Abstract
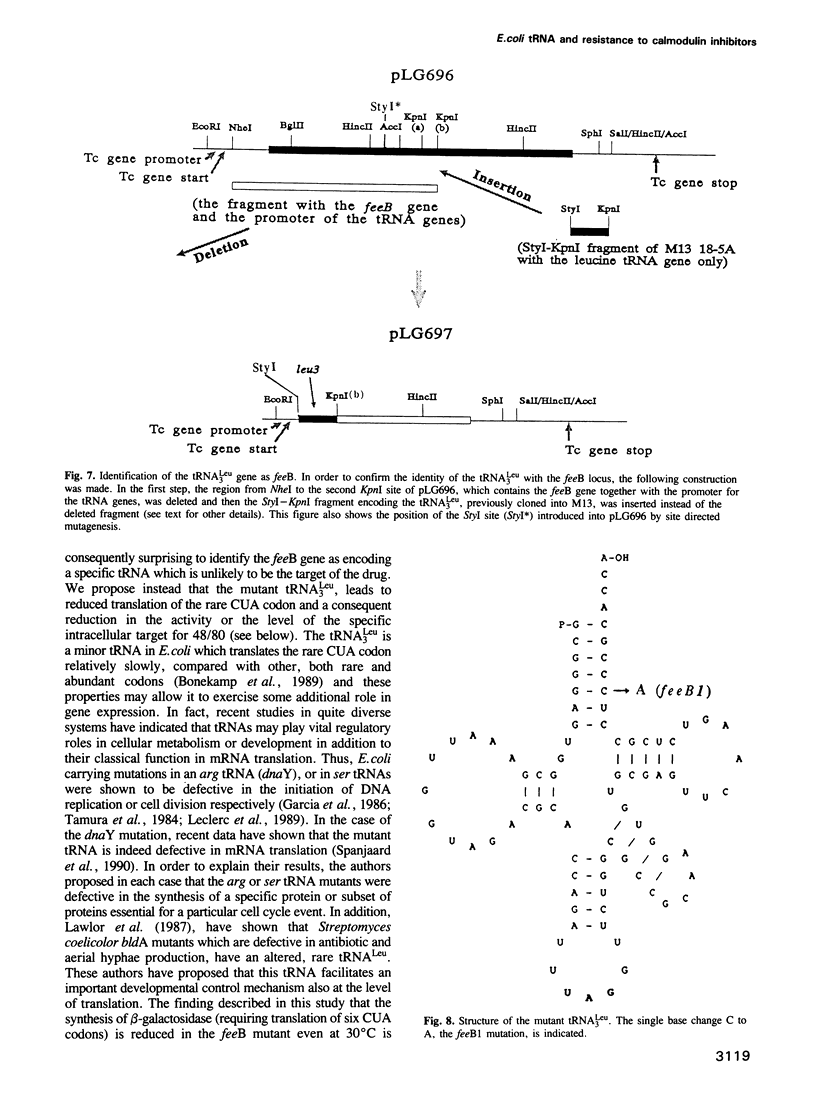
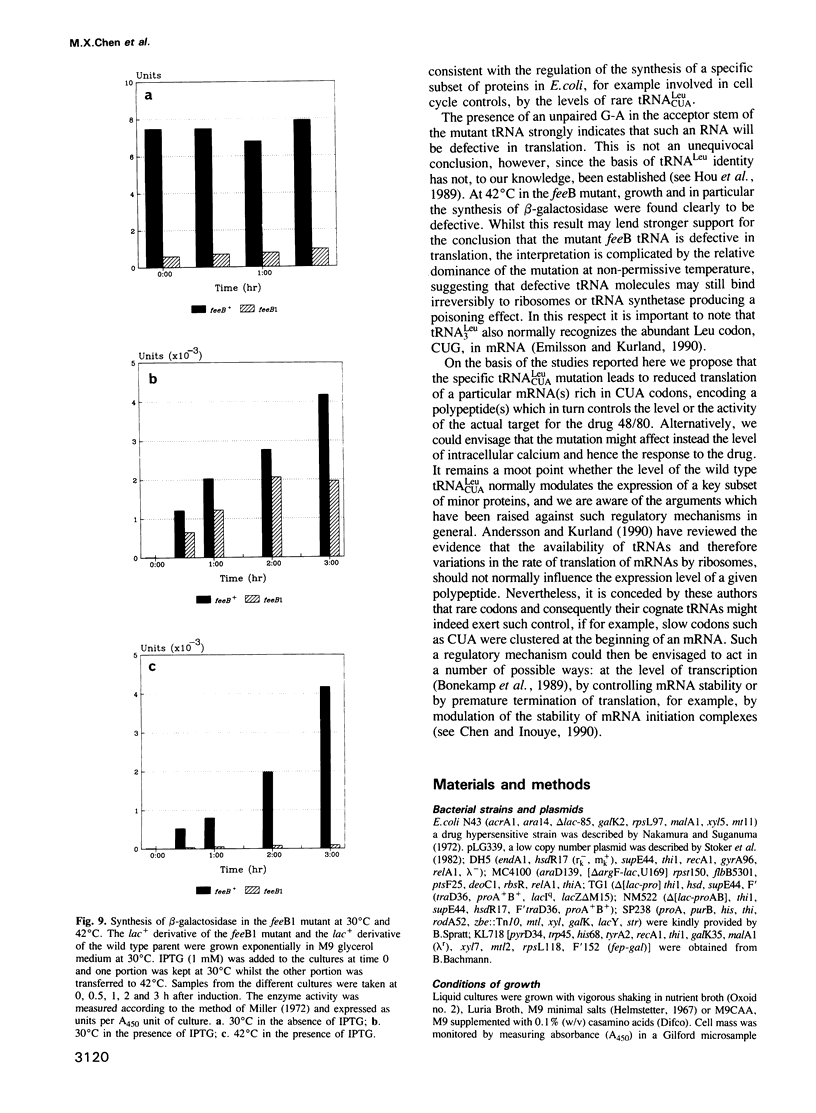
We have isolated several classes of spontaneous mutants resistant to the calmodulin inhibitor 48/80 which inhibits cell division in Escherichia coli K12. Several mutants were also temperature sensitive for growth and this property was exploited to clone a DNA fragment from an E. coli gene library restoring growth at 42 degrees C and drug sensitivity at 30 degrees C in one such mutant. Physical and genetic mapping confirmed that both the mutation and the cloned DNA were located at 15.5 min on the E. coli chromosome at a locus designated feeB. By subcloning, complementation analysis and sequencing, the feeB locus was identified as identical to the tRNA(CUALEU) gene. When the mutant locus was isolated and sequenced, the mutation was confirmed as a single base change, C to A, at position 77 in the acceptor stem of this rare Leu tRNA. In other studies we obtained evidence that this mutant tRNA, recognizing the rare Leu codon, CUA, was defective in translation at both permissive and non-permissive temperatures. The feeB1 mutant is defective in division and shows a reduced growth rate at non-permissive temperature. We discuss the possibility that the mutant tRNA(3Leu) is limiting for the synthesis of a polypeptide(s), requiring several CUA codons for translation which in turn regulates in some way the level or activity of the drug target, a putative cell cycle protein.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adamczyk-Engelmann P., Gietzen K. Induction of histamine release and calmodulin antagonism are two distinct properties of compound 48/80. Cell Calcium. 1989 Feb-Mar;10(2):93–99. doi: 10.1016/0143-4160(89)90049-3. [DOI] [PubMed] [Google Scholar]
- Andersson S. G., Kurland C. G. Codon preferences in free-living microorganisms. Microbiol Rev. 1990 Jun;54(2):198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum P., Furlong C., Byers B. Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5512–5516. doi: 10.1073/pnas.83.15.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonekamp F., Dalbøge H., Christensen T., Jensen K. F. Translation rates of individual codons are not correlated with tRNA abundances or with frequencies of utilization in Escherichia coli. J Bacteriol. 1989 Nov;171(11):5812–5816. doi: 10.1128/jb.171.11.5812-5816.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegielska A., Georgopoulos C. Functional domains of the Escherichia coli dnaK heat shock protein as revealed by mutational analysis. J Biol Chem. 1989 Dec 15;264(35):21122–21130. [PubMed] [Google Scholar]
- Chang C. F., Shuman H., Somlyo A. P. Electron probe analysis, X-ray mapping, and electron energy-loss spectroscopy of calcium, magnesium, and monovalent ions in log-phase and in dividing Escherichia coli B cells. J Bacteriol. 1986 Sep;167(3):935–939. doi: 10.1128/jb.167.3.935-939.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. F., Inouye M. Suppression of the negative effect of minor arginine codons on gene expression; preferential usage of minor codons within the first 25 codons of the Escherichia coli genes. Nucleic Acids Res. 1990 Mar 25;18(6):1465–1473. doi: 10.1093/nar/18.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C. T., Miller R. H. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res. 1988 Apr 25;16(8):3580–3580. doi: 10.1093/nar/16.8.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T. N., Urdea M. S., Masiarz F. R., Thorner J. Isolation of the yeast calmodulin gene: calmodulin is an essential protein. Cell. 1986 Nov 7;47(3):423–431. doi: 10.1016/0092-8674(86)90599-4. [DOI] [PubMed] [Google Scholar]
- Douglas W. W., Nemeth E. F. On the calcium receptor activating exocytosis: inhibitory effects of calmodulin-interacting drugs on rat mast cells. J Physiol. 1982 Feb;323:229–244. doi: 10.1113/jphysiol.1982.sp014070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V., Kurland C. G. Growth rate dependence of transfer RNA abundance in Escherichia coli. EMBO J. 1990 Dec;9(13):4359–4366. doi: 10.1002/j.1460-2075.1990.tb07885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry I. J., Becker-Hapak M., Hageman J. H. Purification and properties of an intracellular calmodulinlike protein from Bacillus subtilis cells. J Bacteriol. 1991 Apr;173(8):2506–2513. doi: 10.1128/jb.173.8.2506-2513.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry I. J., Villa L., Kuehn G. D., Hageman J. H. Calmodulin-like protein from Bacillus subtilis. Biochem Biophys Res Commun. 1986 Jan 14;134(1):212–217. doi: 10.1016/0006-291x(86)90549-8. [DOI] [PubMed] [Google Scholar]
- Gangola P., Rosen B. P. Maintenance of intracellular calcium in Escherichia coli. J Biol Chem. 1987 Sep 15;262(26):12570–12574. [PubMed] [Google Scholar]
- Garcia G. M., Mar P. K., Mullin D. A., Walker J. R., Prather N. E. The E. coli dnaY gene encodes an arginine transfer RNA. Cell. 1986 May 9;45(3):453–459. doi: 10.1016/0092-8674(86)90331-4. [DOI] [PubMed] [Google Scholar]
- Gietzen K. Comparison of the calmodulin antagonists compound 48/80 and calmidazolium. Biochem J. 1983 Dec 15;216(3):611–616. doi: 10.1042/bj2160611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Holland I. B., Casaregola S., Norris V. Cytoskeletal elements and calcium: do they play a role in the Escherichia coli cell cycle? Res Microbiol. 1990 Jan;141(1):131–136. doi: 10.1016/0923-2508(90)90104-x. [DOI] [PubMed] [Google Scholar]
- Hou Y. M., Francklyn C., Schimmel P. Molecular dissection of a transfer RNA and the basis for its identity. Trends Biochem Sci. 1989 Jun;14(6):233–237. doi: 10.1016/0968-0004(89)90033-9. [DOI] [PubMed] [Google Scholar]
- Iida H., Sakaguchi S., Yagawa Y., Anraku Y. Cell cycle control by Ca2+ in Saccharomyces cerevisiae. J Biol Chem. 1990 Dec 5;265(34):21216–21222. [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981 Sep 25;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Inokuchi H., Yamao F., Sakano H., Ozeki H. Identification of transfer RNA suppressors in Escherichia coli. I. Amber suppressor su+2, an anticodon mutant of tRNA2Gln. J Mol Biol. 1979 Aug 25;132(4):649–662. doi: 10.1016/0022-2836(79)90380-2. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kurn N., Sela B. A. Altered calmodulin activity in fluphenazine-resistant mutant strains. Pleiotropic effect on development and cellular organization in Volvox carteri. Eur J Biochem. 1981 Dec;121(1):53–57. doi: 10.1111/j.1432-1033.1981.tb06428.x. [DOI] [PubMed] [Google Scholar]
- Lawlor E. J., Baylis H. A., Chater K. F. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev. 1987 Dec;1(10):1305–1310. doi: 10.1101/gad.1.10.1305. [DOI] [PubMed] [Google Scholar]
- Leclerc G., Sirard C., Drapeau G. R. The Escherichia coli cell division mutation ftsM1 is in serU. J Bacteriol. 1989 Apr;171(4):2090–2095. doi: 10.1128/jb.171.4.2090-2095.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means A. R. Molecular mechanisms of action of calmodulin. Recent Prog Horm Res. 1988;44:223–262. doi: 10.1016/b978-0-12-571144-9.50012-0. [DOI] [PubMed] [Google Scholar]
- Molnár J., Holland I. B., Mándi Y. Selection of ion mutants in Escherichia coli by treatment with phenothiazines. Genet Res. 1977 Aug;30(1):13–20. doi: 10.1017/s0016672300017420. [DOI] [PubMed] [Google Scholar]
- Mándi T. Y., molnár J., Holland I. B., Béládi I. Efficient curing of an Escherichia coli F-prime plasmid by phenothiazines. Genet Res. 1975 Aug;26(1):109–111. doi: 10.1017/s0016672300015895. [DOI] [PubMed] [Google Scholar]
- Nakajima N., Ozeki H., Shimura Y. In vitro transcription of the supB-E tRNA operon of Escherichia coli. Characterization of transcription products. J Biol Chem. 1982 Sep 25;257(18):11113–11120. [PubMed] [Google Scholar]
- Nakamura H., Suganuma A. Membrane mutation associated with sensitivity to acriflavine in Escherichia coli. J Bacteriol. 1972 Apr;110(1):329–335. doi: 10.1128/jb.110.1.329-335.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H., Jaffé A., Imamura R., Ogura T., Hiraga S. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 1991 Jan;10(1):183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris V., Seror S. J., Casaregola S., Holland I. B. A single calcium flux triggers chromosome replication, segregation and septation in bacteria: a model. J Theor Biol. 1988 Oct 7;134(3):341–350. doi: 10.1016/s0022-5193(88)80065-1. [DOI] [PubMed] [Google Scholar]
- Ohya Y., Miyamoto S., Ohsumi Y., Anraku Y. Calcium-sensitive cls4 mutant of Saccharomyces cerevisiae with a defect in bud formation. J Bacteriol. 1986 Jan;165(1):28–33. doi: 10.1128/jb.165.1.28-33.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen C. D., Means A. R. Calmodulin is involved in regulation of cell proliferation. EMBO J. 1987 Dec 20;6(13):3961–3968. doi: 10.1002/j.1460-2075.1987.tb02738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen C. D., Means A. R. Calmodulin is required for cell-cycle progression during G1 and mitosis. EMBO J. 1989 Jan;8(1):73–82. doi: 10.1002/j.1460-2075.1989.tb03350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra-Molina A., Villalobos R., Borbolla M. Calcium uptake during the cell cycle of Saccharomyces cerevisiae. FEBS Lett. 1983 Aug 22;160(1-2):195–197. doi: 10.1016/0014-5793(83)80965-x. [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr, Wu L. F., Reizer J. Regulation of bacterial physiological processes by three types of protein phosphorylating systems. Trends Biochem Sci. 1990 Oct;15(10):391–395. doi: 10.1016/0968-0004(90)90238-7. [DOI] [PubMed] [Google Scholar]
- Sakakibara Y. The dnaK gene of Escherichia coli functions in initiation of chromosome replication. J Bacteriol. 1988 Feb;170(2):972–979. doi: 10.1128/jb.170.2.972-979.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanjaard R. A., Chen K., Walker J. R., van Duin J. Frameshift suppression at tandem AGA and AGG codons by cloned tRNA genes: assigning a codon to argU tRNA and T4 tRNA(Arg). Nucleic Acids Res. 1990 Sep 11;18(17):5031–5036. doi: 10.1093/nar/18.17.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speaker M. G., Orlow S. J., Sturgill T. W., Rosen O. M. Characterization of a calmodulin-binding protein that is deficient in trifluoperazine-resistant variants of the macrophage-like cell line J774. Proc Natl Acad Sci U S A. 1983 Jan;80(2):329–333. doi: 10.1073/pnas.80.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoker N. G., Fairweather N. F., Spratt B. G. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene. 1982 Jun;18(3):335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]
- Swan D. G., Hale R. S., Dhillon N., Leadlay P. F. A bacterial calcium-binding protein homologous to calmodulin. Nature. 1987 Sep 3;329(6134):84–85. doi: 10.1038/329084a0. [DOI] [PubMed] [Google Scholar]
- Takeda T., Yamamoto M. Analysis and in vivo disruption of the gene coding for calmodulin in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3580–3584. doi: 10.1073/pnas.84.11.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura F., Nishimura S., Ohki M. The E. coli divE mutation, which differentially inhibits synthesis of certain proteins, is in tRNASer1. EMBO J. 1984 May;3(5):1103–1107. doi: 10.1002/j.1460-2075.1984.tb01936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- Whitaker M., Patel R. Calcium and cell cycle control. Development. 1990 Apr;108(4):525–542. doi: 10.1242/dev.108.4.525. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]