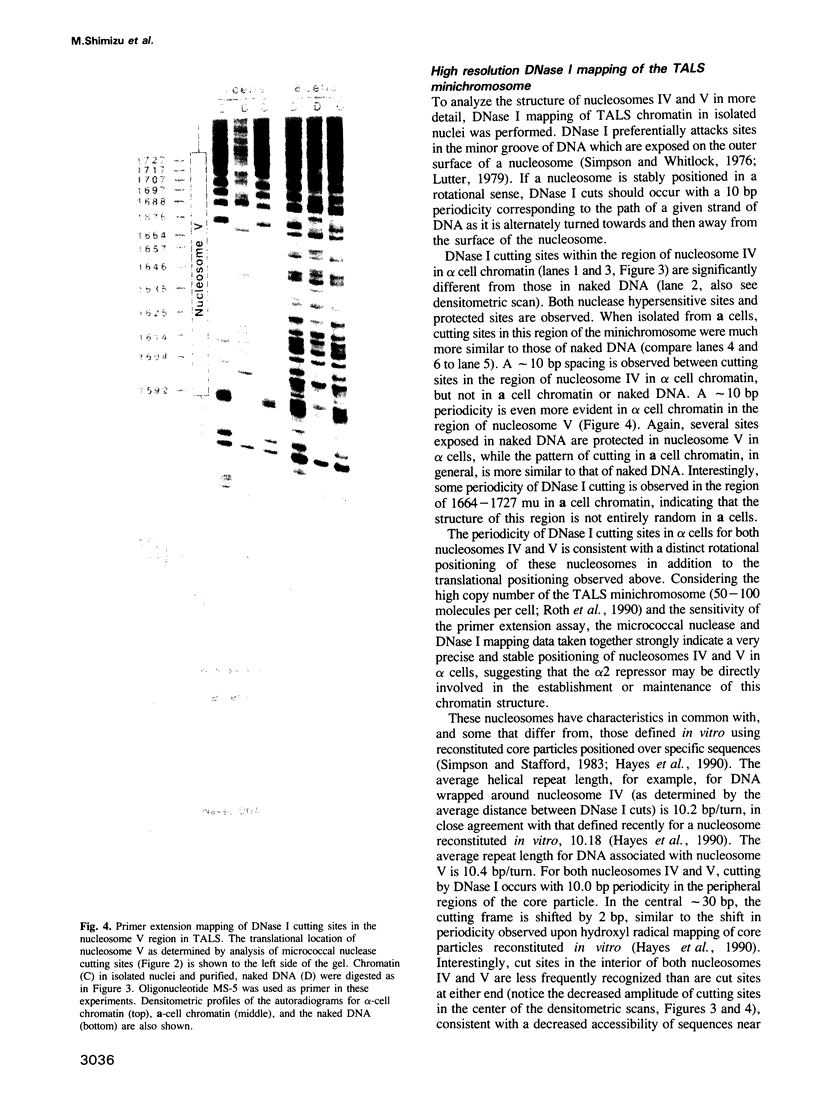
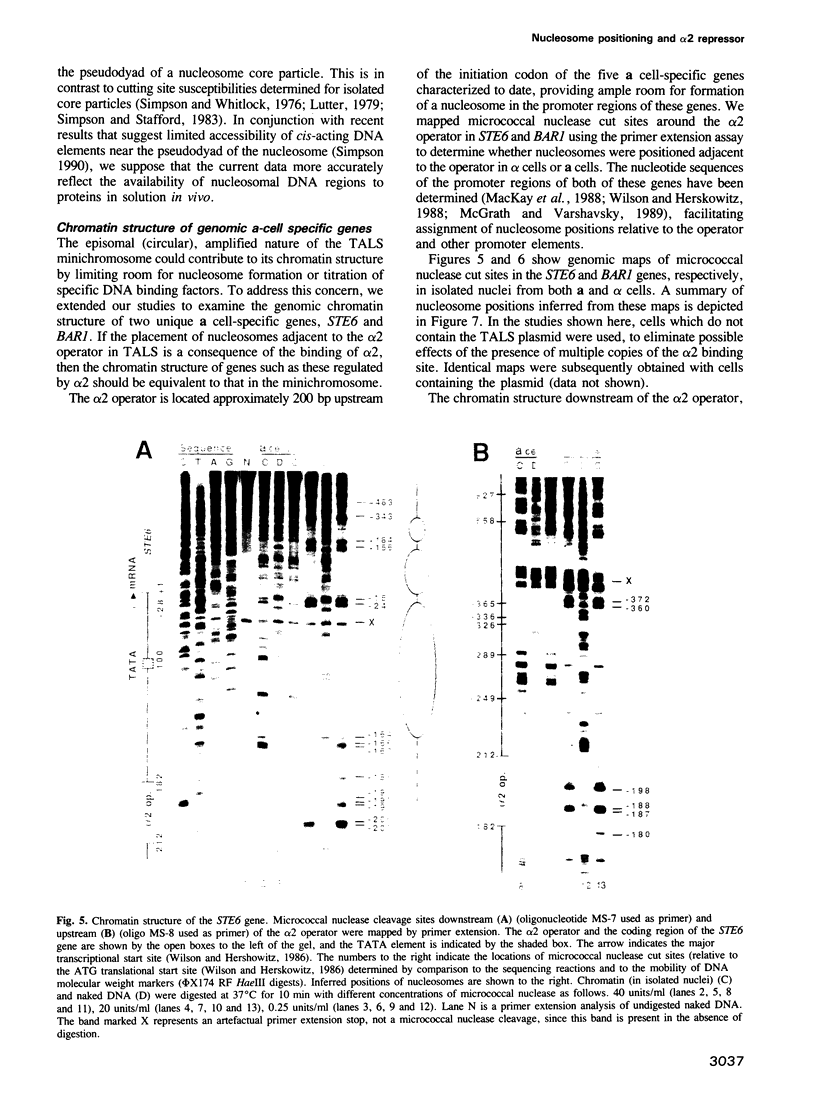
Abstract
Analysis of the chromatin structure of minichromosomes containing the binding site for the yeast alpha 2 repressor protein by indirect end-labeling has previously indicated that nucleosomes are stably positioned over sequences adjacent to the alpha 2 operator in the presence of the repressor. Development of a primer extension assay for nucleosome position now allows a more detailed examination of the location of these nucleosomes relative to the operator sequence, and indicates that nucleosomes are precisely and stably positioned both translationally and rotationally over sequences adjoining the operator. In addition, this assay enables analysis of the chromatin structure of single copy, genomic sequences. Chromatin structures determined for two genes regulated by alpha 2, STE6 and BAR1, are consistent with nucleosomes precisely positioned downstream of the operator sequence, incorporating promoter elements, in alpha cells but not in a-cells. The location of these nucleosomes relative to the operator sequence is highly analogous to that observed in the minichromosome. The stability of the nucleosomes adjacent to the operator together with the precision of their location suggests that they may play a role in repression of a specific gene expression by alpha 2. Further, the primer extension assay allows a comparison of the structure of these positioned nucleosomes formed in vivo to that previously described for core particles reconstituted in vitro.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almer A., Rudolph H., Hinnen A., Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986 Oct;5(10):2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerer G. Identification, purification, and cloning of a polypeptide (PRTF/GRM) that binds to mating-specific promoter elements in yeast. Genes Dev. 1990 Feb;4(2):299–312. doi: 10.1101/gad.4.2.299. [DOI] [PubMed] [Google Scholar]
- Axelrod J. D., Majors J. An improved method for photofootprinting yeast genes in vivo using Taq polymerase. Nucleic Acids Res. 1989 Jan 11;17(1):171–183. doi: 10.1093/nar/17.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R., Cantor C. R., Axel R. Nucleosomes are phased along the mouse beta-major globin gene in erythroid and nonerythroid cells. Cell. 1986 Mar 14;44(5):697–704. doi: 10.1016/0092-8674(86)90835-4. [DOI] [PubMed] [Google Scholar]
- Burkholder A. C., Hartwell L. H. The yeast alpha-factor receptor: structural properties deduced from the sequence of the STE2 gene. Nucleic Acids Res. 1985 Dec 9;13(23):8463–8475. doi: 10.1093/nar/13.23.8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Fedor M. J., Lue N. F., Kornberg R. D. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol. 1988 Nov 5;204(1):109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. A regulatory hierarchy for cell specialization in yeast. Nature. 1989 Dec 14;342(6251):749–757. doi: 10.1038/342749a0. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M., Varshavsky A. In vivo degradation of a transcriptional regulator: the yeast alpha 2 repressor. Cell. 1990 May 18;61(4):697–708. doi: 10.1016/0092-8674(90)90481-s. [DOI] [PubMed] [Google Scholar]
- Huibregtse J. M., Engelke D. R. Genomic footprinting of a yeast tRNA gene reveals stable complexes over the 5'-flanking region. Mol Cell Biol. 1989 Aug;9(8):3244–3252. doi: 10.1128/mcb.9.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis M. A., Myambo K. B., Gelfand D. H., Brow M. A. DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis E. E., Clark K. L., Sprague G. F., Jr The yeast transcription activator PRTF, a homolog of the mammalian serum response factor, is encoded by the MCM1 gene. Genes Dev. 1989 Jul;3(7):936–945. doi: 10.1101/gad.3.7.936. [DOI] [PubMed] [Google Scholar]
- Jarvis E. E., Hagen D. C., Sprague G. F., Jr Identification of a DNA segment that is necessary and sufficient for alpha-specific gene control in Saccharomyces cerevisiae: implications for regulation of alpha-specific and a-specific genes. Mol Cell Biol. 1988 Jan;8(1):309–320. doi: 10.1128/mcb.8.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Herskowitz I. A repressor (MAT alpha 2 Product) and its operator control expression of a set of cell type specific genes in yeast. Cell. 1985 Aug;42(1):237–247. doi: 10.1016/s0092-8674(85)80119-7. [DOI] [PubMed] [Google Scholar]
- Keleher C. A., Goutte C., Johnson A. D. The yeast cell-type-specific repressor alpha 2 acts cooperatively with a non-cell-type-specific protein. Cell. 1988 Jun 17;53(6):927–936. doi: 10.1016/s0092-8674(88)90449-7. [DOI] [PubMed] [Google Scholar]
- Keleher C. A., Passmore S., Johnson A. D. Yeast repressor alpha 2 binds to its operator cooperatively with yeast protein Mcm1. Mol Cell Biol. 1989 Nov;9(11):5228–5230. doi: 10.1128/mcb.9.11.5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D., Stryer L. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 1988 Jul 25;16(14A):6677–6690. doi: 10.1093/nar/16.14.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad J. W., Holly J. A., MacKay V. L. A yeast operator overlaps an upstream activation site. Cell. 1987 Jul 31;50(3):369–377. doi: 10.1016/0092-8674(87)90491-0. [DOI] [PubMed] [Google Scholar]
- Levine M., Manley J. L. Transcriptional repression of eukaryotic promoters. Cell. 1989 Nov 3;59(3):405–408. doi: 10.1016/0092-8674(89)90024-x. [DOI] [PubMed] [Google Scholar]
- Lohr D., Kovacic R. T., Van Holde K. E. Quantitative analysis of the digestion of yeast chromatin by staphylococcal nuclease. Biochemistry. 1977 Feb 8;16(3):463–471. doi: 10.1021/bi00622a020. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- MacKay V. L., Welch S. K., Insley M. Y., Manney T. R., Holly J., Saari G. C., Parker M. L. The Saccharomyces cerevisiae BAR1 gene encodes an exported protein with homology to pepsin. Proc Natl Acad Sci U S A. 1988 Jan;85(1):55–59. doi: 10.1073/pnas.85.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J. P., Varshavsky A. The yeast STE6 gene encodes a homologue of the mammalian multidrug resistance P-glycoprotein. Nature. 1989 Aug 3;340(6232):400–404. doi: 10.1038/340400a0. [DOI] [PubMed] [Google Scholar]
- Passmore S., Elble R., Tye B. K. A protein involved in minichromosome maintenance in yeast binds a transcriptional enhancer conserved in eukaryotes. Genes Dev. 1989 Jul;3(7):921–935. doi: 10.1101/gad.3.7.921. [DOI] [PubMed] [Google Scholar]
- Passmore S., Maine G. T., Elble R., Christ C., Tye B. K. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MAT alpha cells. J Mol Biol. 1988 Dec 5;204(3):593–606. doi: 10.1016/0022-2836(88)90358-0. [DOI] [PubMed] [Google Scholar]
- Piña B., Brüggemeier U., Beato M. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990 Mar 9;60(5):719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- Rahmouni A. R., Wells R. D. Stabilization of Z DNA in vivo by localized supercoiling. Science. 1989 Oct 20;246(4928):358–363. doi: 10.1126/science.2678475. [DOI] [PubMed] [Google Scholar]
- Ramsay N., Felsenfeld G., Rushton B. M., McGhee J. D. A 145-base pair DNA sequence that positions itself precisely and asymmetrically on the nucleosome core. EMBO J. 1984 Nov;3(11):2605–2611. doi: 10.1002/j.1460-2075.1984.tb02181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkawitz R. Transcriptional repression in eukaryotes. Trends Genet. 1990 Jun;6(6):192–197. doi: 10.1016/0168-9525(90)90176-7. [DOI] [PubMed] [Google Scholar]
- Roth S. Y., Dean A., Simpson R. T. Yeast alpha 2 repressor positions nucleosomes in TRP1/ARS1 chromatin. Mol Cell Biol. 1990 May;10(5):2247–2260. doi: 10.1128/mcb.10.5.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer R. T., Smith D. L., Johnson A. D. Flexibility of the yeast alpha 2 repressor enables it to occupy the ends of its operator, leaving the center free. Genes Dev. 1988 Jul;2(7):807–816. doi: 10.1101/gad.2.7.807. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990 Jan 25;343(6256):387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog Nucleic Acid Res Mol Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Stafford D. W. Structural features of a phased nucleosome core particle. Proc Natl Acad Sci U S A. 1983 Jan;80(1):51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P. Mapping DNAase l-susceptible sites in nucleosomes labeled at the 5' ends. Cell. 1976 Oct;9(2):347–353. doi: 10.1016/0092-8674(76)90124-0. [DOI] [PubMed] [Google Scholar]
- Szent-Gyorgyi C., Isenberg I. The organization of oligonucleosomes in yeast. Nucleic Acids Res. 1983 Jun 11;11(11):3717–3736. doi: 10.1093/nar/11.11.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Györgyi C., Finkelstein D. B., Garrard W. T. Sharp boundaries demarcate the chromatin structure of a yeast heat-shock gene. J Mol Biol. 1987 Jan 5;193(1):71–80. doi: 10.1016/0022-2836(87)90628-0. [DOI] [PubMed] [Google Scholar]
- Thoma F. Protein-DNA interactions and nuclease-sensitive regions determine nucleosome positions on yeast plasmid chromatin. J Mol Biol. 1986 Jul 20;190(2):177–190. doi: 10.1016/0022-2836(86)90291-3. [DOI] [PubMed] [Google Scholar]
- Thoma F., Simpson R. T. Local protein-DNA interactions may determine nucleosome positions on yeast plasmids. Nature. 1985 May 16;315(6016):250–252. doi: 10.1038/315250a0. [DOI] [PubMed] [Google Scholar]
- Thoma F., Zatchej M. Chromatin folding modulates nucleosome positioning in yeast minichromosomes. Cell. 1988 Dec 23;55(6):945–953. doi: 10.1016/0092-8674(88)90240-1. [DOI] [PubMed] [Google Scholar]
- Wilson K. L., Herskowitz I. Sequences upstream of the STE6 gene required for its expression and regulation by the mating type locus in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2536–2540. doi: 10.1073/pnas.83.8.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P., Drew H. R. Initiation of transcription on nucleosomal templates. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9817–9821. doi: 10.1073/pnas.86.24.9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P. New approaches to chromatin function. New Biol. 1990 Mar;2(3):211–218. [PubMed] [Google Scholar]