inflammatory responses. By extension, prolonged overexpression of MUC1-C with repetitive episodes of intestinal inflammation results in constitutive activation of the TAK1→NF-κB auto-inductive loop as found here in colon cancer cell lines (Fig. 7H).
Based on our findings in colon cancer cells in vitro and in the MUC1 transgeneic model of colitis, we sought to determine whether MUC1-C and the TAK1→NF-κB pathway are linked in primary colon tumors. A tumorigenesis-associated gene expression pattern induced by the oncogenic MUC1-C subunit was therefore applied to the analysis of colon cancer datasets. The results show that the MUC1-C contributes to the regulation of genes that are highly predictive of poor clinical outcome in colon cancer patients. The data further demonstrate a strong association between MUC1-C-induced gene expression and co-expression of MUC1, TAK1, and TRAF6 in human colon cancers. In an independent dataset of KRAS mutant tumors, patients over-expressing MUC1-C-induced genes were similarly at a significantly higher risk for disease relapse. A positive correlation between MUC1-C-induced gene expression and the NF-κB signaling pathway provided further support for a mutual regulation of both pathways by MUC1-C, indicating that the auto-inductive model derived from in vitro studies of colon cancer cells may extend to colon tumors (Fig. 7H). These findings also provide support for the targeting of MUC1-C in the potential treatment of colon cancers. In this context, a Phase I trial of GO-203 has recently been completed in patients with refractory solid tumors with definition of a maximum tolerated dose for Phase II studies. Therefore, based on the present results that targeting MUC1-C decreases TAK1 expression and function, certain patients with colon cancer may be candidates for GO-203 treatment. The present results further show that targeting MUC1-C decreases BCL-XL expression, which sensitizes KRAS mutant cancers to MEK inhibition (29). Indeed, we found that GO-203 is highly synergistic with different MEK inhibitors that are under clinical development. Therefore, an additional strategy would be to combine GO-203 with a MEK inhibitor for the treatment of certain colon cancers that overexpress MUC1-C and exhibit activation of the TAK1→NF-κB pathway.
Experimental Procedures
Cell culture
Human SK-CO-1 colon cancer cells (ATCC HTB-39) were cultured in EMEM medium (ATCC) containing 10% heat-inactivated fetal bovine serum (HI-FBS). Human SW620 colon cancer cells (ATCC CCL-227) were grown in RPMI1640 medium (ATCC) containing 10% HI-FBS. Human HCT116 (ATCC CCL-247) and LoVo (ATCC CCL-229) colon cancer cells were cultured in DMEM (Corning) and F-12K (ATCC) medium, respectively, each containing 10% HI-FBS. Cells were infected with lentiviruses expressing a MUC1 shRNA (Sigma), a control scrambled CshRNA (Sigma), or MUC1-C (47). Cells were treated with (i) the MUC1-C inhibitor GO-203 or a control peptide CP-2 (48), and (ii) the NF-κB inhibitor BAY11-7085 (Santa Cruz Biotechnology).
Immunoprecipitation and immunoblotting
Total cell lysates were prepared in NP-40 lysis buffer as described (48). Soluble proteins were subjected to immunoprecipitation with anti-MUC1-C (49). Precipitates and cell lysates were analyzed by immunoblotting with anti-MUC1-C (49), anti-β-actin (Sigma), anti-TAK1 (Cell Signaling Technology), anti-phospho-IKKβ, anti-IKKβ, anti-phospho-NF-κB p65 (Cell Signaling Technology), anti-NF-κB p65 (Santa Cruz Biotechnology), anti-TRAF6, anti-BCL-XL and anti-AXIN2 (Cell Signaling Technology). Immune complexes were detected using horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (GE Healthcare).
Quantitative RT-PCR
For qRT-PCR, cDNA synthesis was performed with 1 μg of total RNA using the SuperScript III First-Strand Synthesis System (Invitrogen). The Power SYBR Green PCR Master Mix (Applied Biosystems) was used with 1 μl of diluted cDNA for each sample. The samples were amplified using the 7300 Realtime PCR System (Applied Biosystems). Primers used for RT-PCR analysis are included in Supplemental Table S1.
Promoter-luciferase Reporter Assays
Cells (1 × 105) growing in 24-well plates were transfected with 500 ng of the pGL4.32-Luc plasmid containing NF-κB-activated sequences upstream to luciferase (pGL4.32/luc2P/NF-κB-RE/Hygro; Promega) or pGL3-basic vector (Promega) and 1 ng of SV-40-Renilla-Luc in the presence of Lipofectamine LTX (Invitrogen). The TAK1 promoter was generated by PCR using MCF-10A genomic DNA as a template. The PCR product was digested with XhoI/NcoI and then cloned into corresponding site of pGL3-basic to generate pTAK1-Luc. Site directed mutagenesis PCR (Agilent) was performed to generate a mutant pTAK1-Luc. Primer pairs used in PCR amplification are included in Supplemental Table S1. Cells (1 × 105) growing in 24-well plates were also transfected with 500 ng TOPFlash or FOPFlash (Addgene) and 1 ng of SV-40-Renilla-Luc in the presence of Lipofectamine LTX. At 48 h after transfection, the cells were lysed in passive lysis buffer. Lysates were analyzed for fire-fly and Renilla luciferase activities with the Dual-Luciferase assay kit (Promega).
In vitro binding assays
GST, GST-MUC1-CD, GST-MUC1-CD(1-45), GST-MUC1-CD(46-72) GST-MUC1-CD(20-72) and GST-MUC1-CD(AQA) were prepared as described (37; 11). TAK1 (FL) and TRAF6 (FL) were generated by PCR using SK-CO-1 cDNA as a template. The PCR products were digested with EcoRI/XhoI and then cloned into corresponding sites of pGEX-5X to generate GST fusion proteins. The TAK1 and TRAF6 PCR products were also digested with EcoRI/XhoI and cloned into corresponding sites of pET-28b to generate His-tagged proteins. Purified GST-MUC1-CD was cleaved with thrombin to remove the GST moiety. For bindings assays, purified proteins were incubated for 2 h at room temperature. Adsorbates to glutathione-conjugated beads were analyzed by immunoblotting.
Chromatin immunoprecipitation (ChIP) assays
Soluble chromatin was prepared from 2–3 × 106 cells as described (47) and precipitated with anti-NF-κB or a control nonimmune IgG. For re-ChIP assays, NF-κB complexes from the primary ChIP were eluted and reimmunoprecipitated with anti-MUC1-C as described (47). The SYBR green qPCR kit was used for ChIP qPCRs with the ABI Prism 7000 Sequence Detector (Applied Biosystems). Relative fold enrichment was calculated as described (50). Primers used for qPCR of the TAK1 promoter and control region (CR) are listed in Supplemental Table S1.
MUC1 transgenic mouse model of inflammatory bowel disease and colon tumorigenesis
Human MUC1 transgenic C57BL/6 mice were purchased from Dr. S. Gendler (Mayo Clinic, Scottsdale, AZ). IL-10−/− C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). The MUC1+/− mice were crossed with IL-10−/− mice and then the F1 MUC1+/−/IL-10+/− mice were bred with IL-10−/− mice. Analysis of colon tissue was performed in the Mouse Histopathology Core, Dana-Farber/Harvard Cancer Center (R. T. Bronson, pathologist). Mucosa samples were disrupted into cell suspensions using the Tissue Tearor (BioSpec Products). These studies were performed under animal protocol number 12-029, approved by the Dana-Farber Institutional Animal Care and Use Committee (IACUC).
Clonogenic assays
Cells were seeded into 6-well plates at 100 cells/well and treated with (i) GO-203 each day for 3 days; (ii) AS703026 on day 0; and (iii) GSK1120212 on day 0. After 14 days, the cells were fixed with ice-cold 100% methanol and stained with crystal violet (0.5% w/v).
Statistical analysis of colon cancer databases
A clinical dataset of ulcerative colitis tissue samples (n=62) with corresponding uninflamed (n=61) or normal tissue controls (n=63) was downloaded from GEO under accession number GSE11223. Samples derived from the terminal ileum were removed from analysis. Gene expression values were calculated relative to the median value of uninflamed/normal controls.
Clinical datasets of colon cancers were downloaded from GEO under accession numbers GSE17538 and GSE39582. NF-κB target genes were collected from the following sources: http://www.bu.edu/nf-kb/gene-resources/target-genes/ and http://www.broadinstitute.org/mpr/publications/projects/Lymphoma/FF_NFKB_suppl_revised.pdf. Human orthologs of MUC1-C-induced genes were identified using NetAffx Analysis Center (Affymetrix). Data were median- (GSE17538) or RMA- (GSE39582) normalized across patients. Gene expression was calculated relative to the median value across patient samples. Multiple probe set IDs for a given gene were averaged for each patient sample to obtain a representative expression value for each gene. Probe set IDs derived from exemplar sequences were examined for MUC1, TAK1, and TRAF6 gene expression. Significance Analysis of Microarrays (SAM) version 3.0 was used to identify differentially expressed probe set IDs with a false discovery ratio of 0.74% and a delta value of 1.36. Hierarchical clustering via Ward’s method was used to display gene expression patterns. K-means clustering was used to define patient groups based on gene expression differences. Kaplan-Meier and Cox proportional hazard analyses were utilized to determine survival differences between patient cohorts. Statistical analysis was performed using JMP 9.0 (SAS Institute, Inc).
Supplementary Material
Acknowledgments
Research reported in this publication was supported by Grant CDMRP CA110495 awarded by the Department of Defense and by the National Cancer Institute of the National Institutes of Health under award numbers CA97098 and CA166480.
Abbreviations
- TAK1
TGF-β-activated kinase 1
- TRAF6
TNF receptor-associated factor 6
- MUC1
mucin 1
- MUC1-C
MUC1 C-terminal subunit
- MUC1-CD
MUC1 cytoplasmic domain
Footnotes
Conflict of Interest
D.K. holds equity in Genus Oncology and is a consultant to the company. The other authors disclosed no potential conflicts of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Kufe D. Mucins in cancer: function, prognosis and therapy. Nature Reviews Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrd JC, Bresalier RS. Mucins and mucin binding proteins in colorectal cancer. Cancer Metastasis Rev. 2004;23:77–99. doi: 10.1023/a:1025815113599. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki H, Shoda J, Kawamoto T, Shinozaki E, Miyahara N, Hotta S, et al. Expression of MUC1 recognized by monoclonal antibody MY.1E12 is a useful biomarker for tumor aggressiveness of advanced colon carcinoma. Clin Exp Metastasissis. 2004;21:321–329. doi: 10.1023/b:clin.0000046133.35133.cc. [DOI] [PubMed] [Google Scholar]
- 4.Niv Y. MUC1 and colorectal cancer pathophysiology considerations. World J Gastroenterol. 2008;14:2139–2141. doi: 10.3748/wjg.14.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lugli A, Zlobec I, Baker K, Minoo P, Tornillo L, Terracciano L, et al. Prognostic significance of mucins in colorectal cancer with different DNA mismatch-repair status. J Clin Pathol. 2007;60:534–539. doi: 10.1136/jcp.2006.039552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamori S, Ota DM, Cleary KR, Shirotani K, Irimura T. MUC1 mucin expression as a marker of progression and metastasis of human colorectal carcinoma. Gastroenterology. 1994;106:353–361. doi: 10.1016/0016-5085(94)90592-4. [DOI] [PubMed] [Google Scholar]
- 7.Baldus SE, Monig SP, Hanisch FG, Zirbes TK, Flucke U, Oelert S, et al. Comparative evaluation of the prognostic value of MUC1, MUC2, sialyl-Lewis(a) and sialyl-Lewis(x) antigens in colorectal adenocarcinoma. Histopathology. 2002;40:440–449. doi: 10.1046/j.1365-2559.2002.01389.x. [DOI] [PubMed] [Google Scholar]
- 8.Duncan TJ, Watson NF, Al-Attar AH, Scholefield JH, Durrant LG. The role of MUC1 and MUC3 in the biology and prognosis of colorectal cancer. World J Surg Oncol. 2007;5:31. doi: 10.1186/1477-7819-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 10.Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein blocks GSK3β-mediated phosphorylation and degradation of β-catenin. Cancer Res. 2005;65:10413–10422. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 11.Rajabi H, Ahmad R, Jin C, Kosugi M, Alam M, Joshi M, et al. MUC1-C oncoprotein induces TCF7L2 activation and promotes cyclin D1 expression in human breast cancer cells. J Biol Chem. 2012;287:10703–10713. doi: 10.1074/jbc.M111.323311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmad R, Raina D, Joshi MD, Kawano T, Kharbanda S, Kufe D. MUC1-C oncoprotein functions as a direct activator of the NF-κB p65 transcription factor. Cancer Res. 2009;69:7013–7021. doi: 10.1158/0008-5472.CAN-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Liu D, Chen D, Kharbanda S, Kufe D. Human DF3/MUC1 carcinoma-associated protein functions as an oncogene. Oncogene. 2003;22:6107–6110. doi: 10.1038/sj.onc.1206732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raina D, Kharbanda S, Kufe D. The MUC1 oncoprotein activates the anti-apoptotic PI3K/Akt and Bcl-xL pathways in rat 3Y1 fibroblasts. J Biol Chem. 2004;279:20607–20612. doi: 10.1074/jbc.M310538200. [DOI] [PubMed] [Google Scholar]
- 15.Ahmad R, Raina D, Trivedi V, Ren J, Rajabi H, Kharbanda S, et al. MUC1 oncoprotein activates the IκB kinase β complex and constitutive NF-κB signaling. Nat Cell Biol. 2007;9:1419–1427. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajabi H, Alam M, Takahashi H, Kharbanda A, Guha M, Ahmad R, et al. MUC1-C oncoprotein activates the ZEB1/miR-200c regulatory loop and epithelial-mesenchymal transition. Oncogene. 2013;33:1680–1689. doi: 10.1038/onc.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakurai H. Targeting of TAK1 in inflammatory disorders and cancer. Trends Pharmacol Sci. 2012;33:522–530. doi: 10.1016/j.tips.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Ajibade AA, Wang HY, Wang RF. Cell type-specific function of TAK1 in innate immune signaling. Trends Immunol. 2013;34:307–316. doi: 10.1016/j.it.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Omori E, Matsumoto K, Zhu S, Smart RC, Ninomiya-Tsuji J. Ablation of TAK1 upregulates reactive oxygen species and selectively kills tumor cells. Cancer Res. 2010;70:8417–8425. doi: 10.1158/0008-5472.CAN-10-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ray DM, Myers PH, Painter JT, Hoenerhoff MJ, Olden K, Roberts JD. Inhibition of transforming growth factor-beta-activated kinase-1 blocks cancer cell adhesion, invasion, and metastasis. Br J Cancer. 2012;107:129–136. doi: 10.1038/bjc.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh A, Sweeney MF, Yu M, Burger A, Greninger P, Benes C, et al. TAK1 inhibition promotes apoptosis in KRAS-dependent colon cancers. Cell. 2012;148:639–650. doi: 10.1016/j.cell.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mihaly SR, Ninomiya-Tsuji J, Morioka S. TAK1 control of cell death. Cell Death Differ. 2014;21:1667–1676. doi: 10.1038/cdd.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Relic B, Benoit V, Franchimont N, Ribbens C, Kaiser MJ, Gillet P, et al. 15-deoxy-delta12,14-prostaglandin J2 inhibits Bay 11-7085-induced sustained extracellular signal-regulated kinase phosphorylation and apoptosis in human articular chondrocytes and synovial fibroblasts. J Biol Chem. 2004;279:22399–22403. doi: 10.1074/jbc.M314118200. [DOI] [PubMed] [Google Scholar]
- 24.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 25.Takaesu G, Surabhi RM, Park KJ, Ninomiya-Tsuji J, Matsumoto K, Gaynor RB. TAK1 is critical for IκB kinase-mediated activation of the NF-κB pathway. J Mol Biol. 2003;326:105–115. doi: 10.1016/s0022-2836(02)01404-3. [DOI] [PubMed] [Google Scholar]
- 26.Yamazaki K, Gohda J, Kanayama A, Miyamoto Y, Sakurai H, Yamamoto M, et al. Two mechanistically and temporally distinct NF-κB activation pathways in IL-1 signaling. Sci Signal. 2009;2:ra66. doi: 10.1126/scisignal.2000387. [DOI] [PubMed] [Google Scholar]
- 27.Starczynowski DT, Lockwood WW, Delehouzee S, Chari R, Wegrzyn J, Fuller M, et al. TRAF6 is an amplified oncogene bridging the RAS and NF-κB pathways in human lung cancer. J Clin Invest. 2011;121:4095–4105. doi: 10.1172/JCI58818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raina D, Ahmad R, Rajabi H, Panchamoorthy G, Kharbanda S, Kufe D. Targeting cysteine-mediated dimerization of the MUC1-C oncoprotein in human cancer cells. Int J Oncol. 2012;40:1643–1649. doi: 10.3892/ijo.2011.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corcoran RB, Cheng KA, Hata AN, Faber AC, Ebi H, Coffee EM, et al. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell. 2013;23:121–128. doi: 10.1016/j.ccr.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ullman TA, Itzkowitz SH. Intestinal inflammation and cancer. Gastroenterology. 2011;140:1807–1816. doi: 10.1053/j.gastro.2011.01.057. [DOI] [PubMed] [Google Scholar]
- 31.Campbell BJ, Yu LG, Rhodes JM. Altered glycosylation in inflammatory bowel disease: a possible role in cancer development. Glycoconj J. 2001;18:851–858. doi: 10.1023/a:1022240107040. [DOI] [PubMed] [Google Scholar]
- 32.Rhodes JM. Unifying hypothesis for inflammatory bowel disease and associated colon cancer: sticking the pieces together with sugar. Lancet. 1996;347:40–44. doi: 10.1016/s0140-6736(96)91563-9. [DOI] [PubMed] [Google Scholar]
- 33.Furr AE, Ranganathan S, Finn OJ. Aberrant expression of MUC1 mucin in pediatric inflammatory bowel disease. Pediatr Dev Pathol. 2010;13:24–31. doi: 10.2350/08-06-0479.1. [DOI] [PubMed] [Google Scholar]
- 34.Rowse GJ, Tempero RM, VanLith ML, Hollingsworth MA, Gendler SJ. Tolerance and immunity to MUC1 in a human MUC1 transgenic murine model. Cancer Res. 1998;58:315–321. [PubMed] [Google Scholar]
- 35.Berg DJ, Davidson N, Kuhn R, Muller W, Menon S, Holland G, et al. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J Clin Invest. 1996;98:1010–1020. doi: 10.1172/JCI118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beatty PL, Plevy SE, Sepulveda AR, Finn OJ. Cutting edge: transgenic expression of human MUC1 in IL-10−/− mice accelerates inflammatory bowel disease and progression to colon cancer. J Immunol. 2007;179:735–739. doi: 10.4049/jimmunol.179.2.735. [DOI] [PubMed] [Google Scholar]
- 37.Ahmad R, Rajabi H, Kosugi M, Joshi M, Alam M, Vasir B, et al. MUC1-C oncoprotein promotes STAT3 activation in an auto–inductive regulatory loop. Science Signaling. 2011;4:ra9. doi: 10.1126/scisignal.2001426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khodarev N, Pitroda S, Beckett M, MacDermed D, Huang L, Kufe D, et al. MUC1-induced transcriptional programs associated with tumorigenesis predict outcome in breast and lung cancer. Cancer Res. 2009;69:2833–2837. doi: 10.1158/0008-5472.CAN-08-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai PC, Shi L, Liu VW, Tang HW, Liu IJ, Leung TH, et al. Elevated TAK1 augments tumor growth and metastatic capacities of ovarian cancer cells through activation of NF-κB signaling. Oncotarget. 2014;5:7549–7562. doi: 10.18632/oncotarget.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Omori E, Inagaki M, Mishina Y, Matsumoto K, Ninomiya-Tsuji J. Epithelial transforming growth factor beta-activated kinase 1 (TAK1) is activated through two independent mechanisms and regulates reactive oxygen species. Proc Natl Acad Sci USA. 2012;109:3365–3370. doi: 10.1073/pnas.1116188109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kufe D. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene. 2013;32:1073–1081. doi: 10.1038/onc.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McAuley JL, Linden SK, Png CW, King RM, Pennington HL, Gendler SJ, et al. MUC1 cell surface mucin is a critical element of the mucosal barrier to infection. J Clin Invest. 2007;117:2313–2324. doi: 10.1172/JCI26705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoebler C, Gaudier E, De Coppet P, Rival M, Cherbut C. MUC genes are differently expressed during onset and maintenance of inflammation in dextran sodium sulfate-treated mice. Dig Dis Sci. 2006;51:381–389. doi: 10.1007/s10620-006-3142-y. [DOI] [PubMed] [Google Scholar]
- 44.Petersson J, Schreiber O, Hansson GC, Gendler SJ, Velcich A, Lundberg JO, et al. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am J Physiol Gastrointest Liver Physiol. 2011;300:G327–333. doi: 10.1152/ajpgi.00422.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, et al. IKKβ links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 46.Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alam M, Ahmad R, Rajabi H, Kharbanda A, Kufe D. MUC1-C oncoprotein activates ERK→C/EBPβ-mediated induction of aldehyde dehydrogenase activity in breast cancer cells. J Biol Chem. 2013;288:30829–30903. doi: 10.1074/jbc.M113.477158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raina D, Kosugi M, Ahmad R, Panchamoorthy G, Rajabi H, Alam M, et al. Dependence on the MUC1-C oncoprotein in non-small cell lung cancer cells. Mol Cancer Therapeutics. 2011;10:806–816. doi: 10.1158/1535-7163.MCT-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panchamoorthy G, Rehan H, Kharbanda A, Ahmad R, Kufe D. A monoclonal antibody against the oncogenic mucin 1 cytoplasmic domain. Hybridoma. 2011;30:531–535. doi: 10.1089/hyb.2011.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.