Abstract
Liver fat accumulation generally related to systemic insulin resistance characterizes nonalcoholic fatty liver disease (NAFLD), which in the presence of nonalcoholic steatohepatitis (NASH) can progress towards cirrhosis and hepatocellular carcinoma. Due to the epidemic of obesity, NAFLD is now the most frequent liver disease in Western countries. Epidemiological, familial, and twin studies provide evidence for a strong genetic component of NAFLD susceptibility. Recently, genome-wide association studies led to the identification of the major inherited determinants of hepatic fat accumulation: patatin-like phospholipase domain-containing 3 (PNPLA3) I148M gene and transmembrane 6 superfamily member 2 (TM6SF2) E167K gene variants, involved in lipid droplets remodelling and very low-density lipoproteins secretion, are the major determinants of interindividual differences in liver steatosis, and susceptibility to progressive NASH. In this review, we aimed to provide an overview of recent insights into the genetics of hepatic fat accumulation and steatohepatitis.
1. Introduction
Nonalcoholic fatty liver disease (NAFLD) is defined by liver fat deposition in the absence of excessive alcohol intake. NAFLD is frequently present in obese individuals and it is also related to metabolic changes such as systemic insulin resistance [1]. In susceptible individuals, hepatic steatosis may result in oxidative hepatocellular damage, inflammation, and activation of fibrogenesis, namely, nonalcoholic steatohepatitis (NASH) [2]. This condition potentially progresses towards liver cirrhosis and hepatocellular carcinoma [3]. Due to the epidemic of obesity and the advances in the prevention and treatment of viral hepatitis NAFLD is now the most frequent liver disease and the leading cause of altered liver enzymes in Western countries [4, 5]. Subsequently, NASH will become the leading cause of end-stage liver disease, liver transplantation, and hepatocellular carcinoma within the next decades.
Epidemiological, familial, and twin studies provide evidence for heritability of hepatic fat content, NAFLD, and bona fide metabolic cirrhosis [6, 7]. In the last years, the genetic determinants of steatosis are being unrevealed using genome-wide association studies. These studies have identified patatin-like phospholipase domain-containing 3 (PNPLA3) gene variant, involved in hepatocellular lipid droplets remodelling and very low-density lipoprotein (VLDL) secretion, as a major determinant of interindividual and ethnicity-related differences in hepatic fat content [8, 9]. The transmembrane 6 superfamily member 2 (TM6SF2) E167K gene variant, interfering with VLDL secretion, which may be stimulated in early NAFLD [10], has recently been shown to increase susceptibility to progressive NASH by compartmentalization of lipids within hepatocytes [11, 12]. Furthermore, case-control studies demonstrated a role of other genetic variants implicated in inflammation [13], insulin signaling [14], oxidative stress, iron metabolism [15–18], and fibrogenesis [19] in the progression of fatty liver towards NASH associated fibrosis.
Newly identified genetic risk variants could provide a useful tool for the clinical management and prognosis of patients with NAFLD. They may also lead to the identification of drugs for treating NASH, a condition for which specific pharmacological treatment is still lacking.
The main purpose of this narrative review is to provide an overview of recent insights into the genetics of hepatic fat accumulation and steatohepatitis, specifically focusing on inherited variants regulating lipid metabolism. These recent advances have contributed in a short time to unravel the pathogenesis of NAFLD and may soon translate into therapeutic advances. For a comprehensive overview of all studies related to NAFLD and NASH genetics see the recent review by our group [20].
2. Pathophysiology of NAFLD and NASH: The Current View
The acronym NAFLD defines a wide spectrum of liver disease characterized by hepatic fat accumulation in the form of triglycerides exceeding 5% of liver mass in the absence of significant alcohol consumption. NAFLD may remain uncomplicated or progress into severe hepatitis characterized by severe steatosis, lobular inflammation, and hepatocellular damage and apoptosis with activation of fibrogenesis [21]. The final outcome of NAFLD is life threatening conditions, namely, cirrhosis and hepatocellular carcinoma [3]. Hepatic fat accumulation results from an unbalance between triglycerides acquisition, synthesis, utilization, and secretion [22, 23] and it represents the safest way to store fatty acids (FFAs) in the liver [24]. Several lines of evidence support a model where the excess in hepatocellular triglycerides derives from increased peripheral lipolysis [25] due to adipose tissue insulin resistance [26], an increased hepatic lipogenesis due to hyperinsulinemia, and an excessive food intake (Figure 1(a)). Indeed, the major determinant of NAFLD is systemic insulin resistance [1, 27]. Steatosis per se may then in turn worsen hepatic insulin resistance contributing to metabolic disturbances and cardiovascular damage [28, 29]. Reduction in hepatocellular triglycerides secretion through VLDL [23] and in utilization due to mitochondrial damage is also involved in hepatic fat accumulation.
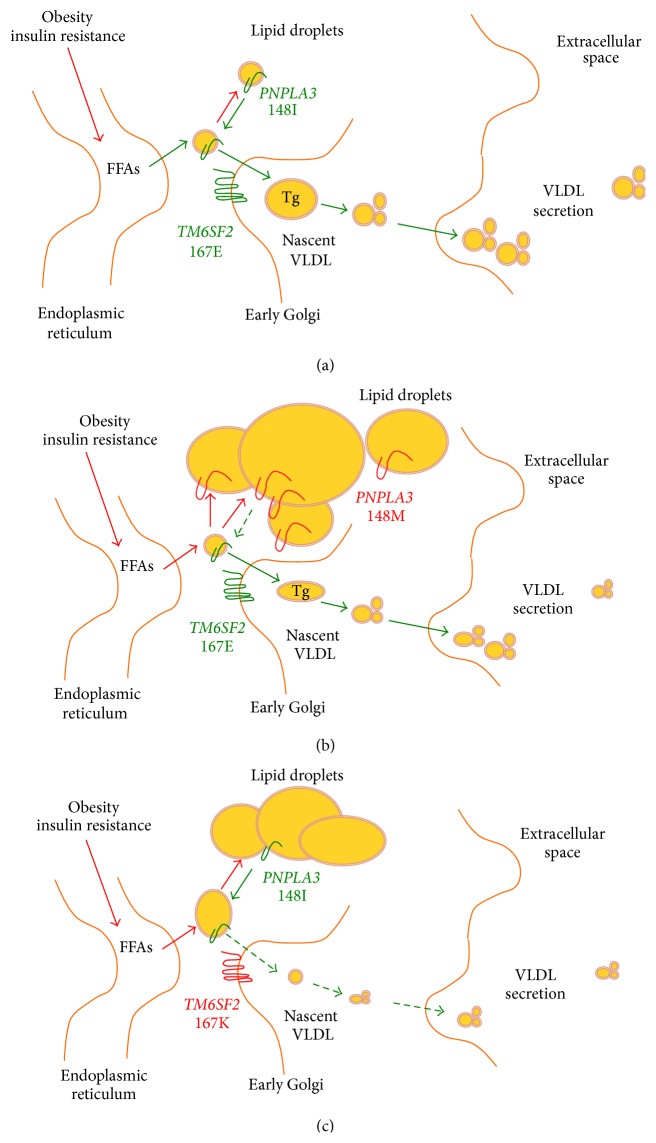
Figure 1.
Molecular genetics of NASH. (a) NAFLD is characterized by the hepatic fat accumulation in lipid droplets resulting from an unbalance between triglycerides acquisition and secretion. FFA stored as triglycerides during hepatic steatosis derive from peripheral lipolysis related to adipose tissue insulin resistance, followed by de novo lipogenesis induced by hyperinsulinemia, and excessive food intake. In the liver, FFA can be catabolized through β-oxidation and reesterification to triglycerides and stored as lipid droplets or exported as VLDL. (b) PNPLA3 I148M variant is attached on the surface of lipid droplets reducing triglyceride breakdown leading to lipid retention in the hepatocyte lipid droplet. (c) TM6SF2 E167K variant reduces triglycerides secretion through VLDL, leading to hepatocellular retention of lipids.
The development of NASH has been classically ascribed to the occurrence of multiple parallel “second-hits,” leading to the activation of inflammation, in the context of hepatic steatosis [30, 31]. This second insult may be related to a variety of conditions: (a) direct hepatic lipotoxicity, (b) hepatocellular oxidative stress secondary to free radicals produced during β- and omega-oxidation of FFAs, (c) inflammation triggered by endotoxin engaging Toll-like receptor-4 in Kupffer cells and hepatocytes due to increased intestinal permeability, (d) qualitative and quantitative changes in gut microbiota [32–34], (e) release of cytokines by the hepatic stellate cells, and (f) endoplasmic reticulum stress. All these conditions lead in the end to inflammation, cellular damage, and activation of fibrogenesis in the liver [35].
3. Heritability of NAFLD and NASH
Epidemiological, familial, and twin studies [6, 7] and clinical case series showing familial clustering support a strong heritability component in NAFLD and NASH [36]. Twin studies show that ALT levels, mostly reflecting liver fat content in the absence of alcohol abuse or viral hepatitis, are a heritable trait explained up to 60% by genetic factors [37]. A study population among Danish twins identified substantial heritability (35–61%) for levels of aminotransferases [38].
Ethnic differences have been reported in the prevalence of NAFLD and NASH [4, 39]. Hispanics are at higher risk than individuals of European descent, whereas those from African descent are protected from these conditions irrespective of diabetes and excess body weight [7, 40].
4. PNPLA3 I148M Mutation Is the Major Genetic Determinant of NAFLD and NASH
The major determinant of the interindividual and ethnicity-related differences in hepatic fat content was identified by an exome wide association study. This is the rs738409 C>G single nucleotide polymorphism (SNP) in the PNPLA3 gene, encoding for the isoleucine to methionine substitution at position 148 (I148M) [8]. In humans PNPLA3, also called adiponutrin, encodes a 481 amino acid membrane protein localized in the endoplasmic reticulum and at the surface of lipid droplets. In human this protein has the highest expression in hepatic stellate cells, retina, and hepatocytes. In mice Pnpla3 is upregulated in the liver after feeding and during insulin resistance by fatty acids and the master regulator of lipogenesis SREBP-1c [41]. Although the mechanism underlying the progression to liver disease remains an area of active research, PNPLA3 has a triglyceride and retinyl-palmitate esterase activity [42–44]. The isoleucine to methionine substitution leads to a loss of function of these activities, of the enzyme, leading to changes in an impairment of lipid catabolism, lipid droplets remodelling, and VLDL secretions [42, 45, 46]. This would favour hepatocellular accumulation of triglycerides during insulin resistance (Figure 1(b)).
A robust association of the I148M variant with hepatic fat content has been confirmed in several studies both in adults [8, 47–58] and in developmental age [59–62]. Most importantly, in carriers of the I148M mutant protein environmental stressors, namely, obesity [63], abdominal fat [64, 65], excessive alcohol consumption [66], chronic viral hepatitis [67, 68], or iron overload [69], trigger progressive liver damage [58]. Dietary habits are also relevant; indeed, the magnitude of the increase in liver enzymes in I148M carriers is correlated to high dietary carbohydrate and sugar consumption [70–72] and increased omega6/omega3 polyunsaturated fatty acids ratio [73, 74]. Interestingly, the carriers of the PNPLA3 I148M variant have a substantial increased risk in cirrhosis and hepatocellular carcinoma [66, 67, 75–84] independently of the predisposition to steatosis. This suggests that PNPLA3 contributes directly to the fibrogenesis and carcinogenesis [9, 57, 61, 83, 85]. Indeed, PNPLA3 retinyl-palmitate activity in hepatic stellate cells may influence hepatic regeneration and differentiation by altering availability of retinol, potent regulators of these phenomena [44].
To summarize the PNPLA3 I148M variant is a robust genetic determinant of hepatic steatosis triggered by a number of environmental factors [9, 85] and the PNPLA3 associated steatohepatitis (PASH) may be mediated by a direct effect on hepatocyte and on hepatic stellate cells [86]. The mechanisms linking the I148M PNPLA3 variant with liver disease progression and hepatocellular carcinoma development have recently been reviewed by our group [9, 83].
5. TM6SF2 E167K and Very Low-Density Lipoproteins Secretion
In 2014, two exome and genome wide association studies were reported identifying the rs58542926 C > T genetic variant of the transmembrane 6 superfamily member 2 gene (TM6SF2), which encodes the loss-of-function lysine (E) to glutamic acid (K) at position 167 substitution (E167K), as a determinant of hepatic triglyceride content, serum aminotransferases, and lower serum lipoproteins [11, 87]. The same studies demonstrated that silencing of TM6SF2 reduces secretion of VLDL resulting in intrahepatic retention of triglycerides and steatosis in mice and in hepatocytes in vitro [11, 87, 88].
Very recently, in large collaborative European study evaluating a large cross-sectional cohort of 1,201 individuals at risk of NASH it was demonstrated that the E167K variant is associated with the full spectrum of liver damage associated with hepatic fat accumulation, including NASH, hepatocellular ballooning, and necroinflammation. Importantly, the association between the E167K variant and advanced fibrosis was abolished after conditioning for NASH, suggesting that fibrosis progression is mediated by the effect of the genetic variant on intracellular retention of lipids, mainly triglycerides and cholesterol, within hepatocytes. In keeping with this interpretation, the severity of liver damage was found to be correlated with the amount of hepatic triglycerides accumulation in patients with NAFLD [89].
The link between the NCAN locus and NAFLD severity [90, 91] and an independent study reporting an association between TM6SF2 and moderate/severe fibrosis [92] support the association between TM6SF2 and liver damage in NAFLD. Indeed, the E167K is the causal variant explaining the association of the NCAN locus with altered lipid metabolism [11, 87].
The association of E167K with NASH and advanced fibrosis contradicts the notion that long-term storage of fatty acids in hepatocytes in triglycerides is benign [93]. In addition, it has been demonstrated that also the PNPLA3 I148M variant by altering lipid droplets remodelling impairs the ability to export triglycerides to secreted VLDL [45, 46]. The deleterious effect on liver damage of the impaired ability to secrete triglycerides in VLDL is also supported by the association between progressive liver disease and rare apolipoprotein B (APOB) and microsomal triglyceride transfer protein (MTTP) mutations directly causing VLDL retention [93, 94]. A possible alternative mechanism may be related to the toxicity of excessive hepatocellular cholesterol and the consequent mitochondrial damage in carriers of the E167K variant [95, 96]. All in all, these novel findings suggest that compartmentalization of neutral lipids within hepatocytes is harmful for the liver.
6. GCKR and Lipogenesis
In a meta-analysis of combined GWAS datasets, besides PNPLA3 I148M, other genetic loci were associated with liver fat content. These included NCAN (explained by TM6SF2, as mentioned previously), glucokinase regulator (GCKR, SNP rs780094), and lysophospholipase-like 1 (LYPLAL1, SNP rs12137855). Both variants in GCKR, a regulator of glucose metabolism and LYPLAL1, involved in triglycerides catabolism, were also shown to influence liver damage [97, 98]. It has been hypothesized that the association of the rs780094 GCKR polymorphism with hepatic fat accumulation can be explained by the linkage disequilibrium with rs1260326, encoding for the P446L protein variant. The P446L variant indeed affects GCKR ability to negatively regulate glucokinase in response to fructose-6-phosphate, thereby determining constitutive activation of hepatic glucose uptake [99]. This would lead to decreased circulating fasting glucose and insulin levels, but on the other hand it would lead to increased glycolysis and production of malonyl-CoA. Malonyl-CoA is a key metabolite, because it favors hepatic fat accumulation by serving as a substrate for lipogenesis and blocks fatty acid oxidation through the inhibition of carnitine-palmitoyl transferase-1. The combined effects of PNPLA3 I148M and GCKR P446L polymorphisms explained up to one-third of variability in liver fat content in a recently reported series of obese children [100, 101]. All this body of evidence shows that genetic variants influencing hepatocellular lipid accumulation predispose both to fatty liver and to progressive NASH.
7. Other Variants Regulating Lipid Metabolism
According to the aforementioned hypothesis, other genes involved in hepatic fat uptake, synthesis, storage, and mobilization are therefore candidates to influence the development and progression of NAFLD. Among these are variants in peroxisome proliferator-activated nuclear receptors (PPAR). PPARα, a molecular target of long chain fatty acids, eicosanoids, and fibrates [102], is highly expressed in tissues that catabolize fatty acids such as the liver, where, under condition of increased hepatic fatty acid influx or decreased fatty acid efflux, PPARα activation prevents the accumulation of triglycerides by increasing the rate of fatty acid catabolism. However, the Leu162Val loss-of-function PPARα variant did not influence the risk of NAFLD and the severity of liver disease [103, 104].
PPARγ is highly expressed in adipose tissue and regulates adipocyte differentiation, FFA uptake, and storage. Pharmacological activation of PPARγ improves insulin resistance in diabetes and decreases steatosis in NAFLD patients by restoring adipose tissue insulin sensitivity and adiponectin release [105, 106]. The Pro12Ala loss-of-function SNP in PPARγ2, inducing a modest impairment of transcriptional activity due to decreased DNA-binding affinity, was associated with a reduction of PPARγ activity in adipose tissue as well as decreased insulin resistance [107]. However, inconsistent results have been reported concerning the association of this variant with the severity of liver damage in NAFLD [103, 104].
Another interesting candidate is represented by Lipin1 (LPIN1), a phosphatidate phosphatase that is highly expressed in the liver and adipose tissue, and is involved in the synthesis of phospholipids downstream of the step catalysed by PNPLA3 [108] and of triglycerides. LPIN1 is required for adipogenesis and the normal metabolic flux between adipose tissue and liver, where it also acts as an inducible transcriptional coactivator to regulate fatty acid metabolism [56]. LPIN1 variants have been associated with several components of the metabolic syndrome, including body mass, insulin levels, resting metabolic rate, and responsiveness to insulin sensitizers [109, 110]. In a pediatric population, LPIN1 rs13412852 TT genotype was protective towards NAFLD [111]. Although independent validation of these results is required, these data suggest that LPIN1 variants predispose to progressive NASH at early age by influencing lipogenesis and lipid metabolism.
Fatty acid transport proteins (FATP) hold a crucial role in mediating cellular FFA uptake. In the liver FATP2 and FATP5 are predominantly expressed [112]. FATP5 encodes a multifunctional protein which increases the hepatic FFA uptake and activates very long-chain fatty acids and has bile-CoA ligase activity [113, 114]. FATP5 silencing reversed diet-induced NAFLD and improved hyperglycemia in mice [115]. Interestingly, the rs56225452 FATP5 promoter variant linked to transcriptional activity has been associated with ALT levels in a population study and with steatosis severity in NAFLD patients [116].
Interestingly, a genetic variant in the promoter region regulating the expression of uncoupling protein 2 (UCP2) has recently been associated with altered lipoprotein metabolism and reduced susceptibility to NASH [117]. Besides regulating mitochondrial redox status and energy dissipation, UCP2 is involved in the export of fatty acids from mitochondria in hepatocytes and is induced during steatosis. It is therefore possible that it may modulate the risk of NASH by altering mitochondrial fluxes of lipids during NAFLD.
Apolipoprotein C3 (APOC3) is a major constituent of VLDL, chylomicrons, and HDL cholesterol, which inhibits lipoprotein lipase and triglyceride clearance [118]. Petersen et al. reported that two common APOC3 T-455C and C-482T promoter variants predispose to liver fat accumulation in Indian individuals. However, these data have not been confirmed in large population studies and in other ethnic groups. Furthermore, APOC3 variants were not associated with the histological severity of liver damage in NAFLD [119–121]. These data suggest that genetic factors influencing triglyceride metabolism outside the liver are not involved in the pathogenesis of progressive NAFLD.
8. Other Variants Not Implicated in Lipid Metabolism
Although recent genetic evidence concordantly points to a major role of variants influencing intrahepatocellular lipid metabolism in the pathogenesis of NAFLD and progressive NASH, as recently reviewed by our group [20], other genetic variants may be implicated in the progression of liver disease.
The most solid associations have been collected for genetic variants involved in the regulation of inflammation, such as TNFalpha and IL28B polymorphisms [13, 122], oxidative stress [13, 122], iron metabolism [15, 17, 18], which is frequently altered in NAFLD [124], and fibrogenesis [19]. However, the overall evidence supporting these associations is weaker than that already obtained for the PNPLA3 I148M and TM6SF2 E167K genetic variants, as these results still need independent validation in independent cohorts.
9. Conclusions
Genes play a key role in the susceptibility and progression of NAFLD. To date, the PNPLA3 I148M and TM6SF2 E167K gene variants are the major determinants of interindividual differences in liver steatosis and susceptibility to progressive NASH. Both of these genes determine liver fat retention through lipid droplets and very low-density lipoproteins modifications. PNPLA3 affects also directly hepatic stellate cells and retinol metabolism. These novel findings suggest that hepatocellular accumulation of neutral lipids is harmful for the liver. Several other genetic variants, including rare mutations, involved in the regulation of hepatocellular lipid metabolism, are being scrutinized.
Future challenges will be (a) to understand the molecular mechanisms underlying the association between gene variants and progressive liver disease, (b) to evaluate the impact of gene variants in the clinical practice to stratify individual risk, and (c) to examine pharmacogenetic response to available therapies. This knowledge will offer insights into pathogenesis of NASH and importantly suggest novel therapeutic targets.
Acknowledgment
Paola Dongiovanni was financed by Italian fiscal contribution 5X1000-2010 devolved to Fondazione IRCCS Cà Granda Ospedale Maggiore Policlinico.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Marchesini G., Brizi M., Blanchi G., et al. Nonalcoholic fatty liver disease: a feature of the metabolic syndrome. Diabetes. 2001;50(8):1844–1850. doi: 10.2337/diabetes.50.8.1844. [DOI] [PubMed] [Google Scholar]
- 2.Day C. P. From fat to inflammation. Gastroenterology. 2006;130(1):207–210. doi: 10.1053/j.gastro.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 3.Bugianesi E., Leone N., Vanni E., et al. Expanding the natural history of nonalcoholic steatohepatitis: from cryptogenic cirrhosis to hepatocellular carcinoma. Gastroenterology. 2002;123(1):134–140. doi: 10.1053/gast.2002.34168. [DOI] [PubMed] [Google Scholar]
- 4.Browning J. D., Szczepaniak L. S., Dobbins R., et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40(6):1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 5.Bellentani S., Saccoccio G., Masutti F., et al. Prevalence of and risk factors for hepatic steatosis in northern Italy. Annals of Internal Medicine. 2000;132(2):112–117. doi: 10.7326/0003-4819-132-2-200001180-00004. [DOI] [PubMed] [Google Scholar]
- 6.Schwimmer J. B., Celedon M. A., Lavine J. E., et al. Heritability of nonalcoholic fatty liver disease. Gastroenterology. 2009;136(5):1585–1592. doi: 10.1053/j.gastro.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerrero R., Vega G. L., Grundy S. M., Browning J. D. Ethnic differences in hepatic steatosis: an insulin resistance paradox? Hepatology. 2009;49(3):791–801. doi: 10.1002/hep.22726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romeo S., Kozlitina J., Xing C., et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nature Genetics. 2008;40(12):1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dongiovanni P., Donati B., Fares R., et al. PNPLA3 I148M polymorphism and progressive liver disease. World Journal of Gastroenterology. 2013;19(41):6969–6978. doi: 10.3748/wjg.v19.i41.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zelber-Sagi S., Salomone F., Yeshua H., et al. Non-high-density lipoprotein cholesterol independently predicts new onset of non-alcoholic fatty liver disease. Liver International. 2014;34(6):e128–e135. doi: 10.1111/liv.12318. [DOI] [PubMed] [Google Scholar]
- 11.Kozlitina J., Smagris E., Stender S., et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nature Genetics. 2014;46(4):352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dongiovanni P., Petta S., Maglio C., et al. TM6SF2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology. 2014 doi: 10.1002/hep.27490. [DOI] [PubMed] [Google Scholar]
- 13.Valenti L., Fracanzani A. L., Dongiovanni P., et al. Tumor necrosis factor α promoter polymorphisms and insulin resistance in nonalcoholic fatty liver disease. Gastroenterology. 2002;122(2):274–280. doi: 10.1053/gast.2002.31065. [DOI] [PubMed] [Google Scholar]
- 14.Dongiovanni P., Valenti L., Rametta R., et al. Genetic variants regulating insulin receptor signalling are associated with the severity of liver damage in patients with non-alcoholic fatty liver disease. Gut. 2010;59(2):267–273. doi: 10.1136/gut.2009.190801. [DOI] [PubMed] [Google Scholar]
- 15.Valenti L., Canavesi E., Galmozzi E., et al. Beta-globin mutations are associated with parenchymal siderosis and fibrosis in patients with non-alcoholic fatty liver disease. Journal of Hepatology. 2010;53(5):927–933. doi: 10.1016/j.jhep.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 16.Al-Serri A., Anstee Q. M., Valenti L., et al. The SOD2 C47T polymorphism influences NAFLD fibrosis severity: evidence from case-control and intra-familial allele association studies. Journal of Hepatology. 2012;56(2):448–454. doi: 10.1016/j.jhep.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 17.Valenti L., Rametta R., Dongiovanni P., et al. The A736V TMPRSS6 polymorphism influences hepatic iron overload in nonalcoholic fatty liver disease. PLoS ONE. 2012;7(11) doi: 10.1371/journal.pone.0048804.e48804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valenti L., Fracanzani A. L., Bugianesi E., et al. HFE genotype, parenchymal iron accumulation, and liver fibrosis in patients with nonalcoholic fatty liver disease. Gastroenterology. 2010;138(3):905–912. doi: 10.1053/j.gastro.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 19.Miele L., Beale G., Patman G., et al. The Kruppel-like factor 6 genotype is associated with fibrosis in nonalcoholic fatty liver disease. Gastroenterology. 2008;135(1):282–291. doi: 10.1053/j.gastro.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dongiovanni P., Anstee Q. M., Valenti L. Genetic predisposition in NAFLD and NASH: impact on severity of liver disease and response to treatment. Current Pharmaceutical Design. 2013;19(29):5219–5238. doi: 10.2174/13816128113199990381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleiner D. E., Brunt E. M., van Natta M., et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 22.Cohen J. C., Horton J. D., Hobbs H. H. Human fatty liver disease: old questions and new insights. Science. 2011;332(6037):1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabbrini E., Mohammed B. S., Magkos F., Korenblat K. M., Patterson B. W., Klein S. Alterations in adipose tissue and hepatic lipid kinetics in obese men and women with nonalcoholic fatty liver disease. Gastroenterology. 2008;134(2):424–431. doi: 10.1053/j.gastro.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi K., Yang L., McCall S., et al. Inhibiting triglyceride synthesis improves hepatic steatosis but exacerbates liver damage and fibrosis in obese mice with nonalcoholic steatohepatitis. Hepatology. 2007;45(6):1366–1374. doi: 10.1002/hep.21655. [DOI] [PubMed] [Google Scholar]
- 25.Donnelly K. L., Smith C. I., Schwarzenberg S. J., Jessurun J., Boldt M. D., Parks E. J. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. Journal of Clinical Investigation. 2005;115(5):1343–1351. doi: 10.1172/JCI200523621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bugianesi E., Gastaldelli A., Vanni E., et al. Insulin resistance in non-diabetic patients with non-alcoholic fatty liver disease: sites and mechanisms. Diabetologia. 2005;48(4):634–642. doi: 10.1007/s00125-005-1682-x. [DOI] [PubMed] [Google Scholar]
- 27.Marchesini G., Brizi M., Morselli-Labate A. M., et al. Association of nonalcoholic fatty liver disease with insulin resistance. The American Journal of Medicine. 1999;107(5):450–455. doi: 10.1016/s0002-9343(99)00271-5. [DOI] [PubMed] [Google Scholar]
- 28.Korenblat K. M., Fabbrini E., Mohammed B. S., Klein S. Liver, muscle, and adipose tissue insulin action is directly related to intrahepatic triglyceride content in obese subjects. Gastroenterology. 2008;134(5):1369–1375. doi: 10.1053/j.gastro.2008.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Targher G., Day C. P., Bonora E. Risk of cardiovascular disease in patients with nonalcoholic fatty liver disease. The New England Journal of Medicine. 2010;363(14):1341–1350. doi: 10.1056/nejmra0912063. [DOI] [PubMed] [Google Scholar]
- 30.Day C. P., James O. F. W. Steatohepatitis: a tale of two ‘Hits’? Gastroenterology. 1998;114(4):842–845. doi: 10.1016/s0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- 31.Tilg H., Moschen A. R. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology. 2010;52(5):1836–1846. doi: 10.1002/hep.24001. [DOI] [PubMed] [Google Scholar]
- 32.Valenti L., Fracanzani A. L., Fargion S. The immunopathogenesis of alcoholic and nonalcoholic steatohepatitis: two triggers for one disease? Seminars in Immunopathology. 2009;31(3):359–369. doi: 10.1007/s00281-009-0152-9. [DOI] [PubMed] [Google Scholar]
- 33.Miele L., Valenza V., la Torre G., et al. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology. 2009;49(6):1877–1887. doi: 10.1002/hep.22848. [DOI] [PubMed] [Google Scholar]
- 34.Bardella M. T., Valenti L., Pagliari C., et al. Searching for coeliac disease in patients with non-alcoholic fatty liver disease. Digestive and Liver Disease. 2004;36(5):333–336. doi: 10.1016/j.dld.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 35.Anstee Q. M., Daly A. K., Day C. P. Genetics of alcoholic and nonalcoholic fatty liver disease. Seminars in Liver Disease. 2011;31(2):128–146. doi: 10.1055/s-0031-1276643. [DOI] [PubMed] [Google Scholar]
- 36.Willner I. R., Waters B., Patil S. R., Reuben A., Morelli J., Riely C. A. Ninety patients with nonalcoholic steatohepatitis: insulin resistance, familial tendency, and severity of disease. The American Journal of Gastroenterology. 2001;96(10):2957–2961. doi: 10.1016/s0002-9270(01)03229-4. [DOI] [PubMed] [Google Scholar]
- 37.Makkonen J., Pietiläinen K. H., Rissanen A., Kaprio J., Yki-Järvinen H. Genetic factors contribute to variation in serum alanine aminotransferase activity independent of obesity and alcohol: a study in monozygotic and dizygotic twins. Journal of Hepatology. 2009;50(5):1035–1042. doi: 10.1016/j.jhep.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 38.Takehara T., Tatsumi T., Suzuki T., et al. Hepatocyte-specific disruption of Bcl-x L leads to continuous hepatocyte apoptosis and liver fibrotic responses. Gastroenterology. 2004;127(4):1189–1197. doi: 10.1053/j.gastro.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 39.Minamiyama Y., Takemura S., Kodai S., et al. Iron restriction improves type 2 diabetes mellitus in Otsuka Long-Evans Tokushima fatty rats. The American Journal of Physiology—Endocrinology and Metabolism. 2010;298(6):E1140–E1149. doi: 10.1152/ajpendo.00620.2009. [DOI] [PubMed] [Google Scholar]
- 40.Kirsch R., Sijtsema H. P., Tlali M., Marais A. D., Hall P. D. L. M. Effects of iron overload in a rat nutritional model of non-alcoholic fatty liver disease. Liver International. 2006;26(10):1258–1267. doi: 10.1111/j.1478-3231.2006.01329.x. [DOI] [PubMed] [Google Scholar]
- 41.Huang Y., He S., Li J. Z., et al. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(17):7892–7897. doi: 10.1073/pnas.1003585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He S., McPhaul C., Li J. Z., et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. The Journal of Biological Chemistry. 2010;285(9):6706–6715. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pingitore P., Pirazzi C., Mancina R. M., et al. Recombinant PNPLA3 protein shows triglyceride hydrolase activity and its I148M mutation results in loss of function. Biochimica et Biophysica Acta (BBA)—Molecular and Cell Biology of Lipids. 2014;1841(4):574–580. doi: 10.1016/j.bbalip.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 44.Pirazzi C., Valenti L., Motta B. M., et al. PNPLA3 has retinyl-palmitate lipase activity in human hepatic stellate cells. Human Molecular Genetics. 2014;23(15):4077–4085. doi: 10.1093/hmg/ddu121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pirazzi C., Adiels M., Burza M. A., et al. Patatin-like phospholipase domain-containing 3 (PNPLA3) I148M (rs738409) affects hepatic VLDL secretion in humans and in vitro. Journal of Hepatology. 2012;57:1257–1262. doi: 10.1016/j.jhep.2012.07.030. [DOI] [PubMed] [Google Scholar]
- 46.Ruhanen H., Perttilä J. D., Hölttä-Vuori M. D., et al. PNPLA3 mediates hepatocyte triacylglycerol remodeling. Journal of Lipid Research. 2014;55(4):739–746. doi: 10.1194/jlr.m046607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan X., Waterworth D., Perry J. R. B., et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. American Journal of Human Genetics. 2008;83(4):520–528. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johansson L. E., Lindblad U., Larsson C. A., Råstam L., Ridderstråle M. Polymorphisms in the adiponutrin gene are associated with increased insulin secretion and obesity. European Journal of Endocrinology. 2008;159(5):577–583. doi: 10.1530/EJE-08-0426. [DOI] [PubMed] [Google Scholar]
- 49.Kotronen A., Johansson L. E., Johansson L. M., et al. A common variant in PNPLA3, which encodes adiponutrin, is associated with liver fat content in humans. Diabetologia. 2009;52(6):1056–1060. doi: 10.1007/s00125-009-1285-z. [DOI] [PubMed] [Google Scholar]
- 50.Kotronen A., Peltonen M., Hakkarainen A., et al. Prediction of non-alcoholic fatty liver disease and liver fat using metabolic and genetic factors. Gastroenterology. 2009;137(3):865–872. doi: 10.1053/j.gastro.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 51.Kollerits B., Coassin S., Beckmann N. D., et al. Genetic evidence for a role of adiponutrin in the metabolism of apolipoprotein B-containing lipoproteins. Human Molecular Genetics. 2009;18(23):4669–4676. doi: 10.1093/hmg/ddp424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sookoian S., Castaño G. O., Burgueño A. L., Gianotti T. F., Rosselli M. S., Pirola C. J. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. The Journal of Lipid Research. 2009;50(10):2111–2116. doi: 10.1194/jlr.p900013-jlr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kantartzis K., Peter A., Machicao F., et al. Dissociation between fatty liver and insulin resistance in humans carrying a variant of the patatin-like phospholipase 3 gene. Diabetes. 2009;58(11):2616–2623. doi: 10.2337/db09-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Romeo S., Sentinelli F., Dash S., et al. Morbid obesity exposes the association between PNPLA3 I148M (rs738409) and indices of hepatic injury in individuals of European descent. International Journal of Obesity. 2010;34(1):190–194. doi: 10.1038/ijo.2009.216. [DOI] [PubMed] [Google Scholar]
- 55.Rotman Y., Koh C., Zmuda J. M., Kleiner D. E., Liang T. J. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology. 2010;52(3):894–903. doi: 10.1002/hep.23759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Speliotes E. K., Butler J. L., Palmer C. D., Voight B. F., Hirschhorn J. N. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 2010;52(3):904–912. doi: 10.1002/hep.23768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valenti L., Al-Serri A., Daly A. K., et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin i148m polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51(4):1209–1217. doi: 10.1002/hep.23622. [DOI] [PubMed] [Google Scholar]
- 58.Sookoian S., Pirola C. J. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53(6):1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 59.Romeo S., Sentinelli F., Cambuli V. M., et al. The 148M allele of the PNPLA3 gene is associated with indices of liver damage early in life. Journal of Hepatology. 2010;53(2):335–338. doi: 10.1016/j.jhep.2010.02.034. [DOI] [PubMed] [Google Scholar]
- 60.Santoro N., Kursawe R., D'Adamo E., et al. A common variant in the patatin-like phospholipase 3 gene (PNPLA3) is associated with fatty liver disease in obese children and adolescents. Hepatology. 2010;52(4):1281–1290. doi: 10.1002/hep.23832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valenti L., Alisi A., Galmozzi E., et al. I148M patatin-like phospholipase domain-containing 3 gene variant and severity of pediatric nonalcoholic fatty liver disease. Hepatology. 2010;52(4):1274–1280. doi: 10.1002/hep.23823. [DOI] [PubMed] [Google Scholar]
- 62.Viitasalo A., Pihlajamaki J., Lindi V., et al. Associations of I148M variant in PNPLA3 gene with plasma ALT levels during 2-year follow-up in normal weight and overweight children: the PANIC study. Pediatric Obesity. 2014 doi: 10.1111/ijpo.234. [DOI] [PubMed] [Google Scholar]
- 63.Romeo S., Sentinelli F., Dash S., et al. Morbid obesity exposes the association between PNPLA3 I148M (rs738409) and indices of hepatic injury in individuals of European descent>. International Journal of Obesity. 2010;34(1):190–194. doi: 10.1038/ijo.2009.216. [DOI] [PubMed] [Google Scholar]
- 64.Miraglia del Giudice E., Grandone A., Cirillo G., et al. The association of PNPLA3 variants with liver enzymes in childhood obesity is driven by the interaction with abdominal fat. PLoS ONE. 2011;6(11) doi: 10.1371/journal.pone.0027933.e27933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Graff M., North K. E., Franceschini N., et al. PNPLA3 gene-by-visceral adipose tissue volume interaction and the pathogenesis of fatty liver disease: the NHLBI Family Heart Study. International Journal of Obesity. 2013;37(3):432–438. doi: 10.1038/ijo.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tian C., Stokowski R. P., Kershenobich D., Ballinger D. G., Hinds D. A. Variant in PNPLA3 is associated with alcoholic liver disease. Nature Genetics. 2010;42(1):21–23. doi: 10.1038/ng.488. [DOI] [PubMed] [Google Scholar]
- 67.Valenti L., Rumi M., Galmozzi E., et al. Patatin-Like phospholipase domain-containing 3 I148M polymorphism, steatosis, and liver damage in chronic hepatitis C. Hepatology. 2011;53(3):791–799. doi: 10.1002/hep.24123. [DOI] [PubMed] [Google Scholar]
- 68.Viganò M., Valenti L., Lampertico P., et al. Patatin-like phospholipase domain-containing 3 I148M affects liver steatosis in patients with chronic hepatitis B. Hepatology. 2013;58(4):1245–1252. doi: 10.1002/hep.26445. [DOI] [PubMed] [Google Scholar]
- 69.Valenti L., Maggioni P., Piperno A., et al. Patatin-like phospholipase domain containing-3 gene I148M polymorphism, steatosis, and liver damage in hereditary hemochromatosis. World Journal of Gastroenterology. 2012;18(22):2813–2820. doi: 10.3748/wjg.v18.i22.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davis J. N., Lê K.-A., Walker R. W., et al. Increased hepatic fat in overweight Hispanic youth influenced by interaction between genetic variation in PNPLA3 and high dietary carbohydrate and sugar consumption. The American Journal of Clinical Nutrition. 2010;92(6):1522–1527. doi: 10.3945/ajcn.2010.30185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nobili V., Liccardo D., Bedogni G., et al. Influence of dietary pattern, physical activity, and I148M PNPLA3 on steatosis severity in at-risk adolescents. Genes and Nutrition. 2014;9(3) doi: 10.1007/s12263-014-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sevastianova K., Santos A., Kotronen A., et al. Effect of short-term carbohydrate overfeeding and long-term weight loss on liver fat in overweight humans. American Journal of Clinical Nutrition. 2012;96(4):727–734. doi: 10.3945/ajcn.112.038695. [DOI] [PubMed] [Google Scholar]
- 73.Santoro N., Savoye M., Kim G., et al. Hepatic fat accumulation is modulated by the interaction between the rs738409 variant in the PNPLA3 gene and the dietary omega6/omega3 PUFA intake. PLoS ONE. 2012;7(5) doi: 10.1371/journal.pone.0037827.e37827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nobili V., Bedogni G., Donati B., Alisi A., Valenti L. The I148M variant of PNPLA3 reduces the response to docosahexaenoic acid in children with non-Alcoholic fatty liver disease. Journal of Medicinal Food. 2013;16(10):957–960. doi: 10.1089/jmf.2013.0043. [DOI] [PubMed] [Google Scholar]
- 75.Trépo E., Pradat P., Potthoff A., et al. Impact of patatin-like phospholipase-3 (rs738409 C>G) polymorphism on fibrosis progression and steatosis in chronic hepatitis C. Hepatology. 2011;54(1):60–69. doi: 10.1002/hep.24350. [DOI] [PubMed] [Google Scholar]
- 76.Valenti L., Aghemo A., Stättermayer A. F., et al. Implications of PNPLA3 polymorphism in chronic hepatitis C patients receiving peginterferon plus ribavirin. Alimentary Pharmacology and Therapeutics. 2012;35(12):1434–1442. doi: 10.1111/j.1365-2036.2012.05109.x. [DOI] [PubMed] [Google Scholar]
- 77.Valenti L., Colombo M., Fargion S. Modulation of the effect of PNPLA3 I148M mutation on steatosis and liver damage by alcohol intake in patients with chronic hepatitis C. Journal of Hepatology. 2011;55(6):1470–1471. doi: 10.1016/j.jhep.2011.04.032. [DOI] [PubMed] [Google Scholar]
- 78.Stickel F., Buch S., Lau K., et al. Genetic variation in the PNPLA3 gene is associated with alcoholic liver injury in caucasians. Hepatology. 2011;53(1):86–95. doi: 10.1002/hep.24017. [DOI] [PubMed] [Google Scholar]
- 79.Falleti E., Fabris C., Cmet S., et al. PNPLA3 rs738409C/G polymorphism in cirrhosis: relationship with the aetiology of liver disease and hepatocellular carcinoma occurrence. Liver International. 2011;31(8):1137–1143. doi: 10.1111/j.1478-3231.2011.02534.x. [DOI] [PubMed] [Google Scholar]
- 80.Trepo E., Guyot E., Ganne-Carrie N., et al. PNPLA3 (rs738409 C>G) is a common risk variant associated with hepatocellular carcinoma in alcoholic cirrhosis. Hepatology. 2012;55(4):1307–1308. doi: 10.1002/hep.25518. [DOI] [PubMed] [Google Scholar]
- 81.Nischalke H. D., Berger C., Luda C., et al. The PNPLA3 rs738409 148M/M genotype is a risk factor for liver cancer in alcoholic cirrhosis but shows no or weak association in hepatitis C cirrhosis. PLoS ONE. 2011;6(11) doi: 10.1371/journal.pone.0027087.e27087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stickel F., Hampe J. Genetic determinants of alcoholic liver disease. Gut. 2012;61(1):150–159. doi: 10.1136/gutjnl-2011-301239. [DOI] [PubMed] [Google Scholar]
- 83.Valenti L., Dongiovanni P., Ginanni Corradini S., Burza M. A., Romeo S. PNPLA3 I148M variant and hepatocellular carcinoma: a common genetic variant for a rare disease. Digestive and Liver Disease. 2013;45(8):619–624. doi: 10.1016/j.dld.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 84.Trépo E., Nahon P., Bontempi G., et al. Association between the PNPLA3 (rs738409 C>G) variant and hepatocellular carcinoma: evidence from a meta-analysis of individual participant data. Hepatology. 2014;59(6):2170–2177. doi: 10.1002/hep.26767. [DOI] [PubMed] [Google Scholar]
- 85.Valenti L., Alisi A., Nobili V. I148M PNPLA3 variant and progressive liver disease: a new paradigm in hepatology. Hepatology. 2012;56(2):1883–1889. doi: 10.1002/hep.25830. [DOI] [PubMed] [Google Scholar]
- 86.Krawczyk M., Portincasa P., Lammert F. PNPLA3-associated steatohepatitis: toward a gene-based classification of fatty liver disease. Seminars in Liver Disease. 2013;33(4):369–379. doi: 10.1055/s-0033-1358525. [DOI] [PubMed] [Google Scholar]
- 87.Holmen O. L., Zhang H., Fan Y., et al. Systematic evaluation of coding variation identifies a candidate causal variant in TM6SF2 influencing total cholesterol and myocardial infarction risk. Nature Genetics. 2014;46(4):345–351. doi: 10.1038/ng.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mahdessian H., Taxiarchis A., Popov S., et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(24):8913–8918. doi: 10.1073/pnas.1323785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chalasani N., Wilson L., Kleiner D. E., Cummings O. W., Brunt E. M., Ünalp A. Relationship of steatosis grade and zonal location to histological features of steatohepatitis in adult patients with non-alcoholic fatty liver disease. Journal of Hepatology. 2008;48(5):829–834. doi: 10.1016/j.jhep.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Speliotes E. K., Yerges-Armstrong L. M., Wu J., et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genetics. 2011;7(3) doi: 10.1371/journal.pgen.1001324.e1001324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gorden A., Yang R., Yerges-Armstrong L. M., et al. Genetic variation at NCAN locus is associated with inflammation and fibrosis in non-alcoholic fatty liver disease in morbid obesity. Human Heredity. 2013;75(1):34–43. doi: 10.1159/000346195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu Y.-L., Reeves H. L., Burt A. D., et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nature Communications. 2014;5, article 4309 doi: 10.1038/ncomms5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cefalu A. B., Pirruccello J. P., Noto D., et al. A novel APOB mutation identified by exome sequencing cosegregates with steatosis, liver cancer, and hypocholesterolemia. Arteriosclerosis, Thrombosis, and Vascular Biology. 2013;33(8):2021–2025. doi: 10.1161/atvbaha.112.301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Di Filippo M., Moulin P., Roy P., et al. Homozygous MTTP and APOB mutations may lead to hepatic steatosis and fibrosis despite metabolic differences in congenital hypocholesterolemia. Journal of Hepatology. 2014;61:891–902. doi: 10.1016/j.jhep.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 95.Marí M., Caballero F., Colell A., et al. Mitochondrial free cholesterol loading sensitizes to TNF- and Fas-mediated steatohepatitis. Cell Metabolism. 2006;4(3):185–198. doi: 10.1016/j.cmet.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 96.Musso G., Gambino R., Cassader M. Cholesterol metabolism and the pathogenesis of non-alcoholic steatohepatitis. Progress in Lipid Research. 2013;52(1):175–191. doi: 10.1016/j.plipres.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 97.Morita T. Heme oxygenase and atherosclerosis. Arteriosclerosis, Thrombosis, and Vascular Biology. 2005;25(9):1786–1795. doi: 10.1161/01.ATV.0000178169.95781.49. [DOI] [PubMed] [Google Scholar]
- 98.Petta S., Miele L., Bugianesi E., et al. Glucokinase regulatory protein gene polymorphism affects liver fibrosis in non-alcoholic fatty liver disease. PLoS ONE. 2014;9(2) doi: 10.1371/journal.pone.0087523.e87523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Beer N. L., Tribble N. D., McCulloch L. J., et al. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Human Molecular Genetics. 2009;18(21):4081–4088. doi: 10.1093/hmg/ddp357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Santoro N., Zhang C. K., Zhao H., et al. Variant in the glucokinase regulatory protein (GCKR) gene is associated with fatty liver in obese children and adolescents. Hepatology. 2012;55(3):781–789. doi: 10.1002/hep.24806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Valenti L., Alisi A., Nobili V. Unraveling the genetics of fatty liver in obese children: additive effect of P446L GCKR and I148M PNPLA3 polymorphisms. Hepatology. 2012;55(3):661–663. doi: 10.1002/hep.25617. [DOI] [PubMed] [Google Scholar]
- 102.Acton R. T., Barton J. C., Passmore L. V., et al. Relationships of serum ferritin, transferrin saturation, and HFE mutations and self-reported diabetes in the hemochromatosis and iron overload screening (HEIRS) study. Diabetes Care. 2006;29(9):2084–2089. doi: 10.2337/dc05-1592. [DOI] [PubMed] [Google Scholar]
- 103.Dongiovanni P., Rametta R., Fracanzani A. L., et al. Lack of association between peroxisome proliferator-activated receptors alpha and gamma2 polymorphisms and progressive liver damage in patients with non-alcoholic fatty liver disease: a case control study. BMC Gastroenterology. 2010;10, article 102 doi: 10.1186/1471-230x-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dongiovanni P., Valenti L. Peroxisome proliferator-activated receptor genetic polymorphisms and nonalcoholic fatty liver disease: any role in disease susceptibility? PPAR Research. 2013;2013:8. doi: 10.1155/2013/452061.452061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Belfort R., Harrison S. A., Brown K., et al. A placebo-controlled trial of pioglitazone in subjects with nonalcoholic steatohepatitis. The New England Journal of Medicine. 2006;355(22):2297–2307. doi: 10.1056/nejmoa060326. [DOI] [PubMed] [Google Scholar]
- 106.Li J. H., Lao X. Q., Tillmann H. L., et al. Interferon-lambda genotype and low serum low-density lipoprotein cholesterol levels in patients with chronic hepatitis C infection. Hepatology. 2010;51(6):1904–1911. doi: 10.1002/hep.23592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tönjes A., Scholz M., Loeffler M., Stumvoll M. Association of Pro12Ala polymorphism in peroxisome proliferator-activated receptor γ with pre-diabetic phenotypes: meta-analysis of 57 studies on nondiabetic individuals. Diabetes Care. 2006;29(11):2489–2497. doi: 10.2337/dc06-0513. [DOI] [PubMed] [Google Scholar]
- 108.Kumari M., Schoiswohl G., Chitraju C., et al. Adiponutrin functions as a nutritionally regulated lysophosphatidic acid acyltransferase. Cell Metabolism. 2012;15(5):691–702. doi: 10.1016/j.cmet.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Olivieri O., Martinelli N., Sandri M., et al. Apolipoprotein C-III, n-3 polyunsaturated fatty acids, and “insulin-resistant” T-455C APOC3 gene polymorphism in heart disease patients: example of gene-diet interaction. Clinical Chemistry. 2005;51(2):360–367. doi: 10.1373/clinchem.2004.040477. [DOI] [PubMed] [Google Scholar]
- 110.Pollin T. I., Damcott C. M., Shen H., et al. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science. 2008;322(5908):1702–1705. doi: 10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gattu S., Becker E. M., Koo J. Y. M. A rare case of non-Hodgkins B-cell lymphoma in a psoriatic patient: a case report and literature review. Journal of Drugs in Dermatology. 2010;9(10):1277–1281. [PubMed] [Google Scholar]
- 112.Hirsch D., Stahl A., Lodish H. F. A family of fatty acid transporters conserved from mycobacterium to man. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(15):8625–8629. doi: 10.1073/pnas.95.15.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Steinberg S. J., Mihalik S. J., Kim D. G., Cuebas D. A., Watkins P. A. The human liver-specific homolog of very long-chain acyl-CoA synthetase is cholate: CoA ligase. The Journal of Biological Chemistry. 2000;275(21):15605–15608. doi: 10.1074/jbc.c000015200. [DOI] [PubMed] [Google Scholar]
- 114.Hubbard B., Doege H., Punreddy S., et al. Mice deleted for fatty acid transport protein 5 have defective bile acid conjugation and are protected from obesity. Gastroenterology. 2006;130(4):1259–1269. doi: 10.1053/j.gastro.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 115.Doege H., Grimm D., Falcon A., et al. Silencing of hepatic fatty acid transporter protein 5 in vivo reverses diet-induced non-alcoholic fatty liver disease and improves hyperglycemia. The Journal of Biological Chemistry. 2008;283(32):22186–22192. doi: 10.1074/jbc.m803510200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Auinger A., Valenti L., Pfeuffer M., et al. A promoter polymorphism in the liver-specific fatty acid transport protein 5 is associated with features of the metabolic syndrome and steatosis. Hormone and Metabolic Research. 2010;42(12):854–859. doi: 10.1055/s-0030-1267186. [DOI] [PubMed] [Google Scholar]
- 117.Fares R., Petta S., Lombardi R., et al. The UCP2 -866 G>A promoter region polymorphism is associated with nonalcoholic steatohepatitis. Liver International. 2014 doi: 10.1111/liv.12707. [DOI] [PubMed] [Google Scholar]
- 118.Feldstein A. E., Werneburg N. W., Canbay A., et al. Free fatty acids promote hepatic lipotoxicity by stimulating TNF-α expression via a lysosomal pathway. Hepatology. 2004;40(1):185–194. doi: 10.1002/hep.20283. [DOI] [PubMed] [Google Scholar]
- 119.Petersen K. F., Dufour S., Hariri A., et al. Apolipoprotein C3 gene variants in nonalcoholic fatty liver disease. New England Journal of Medicine. 2010;362(12):1082–1089. doi: 10.1056/NEJMoa0907295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kozlitina J., Boerwinkle E., Cohen J. C., Hobbs H. H. Dissociation between APOC3 variants, hepatic triglyceride content and insulin resistance. Hepatology. 2011;53(2):467–474. doi: 10.1002/hep.24072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Valenti L., Nobili V., Al-Serri A., et al. The APOC3 T-455C and C-482T promoter region polymorphisms are not associated with the severity of liver damage independently of PNPLA3 I148M genotype in patients with nonalcoholic fatty liver. Journal of Hepatology. 2011;55(6):1409–1414. doi: 10.1016/j.jhep.2011.03.035. [DOI] [PubMed] [Google Scholar]
- 122.Petta S., Grimaudo S., Cammà C., et al. IL28B and PNPLA3 polymorphisms affect histological liver damage in patients with non-alcoholic fatty liver disease. Journal of Hepatology. 2012;56(6):1356–1362. doi: 10.1016/j.jhep.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 123.Salomone F., li Volti G., Rosso C., Grosso G., Bugianesi E. Unconjugated bilirubin, a potent endogenous antioxidant, is decreased in patients with non-alcoholic steatohepatitis and advanced fibrosis. Journal of Gastroenterology and Hepatology. 2013;28(7):1202–1208. doi: 10.1111/jgh.12155. [DOI] [PubMed] [Google Scholar]
- 124.Dongiovanni P., Fracanzani A. L., Fargion S., Valenti L. Iron in fatty liver and in the metabolic syndrome: a promising therapeutic target. Journal of Hepatology. 2011;55(4):920–932. doi: 10.1016/j.jhep.2011.05.008. [DOI] [PubMed] [Google Scholar]