Abstract
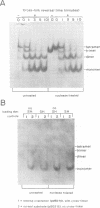
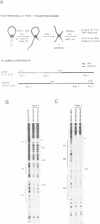
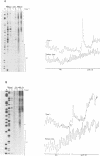
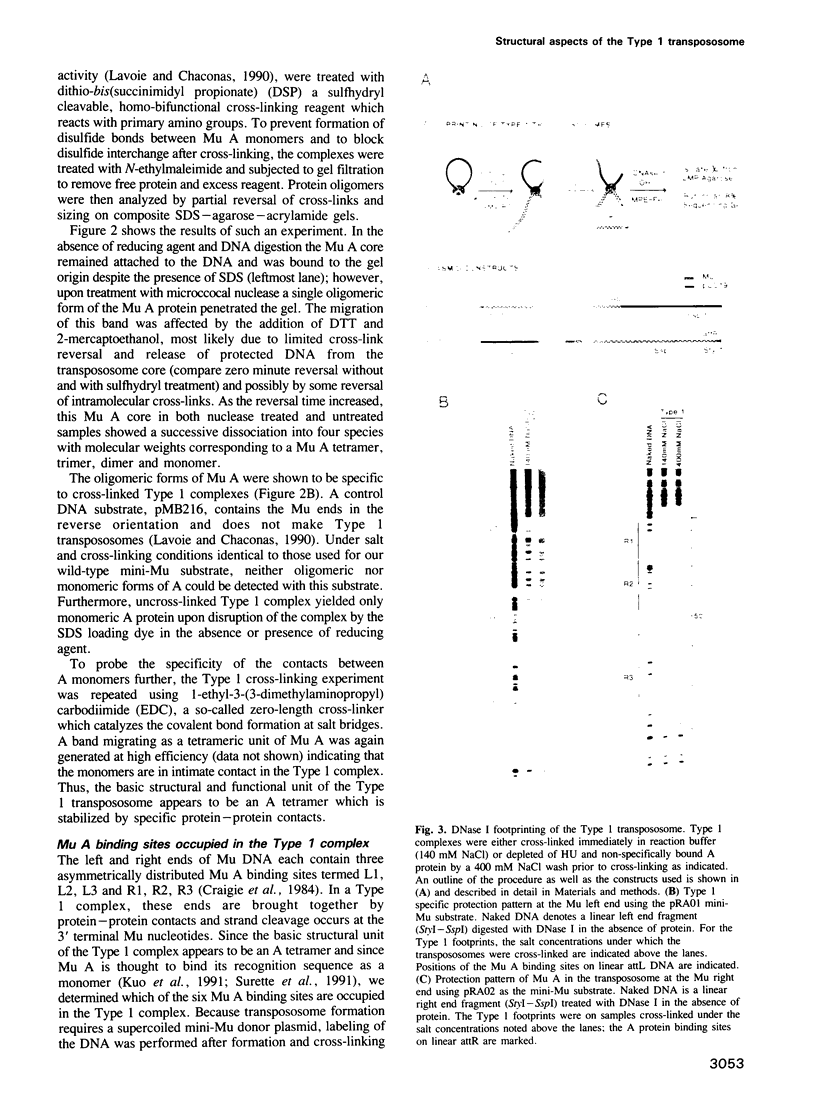
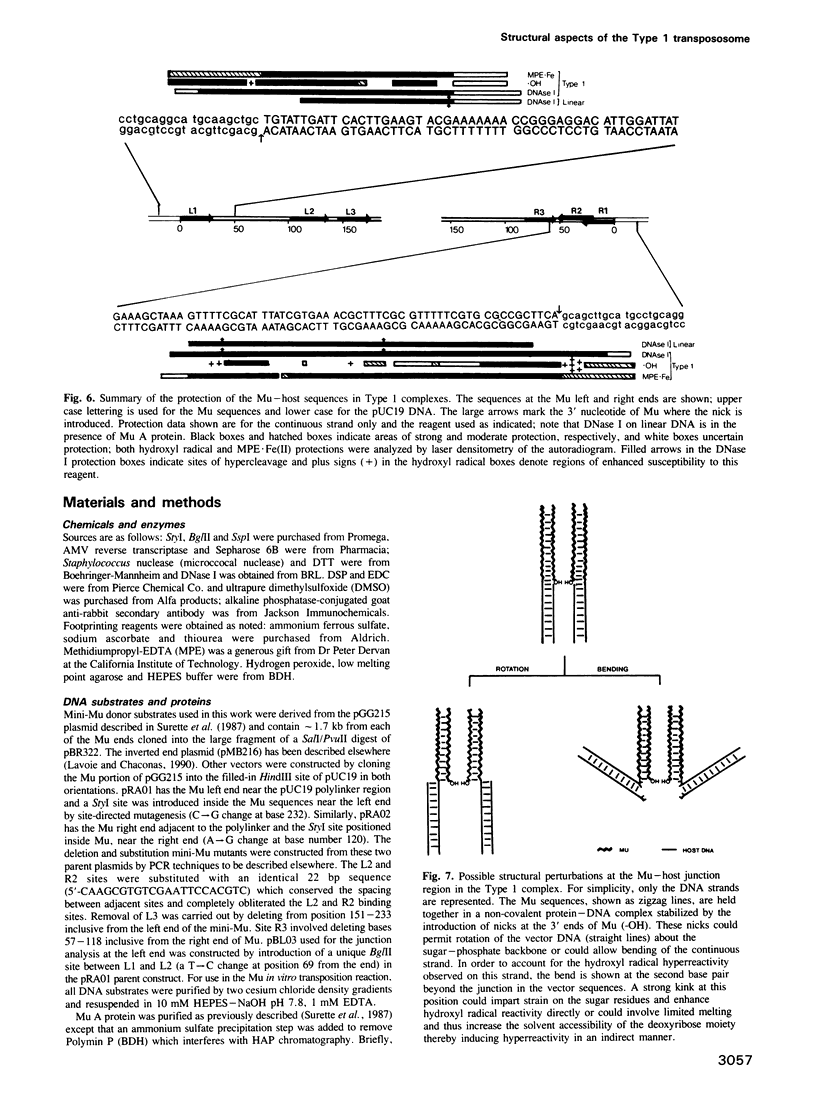
The Mu in vitro strand transfer reaction proceeds via two stable higher order nucleoprotein complexes, the Type 1 and Type 2 transpososomes. The Mu A protein is responsible for the structural and functional integrity of the Type 1 transpososome. We have investigated the quaternary structure of the Mu A protein within this complex by chemical cross-linking experiments and found that the basic structural unit is an A tetramer. Three Mu A binding sites in the transpososome are protected by DNase I footprinting: the outermost A binding sites L1 and R1, as well as R2. Genetic evidence is also presented which corroborates this result. Efficient formation of Type 1 complexes occurs in mini-Mus with the L3 or R3 sites deleted or when the L2 site has been substituted; but no reaction occurs in the absence of R2. The protection at the L1 and R1 sites extends 12-13 bp beyond the Mu-host junctions as seen by DNase I and methidiumpropyl-EDTA.Fe(II) [MPE.Fe(II)] foot-printing, indicating Mu A contacts with the flanking host sequences in the transpososome but not on linear DNA; furthermore, hydroxyl radical footprinting shows an unprecedentedly large enhancement on the continuous strand, 2 bp beyond the nick site outside the Mu right end, which suggests that an altered DNA structure is induced upon Type 1 complex formation.
Full text
PDF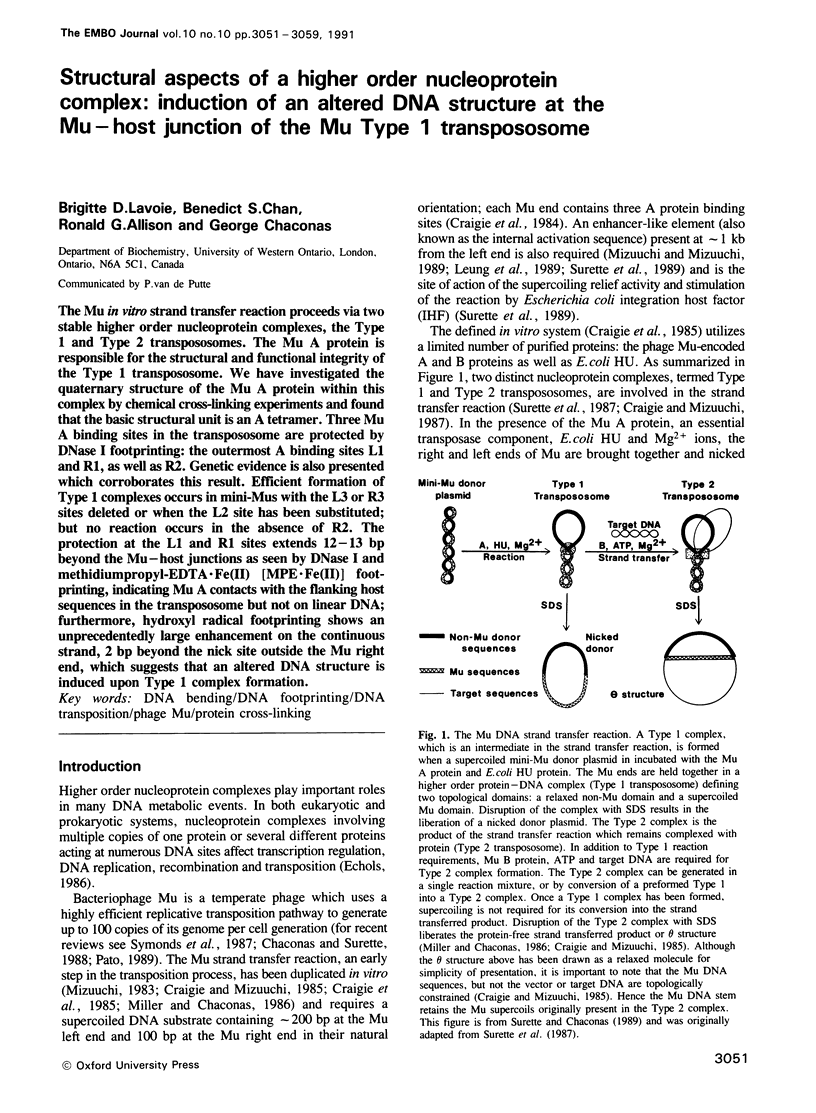





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Betermier M., Alazard R., Ragueh F., Roulet E., Toussaint A., Chandler M. Phage Mu transposase: deletion of the carboxy-terminal end does not abolish DNA-binding activity. Mol Gen Genet. 1987 Nov;210(1):77–85. doi: 10.1007/BF00337761. [DOI] [PubMed] [Google Scholar]
- Burlingame R. P., Obukowicz M. G., Lynn D. L., Howe M. M. Isolation of point mutations in bacteriophage Mu attachment regions cloned in a lambda::mini-Mu phage. Proc Natl Acad Sci U S A. 1986 Aug;83(16):6012–6016. doi: 10.1073/pnas.83.16.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bétermier M., Alazard R., Lefrère V., Chandler M. Functional domains of bacteriophage Mu transposase: properties of C-terminal deletions. Mol Microbiol. 1989 Sep;3(9):1159–1171. doi: 10.1111/j.1365-2958.1989.tb00266.x. [DOI] [PubMed] [Google Scholar]
- Chaconas G., Surette M. G. Mechanism of Mu DNA transposition. Bioessays. 1988 Dec;9(6):205–208. doi: 10.1002/bies.950090606. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Hayes J. J., Tullius T. D. Detection of drug binding to DNA by hydroxyl radical footprinting. Relationship of distamycin binding sites to DNA structure and positioned nucleosomes on 5S RNA genes of Xenopus. Biochemistry. 1990 Jun 26;29(25):6043–6050. doi: 10.1021/bi00477a023. [DOI] [PubMed] [Google Scholar]
- Craigie R., Arndt-Jovin D. J., Mizuuchi K. A defined system for the DNA strand-transfer reaction at the initiation of bacteriophage Mu transposition: protein and DNA substrate requirements. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7570–7574. doi: 10.1073/pnas.82.22.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Mechanism of transposition of bacteriophage Mu: structure of a transposition intermediate. Cell. 1985 Jul;41(3):867–876. doi: 10.1016/s0092-8674(85)80067-2. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Transposition of Mu DNA: joining of Mu to target DNA can be uncoupled from cleavage at the ends of Mu. Cell. 1987 Nov 6;51(3):493–501. doi: 10.1016/0092-8674(87)90645-3. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi M., Mizuuchi K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984 Dec;39(2 Pt 1):387–394. doi: 10.1016/0092-8674(84)90017-5. [DOI] [PubMed] [Google Scholar]
- Dervan P. B. Design of sequence-specific DNA-binding molecules. Science. 1986 Apr 25;232(4749):464–471. doi: 10.1126/science.2421408. [DOI] [PubMed] [Google Scholar]
- Desmet L., Faelen M., Gama M. J., Ferhat A., Toussaint A. Characterization of amber mutations in bacteriophage Mu transposase: a functional analysis of the protein. Mol Microbiol. 1989 Sep;3(9):1145–1158. doi: 10.1111/j.1365-2958.1989.tb00265.x. [DOI] [PubMed] [Google Scholar]
- Dunn S. D. Effects of the modification of transfer buffer composition and the renaturation of proteins in gels on the recognition of proteins on Western blots by monoclonal antibodies. Anal Biochem. 1986 Aug 15;157(1):144–153. doi: 10.1016/0003-2697(86)90207-1. [DOI] [PubMed] [Google Scholar]
- Echols H. Multiple DNA-protein interactions governing high-precision DNA transactions. Science. 1986 Sep 5;233(4768):1050–1056. doi: 10.1126/science.2943018. [DOI] [PubMed] [Google Scholar]
- Groenen M. A., van de Putte P. Analysis of the ends of bacteriophage Mu using site-directed mutagenesis. J Mol Biol. 1986 Jun 20;189(4):597–602. doi: 10.1016/0022-2836(86)90490-0. [DOI] [PubMed] [Google Scholar]
- Hatfull G. F., Noble S. M., Grindley N. D. The gamma delta resolvase induces an unusual DNA structure at the recombinational crossover point. Cell. 1987 Apr 10;49(1):103–110. doi: 10.1016/0092-8674(87)90760-4. [DOI] [PubMed] [Google Scholar]
- Hertzberg R. P., Dervan P. B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984 Aug 14;23(17):3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- Kiehm D. J., Ji T. H. Photochemical cross-linking of cell membranes. A test for natural and random collisional cross-links by millisecond cross-linking. J Biol Chem. 1977 Dec 10;252(23):8524–8531. [PubMed] [Google Scholar]
- Kuo C. F., Zou A. H., Jayaram M., Getzoff E., Harshey R. DNA-protein complexes during attachment-site synapsis in Mu DNA transposition. EMBO J. 1991 Jun;10(6):1585–1591. doi: 10.1002/j.1460-2075.1991.tb07679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B. D., Chaconas G. Immunoelectron microscopic analysis of the A, B, and HU protein content of bacteriophage Mu transpososomes. J Biol Chem. 1990 Jan 25;265(3):1623–1627. [PubMed] [Google Scholar]
- Leung P. C., Teplow D. B., Harshey R. M. Interaction of distinct domains in Mu transposase with Mu DNA ends and an internal transpositional enhancer. Nature. 1989 Apr 20;338(6217):656–658. doi: 10.1038/338656a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller J. L., Chaconas G. Electron microscopic analysis of in vitro transposition intermediates of bacteriophage Mu DNA. Gene. 1986;48(1):101–108. doi: 10.1016/0378-1119(86)90356-2. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K. In vitro transposition of bacteriophage Mu: a biochemical approach to a novel replication reaction. Cell. 1983 Dec;35(3 Pt 2):785–794. doi: 10.1016/0092-8674(83)90111-3. [DOI] [PubMed] [Google Scholar]
- Mizuuchi M., Mizuuchi K. Efficient Mu transposition requires interaction of transposase with a DNA sequence at the Mu operator: implications for regulation. Cell. 1989 Jul 28;58(2):399–408. doi: 10.1016/0092-8674(89)90854-4. [DOI] [PubMed] [Google Scholar]
- Nakayama C., Teplow D. B., Harshey R. M. Structural domains in phage Mu transposase: identification of the site-specific DNA-binding domain. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1809–1813. doi: 10.1073/pnas.84.7.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A., Flamm E., Weisberg R. A., Miller H. I. Overproduction of Escherichia coli integration host factor, a protein with nonidentical subunits. J Bacteriol. 1987 Sep;169(9):4124–4127. doi: 10.1128/jb.169.9.4124-4127.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette M. G., Buch S. J., Chaconas G. Transpososomes: stable protein-DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell. 1987 Apr 24;49(2):253–262. doi: 10.1016/0092-8674(87)90566-6. [DOI] [PubMed] [Google Scholar]
- Surette M. G., Chaconas G. A protein factor which reduces the negative supercoiling requirement in the Mu DNA strand transfer reaction is Escherichia coli integration host factor. J Biol Chem. 1989 Feb 15;264(5):3028–3034. [PubMed] [Google Scholar]
- Surette M. G., Harkness T., Chaconas G. Stimulation of the Mu A protein-mediated strand cleavage reaction by the Mu B protein, and the requirement of DNA nicking for stable type 1 transpososome formation. In vitro transposition characteristics of mini-Mu plasmids carrying terminal base pair mutations. J Biol Chem. 1991 Feb 15;266(5):3118–3124. [PubMed] [Google Scholar]
- Surette M. G., Lavoie B. D., Chaconas G. Action at a distance in Mu DNA transposition: an enhancer-like element is the site of action of supercoiling relief activity by integration host factor (IHF). EMBO J. 1989 Nov;8(11):3483–3489. doi: 10.1002/j.1460-2075.1989.tb08513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint A., Desmet L., Faelen M., Alazard R., Chandler M., Pato M. In vivo mutagenesis of bacteriophage Mu transposase. J Bacteriol. 1987 Dec;169(12):5700–5707. doi: 10.1128/jb.169.12.5700-5707.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A., Churchill M. E., Kam L. Hydroxyl radical footprinting: a high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 1987;155:537–558. doi: 10.1016/0076-6879(87)55035-2. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. C., Nash H. A. The interaction of E. coli IHF protein with its specific binding sites. Cell. 1989 Jun 2;57(5):869–880. doi: 10.1016/0092-8674(89)90801-5. [DOI] [PubMed] [Google Scholar]