Abstract
Breast cancer is a hormone-dependent cancer and usually treated with endocrine therapy using aromatase inhibitors or anti-estrogens such as tamoxifen. A majority of breast cancer, however, will often fail to respond to endocrine therapy. In the present study, we explored miRNAs associated with endocrine therapy resistance in breast cancer. High-throughput miRNA sequencing was performed using RNAs prepared from breast cancer MCF-7 cells and their derivative clones as endocrine therapy resistant cell models, including tamoxifen-resistant (TamR) and long-term estrogen-deprived (LTED) MCF-7 cells. Notably, miR-21 was the most abundantly expressed miRNA in MCF-7 cells and overexpressed in TamR and LTED cells. We found that miR-378a-3p expression was downregulated in TamR and LTED cells as well as in clinical breast cancer tissues. Additionally, lower expression levels of miR-378a-3p were associated with poor prognosis for tamoxifen-treated patients with breast cancer. GOLT1A was selected as one of the miR-378a-3p candidate target genes by in silico analysis. GOLT1A was overexpressed in breast cancer specimens and GOLT1A-specific siRNAs inhibited the growth of TamR cells. Low GOLT1A levels were correlated with better survival in patients with breast cancer. These results suggest that miR-378a-3p-dependent GOLT1A expression contributes to the mechanisms underlying breast cancer endocrine resistance.
Estrogen is an important endocrine hormone that regulates the growth and differentiation of the normal mammary gland1,2. The hormone also plays a critical role in the development and progression of breast cancer, which is one of the most common cancers among women1,2. The estrogen receptor α (ERα) is a member of the nuclear receptor superfamily that functions as transcription factors3,4,5. ERα mediates various functions of estrogen in its normal and malignant target tissues, including breast cancer3,4,5. Determination of ERα status is clinically used as a prognostic and predictive factor in the management of breast cancer. Approximately 85% of breast cancers are ERα positive and can be treated with endocrine therapy using anti-estrogens such as tamoxifen or aromatase inhibitors6,7,8. Tamoxifen has been used for years for adjuvant treatment, which significantly reduces the risk of recurrence of breast cancer9. Despite the obvious benefits of tamoxifen, approximately 40% of patients with early-stage breast cancer treated with tamoxifen as adjuvant therapy would eventually suffer from the relapse with tamoxifen-resistant disease10. Thus, studies have been performed to elucidate the molecular mechanisms underlying endocrine resistance; however, only several potential targets and signaling pathways have been revealed11,12,13. Therfore, a better understanding of the tamoxifen-resistant mechanism may provide novel strategies to overcome tamoxifen resistance in breast cancer.
MicroRNAs (miRNAs) are small noncoding RNAs consisting of an average of 22 nucleotides. miRNAs can function as post-transcriptional regulators by binding to 3′-untranslated regions (3′-UTRs) of their target mRNAs in sequences that have imperfect or perfect complementarity, repressing the translation or degradation of their target mRNAs14,15. Nowadays, particular attention has been paid to the deregulation of miRNAs in tumor progression and metastasis as one of the new transcriptional regulators involved in cancer biology16,17,18. While the molecular mechanisms underlying tamoxifen resistance in terms of its key regulators and signaling events remain to be elucidated, miRNAs could be novel therapeutic targets for endocrine therapy resistant cancers.
Several studies have recently reported the role of miRNAs in tamoxifen resistance. These studies provide a list of miRNAs potentially involved in tamoxifen resistance, including miR-87319, miRNAs-221/22220,21,22, miR-519a23, miR-126 and miR-10a24, miRNA-200b and miR-200c25, miR-146a, -27a, -145, -21, -155, -15a, -125b, and let-7s26,27, miR-37528, miR-45129, miR-34230, and miR-574-3p31. For example, miR-873 is downregulated in tamoxifen-resistant MCF-7 cells and in breast cancer tissues compared with normal tissue. miR-873 decreases ER transcriptional activity through the modulation of ERα phosphorylation and inhibits the proliferation of breast cancer cells via targeting cyclin-dependent kinase 3 (CDK3)19. Loss of miR-375 expression is reported in tamoxifen-resistant breast cancer cells derived from long-term passage of MCF-7 cells with tamoxifen28. Re-expression of miR-375 sensitized tamoxifen-resistant breast cancer cells to tamoxifen and partly reversed the epithelial-mesenchymal transition (EMT)-like properties. This report showed that MTDH, which encodes metadherin, is a direct target of miR-37528. miR-574-3p has been identified as a tamoxifen response-related miRNAs in breast cancer cells by miRNA library-based functional screening31. miR-574-3p potentially targets clathrin heavy chain (CLTC) whose expression is associated with tamoxifen sensitivity in tamoxifen-resistant breast cancer cells.
In the present study, we performed high-throughput sequencing of miRNAs in human breast cancer MCF-7 cells and its derivative tamoxifen-resistant TamR cells and long-term estrogen-deprived (LTED) cells. Differentially expressed miRNAs were identified by comparing miRNA expression profiles among those cells. miR-21 was determined as an upregulated miRNA in TamR and LTED cells, and let-7s (let-7a and f) and miR-378a-3p were identified as downregulated ones. We further studied the expression of miR-378a-3p in clinical breast cancer specimens and found that the miRNA was downregulated in cancer tissues compared to adjacent normal tissues. Additionally, lower miR-378a-3p expression levels were associated with poor prognosis for tamoxifen-treated patients with breast cancer. By in silico analysis and luciferase reporter assay for miRNA binding sites, golgi transport 1A (GOLT1A) was identified as a candidate miR-378a-3p target. Loss-of-function experiments for GOLT1A in TamR cells resulted in a decrease of tamoxifen-resistant cell growth. Low GOLT1A levels were also shown to be correlated with better survival in patients with breast cancer. These results show that miR-378a-3p and its target GOLT1A can provide new insights into the signaling pathways associated with tamoxifen resistance in breast cancer and could be applied to the development of alternative diagnostic and therapeutic options for advanced breast cancer.
Results
Identification of miRNAs differentially expressed in breast cancer cells and endocrine therapy-resistant cells using high throughput sequencing
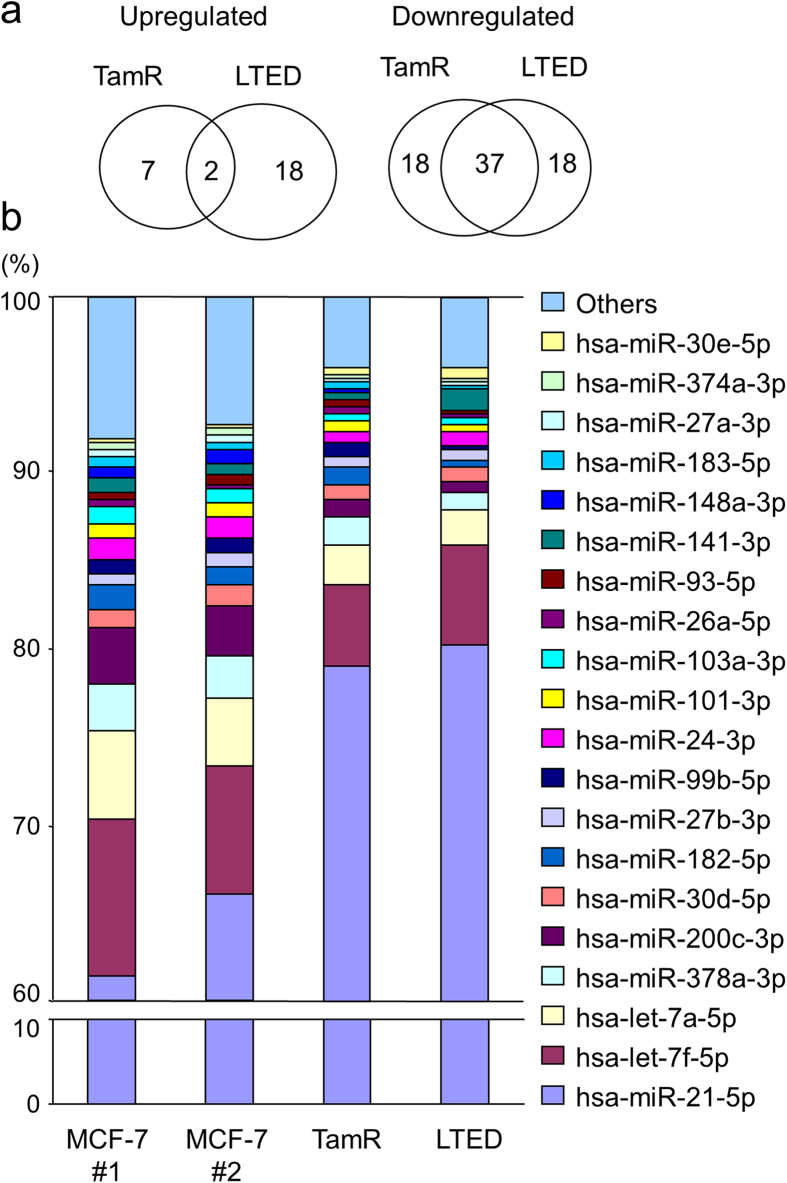
To identify novel miRNAs associated with endocrine therapy-resistance of breast cancer cells, tamoxifen-resistant (TamR) cells32 were obtained from MCF-7 cells by long-term (>3 months) culture with 1 μM tamoxifen and long-term estrogen-deprived (LTED) cells33 were established from MCF-7 cells by culturing them in phenol red-free Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% dextran-charcoal stripped fetal bovine serum (dcc FBS). High-throughput miRNA sequencing was performed using RNAs prepared from parental MCF-7, TamR and LTED cells. Mapping of small RNA reads on the human genome (NCBI35 assembly) and prediction of novel miRNAs were performed as described previously34. An average of 4.6 million reads per sample was mapped on miRNAs. To compare miRNA expression profiles of these cells, a read count for each miRNA was shown as a frequency against the total mapped read count per sample. miRNAs exhibiting >10 reads were selected and resulted in a list of 94 annotated miRNAs in MCF-7, TamR, and LTED cells (Supplementary Table 1). No confident novel miRNA was predicted from the data. Among these annotated miRNAs, 9 and 20 miRNAs were upregulated (>1.2-fold) in TamR and LTED cells, respectively, compared with parental MCF-7 cells. Fifty five miRNAs were downregulated (<0.7-fold) in both TamR and LTED cells compared with their parental cells. Two and 37 miRNAs were commonly upregulated and downregulated, respectively, in TamR and LTED cells compared with their parental cells (Fig. 1a). We found that the percentages of miR-21 counts were approximately two-third of the total counts for 94 miRNAs in MCF-7 cells and >75% of the total counts in TamR and LTED cells (Fig. 1b). miR-21 is a known onco-miRNA in cancer, and its overexpression is shown to be associated with advanced clinical stage, lymph node metastasis, and poor prognosis in cancer patients, including breast cancer patients35,36,37,38,39. It has been also shown that miR-21 regulates the expression of target genes, including phosphatase and tensin homolog (PTEN), programmed cell death 4 (PDCD4), and tropomyosin 1 (TPM1) in breast cancer cells40,41,42.
Figure 1. miRNA expression profiles of breast cancer MCF-7 and its derivative endocrine therapy resistant cells.
(a) Differentially expressed miRNAs in tamoxifen-resistant breast cancer cells (TamR) and long-term estrogen-deprived breast cancer cells (LTED) compared to their parental MCF-7 cells. TamR and LTED were established from MCF-7 cells (see Materials and Methods). Small RNA libraries were constructed from MCF-7, TamR, and LTED cells and sequenced using the Illumina GAIIx. (b) Percentages of top 20 miRNAs abundantly expressed in MCF-7 and endocrine therapy resistant cells. Percentages in distinctive cell lines were determined by a read count for each indicated miRNA normalized to the total count of 94 selected miRNAs (>10 reads) listed in Supplementary Table S1.
Let-7f and let-7a, members of let-7 family, were also identified as abundantly and differentially expressed miRNAs among these cells. The expression levels of both let-7f and let-7a were decreased in TamR and LTED cells compared with those in MCF-7 cells. Let-7 family miRNAs have been generally considered as tumor suppressor miRNAs and the expressions of let-7 family members were reported to be downregulated in many cancer types including breast cancer and at times during tumor progression43,44. Let-7a and let-7f expression was inversely correlated with that of LIN28, which inhibits the biogenesis of the let-7 family and is shown to contribute to breast tumorigenesis45,46.
miR-378a-3p was another abundantly and differentially expressed miRNAs among these cells, as its expression in TamR and LTED cells was downregulated compared with that in MCF-7 cells. miR-378a-3p may contribute to the pathophysiology of cancers, which might be suggested by some studies in colorectal cancer and rhabdomyosarcoma47,48. We further studied miR-378a-3p because its functional role has not been yet elucidated in breast cancer.
Knockdown of miR-378a-3p reverses tamoxifen-dependent suppression of cell growth in MCF-7 cells
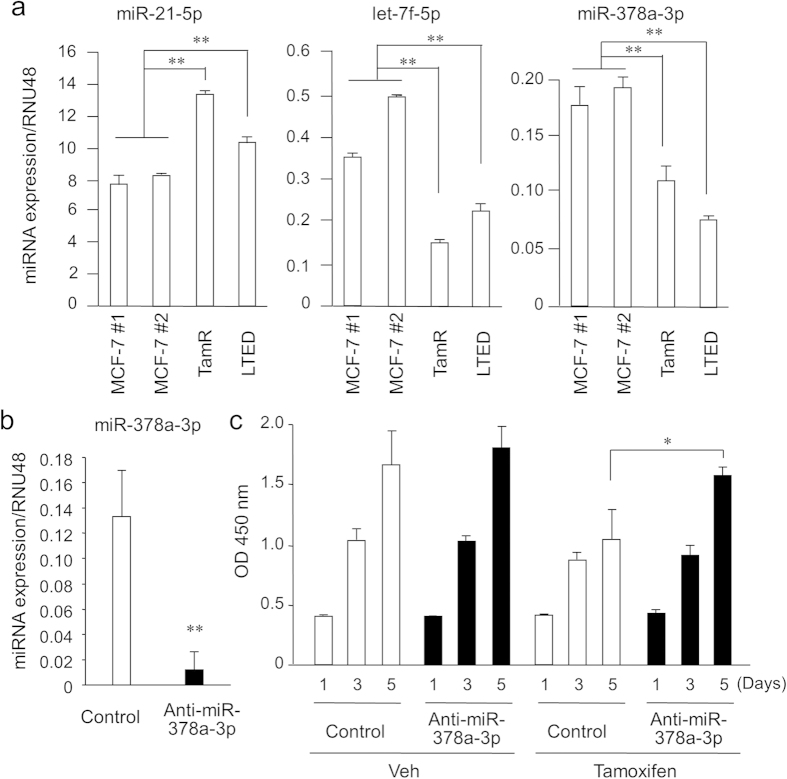
To validate the endogenous expression levels of miR-21-5p, let-7f-5p, and miR-378a-3p, we performed quantitative polymerase chain reaction (qPCR) using cDNAs reversely transcribed from the RNAs prepared from MCF-7, TamR, and LTED cells (Fig. 2a). miR-21 expression levels were significantly upregulated, whereas those of let-7f and miR-378a-3p were significantly downregulated in TamR, and LTED cells compared with MCF-7 cells. To further assess the functional role of miR-378a-3p in the proliferation of breast cancer cells, we performed loss-of-function studies using the miRNA inhibitor, anti-miR-378a-3p. The expression of miR-378a-3p was significantly reduced in MCF-7 cells transfected with anti-miR-378a-3p (Fig. 2b). A cell viability assay showed that tamoxifen treatment significantly repressed the growth of MCF-7 cells transfected with a control miRNA inhibitor. When the cells were transfected with anti-miR-378a-3p, however, tamoxifen-mediated suppression of cell growth was impaired in MCF-7 cells (Fig. 2c). On the other hand, overexpression of miR-378a-3p reduced tamoxifen-resistant growth of TamR cells (Supplementary Fig. S1). These results indicate that miR-378a-3p downregulation will contribute to tamoxifen resistance in breast cancer cells. To assess the downregulation mechanism of miR-378a-3p in TamR cells, we investigated the effect of 5-aza-2′-deoxycytidine (5Aza-dC), an inhibitor of DNA methylation, on the expression of miR-378a-3p (Supplementary Fig. S2). We observed that the expression of miR-378a-3p was increased by the treatment with 5-Aza-dC. Notably, in parental MCF-7 cells, 5Aza-dC was not influential on the expression levels of miR-378a-3p, suggesting that the epigenetic regulation of miR-378a-3p expression would be accompanied with acquired tamoxifen resistance (Supplementary Fig. S3).
Figure 2. miR-21, let-7f, and miR-378a-3p expression determined by qPCR and effect of miR-378a-3p knockdown on MCF-7 cell growth in the presence of tamoxifen.
(a) miR-21, let-7f, and miR-378a-3p expression levels in MCF-7, TamR, and LTED cells were determined by qPCR and normalized to RNU48 levels. Data are presented as mean ± SD. Statistical analysis was performed using the Mann-Whitney U test. **P < 0.01. (b) Knockdown efficiency of anti-miR-378a-3p. MCF-7 cells were transfected with anti-miR-378a-3p or negative control for 48 h. miR-378a-3p levels were determined by qPCR and normalized to RNU48 levels. Data are presented as mean ± SD in triplicates; **P < 0.01. (c) Knockdown of miR-378a-3p significantly increased MCF-7 cell growth in the presence of tamoxifen. Cells were transfected with anti-miR-378a-3p or negative control for 12 h, and then treated with 1 μM tamoxifen or vehicle. Cell viability was analyzed using the WST-8 cell proliferation assay at 1, 3, and 5 days after transfection. Data are presented as mean ± SD, in triplicate; *P < 0.05.
Decreased miR-378a-3p expression in clinical breast cancer tissues is associated with poor prognosis
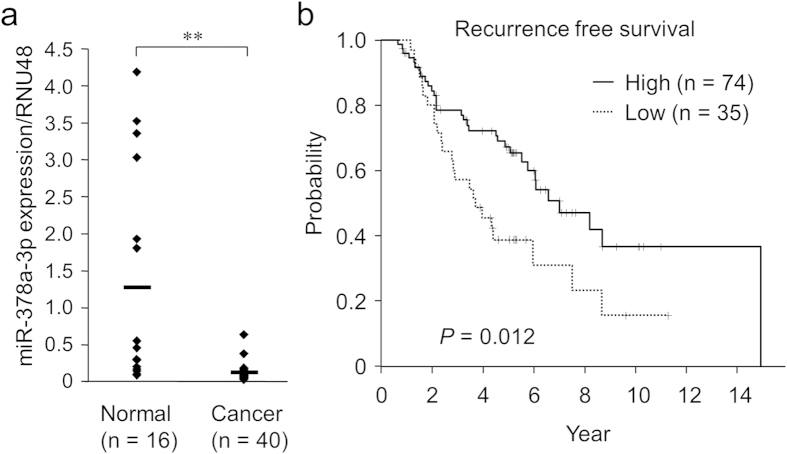
miR-378a-3p expression levels were examined in clinical breast cancer samples and adjacent normal tissues (Fig. 3a). miR-378a-3p expression was significantly lower in breast cancer tissues than in adjacent normal tissues. Moreover, we assessed the association between miR-378a-3p levels and clinical outcomes using a breast cancer cohort with previously reported miRNA microarray data deposited in the Gene Expression Omnibus (GEO) database (GSE37405)(Fig. 3b). This dataset contains the miRNA expression data of 152 ER-positive primary breast cancers from high-risk patients. All patients had received adjuvant tamoxifen therapy as a monotherapy (median clinical follow-up: 4.6 years) and the half of the patients had developed distant recurrence (median time-to-recurrence: 3.5 years)49. Using the data of 109 patients out of 152 patients for whom miR-378a-3p expression data were available, Kaplan-Meier analysis was performed to evaluate survival (time to recurrence) for groups with high and low expression of miR-378a-3p. Kaplan-Meier survival analysis indicates that patients with low miR-378a-3p levels presented poor recurrence-free survival (log-rank, P = 0.012) (Fig. 3b). The results indicate that miR-378a-3p downregulation could be correlated with the development and progression of tamoxifen resistance in breast cancer.
Figure 3. Reduced expression of miR-378a-3p in clinical breast cancer tissues is associated with poor prognosis for tamoxifen-treated patients.
(a) Decreased miR-378a-3p expression levels in breast cancer tissues compared with those in paired adjacent normal tissues. miR-378a-3p expression levels were determined by qPCR and normalized to RNU48 levels. **P < 0.01. (b) Kaplan-Meier survival analysis according to miR-378a-3p levels in tamoxifen-treated breast cancer patients. The recurrence free survival of miR-378a-3p high-expressing group (n = 74) was significantly lower than that of the low-expressing group (n = 35; log-rank test; P = 0.012).
Identification of candidate target genes for miR-378a-3p
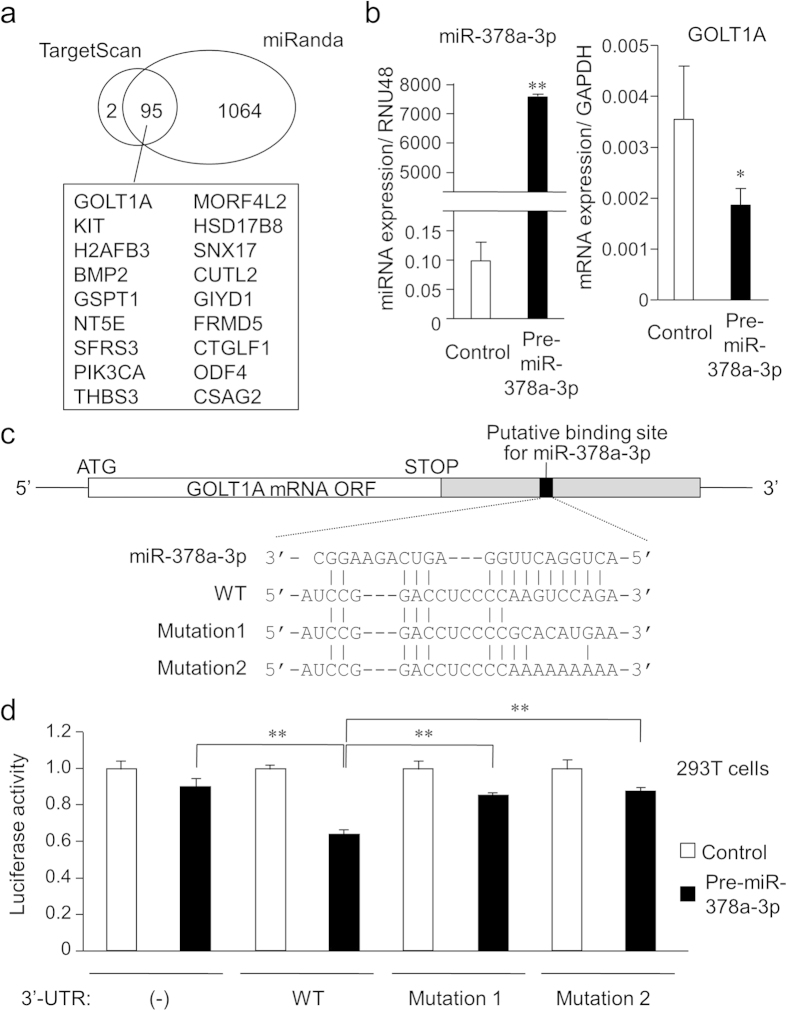
To identify miR-378a-3p candidate targets, we used 2 target gene prediction programs, TargetScan50 and miRanda51 (Fig. 4a). TargetScan and miRanda identified 97 and 1,159 candidate targets for miR-378a-3p, respectively. Notably, 95 genes were common between these programs. Of these candidates, we focused on GOLT1A because it presented one of the highest precision score. To examine whether miR-378a-3p directly regulates GOLT1A expression, we transfected MCF-7 cells with pre-miR-378a-3p for 48 h and then evaluated GOLT1A mRNA expression by quantitative reverse transcription polymerase chain reaction (qRT-PCR). Transfection with pre-miR-378a-3p significantly decreased GOLT1A mRNA expression level in MCF-7 cells compared to that of the control pre-miR (Fig. 4b). We next constructed luciferase reporter vectors containing either the wild-type GOLT1A 3′-UTR sequence with a putative binding site for miR-378a-3p or the altered sequences of the 3′-UTR in which the putative binding site was mutated (mutations 1 and 2) (Fig. 4c). In 293T cells, the luciferase assay demonstrated that miR-378a-3p decreased the activity of luciferase reporter with the wild-type sequence, but not that of the reporters with the mutated sequences (Fig. 4d). This result suggests that miR-378a-3p regulates GOLT1A expression by binding to the 3′-UTR of GOLT1A.
Figure 4.
Identification of miR-378a-3p target genes in breast cancer (a) Schematic presentation of miR-378a-3p target prediction by in silico analyses. Venn diagrams indicating numbers of candidate hits determined by the TargetScan and miRanda prediction algorithms. Candidate genes commonly predicted by the algorithms are described. (b) Overexpression of miR-378a-3p suppressed GOLT1A mRNA expression. MCF-7 cells were transfected with pre-miR-378a-3p or control miR for 48 h and the expression levels of miR-378a-3p and GOLT1A mRNA were then evaluated by qPCR and qRT-PCR, respectively. Data are presented as mean ± SD; *P < 0.05; **P < 0.01. (c) Location of putative miR-378a-3p-binding sequence in the 3′-UTR of GOLT1A gene and its mutated sequences in luciferase reporters. (d) Luciferase reporter assay using vectors containing a putative GOLT1A 3′-UTR binding site for miR-378a-3p and mutated versions of the site. MCF-7 cells were transiently transfected with psiCHECK2 vectors containing either the wild-type or mutated putative binding sites for miR-574-3p, together with pre-miR-378a-3p precursor or control pre-miR for 48 h. The luciferase assay was then performed. Data are presented as mean ± SD; **P < 0.01.
GOLT1A knockdown restores tamoxifen sensitivity and low GOLT1A levels are associated with better survival in patients with breast cancer
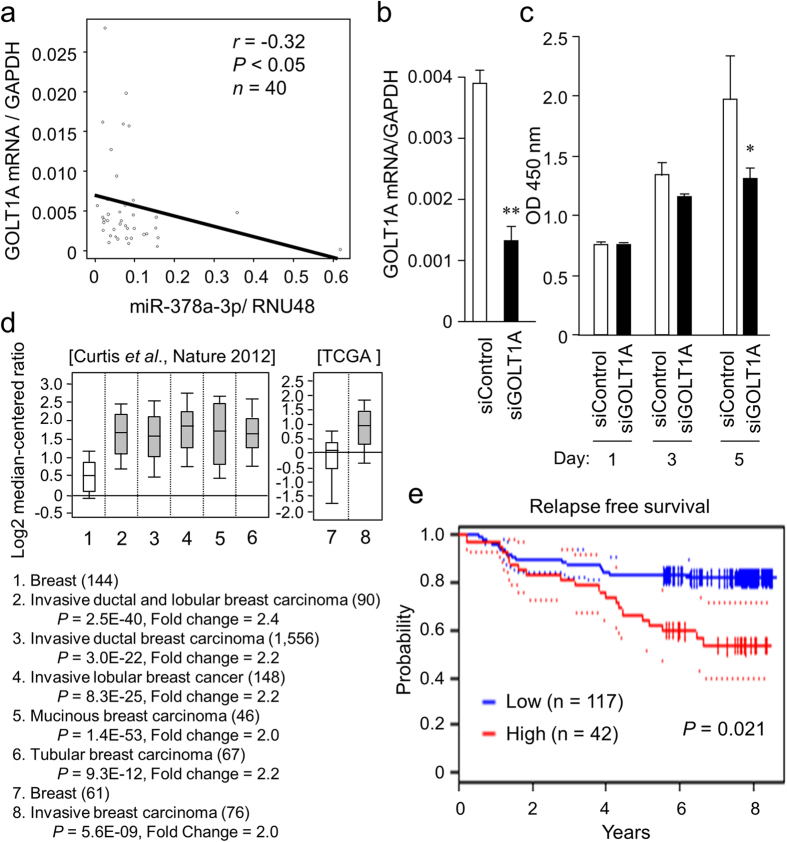
Since GOLT1A was shown as a candidate miR-378a-3p target in breast cancer cells, we investigated whether a negative correlation between miR-378a-3p and GOLT1A expression can be observed in clinical breast cancer specimens (Fig. 5a). A negative correlation was shown between the expression levels of miR-378a-3p and GOLT1A (r = −0.32, P < 0.05). To examine a functional role of GOLT1A in the tamoxifen response, we performed a loss-of-function study in TamR cells using GOLT1A siRNA. We found that GOLT1A mRNA expression was markedly repressed in TamR cells transfected with GOLT1A siRNA (Fig. 5b). Cell proliferation assay showed a significant growth inhibition of cells transfected with GOLT1A siRNA as compared to the control transfectants (Fig. 5c). It was confirmed by using another siRNA (siGOLT1A-2) that the growth inhibitory effect of GOLT1A silencing was observed markedly when TamR cells were treated with tamoxifen (Supplementary Fig. S4). To further examine whether the tumor-suppressive effect of miR-378a-3p is mediated by GOLT1A, MCF-7 cells were transfected with the combinations of anti-miR-378a-3p and siGOLT1A (Supplementary Fig. S5). Anti-miR-378a-3p transfection upregulated the MCF-7 cell growth in the presence of tamoxifen compared with anti-miR negative control transfection; however, the tamoxifen-resistant growth induced by anti-miR-378a-3p was abrogated by siGOLT1A. We then examined the Oncomine microarray database to determine whether GOLT1A expression was altered in other cohorts of clinical breast cancer samples52. Two datasets indicated that GOLT1A was substantially overexpressed (by > 2-fold) in clinical breast cancers compared to normal breast tissues (P < 1e-8, Fig. 5d). Of note, we found that low GOLT1A expression was associated with good prognosis for breast cancer in a breast cancer microarray dataset (Fig. 5e). These results suggest that GOLT1A plays a role in malignant alteration of breast cancer, including the acquisition of tamoxifen resistance.
Figure 5. GOLT1A knockdown restored tamoxifen resistance and low GOLT1A levels correlate with better survival in patients with breast cancer.
(a) Negative correlation between miR-378a-3p and GOLT1A expression levels in breast cancer tissues. The expression levels of miR-378a-3p and GOLT1A mRNA were examined in cancer samples used in Fig. 3a. (b) Knockdown efficiency of GOLT1A siRNA. TamR cells were transfected with siGOLT1A or negative control for 48 h. GOLT1A mRNA levels were determined by qRT-PCR and normalized to GAPDH levels. Data are presented as mean ± SD, in triplicate; **P < 0.01. (c) GOLT1A knockdown significantly reduced TamR cell growth in the presence of tamoxifen. Cells were transfected with siGOLT1A or negative control. Cell viability was then analyzed by WST-8 cell proliferation assay at 1, 3, and 5 days after transfection. Data are presented as mean ± SD, in triplicate; *P < 0.05. (d) GOLT1A mRNA was overexpressed in clinical breast cancer tissues compared to normal mammary tissues, based on the Oncomine cancer profiling database, by >2-fold at P < 1e-8. (e) GOLT1A expression correlates with poor relapse-free survival in patients with breast cancer. Survival curves for high (n = 42) and low (n = 117) expression groups dichotomized at the optimal cut point are plotted using the PrognoScan64. The 95% confidence intervals for each group are also indicated by dotted lines.
Discussion
In the present study, we identified miRNAs that are differentially expressed in endocrine therapy resistant breast cancer cells (TamR and LTED) compared to parental MCF-7 cells. Among them, miR-21 and let-7 (let-7f and let-7a) that are generally known as an onco-miRNA and an anti-onco-miRNA, respectively, were the top three differentially expressed miRNAs in the endocrine therapy resistant breast cancer cells. miR-21 and let-7s (let-7f and let-7a) expression levels are increased and decreased, respectively, in the endocrine therapy resistant breast cancer cells, suggesting their roles that will contribute to aggressive type of breast cancer, including the acquisition of tamoxifen resistance35,43.
As the fourth most differentially expressed miRNA in the endocrine therapy resistant breast cancer cells, we focused on miR-378a-3p whose expression levels were decreased in these cells. miR-378a-3p knockdown facilitated MCF-7 cell growth in the presence of tamoxifen. In addition, miR-378a-3p expression was reduced in clinical breast cancer tissues compared with cancer-surrounding normal tissues and low expression of miR-378a-3p was associated with poor prognosis in ER-positive breast cancer patients treated with adjuvant tamoxifen. In a clinicopathological study of colorectal cancer, miR-378a-3p expression is decreased in cancer tissues compared to that in adjacent normal colorectal tissues47. miR-378a-3p expression was correlated with pathological parameters, including histological differentiation and TNM stage, and low miR-378a-3p expression was significantly associated with shorter survival time. Ectopic miR-378a-3p expression could inhibit colorectal cancer cell growth and colony formation and induce apoptosis and cell cycle arrest. Loss- and gain-of-function experiments of miR-378a-3p in colorectal cancer cells suggest that miR-378a-3p may regulate the expression of the insulin-like growth factor 1 receptor (IGF1R) as a target gene. A significant negative correlation between IGF1R protein and miR-378a-3p expression levels was observed in colorectal cancer tissues. In another study, miR-378a-3p downregulation is observed in rhabdomyosarcoma tissues and cell lines48. miR-378a-3p overexpression suppressed IGF1R expression in rhabdomyosarcoma-derived RH30 cells and decreased the phosphorylation level of AKT protein, which is known as a key signaling molecule in rhabdomyosarcoma. miR-378a-3p overexpression resulted in an increase of apoptosis and a decrease of cell growth in RH30 cells. miR-378a-3p could also induce myogenic differentiation by modulating myogenic marker genes in RH30 cells. In this report, treatment with the DNA methyltransferase inhibitor, 5-Aza-dC, upregulated miR-378a-3p expression. These findings together with our data suggest that miR-378a-3p may play a tumor-suppressive role in malignant tumors. In breast cancer, we assumed that miR-378a-3p downregulation may particularly contribute to the acquisition of endocrine therapy resistance. Epigenetic modification may lead to the deregulation of miR-378a-3p expression in breast cancer cells53,54.
We selected GOLT1A as one of the miR-378a-3p candidate target genes by in silico algorithms. Our computational analysis is consistent with our experimental results that GOLT1A mRNA level was downregulated in MCF-7 cells transfected with miR-378a-3p precursor. Luciferase reporter assay using the 3′-UTR region of GOLT1A gene including a putative binding site for miR-378a-3p suggested that miR-378a-3p could decrease GOLT1A mRNA expression. In addition, GOLT1A knockdown by siRNA restored tamoxifen sensitivity in TamR cells. Moreover, the examination of publicly available microarray datasets revealed that high levels of GOLT1A expression correlated with poor rates of survival for patients with breast cancer. These results suggest that miR-378a-3p and its candidate target gene, GOLT1A, could contribute to the mechanisms underlying the endocrine resistance of breast cancer.
GOLT1A is an evolutionarily conserved protein with a structure including four putative transmembrane domains. Got1p has been identified as a yeast homolog of GOLT1A by screening for mutations that show synthetic lethality with a membrane protein sft2, which is localized in the Golgi apparatus55. The yeast study showed that Got1p protein is present in the early Golgi cisternae. Got1p mutant yeast exhibited reduced endoplasmic reticulum (ER)-to-Golgi transport due to the impairment of SNARE-dependent fusion. Another yeast study revealed that Got1p displayed moderate suppressor activity toward temperature-sensitive mutations in the SEC23 and SEC31 genes, which encode subunits of the coat protein complex II (COPII), a type of vesicle coat protein that transports forward from the rough ER to the Golgi apparatus56. The study further showed that Got1p was efficiently packaged into COPII vesicles and cycled rapidly between the ER and Golgi compartments. As overexpression of yeast Got1p or N-terminal region of human GOLT1A rather impaired the ER-to-Golgi transport56,57, GOLT1A could plausibly saturate or somehow interfere the normal cargo recongnition functions of the COPII vesicles. Further studies will be required to fully assess the role of GOLT1A in the ER-to-Golgi network with differential expression levels, nevertheless, GOLT1A could influence membrane properties and modulate vesicle formation in the ER-to-Golgi network.
Recent evidence suggests that the ER-to-Golgi network regulates cellular processes including stress response, apoptosis, and mitotic checkpoint in cancer cells. In prostate cancer, Golgi is emerging as a new therapeutic target since several Golgi-associated molecules have been demonstrated to be regulated by androgen58. Brefeldin A (BFA) is known as is an inhibitor of ER-to-Golgi protein transport by inhibiting the interaction between ADP-ribosylation factor 1 (Arf1) and guanine nucleotide exchange factor (GEF). Treatment of mammalian cells with BFA induces both ER and Golgi stress due to its ability to cause a cytotoxic accumulation of proteins in the ER that would normally be trafficked through the Golgi to be processed for secretion. The stress to both organelles resulting from BFA treatment can induce apoptosis59,60. The inhibition of the Arf1-GEF interaction will be beneficial for cancer therapy. For example, limited effective therapeutic options are available for patients with chronic lymphocytic leukemia (CLL) refractory to alkylating agents such as fludarabine61. It is notable that BFA could induce apoptosis in primary CLL cells from fludarabine-refractory patients61. Although BFA and its derivatives have not progressed beyond the pre-clinical stage of drug development because of their poor bioavailability, targeting the ER-to-Golgi network will be an attractive therapeutic option for cancers, especially for those that respond poorly to conventional treatments. It remains to be investigated whether new inhibitors for the ER-to-Golgi network can manage tamoxifen-resistant breast cancers, particularly those with high expression of GOLT1A and with altered ER-to-Golgi network.
In summary, we have identified miR-378a-3p as a modulating factor for the tamoxifen response in breast cancer by high throughput sequencing. A combination of in silico and in vitro analyses indicates that GOLT1A is a potential target of miR-378a-3p. These findings could be applied to develop alternative approaches to breast cancer diagnosis and treatment.
Methods
Cell culture
MCF-7 and 293T cells were purchased from ATCC (Manassas, VA, USA) and cultured in Dulbecco’s modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 units/ml penicillin, and 50 μg/ml streptomycin at 37 °C in a humidified atmosphere of 5% CO2 in air. Resistant clones for tamoxifen (TamR) were established from MCF-7 cells by long-term (>3 months) culture with 1 μM tamoxifen32. Tamoxifen was purchased from Sigma (St. Louis, MO, USA). Long-term estrogen-deprived (LTED) cells were established from MCF-7 cells by culturing them in phenol red-free DMEM supplemented with 10% dextran-charcoal stripped FBS (dccFBS).
RNA extraction and high-throughput sequencing
Total RNA was isolated from MCF-7, TamR and LTED cells using the ISOGEN reagent (Nippon Gene, Toyama, Japan) in accordance with the manufacturer’s instructions. Small RNA cDNA library was generated from the total RNAs and high-throughput sequencing was performed using an Illumina GAIIx sequencer (Illumina, San Diego, CA, USA)62. Mapping of small RNA reads on human genomes (NCBI35 assembly) and prediction of novel miRNAs were performed as describes previously34.
Transfection of miRNA precursors/inhibitors and siRNA
Pre-miR-378a-3p and its positive control as well as anti-miR-378a-3p and its negative control were purchased from Ambion (Carlsbad, CA, USA). Transfection of miRNA precursors or inhibitors was carried out using Lipofectamine RNAiMAX transfection reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction. siRNA duplexes, siGOLT1A (5′-GAUUCUUCAGCCUCUUUAAGG-3′ and 5′-UUAAAGAGGCUGAAGAAUCCG-3′) and siGOLT1A-2 (5′-GAAACCUACGGAUUCUUCA-3′ and 5′-UGAAGAAUCCGUAGGUUUCCA-3′) both of which target GOLT1A was synthesized using an algorithm that significantly improves the target specificity of siRNA, in particular, by efficiently estimating off-target sequences63. A non-targeting control siRNA (siControl) with no homology to known gene targets in mammalian cells was obtained from RNAi Inc. (Tokyo, Japan)63. Cells were cultured overnight and transfected with siRNA at a final concentration of 10 nM using Lipofectamine RNAiMAX. Knockdown efficiency of siRNA was determined by qRT-PCR using RNA prepared from the cells 48 h after transfection and normalized to that of siControl.
qPCR and qRT-PCR
Total RNA was extracted from cells or tissues using the ISOGEN reagent. For treatment of 5-aza-2′-deoxycytidine (5-Aza-dC), cells were incubated with 1 or 10 μM 5-Aza-dC or vehicle for 72 h. miRNA levels were determined by qPCR using triplicate TaqMan microRNA assays (Applied Biosystems, CA, USA). The target gene mRNA levels were evaluated by the StepOne Real-time PCR System (Applied Biosystems) using cDNAs converted from total RNA with SuperScript III Reverse Transcriptase (Invitrogen). Results from 3 independent experiments were normalized to the expression of endogenous RNU48 for miRNA or that of GAPDH for mRNA, respectively. Primers for GOLT1A and GAPDH were as follows: GOLT1A forward: 5′- GGGCCTGTCCCTCATCATT -3′,
GOLT1A reverse: 5′- TTTGTGCCGTTGGAAGAAGAA -3′,
GAPDH forward: 5′-GGTGGTCTCCTCTGACTTCAACA-3′, and
GAPDH reverse: 5′-GTGGTCGTTGAGGGCAATG-3′.
miRNA target prediction
Candidate targeted genes by miR-378a-3p were examined using 2 online database algorithms for miRNA target prediction: TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/microrna/getGeneForm.do)50,51.
Cell growth assay
Cell proliferation was evaluated using the WST-8 assay kit (Nacalai Tesque, Kyoto, Japan). Two thousands of MCF-7 or TamR cells per well were cultured in 96-well plates and 10 μL of WST-8 solution was added to each well at the indicated time points after transfection. Cells were further incubated for 2 h at 37 °C in a 5% CO2 incubator. The absorbance was measured at 450 nm with Multiscan FC Microplate Photometer (Thermo Fisher Scientific, Rochester, NY, USA).
Luciferase reporter assay
The sequence of GOLT1A 3′-UTR containing a putative binding site for miR-378a-3p was amplified by specific primers using MCF-7 cDNA and inserted into the psiCHECK-2 vector (Promega, Madison, WI, USA). Two mutants for the putative binding site for miR-378a-3p were constructed by PCR with the mutated primers. For 3′-UTR luciferase assay, 293T cells were transfected with psiCHECK2 vector containing the wild-type or mutated putative binding sites for miR-378a-3p together with pre-miR-378a-3p precursor or control pre-miR using Lipofectamine 2000 transfection reagent (Invitrogen). The psiCHECK2 empty vector was also transfected as a mock control. Luciferase reporter assay was performed using the Dual-Luciferase Reporter Assay System (Promega) 48 hours after transfection. Firefly luciferase activities for GOLT1A 3-UTR reporters were normalized to those for corresponding Renilla luciferase activities. Experiments were performed in triplicate, and the results were expressed as mean ± SD.
Clinical specimens
All clinical breast cancer tissues (n = 40) and the adjacent normal tissues (n = 16) were resected from patients those received surgery at Saitama Medical University. All procedures were performed under a protocol approved by the institutional Ethics Committee, and written informed consent was obtained from all patients. The methods were carried out in accordance with the approved guidelines. Total RNA was isolated from these dissected samples and subjected to qPCR and qRT-PCR analyses. Oncomine Research Edition46 was used for the evaluation of GOLT1A mRNA expression in clinical breast cancer and normal mammary tissues based on microarray datasets. Kaplan–Meier curves of relapse-free survival times stratifying breast cancer patients by GOLT1A expression were obtained using the Prognoscan (http://www.abren.net/PrognoScan/), which is a database for meta-analysis of the prognostic value of genes64.
We also used the publicly available dataset GSE3740549 from the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc = GSE37405), which contains miRNA expression data of ER-positive breast cancer patients treated with adjuvant tamoxifen. Kaplan–Meier analysis of recurrence-free survival was performed using a statistical software package EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan)65.
Additional Information
How to cite this article: Ikeda, K. et al. miR-378a-3p modulates tamoxifen sensitivity in breast cancer MCF-7 cells through targeting GOLT1A. Sci. Rep. 5, 13170; doi: 10.1038/srep13170 (2015).
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the help of Dr. Daisuke Tanaka and Ms. Akane Kitamura, Research Center for Genomic Medicine, Saitama Medical University for advice regarding experimental procedures. The authors thank RIKEN for sequencing our samples. This work was supported by Grants of the Cell Innovation Program, P-DIRECT, Grants-in-Aid, and Support Project of Strategic Research Center in Private Universities from the Ministry of Education, Culture, Sports, Science, and Technology, Japan; by Grants from the Japan Society for the Promotion of Science, Japan (23249040 and 15K15353); by Grants-in-Aid from the Ministry of Health, Labour, and Welfare, Japan; by the Advanced Research for Medical Products Mining Program of the National Institute of Biomedical Innovation, Japan.
Footnotes
Author Contributions Conceived and designed the experiments: K.I. and S.I. Performed the experiments: K.I., T.U., T.S., W.S. and E.B. Analyzed the data: K.H.-I., E.B. and H.M. Contributed reagents/materials/analysis tools: T.S., A.O., T.S. and K.O. Wrote the paper: K.I., K.H.-I. and S.I. All authors reviewed the manuscript.
References
- Lozano R. et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2095–2128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early Breast Cancer Trialists’ Collaborative Group. Tamoxifen for early breast cancer: an overview of the randomized trials. Lancet 351, 1451–1467 (1998). [PubMed] [Google Scholar]
- Chow J., Tobias J. H., Colston K. W. & Chambers T. J. Estrogen maintains trabecular bone volume in rats not only by suppression of bone resorption but also by stimulation of bone formation. J Clin Invest 89, 74–78 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K., Horie-Inoue K. & Inoue S. Identification of estrogen-responsive genes based on the DNA binding properties of estrogen receptors using high-throughput sequencing technology. Acta Pharmacol Sin 36, 24–31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhat M. Y., Lavigne M. C. & Ramwell P. W. The vascular protective effects of estrogen. FASEB J 10, 615–624 (1996). [PubMed] [Google Scholar]
- Blamey R. W. Guidelines on endocrine therapy of breast cancer EUSOMA. Eur J Cancer 38, 615–634 (2002). [DOI] [PubMed] [Google Scholar]
- Cole M. P., Jones C. T. & Todd I. D. A new anti-oestrogenic agent in late breast cancer. An early clinical appraisal of ICI 46474. Br J Cancer 25, 270–275 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J. et al. Effect of tamoxifen and radiotherapy in women with locally excised ductal carcinoma in situ: long-term results from the UK/ANZ DCIS trial. Lancet Oncol 12, 21–29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet 378, 771–784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring A. & Dowsett M. Mechanisms of tamoxifen resistance. Endocr Relat Cancer 11, 643–658 (2004). [DOI] [PubMed] [Google Scholar]
- Badia E., Oliva J., Balaguer P. & Cavaillès V. Tamoxifen resistance and epigenetic modifications in breast cancer cell lines. Curr Med Chem 14, 3035–3045 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijichi N. et al. Association of double-positive FOXA1 and FOXP1 immunoreactivities with favorable prognosis of tamoxifen-treated breast cancer patients. Horm Cancer 3, 147–159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijichi N. et al. Association of positive EBAG9 immunoreactivity with unfavorable prognosis in breast cancer patients treated with tamoxifen. Clin Breast Cancer 13, 465–470 (2013). [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A. & Slack F. J. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6, 259–269 (2006). [DOI] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S. N. & Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9, 102–114 (2008). [DOI] [PubMed] [Google Scholar]
- Iorio M. V. & Croce C. M. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 4, 143–159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce C. M. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10, 704–714 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn D. M. MicroRNAs and metastasis: little RNAs go a long way. Cancer Res 70, 6401–6406 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J. et al. MiR-873 regulates ERα transcriptional activity and tamoxifen resistance via targeting CDK3 in breast cancer cells. Oncogene 2014. Dec 22. 10.1038/onc.2014.430. [Epub ahead of print] [DOI] [Google Scholar]
- Gan R., Yang Y., Yang X., Zhao L., Lu J. & Meng Q. H. Downregulation of miR-221/222 enhances sensitivity of breast cancer cells to tamoxifen through upregulation of TIMP3. Cancer Gene Ther 21, 290–296 (2014). [DOI] [PubMed] [Google Scholar]
- Miller T. E. et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem 283, 29897–29903 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. J. et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem 283, 31079–31086 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ward A. et al. MicroRNA-519a is a novel oncomir conferring tamoxifen resistance by targeting a network of tumour-suppressor genes in ER+ breast cancer. J Pathol 233, 368–379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe R. et al. Increased expression of miR-126 and miR-10a predict prolonged relapse-free time of primary oestrogen receptor-positive breast cancer following tamoxifen treatment. Eur J Cancer 49, 3598–3608 (2013). [DOI] [PubMed] [Google Scholar]
- Manavalan T. T., Teng Y., Litchfield L. M., Muluhngwi P., Al-Rayyan N. & Klinge C. M. Reduced expression of miR-200 family members contributes to antiestrogen resistance in LY2 human breast cancer cells. PLoS One 8, e62334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S., Long X., Kwon C., Kim S. & Nephew K. P. An integrative analysis of cellular contexts, miRNAs and mRNAs reveals network clusters associated with antiestrogen-resistant breast cancer cells. BMC Genomics 13, 732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. et al. let-7 microRNAs induce tamoxifen sensitivity by downregulation of estrogen receptor α signaling in breast cancer. Mol Med 17, 1233–1241 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A. et al. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene 32, 1173–1182 (2013). [DOI] [PubMed] [Google Scholar]
- Bergamaschi A. & Katzenellenbogen B. S. Tamoxifen downregulation of miR-451 increases 14-3-3ζ and promotes breast cancer cell survival and endocrine resistance. Oncogene 31, 39–47 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cittelly D. M. et al. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol Cancer 9, 317 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujihira T. et al. MicroRNA-574-3p, identified by microRNA library-based functional screening, modulates tamoxifen response in breast cancer. Sci Rep 5, 7641 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama M. et al. Integrated quantitative analysis of the phosphoproteome and transcriptome in tamoxifen-resistant breast cancer. J Biol Chem 286, 818–829 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma K. Y. et al. Association of estrogen receptor alpha and histone deacetylase 6 causes rapid deacetylation of tubulin in breast cancer cells. Cancer Res 69, 2935–2940 (2009). [DOI] [PubMed] [Google Scholar]
- Takada S. et al. Mouse microRNA profiles determined with a new and sensitive cloning method. Nucleic Acids Res 34, e115 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L. X. et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 14, 2348–2360 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. A., Krichevsky A. M. & Kosik K. S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 65, 6029–6033 (2005). [DOI] [PubMed] [Google Scholar]
- Iorio M. V. et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65, 7065–7070 (2005). [DOI] [PubMed] [Google Scholar]
- Volinia S. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103, 2257–2261 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulci V. et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood 109, 4944–4951 (2007). [DOI] [PubMed] [Google Scholar]
- Eto K. et al. The microRNA-21/PTEN pathway regulates the sensitivity of HER2-positive gastric cancer cells to trastuzumab. Ann Surg Oncol 21, 343–350 (2014). [DOI] [PubMed] [Google Scholar]
- Frankel L. B., Christoffersen N. R., Jacobsen A., Lindow M., Krogh A. & Lund A. H. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem 283, 1026–1033 (2008). [DOI] [PubMed] [Google Scholar]
- Zhu S., Si M. L., Wu H. & Mo Y. Y. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem 282, 14328–14336 (2007). [DOI] [PubMed] [Google Scholar]
- Boyerinas B., Park S. M., Hau A., Murmann A. E. & Peter M. E. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer 17, F19–36 (2010). [DOI] [PubMed] [Google Scholar]
- Yu F. et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131, 1109–1123 (2007). [DOI] [PubMed] [Google Scholar]
- Sakurai M. et al. LIN28: a regulator of tumor-suppressing activity of let-7 microRNA in human breast cancer. J Steroid Biochem Mol Biol 131, 101–106 (2012). [DOI] [PubMed] [Google Scholar]
- Faehnle C. R., Walleshauser J. & Joshua-Tor L. Mechanism of Dis3l2 substrate recognition in the Lin28-let-7 pathway. Nature 514, 252–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. Clinical and biological significance of miR-378a-3p and miR-378a-5p in colorectal cancer. Eur J Cancer 50, 1207–1221 (2014). [DOI] [PubMed] [Google Scholar]
- Megiorni F. et al. Deep Sequencing the microRNA profile in rhabdomyosarcoma reveals down-regulation of miR-378 family members. BMC Cancer 14, 880 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyng M. B., Lænkholm A. V., Søkilde R., Gravgaard K. H., Litman T. & Ditzel H. J. Global microRNA expression profiling of high-risk ER+ breast cancers from patients receiving adjuvant tamoxifen mono-therapy: a DBCG study. PLoS One 7, e36170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan C. H., Friedman R. C., Ruby J. G. & Bartel D. P. Formation, regulation and evolution of Caenorhabditis elegans 39 UTRs. Nature 469, 97–101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D., Wilson M., Gabow A., Marks D. S. & Sander C. Targets and expression. Nucleic Acids Res 36, (Database Issue) D149–D153 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D. R. et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9, 166–180 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. E., Anderton D. L., Lee M. P., Pentecost B. T. & Arcaro K. F. High-density array analysis of DNA methylation in Tamoxifen-resistant breast cancer cell lines. Epigenetics 9, 297–307 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q. et al. An epigenomic approach to therapy for tamoxifen-resistant breast cancer. Cell Res 24, 809–819 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conchon S., Cao X., Barlowe C. & Pelham H. R. Got1p and Sft2p: membrane proteins involved in traffic to the Golgi complex. EMBO J 18, 3934–3946 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townley A. K. et al. Efficient coupling of Sec23-Sec24 to Sec13-Sec31 drives COPII-dependent collagen secretion and is essential for normal craniofacial development. J Cell Sci 2008. 121, 3025–3034 (2008). [DOI] [PubMed] [Google Scholar]
- Starkuviene V. et al. High-content screening microscopy identifies novel proteins with a putative role in secretory membrane traffic. Genome Res 14, 1948–1956 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migita T. & Inoue S. Implications of the Golgi apparatus in prostate cancer. Int J Biochem Cell Biol 44, 1872–1876 (2012). [DOI] [PubMed] [Google Scholar]
- Ferri K. F. & Kroemer G. Organelle-specific initiation of cell death pathways. Nat Cell Biol 3, E255–E263 (2001). [DOI] [PubMed] [Google Scholar]
- Hicks S. W. & Machamer C. E. Golgi structure in stress sensing and apoptosis. Biochim Biophys Acta 1744, 406–414 (2005). [DOI] [PubMed] [Google Scholar]
- Carew J. S. et al. Targeting endoplasmic reticulum protein transport: a novel strategy to kill malignant B cells and overcome fludarabine resistance in CLL. Blood 107, 222–231 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs A. M., Kawano M., Ando Y., Daub C. O. & Hayashizaki Y. pre-miRNA profiles obtained through application of locked nucleic acids and deep sequencing reveals complex 5′/3′ arm variation including concomitant cleavage and polyuridylation patterns. Nucleic Acids Res 40, 1424–1437 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueyama K. et al. Knockdown of Efp by DNA-modified small interfering RNA inhibits breast cancer cell proliferation and in vivo tumor growth. Cancer Gene Ther 17, 624–632 (2010). [DOI] [PubMed] [Google Scholar]
- Mizuno H., Kitada K., Nakai K. & Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics 2, 18 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda Y. Investigation of the freely-available easy-to-use software “EZR” (Easy R) for medical statistics. Bone Marrow Transplant 48, 452–458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.