Abstract
High hyperdiploidy defines the largest genetic entity of childhood acute lymphoblastic leukemia (ALL). Despite its relatively low recurrence risk, this subgroup generates a high proportion of relapses. The cause and origin of these relapses remains obscure. We therefore explored the mutational landscape in high hyperdiploid (HD) ALL with whole-exome (n=19) and subsequent targeted deep sequencing of 60 genes in 100 relapsing and 51 non-relapsing cases. We identified multiple clones at diagnosis that were primarily defined by a variety of mutations in receptor tyrosine kinase (RTK)/Ras pathway and chromatin-modifying genes. The relapse clones consisted of reappearing as well as new mutations, and overall contained more mutations. Although RTK/Ras pathway mutations were similarly frequent between diagnosis and relapse, both intergenic and intragenic heterogeneity was essentially lost at relapse. CREBBP mutations, however, increased from initially 18–30% at relapse, then commonly co-occurred with KRAS mutations (P<0.001) and these relapses appeared primarily early (P=0.012). Our results confirm the exceptional susceptibility of HD ALL to RTK/Ras pathway and CREBBP mutations, but, more importantly, suggest that mutant KRAS and CREBBP might cooperate and equip cells with the necessary capacity to evolve into a relapse-generating clone.
Introduction
High hyperdiploidy denotes the largest B-cell precursor acute lymphoblastic leukemia (BCP ALL) subgroup in children and adolescents.1 Affected patients usually present with low-risk features and respond well to treatment. The relative proportion of relapses (up to 15%) is small but nevertheless constitutes the largest genetically homogeneous fraction in the BCP ALL cohort.2, 3, 4
High hyperdiploid (HD) ALL has a modal number of 51–68 non-randomly gained chromosomes and is probably caused by the maldistribution of chromosomes during a single abnormal non-disjunction event already in utero.5, 6, 7, 8 Subsequent additional hits are then required to promote disease development. These apparently consist of recurrent submicroscopic copy number aberrations as well as non-overlapping activating mutations in the receptor tyrosine kinase (RTK)/Ras signaling pathway genes (KRAS, NRAS, PTPN11 and FLT3) in up to 50% of cases.9, 10, 11 RTK/Ras signaling is prominently involved in critical cellular processes, such as proliferation, differentiation and cell survival. Mutations in the respective genes lead to the constitutive activation of downstream pathways and provide oncogenic properties, whose effects strongly depend on the respective genomic context as well as on the differentiation stage of the affected cell, and also on the type of tissue to which it belongs.12, 13
So far, none of the identified gene deletions or mutations does qualify as an indisputable indicator of an elevated relapse risk. Although clones with various recurrent genomic alterations may contribute to the initial clinical manifestation of the leukemia, they may be gone at relapse and replaced by others with different aberrations, which, however, still affect the same cohort of genes. In HD ALL, this phenomenon is particularly pronounced in genes that encode RTK/Ras pathway components.14, 15, 16, 17
Of special interest in the research addressed herein is also CREBBP (or CBP). It encodes the binding protein for the cAMP-response element binding protein and serves as transcriptional coactivator by modulating multiple signaling and developmental pathways. Inherited mutations are responsible for the Rubinstein–Taybi syndrome.18 Acquired mutations in CREBBP occur in high-risk and relapsed childhood ALL, particularly in the HD ones, as well as in B-cell lymphomas.17, 19, 20, 21 They cluster in the HAT domain and attenuate the function of the encoded protein, which leads to a reduction in acetylation of histone and non-histone proteins, deregulation of transcription and resistance to glucocorticoids in vitro.19, 20
Previous HD ALL studies have focused their screening on the qualitative identification of mutations in selected target genes and only a few investigated matched diagnosis and relapse samples.16, 17 We therefore performed whole-exome sequencing (WES) and targeted sequencing in a large cohort of relapsing HD ALL cases to investigate the mutational landscape in a genome-wide, unbiased and quantitative manner. This approach allowed us to infer and compare the clonal composition of diagnostic and relapse samples, and, to some extent, also to deduce the developmental path of the relapse clone. Moreover, it enabled us to examine the impact of the most abundant mutations on in vivo drug resistance and course of disease, as well as to check their potential suitability as biomarkers.
Materials and methods
Patients and samples
Frozen viable cells or DNA were obtained from 151 children and adolescents with HD ALL from Austria (n=66), Germany (n=57) and Italy (n=28) (Supplementary Table S1), who were treated according to AIEOP/BFM-ALL95 and 2000, CoALL-08-09 and ALL-Rez BFM 2002 protocols.3, 22, 23, 24 They comprised 100 cases with a bone marrow relapse, among them 50 with matched diagnosis and relapse samples and another 50 with material from either diagnosis (n=16) or relapse (n=34). Germline controls in the form of remission samples were available from all cases. Another 51 diagnosis samples were obtained from HD ALL cases in long-term remission (mean 141 months, range 68–253). The selection of cases was based on the presence of an HD karyotype (51–68 chromosomes) or a DNA index of ⩾1.16 and on at least 85% blast cells at diagnosis or 50% at relapse. All samples were obtained from the respective study centers upon institutional review approval and with approval of their ethics committees. Informed consent for tissue banking and research studies was obtained from patients, their parents or legal guardians in accordance with the Declaration of Helsinki.
Sample preparation and high-throughput sequencing
Genomic DNA from leukemic and remission bone marrow mononuclear cells was isolated with the QIAamp DNA Blood Extraction Kit (Qiagen, Venlo, The Netherlands). The quantity and quality of genomic double-stranded DNA was assessed with the Qubit Fluorometric Quantitation system (Invitrogen, Carlsbad, CA, USA) and Experion Bioanalyzer (Bio-Rad, Hercules, CA, USA), respectively. For WES, libraries were prepared from DNA of matched diagnosis, remission and relapse samples from 19 cases with the Illumina TruSeq DNA Sample Prep and TruSeq Exome Enrichment Kits (Illumina, San Diego, CA, USA) according to the manufacturer's recommendations. The 100 bp paired-end sequencing was performed on a HiSeq 2000 (Illumina). Further details regarding key performance metrics of WES (including coverage) are provided in Supplementary Methods and Supplementary Figure S1.
DNA from these 19 cases submitted to WES and additional 132 HD ALL cases was subjected to deep targeted sequencing of the entire coding regions of 60 selected genes. Sample and library preparation was performed with Illumina Nextera Rapid Capture Custom Kit (Illumina) and sequenced on a HiSeq 2000 (Figure 1a, Supplementary Methods and Supplementary Figures S1 and S2).
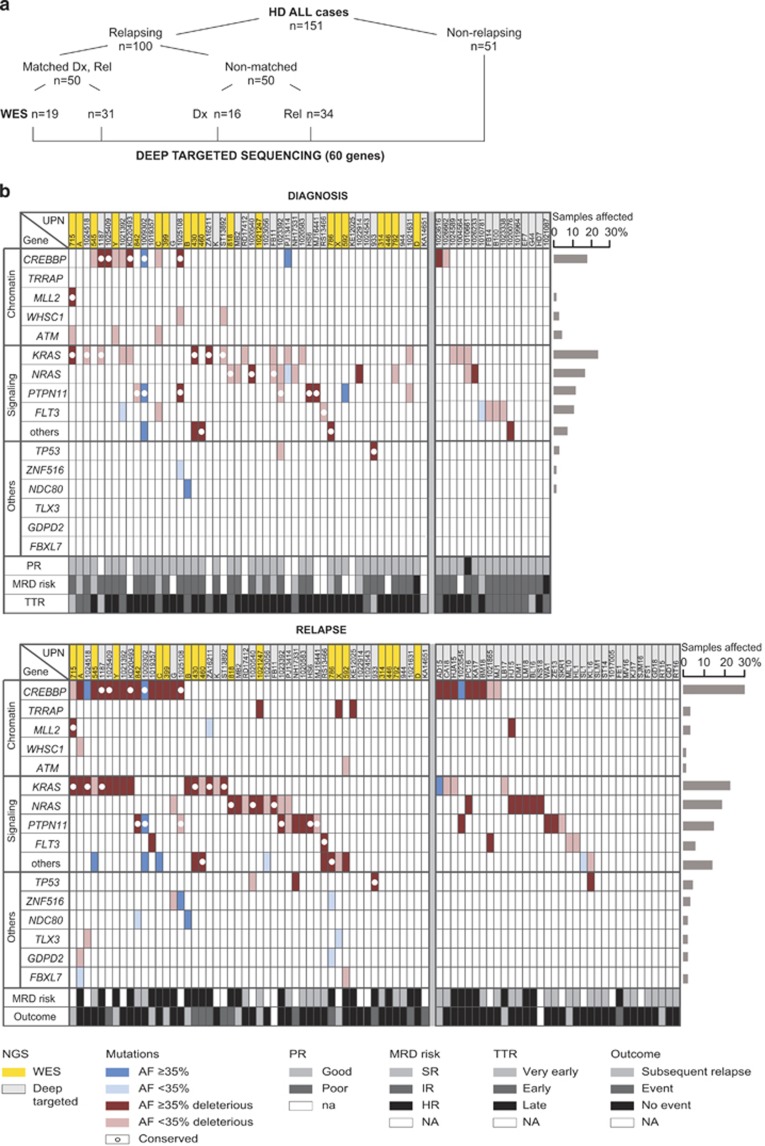
Figure 1.
NGS design and genomic sequence alterations in HD ALL relapsing cases. (a) Schematic representation of the experimental approach for NGS of 151 HD ALL cases. (b) Recurrently mutated genes in relapsing cases. Non-silent mutations in genes are listed according to functional groups in diagnostic (top) and relapse (bottom) samples. Left part shows 50 cases with matched diagnosis/relapse material, right part displays cases lacking relapse (top, 16 cases) or diagnosis (bottom, 34 cases) samples. Mutations are marked by color codes (as shown at the bottom of the figure) to indicate their clonal or subclonal nature based on AF, predicted functional effect and conservation from diagnosis to relapse. Mutated genes in the 'signaling others' category comprise JAK2, PI3KCB, USP9X and MAGI1. UPN, unique patient number; highlighted in yellow are cases from the WES cohort. HR, high risk; IR, intermediate risk; na, not applicable; NA, not available; PR, prednisone response; SER, slow early responder; SR, standard risk; TTR, time to relapse.
Sanger sequencing
CREBBP mutations in non-relapsing cases were validated on genomic DNA from diagnosis and remission samples using published primers and conditions.17
Bioinformatics analysis
Sequence reads were aligned against the human reference genome hg19 using BWA25 and postprocessed with GATK.26 Somatic mutations were called with MuTect27 and IndelGenotyper2 (http://www.broadinstitute.org/cancer/cga/indelocator) and filtered by custom scripts to increase specificity. Variant annotation was performed using SnpEff and SnpSift.28 Mutation calling in non-relapsing samples without germline samples was performed with SAMtools and VarScan 2.29 For pathway analysis Genome MuSiC PathScan was used.30
Ras pathway mutations with low allelic frequency (AF) were called by a custom script that inspected aligned reads at known mutational hotspots for the presence of variant alleles (detailed in Supplementary Methods).
Mutation diagrams ('lolliplots') were generated using a modified version of the 'mutation-diagram' module distributed with Genome MuSiC.31
Statistical analysis
Associations between categorical variables were examined using Fisher's exact test. Event-free survival (EFS) and overall survival (OS) were analyzed according to the Kaplan–Meier method and compared by the log-rank test.32 Cumulative incidence of relapse was calculated with R package 'cmprsk' and compared using the Gray test.33 Pathway enrichment P-values were computed with PathScan.30
Results
Acquired mutations in HD ALL relapsing cases
In the discovery cohort of 19 matched diagnosis and relapse samples subjected to WES, we identified altogether 872 somatic mutations impacting coding sequences (Supplementary Table S2). Six hundred and twenty-six (72%) of these were non-silent mutations and comprised 603 nonsynonymous point mutations, 16 splice site mutations and 7 indels. Three hundred and thirty-six (54%) of non-silent mutations were predicted to be deleterious.
There were 225 non-silent mutations (median 12 per case, range 3–23) at diagnosis and 517 (median 25 per case, range 10–90) at relapse, the latter including both conserved and newly acquired ones (Supplementary Figure S3). Mutations were enriched in genes involved in signaling, development and differentiation, cell cycle and receptor/transporter genes (Supplementary Results, Supplementary Table S3 and Supplementary Figures S4 and S5).
Applying a ⩾10% AF cutoff, we found 12 recurrently mutated genes including all those which had been recognized earlier as being commonly involved in HD ALL, namely the RTK/Ras pathway genes KRAS, NRAS, PTPN11 and FLT3, as well as CREBBP.17, 19, 34, 35 Apart from TRRAP,36 none of the others, MAGI1, USP9X, ZNF516, NDC80, GDPD2 and FBXL7, had been previously identified in hematologic malignancies or solid tumors.
Validation of mutation frequency by targeted sequencing
To validate and better quantify both allelic and case frequencies of these potential biologically and clinically relevant mutations, we deep sequenced 60 selected genes in all 151 cases to ≈500 × coverage (Supplementary Table S4 and Supplementary Figure S1). These analyses not only verified the previously observed mutation frequency in RTK/Ras pathway genes (63%), CREBBP (24%), TRAPP (3%), ZNF516 (3%), FBXL7 (2%), GDPD2 (2%), MAGI1 (2%), NDC80 (2%) and USP9X (2%), but also recovered additional recurrently mutated genes, such as TP53 (5%), ATM (4%), WHSC1 (3%), MLL2 (3%), JAK2 (2%), PI3KCB (2%) and TLX3 (2%). A detailed list of these genes and their respective mutations is provided as Supplementary Table S5 and Supplementary Results. Mutated genes were then manually curated according to their biological functions and grouped together into the three categories 'chromatin modifiers', 'signaling' and 'others' (Figure 1b). Sequence data are available at the European Genome-phenome Archive (accession number EGAS00001001113).
Kinetics of leukemic clones and origin of relapses
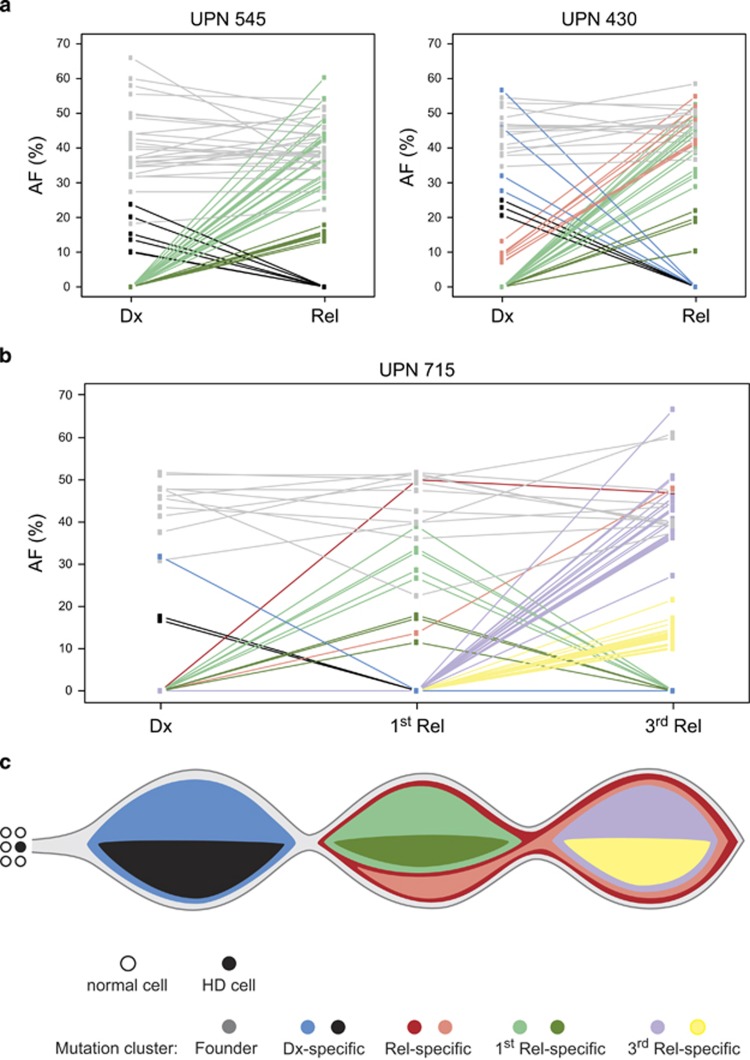
To determine the size of leukemic clones, we normalized AFs of all somatic mutations to single-nucleotide polymorphism array-derived copy numbers of respective genes in eight cases and assumed that mutations affect one allele only. Our analyses revealed two major patterns of clonal diversification (Figure 2a and Supplementary Figure S6). In five cases, we saw a typical clonal progression from diagnosis to relapse with the appearance of new mutations, and in three cases, the relapse clearly evolved from an ancestral clone that was not detectable at diagnosis. Apart from the newly acquired mutations, some of the original ones were still preserved, but some had been lost. Taking into account also single-nucleotide polymorphism array-derived microdeletions, all relapses were apparently derived from an ancestral clone (data not shown). Case UPN 715 experienced three relapses and the resulting mutation patterns enabled us to determine the origin and thereby also to reconstruct the evolution of relapse clones (Figures 2b and c). All but one mutation that defined the major clone at diagnosis were redetected in both relapses, whereas smaller clones were wiped out. Apart from many individual subclonal mutations that were discovered in only one of the relapse samples, these relapse leukemias also shared two distinct mutations, which were absent from the diagnostic sample. This suggests that both relapses emerged from the same ancestral clone and subsequently acquired different mutations.
Figure 2.
Clonal evolution of relapses from HD ALL. (a) The two major patterns of leukemia evolution characterized by the predominant progression of the initial clone (left) and the selection of an ancestral clone (right), exemplarily shown in case UPN 545 and UPN 430. AF denotes the AF of all somatic mutations ⩾10 at diagnosis (Dx) and relapse (Rel) to better illustrate the kinetics of clones. Colored lines connecting individual mutations between diagnosis and relapse visualize the kinetics of major and minor populations; gray, shared mutations; blue, diagnosis-specific mutations in the major clone; black, diagnosis-specific mutations in minor clones; red, initially subclonal mutations developing into the major relapse clone; light and dark green, relapse-specific mutations characterizing major and minor clones, respectively. (b) Kinetics of clones in case UPN 715 inferred from copy number-adjusted AFs of mutations present at diagnosis, first and third relapse. Red and pink, shared relapse-specific mutations; purple and yellow, third relapse-specific mutations in major and minor clones, respectively; remaining color code as in (a). (c) Model for the evolution of initial and two relapse leukemias from case UPN 715 derived from AFs shown in (b). Non-disjunction of chromosomes as the presumed first event occurs in a stem or progenitor cell and generates the HD clone. The founding clone harbors additional mutations and is preserved at both relapses (mutation cluster gray). Within this founder clone, specific major (blue) and minor (black) leukemic populations are present at diagnosis, whereas relapses share two additional mutations, and also comprise distinctive mutations. Affected genes are provided in Supplementary Table S6.
RTK/Ras pathway mutations are highly heterogeneous
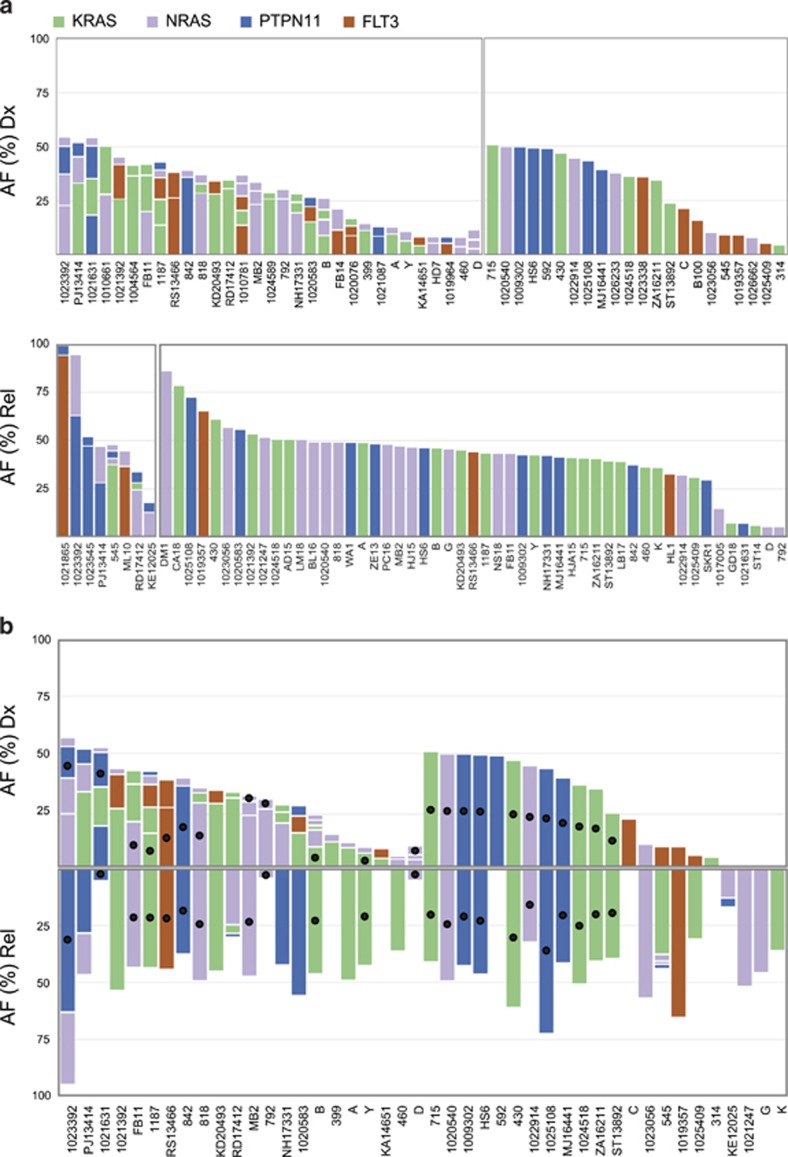
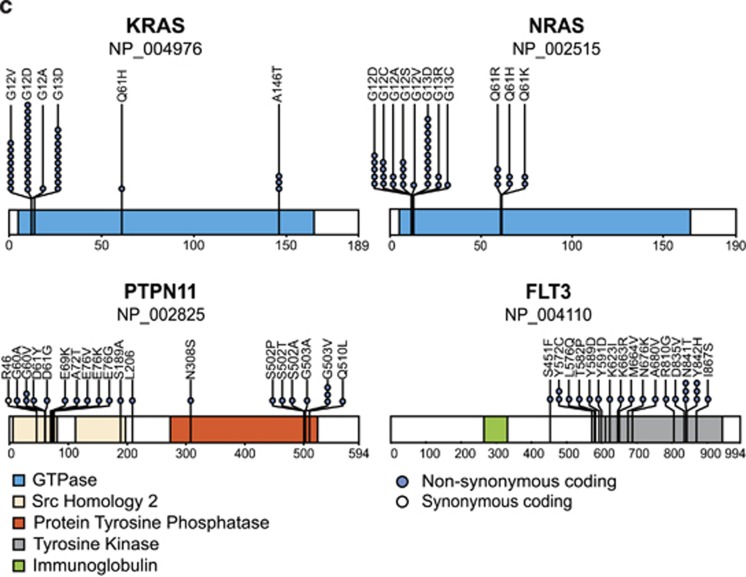
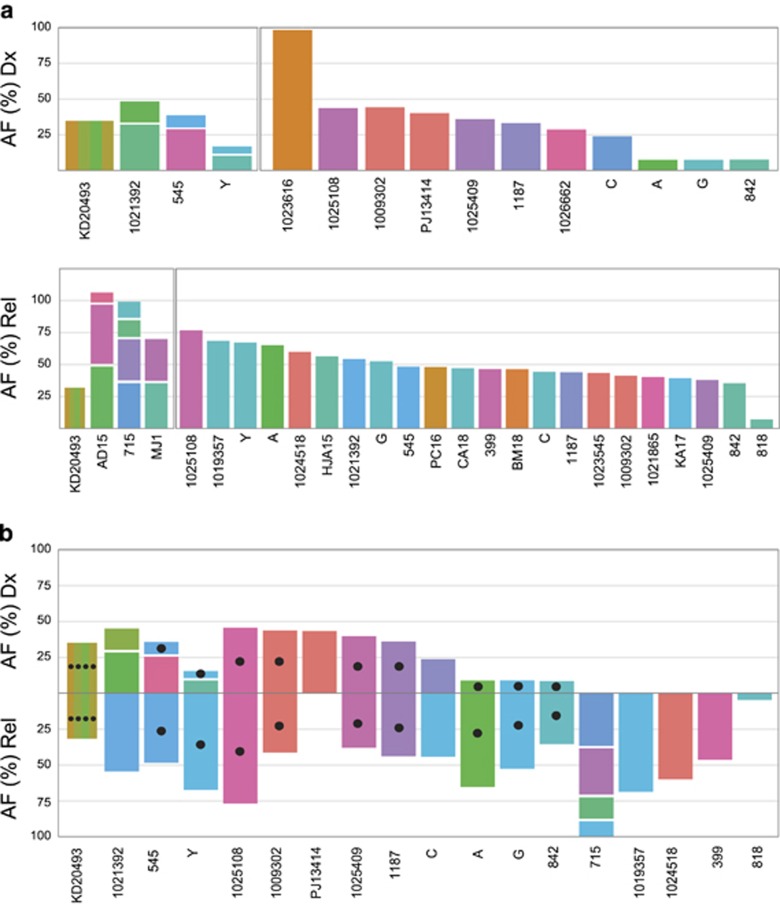
RTK/Ras pathway mutations with an AF⩾10% were equally frequent at diagnosis and relapse (53% and 60%, respectively). The most common ones were KRAS, followed by NRAS, PTPN11 and FLT3. These mutations were usually only present in small clones at diagnosis, and, in the matched cohort, 57% of them were lost at relapse in approximately half of cases. The retained ones are mainly derived from the predominant clone at diagnosis (Figure 1b). Including mutations with AF <10% increases these numbers significantly (Figure 3a and Supplementary Table S7). It provides another 60 mutations in 35 cases at diagnosis and 14 mutations in 11 relapses. At diagnosis, 40% of cases had mutations in two to four different genes, but only 8% at relapse. In addition, a distinctive intragenic heterogeneity was found in 24% of cases exclusively at diagnosis (Figures 3a and b). As reported previously,12 these RTK/Ras pathway mutations were primarily located in signal-transduction domains (Figure 3c).
Figure 3.
Frequencies and patterns of RTK/Ras pathway mutations. Inter- and intragenic heterogeneity of RTK/Ras pathway mutations (including those with low AF) at diagnosis (Dx; top) and relapse (Rel; bottom) in (a) all relapsing cases and (b) cases with matched diagnosis and relapse samples. Cases are grouped according to heterogeneity of mutations (left, heterogeneous; right, homogeneous) and within each group sorted by cumulative AFs. UPN of cases indicated at the x axis; AFs of mutations at the y axis; AF was adjusted to blast counts obtained from bone marrow smears. Mutations with AF<5% not drawn to scale. Color code of mutated genes given at the top of the graph; black filled circles in bars indicate the conservation of the respective mutation at relapse. (c) Schematic diagram of KRAS, NRAS, PTPN11 and FLT3 proteins with alterations present in diagnostic and relapse samples. Color codes for their functional domains and types of mutations given at the bottom of the graph.
In the matched cohort, approximately half of the 50 cases retained merely 28% of all initial RTK/Ras pathway mutations and 16 cases acquired new ones (Figure 3b). There was a high likelihood that larger clones with such particular mutations were conserved at relapse (P=0.001). However, also small clones evolved into the dominant clone. These observations combined with the fact that we observed similar patterns of mutations in leukemic samples from non-relapsing cases (Supplementary Table S8) corroborate their clinically inert behavior. Of note, low-abundant RTK/Ras pathway mutations were not detected in 82 analyzed remission samples.
Taken together, RTK/Ras pathway mutations are present in the vast majority of cases and more abundant at diagnosis. They are dispersed over multiple (sub)clones, located in several RTK/Ras pathway-associated genes and highly instable.
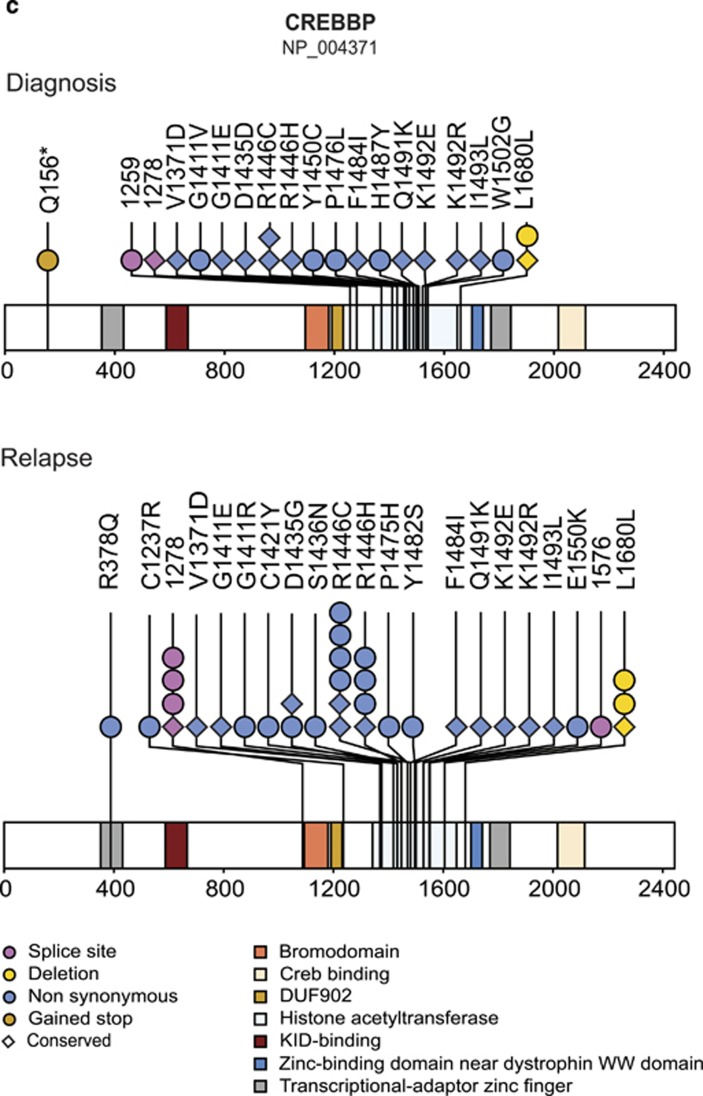
CREBBP mutations cluster in the HAT domain, are generally maintained and accumulate at relapse
CREBBP mutations were present in 18% of cases at diagnosis and in 30% at relapse. If present, they were already part of the major clone in more than half of the cases at diagnosis, but they were virtually always present in the predominant clone at relapse (Figures 1b and 4a).
Figure 4.
Frequency and patterns of CREBBP mutations. Intragenic heterogeneity of CREBBP mutations at diagnosis (Dx; top) and relapse (Rel; bottom) in (a) all cases and (b) cases with matched diagnosis and relapse samples; UPN of cases indicated at the x axis; allelic frequencies of mutations (AF; including those with <10%), adjusted to the percentage of blasts from bone marrow smears, at the y axis; colors denote distinct mutations. Mutations with AF<5% not drawn to scale. Black filled circles indicate conserved mutations. (c) Schematic diagram of CREBBP protein with mutations present at diagnosis (top) and relapse (bottom). Color codes for functional domains of CREBBP and types of mutations given at the bottom of the graph.
In the matched set, 14 CREBBP mutations were found in 10 cases at diagnosis and 22 were discovered in 16 relapses. All but one mutation with AF ⩾35% were conserved, whereas none of the five originally small mutation-carrying clones reoccurred at relapse. Instead, we detected other clones with new distinct CREBBP mutations in these cases, which were successfully backtracked to the diagnostic sample in two of them (UPN 545, Y). A similar reexamination of diagnostic samples from the remaining seven cases with novel predominant relapse-specific CREBBP mutations discovered these mutations also in another three cases (UPN A, G, 842; Figure 4b). Case UPN KD20493 was exceptional because it had four distinct CREBBP mutations on one allele that were also preserved at relapse (Supplementary Figure S7). Such a cluster of regional mutations has been described as kataegis in breast cancer.37
In accordance with previous reports, the vast majority of CREBBP mutations affected the HAT domain mainly at R1446 (Figure 4c).17, 19, 20 These mutations are functionally disruptive, reduce proliferation and confer glucocorticoid resistance in vitro.19, 20 By contrast, none of the CREBBP-mutated cases had a poor prednisone response (Figure 1b).
Co-occurrence of CREBBP and KRAS mutations at relapse
Our most salient finding was the coexistence of CREBBP and KRAS mutations in HD ALL relapse clones, an insight that derived from the quantitative overlap of these two mutations. It originally became evident in our WES data and was subsequently confirmed in the analysis of the targeted sequencing results of the entire cohort including additional 65 relapse samples (P<0.001) (Figure 1b). Although KRAS and NRAS mutations at relapse were similarly frequent (23% and 19%, respectively), only the KRAS ones concurred with the CREBBP mutations. This observation implies that these two mutations exert an interdependent function and cooperate in a RAS isoform-specific and synergistic manner under the selective pressure of the applied chemotherapy.
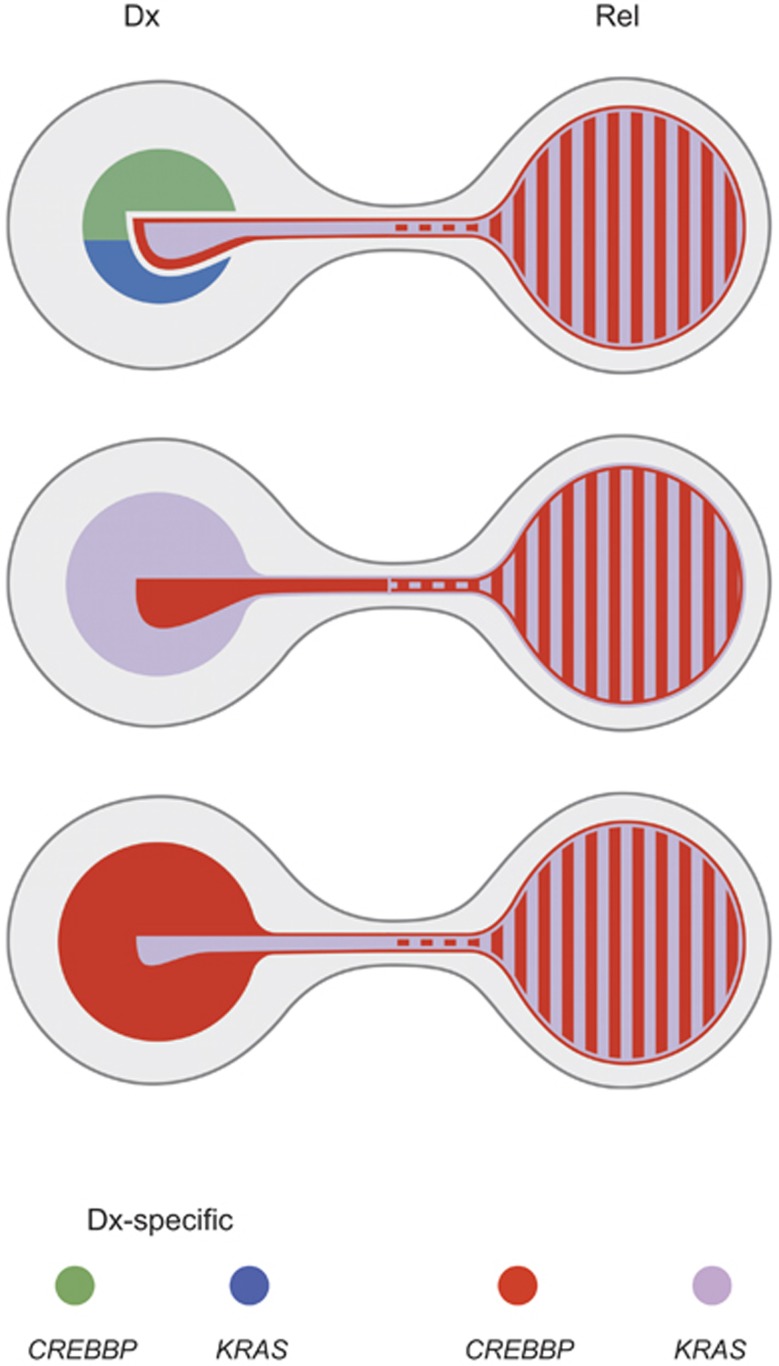
The relapse-associated co-occurrence of CREBBP and KRAS mutations also allowed us to determine from which clones they originally derived and thereby to reconstruct the respective clonal development in nine suitable cases. The three ensuing patterns of relapse evolution are depicted in Figure 5. They clearly highlight the specific selective advantage of CREBBP- and KRAS-double-mutated clones, which eventually culminates in the emergence of a predominant clone at relapse. The unique alliance of these two mutations is further corroborated by the fact that seven of these nine cases initially also had clones with other RTK/Ras signaling gene mutations; however, none of them, persisted.
Figure 5.
Model for the evolution of relapses with concurring CREBBP and KRAS mutations. Shown are the three most frequently observed scenarios representing the diverse composition of initial leukemia populations defined by mutations in CREBBP, KRAS or a combination thereof and their presence at relapse. The clone size was estimated from the means of the AFs of respective mutations present at initial diagnosis (Dx; left) and relapse (Rel; right) in corresponding cases. The first and most frequent scenario indicates the initial coexistence of various clones with two major ones carrying either a CREBBP or KRAS mutation in addition to a minor clone that harbors distinct mutations in both genes. At relapse, this double-mutated population constitutes the entire leukemic clone (upper part; summarizes data from cases UPN A, UPN 545, UPN Y, UPN 1021392). The two other scenarios highlight the initial presence of a KRAS- and CREBBP-mutated major or minor clone, in which a minute population of cells also carries a CREBBP (middle) or KRAS (bottom) mutation, respectively. Again, only clones harboring both mutations prevail at relapse (middle, cases UPN 1187, UPN 1025409, UPN KD20493; and bottom, cases UPN 715, UPN 1024518). Color code of mutations at the bottom of the graph.
Somatic CREBBP mutations in non-relapsing HD ALL cases
Based on the reported virtual absence of CREBBP mutations in non-relapsing ALL,17, 19 we checked whether they might also qualify as a relapse-predicting marker in HD ALL. Targeted sequencing of diagnostic samples from 51 HD ALL cases in long-term remission, which were not included in our previous study,17 and subsequent validation by bidirectional Sanger sequencing revealed, however, that somatic CREBBP mutations were present in four of these cases (Supplementary Figure S8 and Supplementary Table S8), thereby precluding their use as biomarkers.
CREBBP and KRAS mutations are associated with unfavorable clinical features at relapse
Finally, we also checked the clinical consequences of mutations, in particular how they might influence time to relapse, the molecular response to relapse treatment and outcome (Figure 1b and Supplementary Table S9). Overall, RTK/Ras pathway and CREBBP-mutated relapses showed only a slight prevalence of occurring early while KRAS-mutated relapses do develop primarily early; in line, relapses with concurrent CREBBP and KRAS mutations appeared earlier than those in the remaining cases with only one or no mutation in either gene (P=0.012) (Supplementary Table S10). PTPN11-mutated relapses, on the other hand, occur late, and NRAS- and FLT3-mutated ones do not differ from cases without such mutations.
Minimal residual disease standard risk (SR) and high risk (HR) relapses were similarly distributed in RTK/Ras pathway mutated and wild type cases. By contrast, both KRAS- and CREBBP-mutated cases fell mainly into the high-risk group while PTPN11-mutated ones were found in the standard risk group.
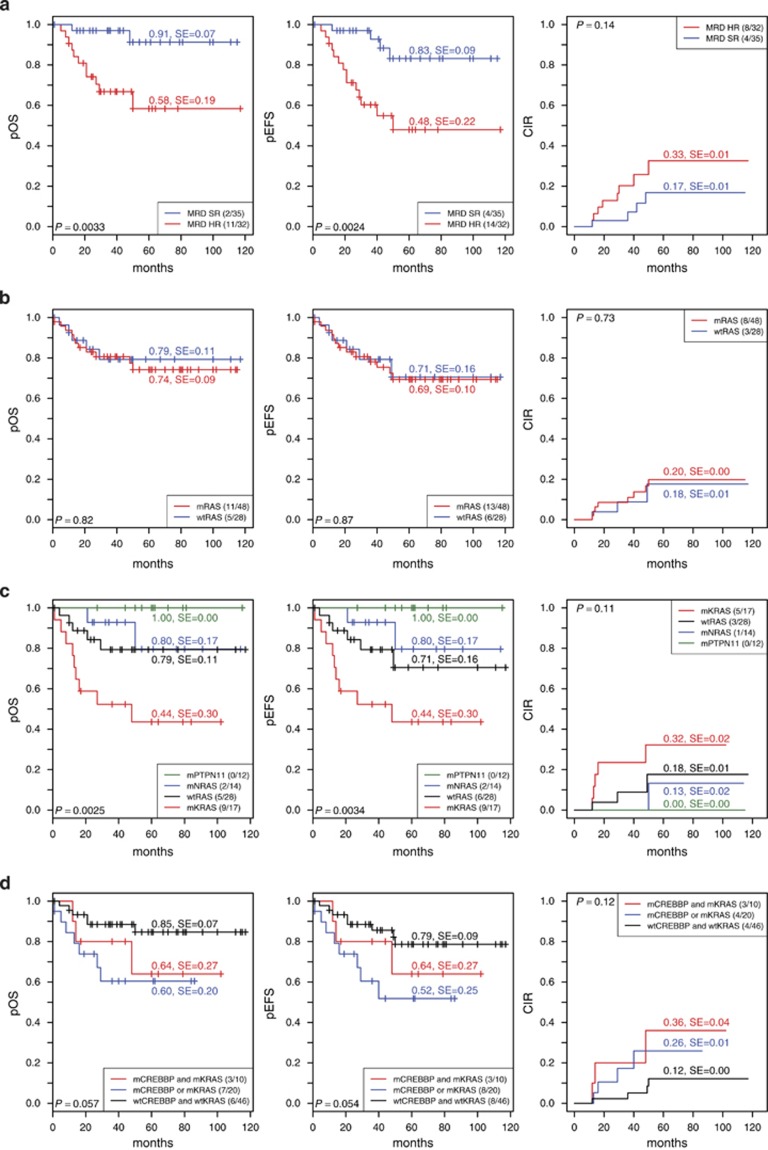
As expected and in line with previous reports,38 high-risk patients had a significantly lower pOS and pEFS at 5 years compared with standard risk patients, and they showed a trend towards a higher cumulative incidence of subsequent relapses (Figure 6a). Although Ras pathway mutations had, in general, no influence on the pOS and pEFS, a more in-depth analysis revealed that especially KRAS mutations were associated with a significantly adverse pOS and pEFS and marginally increased cumulative incidence of relapse. NRAS mutations had no impact and PTPN11 mutations seemed to indicate a more favorable outcome (Figures 6b and c). Although cases with either CREBBP and KRAS mutations or only one of these mutations had a significantly poorer outcome compared with wild-type cases, the cumulative incidence of relapses of the double mutant cases tended to have an increased incidence of subsequent relapses (Figure 6d and Supplementary Table S10).
Figure 6.
Clinical outcome of children with HD ALL after first relapse according to treatment response and mutation status. Kaplan–Maier estimates at 5 years showing the probability of pOS (left), pEFS (middle) and cumulative incidence of relapse (CIR) (right) according to (a) minimal residual disease (MRD) risk at relapse (SR, standard risk; HR, high risk), (b) RTK/Ras pathway (RAS) mutation status, (c) mutated RTK/Ras pathway genes (KRAS, NRAS and PTPN11) or wild-type (wtRAS) and (d) CREBBP and KRAS mutation status.
Discussion
HD ALL is generally perceived as a disease with a low recurrence risk and an excellent prognosis.1, 2, 3, 4 What one might easily overlook in this context is that the small relative proportion of recurrent diseases is, in fact, responsible for a high proportion of relapses that occur in ALL. We consider it thus appropriate to explore the causes and origin of these relapses, especially as all efforts to identify patients at risk in advance have, so far, been unsuccessful.
To achieve our aim of defining the mutational landscape of relapse-prone HD ALL and exploring the potential origin and evolutionary path of relapse clones, we first performed WES in a discovery cohort of 19 matched diagnosis/relapse pairs followed by targeted sequencing of 60 selected genes in these 19 cases and additional 132 ones. We found that at diagnosis the karyotypically homogeneously appearing cell population contains many small subclones whose specific features are abundant mutations that primarily affect RTK/Ras pathway-associated genes. Owing to the fact that relapses arise from highly selected cell populations that survived first-line treatment, the ensuing relapse clones have a more restricted repertoire of mutations (Figures 1 and 3). Although clonal heterogeneity is a typical characteristic of all malignancies and of childhood ALL, in particular,14, 39, 40, 41 the exceptional position of RTK/Ras pathway mutations in this group is a remarkable and noteworthy, albeit not entirely unprecedented, phenomenon.5, 16 Despite the previously suggested apparent mutual exclusive occurrence of RTK/Ras pathway mutations,16 targeted sequencing uncovered an extraordinary inter- and intragenic heterogeneity in the vast majority of cases, particularly at diagnosis (Figures 3a and b). Moreover, RTK/Ras pathway mutations are also quite frequent in the related near-haploid and low hypodiploid ALL forms, and, in rare instances, two distinct Ras pathway mutations and double-mutated RAS genes were found.42 Apart from that, it was shown that this specific pathway might be activated in cases without such mutations.12, 42 Taken together, these findings rank the activation of the Ras pathway as the currently most important disease-relevant functional change in HD ALL.
It comes as a surprise, therefore, that, in contrast to the overall less common CREBBP mutations, which are definitely involved in relapse development,17, 19, 21 RTK/Ras pathway mutations should have only little or no impact at all on the clinical behavior of the disease. At diagnosis, RTK/Ras pathway mutations are as frequent in cases that eventually experience a relapse as in those that stay in long-term remission and they also share a similar distribution. CREBBP mutations are, in contrast, far less common in non-relapsing cases.17, 19 Moreover, at relapse, RTK/Ras pathway mutations remain as frequent as at diagnosis, whereas the frequency of CREBBP mutations increases. Of high interest in this context is, that of all RTK/Ras pathway mutations, only those affecting KRAS pair with CREBBP mutations. The high concordance rate of KRAS and CREBBP mutations at relapse suggests a first circumstantial but nevertheless important clue that these two alterations are functionally linked. The simultaneous deregulation of both proteins may have a synergistic effect, render the affected clone more resistant to therapy and thereby endow it with a considerable proliferative and selective advantage. This notion is further supported by our observation in four cases that already carried two distinct CREBBP-mutated clones at diagnosis (Figure 5). As all these alterations affect the HAT domain and presumably attenuate the acetylation of proteins to a similar extent, it is safe to assume that they are functionally equivalent.19, 20 Nevertheless, only those that also acquired KRAS mutations eventually evolved into predominant relapse clones. CREBBP acetylates both histone and non-histone proteins, such as H3K18, p53 and BCL6, and missense mutations in the HAT domain generally attenuate or destroy the acetylating and coactivator function of the protein.19, 20, 43 As CREBBP is a cofactor for more than 400 transcription factors and most likely acetylates even many more proteins than have yet been identified,43, 44 it is virtually impossible to predict which targets may be affected and are of specific relevance in HD ALL.
Similarly complex and unresolved are the functional impact and consequences of different types of RAS mutations in this specific genetic and cellular context. In transformed cells, oncogenic RAS operates mainly via the RAF-MEK-ERK and PI3K-AKT-mTOR pathways, which prominently control many functions that are essential for cancer.12, 13, 45 Whether and to which extent the deregulation of these particular pathways also contributes to the leukemia development in HD ALL is, so far, not known. Whereas the balanced frequency of KRAS and NRAS mutations at diagnosis implies that they could exert similar functions, only KRAS seems to profit from attenutated CREBBP and thus co-occurs at relapse. This strongly biased selection provides another good example of the fact that individual RAS isoforms can indeed have distinctive and unique properties, which however might only become relevant and obvious under very specific circumstances.12, 13, 46 It is likely that CREBBP modulates or alters not only the function of Ras effectors but also of specific RAS proteins as well.47
KRAS- and CREBBP-double-mutated relapse cases are among those with the highest risk and are therefore prime candidates for novel therapeutic approaches. They could, for instance, combine histone deacetylase inhibitors with Ras pathway inhibitors or drugs that inhibit the connected PI3K-AKT-mTOR pathway, as proposed previously for hypodiploid and HD leukemias with activated Ras signaling.19, 42, 45, 48 Finally, as somatic CREBBP mutations are also present in a small number of non-relapsing HD ALL cases, they unfortunately turn out to be not very useful predictive or prognostic markers.
Acknowledgments
We like to acknowledge the technical support of Maria Morak for single-nucleotide polymorphism array analysis and Aidan Swanton for designing Figures 2c and 5. This study was supported in part by research funding from the Jubiläumsfonds of the Austrian National Bank, OENB 15480 (to GM), the Austrian Science Fund, FWF I-1226-B19 (to RP-G), the FP7-ERA-NET Grant TRANSCALL (to MS) and by a charitable donation of the Kapsch group (http://www.kapsch.net/kapschgroup) (to RP-G).
Author Contributions
KM-O performed wet lab work, analyzed and compiled data and contributed to manuscript writing; CF performed bioinformatics and statistical analyses, and contributed to manuscript writing; AS performed next-generation sequencing and primary sequence analysis; AM performed single-nucleotide polymorphism array analysis and prepared patient samples; M Schuster performed primary next-generation sequence analysis; DK prepared patient samples; CE, GC, M Stanulla, UZS, FL, AVS, M Schrappe, MH and AA provided patient samples and clinical information; CB supervised next-generation sequencing and primary sequence analysis and contributed to the design of the study; GM provided patient samples and clinical data and conceived the study; OAH contributed to the design of the study, interpreted results and wrote the paper; RP-G conceived the study, supervised research, interpreted results and wrote the paper.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- Moorman AV, Ensor HM, Richards SM, Chilton L, Schwab C, Kinsey SE, et al. Prognostic effect of chromosomal abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: results from the UK Medical Research Council ALL97/99 randomised trial. Lancet Oncol. 2010;11:429–438. doi: 10.1016/S1470-2045(10)70066-8. [DOI] [PubMed] [Google Scholar]
- Conter V, Bartram CR, Valsecchi MG, Schrauder A, Panzer-Grumayer R, Moricke A, et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood. 2010;115:3206–3214. doi: 10.1182/blood-2009-10-248146. [DOI] [PubMed] [Google Scholar]
- Tallen G, Ratei R, Mann G, Kaspers G, Niggli F, Karachunsky A, et al. Long-term outcome in children with relapsed acute lymphoblastic leukemia after time-point and site-of-relapse stratification and intensified short-course multidrug chemotherapy: results of trial ALL-REZ BFM 90. J Clin Oncol. 2010;28:2339–2347. doi: 10.1200/JCO.2009.25.1983. [DOI] [PubMed] [Google Scholar]
- Paulsson K, Johansson B. High hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2009;48:637–660. doi: 10.1002/gcc.20671. [DOI] [PubMed] [Google Scholar]
- Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer. 2003;3:639–649. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- Panzer-Grumayer ER, Fasching K, Panzer S, Hettinger K, Schmitt K, Stockler-Ipsiroglu S, et al. Nondisjunction of chromosomes leading to hyperdiploid childhood B-cell precursor acute lymphoblastic leukemia is an early event during leukemogenesis. Blood. 2002;100:347–349. doi: 10.1182/blood-2002-01-0144. [DOI] [PubMed] [Google Scholar]
- Maia AT, van der Velden VH, Harrison CJ, Szczepanski T, Williams MD, Griffiths MJ, et al. Prenatal origin of hyperdiploid acute lymphoblastic leukemia in identical twins. Leukemia. 2003;17:2202–2206. doi: 10.1038/sj.leu.2403101. [DOI] [PubMed] [Google Scholar]
- Paulsson K, Forestier E, Lilljebjorn H, Heldrup J, Behrendtz M, Young BD, et al. Genetic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2010;107:21719–21724. doi: 10.1073/pnas.1006981107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- Kawamata N, Ogawa S, Zimmermann M, Kato M, Sanada M, Hemminki K, et al. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high-resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood. 2008;111:776–784. doi: 10.1182/blood-2007-05-088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuster L, Grausenburger R, Fuka G, Kaindl U, Krapf G, Inthal A, et al. ETV6/RUNX1-positive relapses evolve from an ancestral clone and frequently acquire deletions of genes implicated in glucocorticoid signaling. Blood. 2011;117:2658–2667. doi: 10.1182/blood-2010-03-275347. [DOI] [PubMed] [Google Scholar]
- Davidsson J, Paulsson K, Lindgren D, Lilljebjorn H, Chaplin T, Forestier E, et al. Relapsed childhood high hyperdiploid acute lymphoblastic leukemia: presence of preleukemic ancestral clones and the secondary nature of microdeletions and RTK-RAS mutations. Leukemia. 2010;24:924–931. doi: 10.1038/leu.2010.39. [DOI] [PubMed] [Google Scholar]
- Inthal A, Zeitlhofer P, Zeginigg M, Morak M, Grausenburger R, Fronkova E, et al. CREBBP HAT domain mutations prevail in relapse cases of high hyperdiploid childhood acute lymphoblastic leukemia. Leukemia. 2012;26:1797–1803. doi: 10.1038/leu.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, et al. Rubinstein–Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Mullighan CG, Harvey RC, Wu G, Chen X, Edmonson M, et al. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children's Oncology Group. Blood. 2011;118:3080–3087. doi: 10.1182/blood-2011-03-341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert C, Henze G, Seeger K, Hagedorn N, Mann G, Panzer-Grumayer R, et al. Use of allogeneic hematopoietic stem-cell transplantation based on minimal residual disease response improves outcomes for children with relapsed acute lymphoblastic leukemia in the intermediate-risk group. J Clin Oncol. 2013;31:2736–2742. doi: 10.1200/JCO.2012.48.5680. [DOI] [PubMed] [Google Scholar]
- Escherich G, Zimmermann M, Janka-Schaub G. Co ALLsg. Doxorubicin or daunorubicin given upfront in a therapeutic window are equally effective in children with newly diagnosed acute lymphoblastic leukemia. A randomized comparison in trial CoALL 07-03. Pediatr Blood Cancer. 2013;60:254–257. doi: 10.1002/pbc.24273. [DOI] [PubMed] [Google Scholar]
- Moricke A, Reiter A, Zimmermann M, Gadner H, Stanulla M, Dordelmann M, et al. Risk-adjusted therapy of acute lymphoblastic leukemia can decrease treatment burden and improve survival: treatment results of 2169 unselected pediatric and adolescent patients enrolled in the trial ALL-BFM 95. Blood. 2008;111:4477–4489. doi: 10.1182/blood-2007-09-112920. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM, et al. Using Drosophila melanogaster as a Model for Genotoxic Chemical Mutational Studies with a New Program, SnpSift. Front Genet. 2012;3:35. doi: 10.3389/fgene.2012.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendl MC, Wallis JW, Lin L, Kandoth C, Mardis ER, Wilson RK, et al. PathScan: a tool for discerning mutational significance in groups of putative cancer genes. Bioinformatics. 2011;27:1595–1602. doi: 10.1093/bioinformatics/btr193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dees ND, Zhang Q, Kandoth C, Wendl MC, Schierding W, Koboldt DC, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 2012;22:1589–1598. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan ELMP. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:4. [Google Scholar]
- Gray R. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat. 1988;16:1141–1154. [Google Scholar]
- Paulsson K, Horvat A, Strombeck B, Nilsson F, Heldrup J, Behrendtz M, et al. Mutations of FLT3, NRAS, KRAS, and PTPN11 are frequent and possibly mutually exclusive in high hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2008;47:26–33. doi: 10.1002/gcc.20502. [DOI] [PubMed] [Google Scholar]
- Wiemels JL, Zhang Y, Chang J, Zheng S, Metayer C, Zhang L, et al. RAS mutation is associated with hyperdiploidy and parental characteristics in pediatric acute lymphoblastic leukemia. Leukemia. 2005;19:415–419. doi: 10.1038/sj.leu.2403641. [DOI] [PubMed] [Google Scholar]
- Parry M, Rose-Zerilli MJ, Gibson J, Ennis S, Walewska R, Forster J, et al. Whole exome sequencing identifies novel recurrently mutated genes in patients with splenic marginal zone lymphoma. PLoS One. 2013;8:e83244. doi: 10.1371/journal.pone.0083244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert C, von Stackelberg A, Seeger K, Groeneveld TW, Peters C, Klingebiel T, et al. Minimal residual disease after induction is the strongest predictor of prognosis in intermediate risk relapsed acute lymphoblastic leukaemia - long-term results of trial ALL-REZ BFM P95/96. Eur J Cancer. 2013;49:1346–1355. doi: 10.1016/j.ejca.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morak M, Attarbaschi A, Fischer S, Nassimbeni C, Grausenburger R, Bastelberger S, et al. Small sizes and indolent evolutionary dynamics challenge the potential role of P2RY8-CRLF2-harboring clones as main relapse-driving force in childhood ALL. Blood. 2012;120:5134–5142. doi: 10.1182/blood-2012-07-443218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waanders E, Scheijen B, van der Meer LT, van Reijmersdal SV, van Emst L, Kroeze Y, et al. The origin and nature of tightly clustered BTG1 deletions in precursor B-cell acute lymphoblastic leukemia support a model of multiclonal evolution. PLoS Genet. 2012;8:e1002533. doi: 10.1371/journal.pgen.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford DC, Brindle PK. Is histone acetylation the most important physiological function for CBP and p300. Aging. 2012;4:247–255. doi: 10.18632/aging.100453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JS, Koutelou E, Schibler AC, Dent SY. Histone-modifying enzymes: regulators of developmental decisions and drivers of human disease. Epigenomics. 2012;4:163–177. doi: 10.2217/epi.12.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AF, Braun BS, Shannon KM. Targeting oncogenic Ras signaling in hematologic malignancies. Blood. 2012;120:3397–3406. doi: 10.1182/blood-2012-05-378596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omerovic J, Laude AJ, Prior IA. Ras proteins: paradigms for compartmentalised and isoform-specific signalling. Cell Mol Life Sci. 2007;64:2575–2589. doi: 10.1007/s00018-007-7133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson ML, Kang HS, Lee GM, Blaszczak AG, Lau DK, McIntosh LP, et al. Ras signaling requires dynamic properties of Ets1 for phosphorylation-enhanced binding to coactivator CBP. Proc Natl Acad Sci USA. 2010;107:10026–10031. doi: 10.1073/pnas.0915137107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving J, Matheson E, Minto L, Blair H, Case M, Halsey C, et al. Ras pathway mutations are highly prevalent in relapsed childhood acute lymphoblastic leukaemia, may act as relapse-drivers and confer sensitivity to MEK inhibition. Blood. 2014;124:3420–3430. doi: 10.1182/blood-2014-04-531871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.