Abstract
A sufficient cell source and minimal invasiveness in obtaining human adipose stromal cells (hASCs) hold great promise for their utilization in wound repair. However, little is known about how cell-residing microenvironments regulate the cellular response. In this study we explored the effects of polycaprolactone (PCL)/collagen nanofibers with distinct spatial arrangements (aligned and random) on phenotypic expression of hASCs in vitro. Elongated cell morphology, higher proliferation, and faster migration rate were observed for hASCs cultured on the aligned nanofibers, showing that hASCs could detect the nanofiber spatial arrangement and then distinctively respond. This study on the expression of extracellular matrix (ECM) related genes in hASCs revealed higher synthesis capacity for critical ECM molecules including tropoelastin, collagen I, and matrix metalloproteinase (MMP)-1 on the aligned nanofibers. Integrins α5, β1, β3, β6, and transforming growth factor (TGF)-β1 were differentially regulated by PCL/collagen nanofiber arrangements. Our results indicate that fiber orientation-induced phenotypic change of hASCs may be regulated by integrins and TGF-β signaling synergistically. These findings demonstrate the potential application of hASCs and aligned PCL/collagen nanofibers for accelerated wound repair.
Introduction
In the past two decades, advances in cellular and molecular biology have significantly improved our understanding of wound repair and tissue regeneration and therefore enhanced our ability to heal difficult wounds.1,2 Tissue-engineered skin substitutes, especially those of autologous skin cells, have proved to be effective in treating deep wounds.3–7 However, it remains a big challenge to obtain enough skin cells from patients who have limited intact skin and fabricate sufficient skin substitutes to treat large wounds. In this regard, stem cells, with their unique capability of differentiating into various types of tissue cells, hold great promise in dermatology to compensate the shortage of skin cells. Among different stem cells, adipose-derived stromal cells (ASCs) have drawn particular attention in consideration of their easy access, minimal invasiveness and large quantity for tissue harvest, and self-renewal capacity.8,9 Increasing evidence has shown the advantages of ASCs in wound repair by promoting the proliferation and migration of fibroblasts, stimulating the angiogenesis and accelerating the re-epithelialization from wound edge.10–16 We have also found that 3D dermal substitutes fabricated from ASCs could stimulate the rapid closure of full thickness wounds of athymic mice with proper re-epithelialization.16 The wound healing capacity of ASCs is synergistically regulated by soluble factors as well as the direct interaction with other neighboring cells and extracellular matrix (ECM).10 In this regard, the local environment in which ASCs reside would play a vital role in ASC phenotypic expression and therefore determine their wound healing capacity. Recently, electrospun nanofiber scaffolds have received much attention in wound repair mainly due to their morphological and dimensional similarity to the ECM of native tissues. With the capability of incorporating ECM molecules like collagen into electrospun nanofibers, it is possible for us to formulate a biomimetic microenvironment close to the native cell-growing environment. In this regard, it is reasonable to hypothesize that such nanofiber scaffolds can support cell growth and better maintain cell phenotype.17–19 During electrospinning, electrospun nanofibers can be oriented in the collected fiber scaffolds depending on the fiber collection setup. Studies have shown that oriented nanofibers can guide the cell adhesion and spatial arrangement of intracellular cytoskeletal proteins, and as a result lead to the elongation of cells along the fiber orientation.20–22 However, it is unclear whether fiber orientation can also regulate the cellular functions (e.g., cell proliferation and migration) and the synthesis capacity of new ECM such as collagen and elastin, especially in the case of ASCs, which are critical to wound healing.
In this study, collagen-containing electrospun nanofibers with two types of spatial arrangements (aligned=fibers oriented in the same direction; random=fibers collected without specific orientation) were fabricated and used to culture human adipose stromal cells (hASCs). The morphology of hASCs on aligned on random fibers was examined by immunofluoresent staining of cytoskeletal proteins. The proliferation and migration of hASCs in association with fiber orientation were studied. Genes of various integrins and those involved in ECM synthesis and degradation were investigated to determine the phenotypic expression. It was found that fiber orientation greatly affected the cell morphology, proliferation, and migration by differentially regulating the expression of integrins and the formation of focal adhesion. The synthesis of collagen I, tropoelastin, and matrix metalloproteinase (MMP)-1 by hASCs was also influenced by fiber orientation, in which transforming growth factor (TGF)-β1 might be involved as suggested by the upregulation of TGF-β1 gene.
Materials and Methods
Materials
Poly(epsilon-caprolactone) (PCL, MW=80,000), MCDB201 medium, MCDB131 medium, ITS-X, bovine serum albumin (BSA), linoleic acid-BSA, L-ascorbate-2-phosphate, β-mercaptoethanol, recombinant human epithelial growth factor (rhEGF), recombinant human platelet derived growth factor (rhPDGF)-BB, and dexamethasone were purchased from Sigma-Aldrich (St. Louis, MO). Collagen type I was purchased from Elastin Products Company (Owensville, MO). 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) was obtained from Oakwood Products Inc. (West Columbia, SC). Fetal bovine serum (FBS) was purchased from American Type Culture Collection (ATCC, Manassas, VA). All other reagents and solutions were obtained from Invitrogen (Carlsbad, CA) except as indicated.
Electrospinning and characterization
Nanofibers were similarly prepared using electrospinning technique as described previously.17,23 Briefly, collagen type I solution (8% w/v), and PCL solution (8% w/v) dissolved in HFIP were mixed thoroughly at a volume ratio of 1:3. The solution was transferred to a 3 mL syringe attached with a blunt-tip capillary (inner diameter=0.9mm) and electrospun at 10 kV. Steady flow of the solution from the capillary spinneret was achieved using a syringe pump from KD Scientific (Holliston, MA), operating at a flow rate from 5 μL/min to 15 μL/min. To fabricate random nanofiber scaffolds, the collecting surface consisted of a stationary aluminum foil plate, and to collect the nanofibers with desired alignment, a customized rotating mandrel (φ=10 cm) was used. Nanofibrous scaffolds were collected on square glass cover slips. All fibers were fabricated under a sterile condition. To characterize the electrospun nanofiber using a scanning electron microscope (SEM), fibers were collected on silicon wafer and sputter coated with gold. The coated fibers were examined with a LEO 982 FEG SEM. To determine the diameter of nanofibers, images of five randomly selected areas were captured and analyzed by analysis software (NIS-elements BR 3.10) from Nikon. The orientation of electrospun nanofibers was determined by using the ImageJ program (NIH) along with the Oval Profile Plot plugin. Briefly, representative SEM images of the electrospun fibers were cropped into a square covering as much of the image as possible (the images used were 905×905 pixels in size). These cropped images were then processed with a fast Fourier transform (FFT) and then a circular selection of the image was made maximizing the amount selected (edges of the circle were tangent with the edges of the image). This selection then underwent a radial summation using the Oval Profile Plot plugin with 360 points such that the degree interval was 1°. The raw data were normalized to 0 and data from all sets were rescaled to an arbitrary range between 0 and 0.3. The final results were presented by plotting this arbitrary scale versus degrees, where the height of peaks represents a greater degree of alignment at a given angle.
Cell seeding and culture
Human adipose stromal cells (hASCs) kindly provided by Dr. Moscatello from Coriell Institute for Medical Research (Camden, NJ) were used in this study. Cells were cultured in the proliferation medium, which consisted of Dulbecco's modified Eagle medium (DMEM, low glucose), MCDB201 and MCDB131 at a ratio of 3:1:1, 0.5% FBS, 1× ITSX, 0.2% BSA, 1× linoleic acid-BSA, 100 μM L-ascorbate-2-phosphate, 100 μM β-mercaptoethanol, 10 ng/mL rhEGF, 5 ng/mL rhPDGF-BB, 1nM dexamethasone, 1% penicillin, and 1% streptomycin. Medium was refreshed every 3–4 days. Cell splitting was performed when the monolayer cells reached 70–80% confluence. Cells between the second to fifth passages were used in this study. To seed hASCs onto electrospun fibers, cells were trypsinized, centrifuged, and resuspended in the proliferation medium at a final concentration of 1×105/mL. hASCs were then seeded onto various nanofibrous scaffolds (aligned and random) at a density of 5×103 cells/cm2 and allowed to attach for 60 min before adding more medium to completely immerse the culture. The culture was kept in a humidified incubator at 37°C with 5% carbon dioxide for designated times. Monolayer culture of hASCs on glass cover slips served as controls.
Immunofluorescent staining
The morphology and adhesion of hASCs onto nanofibrous scaffolds were evaluated by immunofluorescently staining the cytoskeletal proteins and vinculin. Briefly, after fixation in 4% paraformaldehyde for 20 minutes the culture was permeabilized with 0.5% Triton X-100 (Fisher Scientific, Fair Lawn, NJ), and then incubated with phalloidin-tetramethyl rhodamine isothiocyanate (TRITC) (Sigma, 1:500), anti-α-tubulin (DSHB, 1:25), anti-vinculin conjugated with fluorescein isothiocyanate (FITC) (Sigma, 1:50), anti-vimentin (Biovision, Mountain View, CA, 1:20) or anti-αSMA (α-smooth muscle actin) conjugated with Cy3 (Sigma, 1:200) for 60 min at room temperature in dark. For those primary antibodies without fluorescent labels, the cells were further incubated with goat anti-mouse IgG-FITC conjugate secondary antibody (Caltag, PAB4971, 1:400) or goat anti-rabbit IgG-Alexa Fluor 594 conjugate secondary antibody (Molecular Probes, A11037, 1:500). Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma, 1:1000). Excessive fluorescent conjugate was removed by washing with phosphate buffered solution (PBS). DAPI was used to stain cell nucleus. The staining was examined under a Nikon Eclipse 80i epifluorescent microscope (Nikon, Japan).
Cell proliferation
Cell proliferation on electrospun nanofibers (n=4) was determined by DNA assay using a CyQUANT® Cell Proliferation Assay Kit (Molecular Probes, Inc., Eugene, OR) as described previously.24 Briefly, the samples harvested at different times were digested with proteinase K solution (Sigma). Two hundred microliters of CyQUANT® GR dye/cell-lysis buffer was added to each sample and incubated at room temperature for 2–5 minutes. The fluorescence intensity was measured with excitation at 480 nm and emission at 520 nm using a Synergy™ HT Multi-Detection Microplate Reader (BioTek Instruments Inc., Winooski, VT).
Cell migration
A collagen gel drop assay was developed and used to analyze cell mobility on nanofibrous scaffolds. In this assay, a small drop of collagen gel suspended with a high density of hASCs was placed on top of the nanofibrous scaffolds and then the cell migratory behavior out of gel was evaluated by optical microscopy. Briefly, hASCs was carefully resuspended in neutralized type I collagen solution (6 mg/mL in 0.01 M HCl) at a final concentration of 2×106 cells/mL and then droplets of 10 μL cell/gel suspension were added onto the scaffold surface. Upon solidification of collagen gel at 37°C for 30 min, cell/gel droplets were completely covered with culture medium. After incubation for 3 and 7 days, samples were fixed in 4% paraformaldehyde, subsequently stained with methylene blue, and then examined under a stereo microscope (Nikon SMZ1500). Cell migration on different scaffolds was determined by measuring the longest distance that the cells migrated away from the boundary of gel drop.
Gene expression
Gene expression (n=4 per group) was measured using reverse transcriptase polymerase chain reaction (RT-PCR) at 1, 3, and 7 days. Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA). The isolated RNA was reverse transcribed into complementary DNA (cDNA) using the SuperScript First-Strand Synthesis System (Promega, Madison, WI), and the cDNA product was then amplified using recombinant Taq DNA polymerase (Promega). Expression of integrins α2, α5, αV, β1, β3, β6, MMP-1, MMP-2, MMP-9, TGF-β1, tropoelastin, type I collagen, type III collagen, vimentin, and α-SMA (ACTA-2) were determined over time. β-actin served as housekeeping gene. Primer sequences for various genes are listed in Table 1. All genes were amplified for 30 cycles in a thermocycler (Eppendorf Mastercycler gradient, Brinkmann, Westbury, NY). Semiquantitative analysis of gene expression was performed, and band intensity was normalized to that of β-actin.
Table 1.
Primer Sequences Used for Reverse Transcription-Polymerase Chain Reaction Gene Expression Analysis
| Gene | 5′–3′ | Primers |
|---|---|---|
| ITGα2 | Sense | 5′-CAGAATTTGGAACGGGACTT-3′ |
| Anti-sense | 5′-CAGGTAGGTCTGCTGGTTCA-3′ | |
| ITGα5 | Sense | 5′-GTGGCCTTCGGTTTACAGTC-3′ |
| Anti-sense | 5′-AATAGCACTGCCTCAGGCTT-3′ | |
| ITGαV | Sense | 5′-GATGGACCAATGAACTGCAC-3′ |
| Anti-sense | 5′-TTGGCAGACAATCTTCAAGC-3′ | |
| ITGβ1 | Sense | 5′-CCAAAAGCTCAGGACAAAGC-3′ |
| Anti-sense | 5′-GCTTGCATCTGTGGTCTTCA-3′ | |
| ITGβ3 | Sense | 5′-ACACTGGCAAGGATGCAGTGAATTGTAC-3′ |
| Anti-sense | 5′-CGTGATATTGGTGAAGGTAGCGTGGC-3′ | |
| ITGβ6 | Sense | 5′-TCTGGAGTTGGCGAAAGG-3′ |
| Anti-sense | 5′-TCCACCGGGTAGTCCTCA-3′ | |
| Collagen I | Sense | 5′-TGCTGGCCAACTATGCCTCT-3′ |
| Anti-sense | 5′-TTGCACAATGCTCTGATC-3′ | |
| Collagen III | Sense | 5′-CCAAACTCTATCTGAAATCC-3′ |
| Anti-sense | 5′-GGACTCATAGAATACAATCT-3′ | |
| MMP1 | Sense | 5′-CTGCTTACGAATTTGCCGACAGA-3′ |
| Anti-sense | 5′-GTTCTAGGGAAGCCAAAGGAGCTG-3′ | |
| MMP2 | Sense | 5′-GGCCCTGTCACTCCTGAGAT-3′ |
| Anti-sense | 5′-GGCATCCAGGTTATCGGGGA-3′ | |
| MMP9 | Sense | 5′-TGTACCGCTATGGTTACAC-3′ |
| Anti-sense | 5′-CCGCGACACCAAACTGGAT-3′ | |
| Vimentin | Sense | 5′ -ATTGAGATTGCCACCTACAGGAAGC-3′ |
| Anti-sense | 5′ -TGATGCTGAGAAGTTTCGTTGATAA-3′ | |
| ACTA2 | Sense | 5′ -TCGTGCTGGACTCTGGAGATGGTGT-3′ |
| Anti-sense | 5′ -TGAAGGATGGCTGGAACAGGGTCTC-3′ | |
| β-actin | Sense | 5′- GAGGAATACAGCCTGTGGGT-3′ |
| Anti-sense | 5′CAGAAGGTGCAGAGATGATGA-3′ |
MMP, matrix metalloproteinase; ACTA2, alpha actin 2.
Statistical analysis
Each experiment was repeated at leastthree3 times on different days and data were expressed as the mean±standard deviation. All the cytotoxicity and cell attachment measurements were collected in triplicate for each group. Unpaired student t-test was used to evaluate the significance between experimental groups. A value of p<0.05 was considered statistically significant.
Results
Nanofiber meshes with various fiber orientations
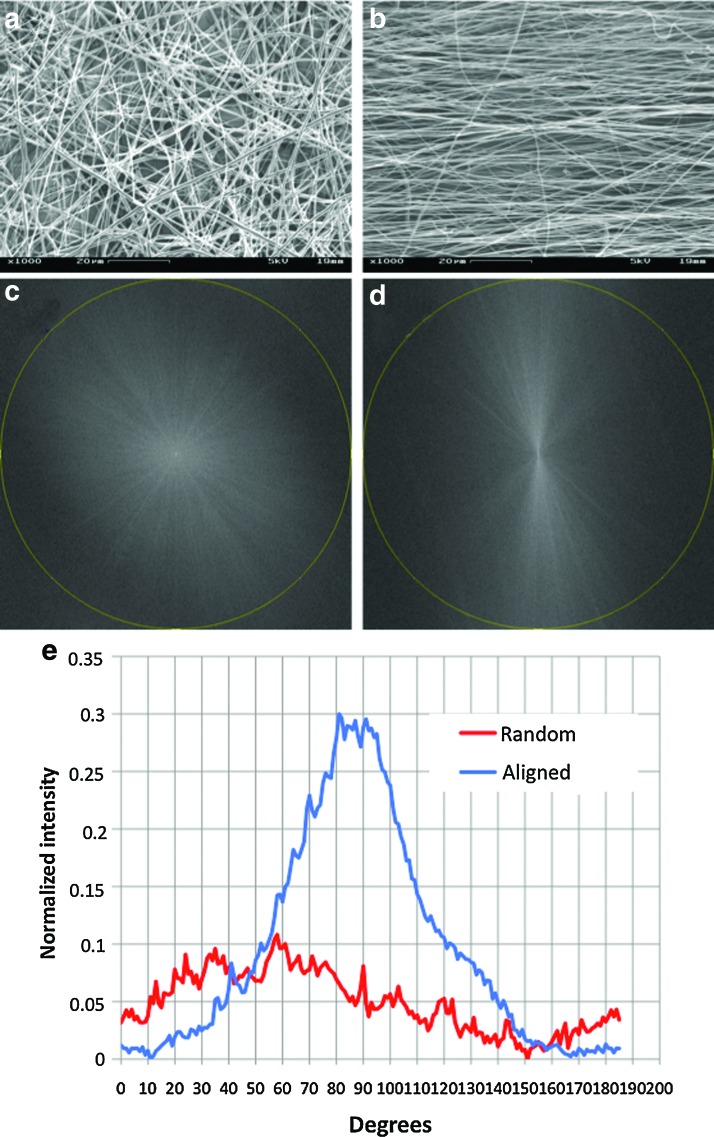
By collecting the nanofibers either on a grounded rotating mandrel or on a grounded stationary flat surface, nanofiber meshes with fibers oriented either in one direction (i.e., aligned) or in all x-y directions (i.e., random) were obtained. SEM examination clearly showed various spatial arrangements of collected fibers (Fig. 1a vs. 1b). To better characterize the orientation, the SEM images of obtained nanofibers were analyzed with a fast Fourier transform (FFT) and the analysis results were shown in Figure1 (c–e). More than 90% electrospun nanofibers aligned in one direction in the aligned scaffolds, and no preferred orientation could be identified in the random scaffolds. Based on the SEM images, the average fiber diameter in the aligned scaffolds seemed slightly smaller than that of the random ones (Table 2), but there was no significant difference. SEM examination at a high magnification revealed that both aligned and random 3:1 PCL/collagen nanofibers had a smooth surface (data not shown).
FIG. 1.
Representative scanning electron microscope images of random (a) and aligned (b) nanofiber meshes. Fast Fourier transform output images of random (c) and aligned (d) nanofiber meshes, where circular line indicates radial projection. (e) Pixel intensity plots against the angle of acquisition for a random (red) and an aligned (blue) nanofiber meshes. Note the distinctive peak in aligned meshes. Color images available online at www.liebertonline.com/tea
Table 2.
Mean Diameters of Nanofiber Meshes
| Fiber group | Composition | Mean diameter (nm) |
|---|---|---|
| Random | PCL/Collagen | 407±193 |
| Aligned | PCL/Collagen | 323±98 |
Attachment and morphology of hASCs on nanofiber meshes
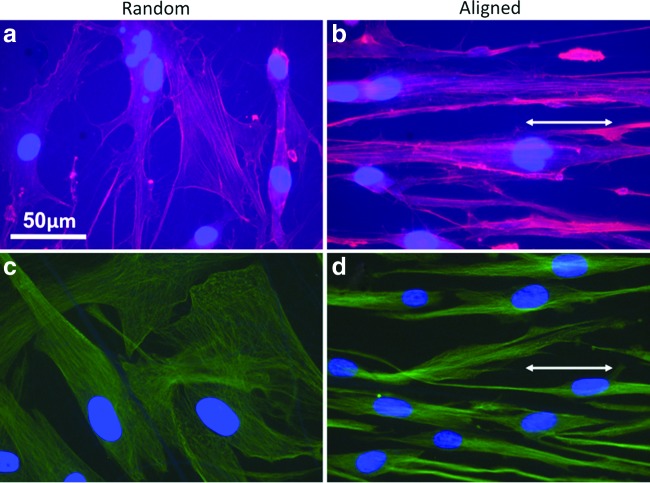
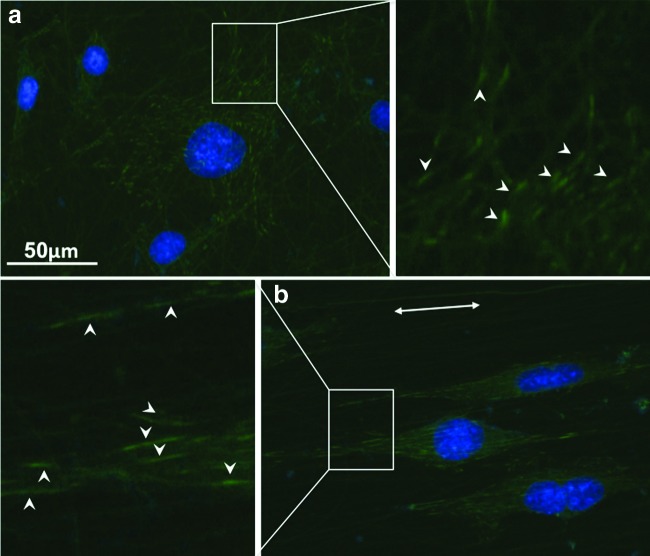
The inclusion of collagen in PCL electrospun nanofibers favored the adhesion and spreading of hASCs. As the cell morphology was defined by the arrangement of intracellular cytoskeletal proteins, immunofluorescent staining for F-actin and α-tubulin was performed on the hASCs cultured on PCL/collagen nanofibers for 3 days. Both F-actin and α-tubulin staining showed that hASCs on random nanofiber surfaces had a polygonal morphology without preferred orientation, while those on aligned nanofiber surfaces elongated along with the nanofiber orientation, showing a typical spindle-like morphology (Fig. 2). The intracellular cytoskeletal protein fibers also oriented in the same direction as the electrospun nanofibers on the aligned PCL/collagen nanofiber surfaces and were more compacted as compared to those on the random nanofiber surfaces. The morphology remained similar for the entire experiment time (up to 7 days). To better quantify the morphology difference of hASCs on aligned and random fibers, fluorescent images of five randomly selected fields were analyzed using the NIH ImageJ analysis software. As shown in Table 3, the average cell area on aligned PCL/collagen nanofibers was consistently lower than that on the random nanofibers for both days 1 and 7 despite no statistical significance (p>0.05). The average cell area increased approximately one fold after cultured for 7 days compared to that from day 1. The long/short axis ratio of hASCs, an indicator of cell polarity, was also calculated (Table 3). Clearly, the cells on aligned fibers remained constantly elongated with the long/short axis ratio as high as 17, more than three times of the random ones. The formation of cell/fiber focal adhesion was evaluated by immunofluorescent staining of vinculin, a protein involved in the connection of focal adhesion point to cytoskeletal actin. It was found that vinculin of hASCs cultured on the aligned fibers showed long spikes with the same orientation as the fibers (Fig. 3b). In contrast, on random fibers vinculin were short strokes without preferred orientation (Fig. 3a).
FIG. 2.
Representative fluorescent images of human adipose stromal cells (hASC) cultured for 3 days on random (a, c) and aligned (b, d) nanofiber meshes. Cells were stained with phalloidin-tetramethyl rhodamine isothiocyanate (TRITC) (red) for filament-actin (a, c) or stained with Cy3-conjugated anti-α-tubulin (green). Cell nuclei were stained blue with 4′,6-diamidino-2-phenylindole (DAPI). Double-head arrow indicates the fiber orientation. Scale bar: 50 μm. Color images available online at www.liebertonline.com/tea
Table 3.
Measurement of Cell Spreading on Nanofiber Meshes
| Random | Aligned | ||
|---|---|---|---|
| Mean cell area (μm2) | Day 1 | 6228±2732 | 4140±1766 |
| Day 7 | 1348 1±4836 | 10969±773 | |
| Long/short axis ratio | Day 1 | 3.4±1.5 | 17.4±6.6* |
| Day 7 | 3.2±1.2 | 11.3±4.0* |
Statistically significant, p<0.001.
FIG. 3.
Representative fluorescent images of the distribution of vinculin in hASCs cultured on random (a) and aligned (b) nanofiber meshes. Vinculin was stained green with fluorescein isothiocyanate (FITC) and cell nuclei were stained blue with DAPI. Arrowheads indicate the vinculin and double-head arrow indicates the fiber orientation. Scale bar: 50 μm. Color images available online at www.liebertonline.com/tea
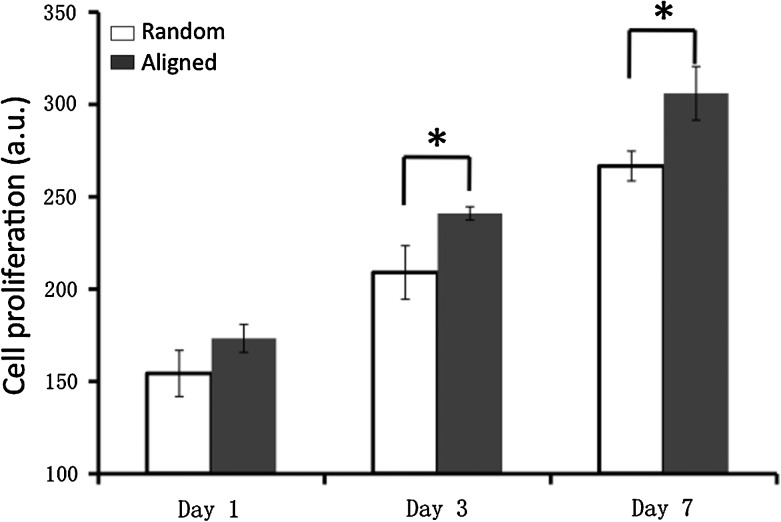
Cell proliferation and migration
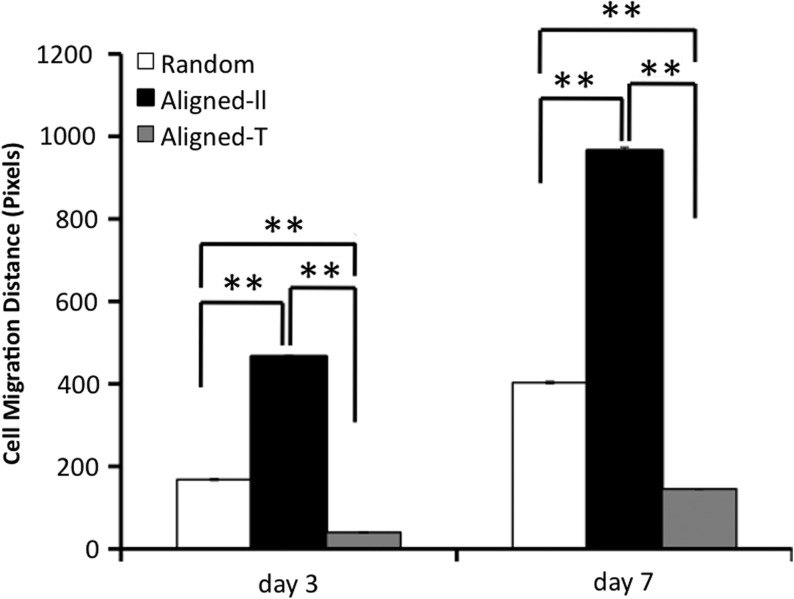
Cell proliferation on aligned and random nanofiber meshes was evaluated by DNA assay. Continuous cell proliferation was observed for hASCs on both aligned and random nanofibers. However, cells on aligned nanofibers proliferated faster than those on random fibers (Fig. 4), and this trend remained for all the times investigated. The migration of hASCs on the aligned and random fiber surfaces was determined by a collagen gel drop assay. Obviously, cells migrated out of the original droplet after 3 days on both aligned and random fiber surfaces. Interestingly, an isotropic hASC migration was observed on the random nanofibers, while an anisotropic migration was observed on the aligned nanofibers (i.e., accelerated migration along the fiber alignment in contrast to an inhibited migration perpendicular to fiber alignment) (Fig. 5). Besides, hASCs migrated in a longer distance on the aligned nanofiber meshes than they did on the random fibers. The migration distance away from collagen gel was measured on the samples stained with methylene blue (n=3 per time point). The results showed that hASC migration along the aligned fibers was more than two times faster than that on the random fibers, while the migration perpendicular to fiber alignment was less than half of that on the random one (Fig. 6).
FIG. 4.
Proliferation of human adipose stromal cells on nanofiber meshes. Cell amount was determined by DNA assay. *Statistically significant, p<0.05.
FIG. 5.
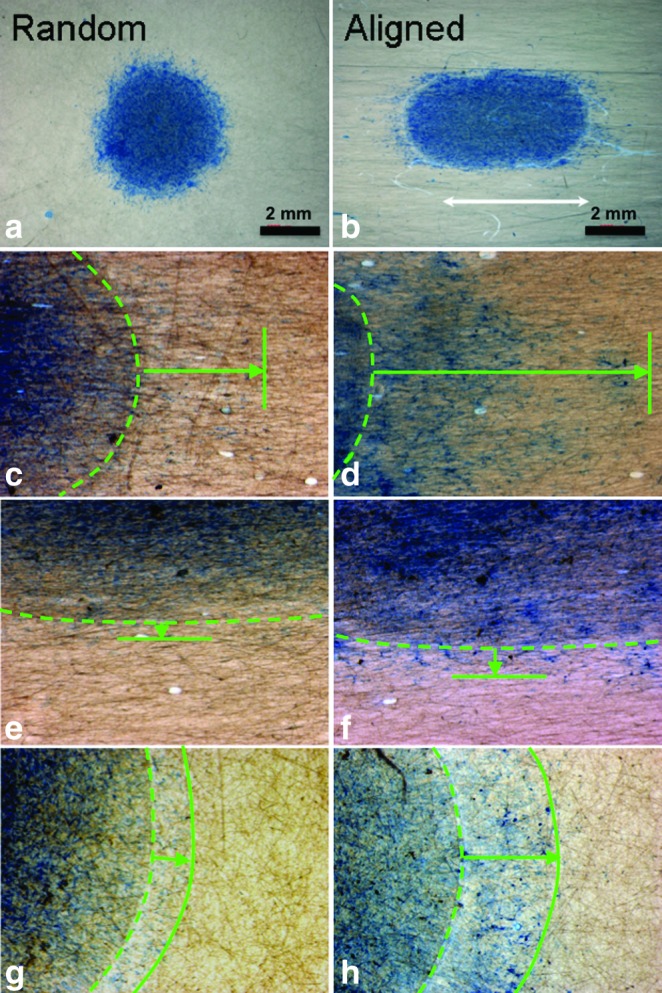
Optical images of hASCs migrated out of the collagen gel borderline (green broken line) on random (a, g, h) or aligned (b, c, d, e, f) nanofiber meshes for 3 days (c, e, g) and 7 days (d, f, h). Cells were fixed and stained blue with methylene blue. White arrow indicates the fiber orientation and green solid lines indicate the frontline of migrated cells. Green arrows show the maximum travel distance. (a) and (b): overview of hASC migration after 7 days; (c) and (d): cell migration parallel to fiber alignment; (e) and (f): cell migration perpendicular to fiber alignment. Color images available online at www.liebertonline.com/tea
FIG. 6.
Quantitative measurement of hASC migration on nanofiber meshes. Aligned-ll: migration parallel to fiber alignment; Aligned-⊥; migration perpendicular to fiber alignment. **Statistically significant, p<0.01.
Expression of integrins
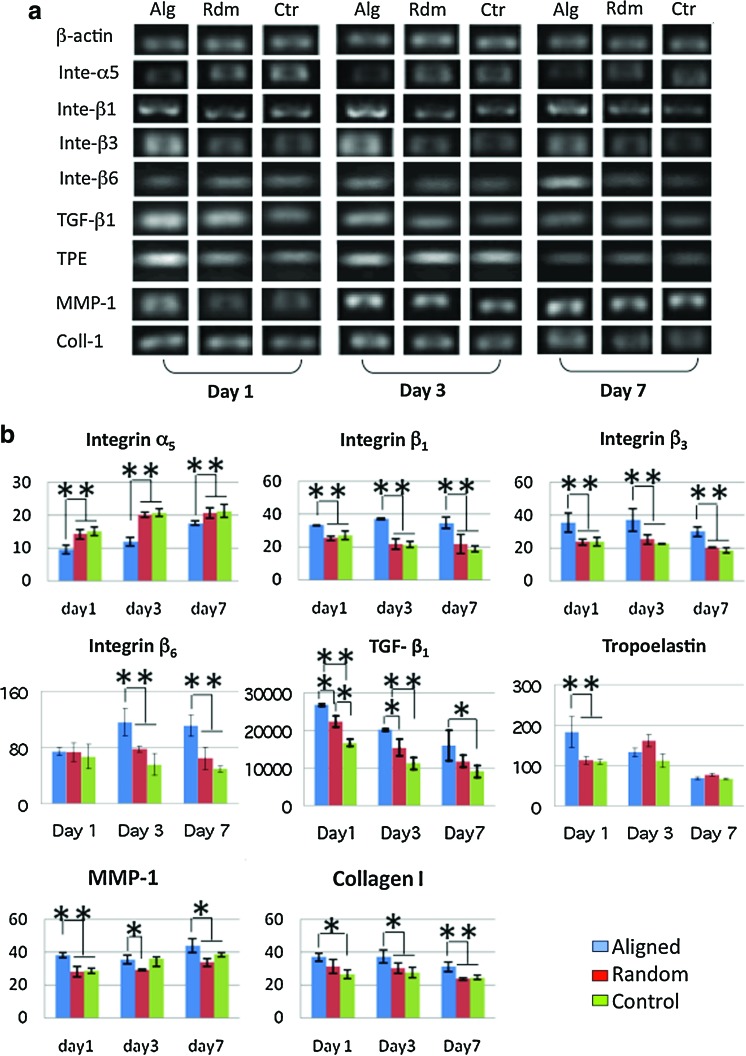
To determine whether integrins were involved in the fiber-orientation induced cell phenotypic change, gene expression for integrins (α2, α5, αv, β1, β3, and β6) in hASCs was analyzed using RT-PCR (Fig. 7). The results showed that gene expression of α2 and αv was not affected by various fiber and glass cover slip (controls) surfaces (data not shown). Western blot was also performed to determine the protein level of α2 and αv integrins on the cell membranes. Again, no noticeable difference was identified (data not shown). However, integrins α5, β1, β3, and β6 were significantly regulated by fiber orientation. A significant upregulation of β6 integrin was observed after 3 days. The fiber orientation upregulated the gene expression of integrins β1 and β3 as early as day 1 and remained for the rest of the experimental times (Fig. 7a, b). Somehow, the fiber orientation down-regulated integrin α5 expression for all the times investigated.
FIG. 7.
Gene expression of hASCs cultured on random (Rdm) and aligned (Alg) nanofiber meshes and glass coverslips (Ctr). (a) Representative electrophoresis gel. (b) Semiquantitative analysis of gene expression. The data presented were normalized with β-actin. *Statistically significant, p<0.05 and **p<0.001. Color images available online at www.liebertonline.com/tea
Expression of ECM related genes
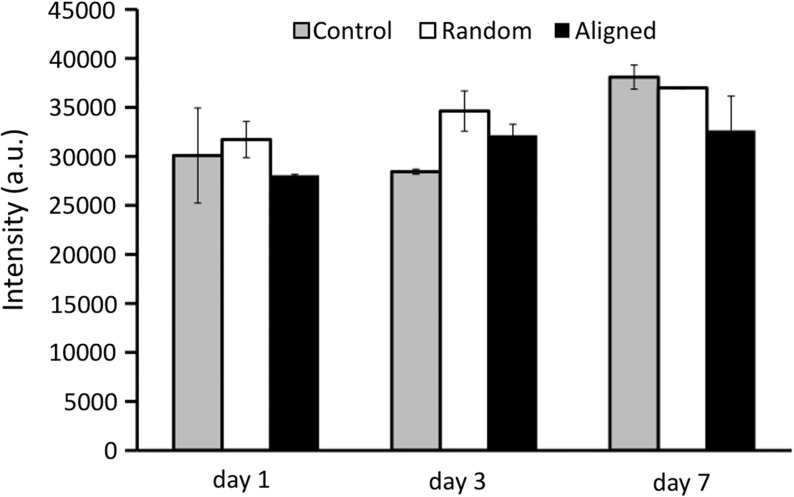
As part of the phenotypic expression of hASCs, the capability of regulating ECM turnover is a crucial measure. In this regard, the gene expression of major ECM proteins related to wound healing (i.e., collagen type I [Coll I], collagen type III [Coll III] and tropoelastin [TPE]) were particularly investigated. Interestingly, higher gene expression of Coll I was observed for the hASCs cultured on aligned nanofibers compared to those on random nanofibers on days 3 and 7 (Fig. 7a, b), while no difference was observed in the expression of Coll III (Supplementary Fig. S1; Supplementary Data are available online at www.liebertonline.com/tea). Significantly higher expression of TPE was found on the aligned nanofibers, but only on day 1 (Fig. 7a, b). ECM degrading matrix metalloproteinase (MMP-1, MMP-2, MMP-9) were also investigated. Gene expression of both MMP-2 and MMP-9 was not significantly different between the aligned and random nanofibers (data not shown). Gelatin zymography further confirmed the gene result with no significant difference in MMP-2 enzymatic activity between the two groups (Fig. 8). In contrast, noticeably high MMP-1 was consistently found in the hASCs cultured on aligned nanofibers (Fig. 7a, b). Gene expression for TGF-β1, an important ECM regulator, was also studied. It was found that TGF-β1 was highly upregulated by aligned nanofibers (Fig. 7a, b).
FIG. 8.
Semiquantification of matrix metalloproteinase (MMP)-2 activity as measured by zymography.
Fibroblastic differentiation of hASCs on nanofibers
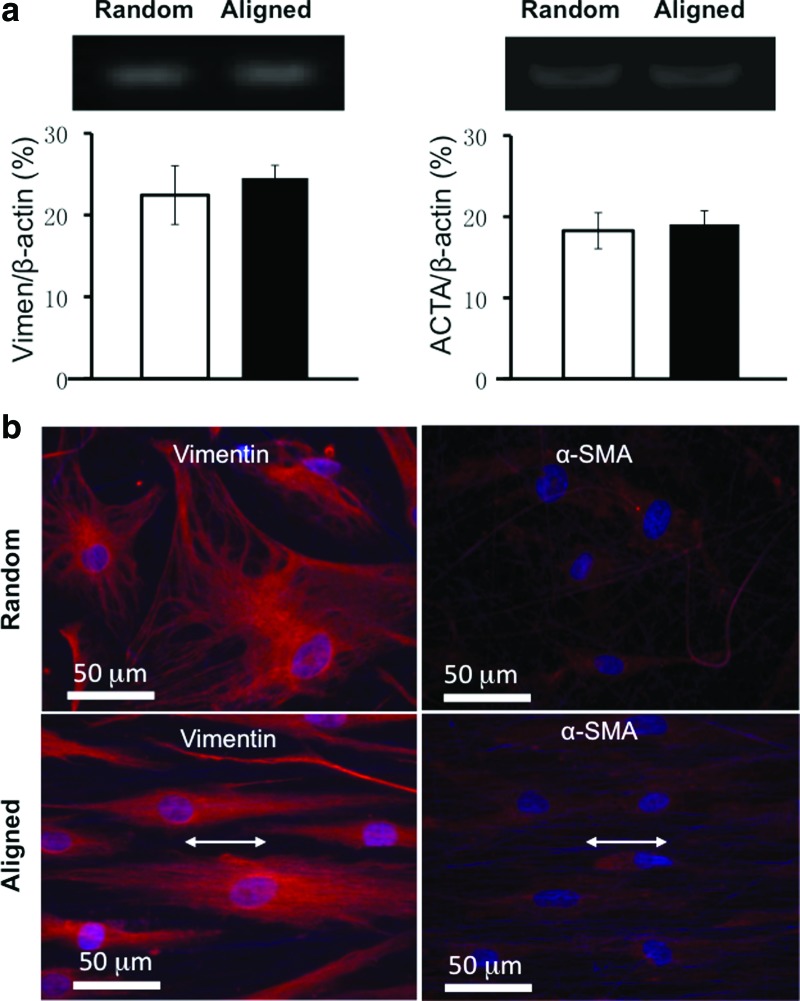
To determine the fibroblastic differentiation of hASCs on nanofibers, gene expression for vimentin, a reliable fibroblast marker, was evaluated using RT-PCR. Semiquantitative analysis showed that gene expression for vimentin was similarly detected on hASCs cultured on aligned or random nanofibers (Fig. 9a). Immunofluorescent staining for vimentin further confirmed the gene expression result. hASCs had a strong fluorescent staining for vimentin on both nanofiber meshes despite a distinct variation in spatial arrangement (spindle [aligned] vs. polygonal [random]; Fig. 9b), suggesting a fibroblastic phenotype. This observation remained consistent for all the time points up to 7 days (data not shown). α-SMA, involved in wound contraction, normally appeared in the wound myofibroblasts. To assess whether nanofiber orientation regulated the expression of α-SMA in hASCs, cells cultured on nanofiber meshes were immunofluorescently stained for α-SMA. No obvious myofibroblastic differentiation was detected in the hASCs either on aligned or random nanofibers (Fig. 9b). The expression level for α-SMA gene, (i.e., ACTA-2) was checked using RT-PCR. A low level expression was detected and it was comparable between aligned and random culture groups (Fig. 9a).
FIG. 9.
Fibroblastic differentiation of hASCs on nanofiber meshes. (a) Gene expression for vimentin (day 1) and alpha-actin-2 (ACTA-2) (day 3). (b) Immunofluorescent images for vimentin (day 1) and alpha-smooth muscle actin (α-SMA) (day 3). Double-head arrow indicates the fiber orientation. Scale bar: 50 μm. Color images available online at www.liebertonline.com/tea
Discussion
The advantages of electrospun nanofibers in promoting cell adhesion and maintaining cell phenotype, which mainly comes from the morphological similarity to native ECM fibrils, have been well recognized.19 As a result, extensive efforts have been made to explore the possible utility of such nanofibers for regenerating a variety of tissues including skin.25–31 It is known that native ECM not only provides a substrate for cells to attach, but also regulates cellular function via cell/matrix interactions. Inclusion of ECM molecules (e.g., type I collagen) into electrospun nanofibers can better recapitulate the bioactivity of ECM32 and therefore lead to cell/nanofiber interactions similar to the cell/ECM interactions in tissues. With the presence of type I collagen in electrospun nanofibers, hASCs can rapidly attach and spread on PCL/collagen nanofibers (Fig. 2). Moreover, fiber arrangements (aligned vs. random; Fig. 1) have a significant impact on the morphology of attached hASCs: cytoskeletal proteins (F-actin and α-tubulin) oriented along with the PCL/collagen nanofibers in the aligned group, but no preferred orientation in the random group (Fig. 2). Meanwhile, the hASCs on aligned PCL/collagen nanofibers are more elongated (long/short axis ratio: 11–17 [aligned] vs. 3 [random], p<0.001) with a lower mean cell area (Table 3). Although higher Young's modulus was measured on aligned PCL/collagen fibers in the orientation in other study compared to that of random ones (8.98±3.72 MPa vs. 2.83±0.60 MPa), the mechanical contribution to fiber-induced hASC response can be neglected in this study due to the use of rigid glass coverslips to collect nanofibers for cell culture, and no fiber detachment from coverslip surface has been observed during the culture. In addition, comparable fiber diameter has been measured between aligned and random nanofibers (Table 2); therefore, it is reasonable to hypothesize that the striking variation in cell morphology and other behaviors mainly comes from fiber arrangement. This fiber organization-induced cell morphology change has been reported for several types of cells, including cardiomyocytes, crucial ligament fibroblasts, and dermal fibroblasts.22,33,34 Although the exact mechanism is not fully understood, studies have shown that various integrins such as α2β1 are involved in fiber-induced cell adhesion.35,36 It is believed that integrins, particularly those involved in focal adhesion formation, are important in regulating cell morphology.37 Indeed, the size and spatial distribution of focal adhesion plaques in hASCs seems well correlated with cell morphology and are controlled by fiber arrangement, as observed from the immunofluorescent staining of vinculin. Compared to short and random focal adhesion plaques formed on random PCL/collagen nanofibers (Fig. 3a), long focal adhesion plaques were observed for hASCs cultured on the aligned fibers and they also oriented in the fiber alignment (Fig. 3b). The distinct vinculin distribution would inevitably lead to corresponding cytoskeletal arrangement as shown in Figure 2.
In this study, we also found that aligned PCL/collagen nanofiber meshes promoted the proliferation and migration of hASCs. Approximately 15% increase in cell number was consistently observed in the aligned group compared to random one (p<0.05) (Fig. 4), contradicting to the results observed with ligament fibroblasts,34 in which no significant difference in cell proliferation was noted between aligned and random nanofibers. We believe that the mitosis of fully differentiated cells like ligament fibroblasts will not be affected by the fiber organization, however, pluoripotent cells like hASCs may have various mitotic responses as a result of the commitment to different cell lineages induced by substrate topography.38,39 Accelerated migration of hASCs was also observed on the aligned nanofibers, however, only in the direction parallel to fiber alignment, but not in the perpendicular direction (Figs. 5, 6). This fiber orientation regulated cell migration was similarly reported with other cells.40 Cell migration is controlled by a highly orchestrated set of events involving the polymerization and depolymerization of actin filaments and the exertion of force through actinomyosin mediated contraction.41,42 The role of integrins in cell migration has been well recognized for both their involvement in binding to ECM and in regulating the turnover of intracellular cytoskeletal proteins.43 Gene expression for several integrin genes was particularly studied in this study and indeed a notable difference was observed between aligned and random nanofibers. Significantly higher expression of integrins β1 and β3 were consistently observed in the aligned group (Fig. 7). It is known that both integrin β1 and β343,44 are involved in cell migration, which suggests that accelerated hASC migration on aligned fibers may be the result of elevated integrins β1 and β3. Our ongoing efforts will further determine whether integrin β1 plays the determinant role by using a gene knockout model. In the gene expression study, significantly lower α5 was observed in the aligned group. Previous studies have shown that integrin α5β1 negatively regulates cell proliferation.45 The down-regulated α5 integrins may partially explain the promoted cell proliferation on the aligned group.
Further studies on several ECM-related genes showed that tropoelastin, collagen I, and MMP-1 were all upregulated by the aligned PCL/collagen nanofibers (Fig. 7). Active ECM synthesis and degradation is crucial to wound healing, especially in the case of a significant loss of tissue. Elevated expression of ECM-related genes implies the activation of ECM turnover, which may favor the migration and proliferation of fibroblast and keratinocytes and therefore facilitate wound healing. Our previous study demonstrated that increased deposition of collagen by hASCs in the skin grafts could significantly promote wound closure with accelerated re-epithelialization.16 TGF-β1 signaling, known for its essential role in regulating ECM turnover and the expression of integrin subunits that facilitate keratinocyte and fibroblast migration, and in inducing myofibroblastic differentiation that is involved in wound contraction,46–50 can come from either new synthesized TGF-β1 or those of activated latent ones.51,52 Although no TGF-β1 was supplemented to the culture in this study, upregulation of TGF-β1 gene was observed for hASCs cultured on both aligned and random nanofibers, and a significantly higher TGF-β1 expression was particularly observed in the aligned group (Fig. 7). Cumulative evidence indicates that integrins β6 can activate latent TGF-β through possible binding to latent association protein (LAP) to release the active TGF-β.53–55 Indeed, elevated gene expression for integrin β6 was observed for hASCs cultured on aligned nanofibers (Fig. 7). Thus, aligned nanofibers may not only promotes the synthesis of new TGF-β1 (most likely latent one) by hASCs, but also elevates the synthesis of integrin β6 to activate the latent TGF-β1 for initiating TGF-β1 signaling, which greatly regulates the production of new ECM molecules (collagen I and tropoelastin52 and MMP-156). However, the increased TGF-β1 did not induce hASCs to further differentiate into myofibroblastic phenotype (Fig. 9b), which instead still retained a high expression of fibroblastic marker protein of vimentin (Fig. 9a). Clearly, the enhanced migration and proliferation of hASCs as well as promoted ECM turnover by aligned PCL/collagen nanofibers would be beneficial to regenerate large wounds in consideration of both abundant hASCs and the biocompatible and biodegradable nanofibers. Our ongoing efforts that focus on the in vivo evaluation of the effect of hASCs and PCL/collagen nanofibers on wound healing will provide critical evidence to the in vitro findings reported in this study.
Taken together, it is clear that hASCs can detect the spatial arrangement of nanofibers by exhibiting various cell morphologies initially upon their attachment and then distinctively respond by showing various cell proliferations, migration, and synthesis of new ECM molecules. The fiber orientation-induced phenotypic change of hASCs is most likely regulated by integrins and TGF-β signaling synergistically.
Conclusions
This study for the first time systematically explored the effects of PCL/collagen nanofibers with distinct spatial fiber arrangements (aligned and random) on cellular response of hASCs in vitro. Our results clearly show the behavioral difference of hASCs on nanofibers in response to various spatial arrangements. hASCs grown on aligned scaffolds have elongated morphology, higher proliferation and migration rate, and higher synthesis of ECM molecules, all of which occur during wound healing process. Moreover, differential expression of integrins and TGF- β1 on various nanofiber scaffolds suggests their involvement in regulating the distinct cell behavior. These findings demonstrate the potential application of hASCs and aligned PCL/collagen nanofibers for accelerated wound repair.
Supplementary Material
Acknowledgments
The project described was supported by Grant Number 1R21 AR056416 from NIAMS. Xiaoling Fu is also supported by an Innovation and Entrepreneurship Doctoral Fellowship from Stevens Institute of Technology. The authors would like to thank Dr. Chengyang Huang for his scientific review and important discussion of this manuscript.
Disclosure Statement
No competing financial interests exist.
References
- 1.Wu S.C. Marston W. Armstrong D.G. Wound care: the role of advanced wound healing technologies. J Vasc Surg. 2010;52:59S. doi: 10.1016/j.jvs.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Shakespeare P. Burn wound healing and skin substitutes. Burns. 2001;27:517. [PubMed] [Google Scholar]
- 3.Ehrenreich M. Ruszczak Z. Update on tissue-engineered biological dressings. Tissue Eng. 2006;12:2407. doi: 10.1089/ten.2006.12.2407. [DOI] [PubMed] [Google Scholar]
- 4.Mansbridge J.N. Tissue-engineered skin substitutes in regenerative medicine. Curr Opin Biotechnol. 2009;20:563. doi: 10.1016/j.copbio.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Mason C. Tissue Engineering Skin: A Paradigm Shift in Wound Care. Med Device Technol. 2005;16:32. [PubMed] [Google Scholar]
- 6.Metcalfe A.D. Ferguson M.W.J. Tissue engineering of replacement skin: the crossroads of biomaterials, wound healing, embryonic development, stem cells and regeneration. J R Soc Interface. 2007;49:413. doi: 10.1098/rsif.2006.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shakespeare P.G. The role of skin substitutes in the treatment of burn injuries. Clin Dermatol. 2005;23:413. doi: 10.1016/j.clindermatol.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 8.Tran T.T. Kahn C.R. Transplantation of adipose tissue and stem cells: role in metabolism and disease. Nat Rev Endocrinol. 2010;6:195. doi: 10.1038/nrendo.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Branski L.K. Gauglitz G.G. Herndon D.N. Jeschke M.G. A review of gene and stem cell therapy in cutaneous wound healing. Burns. 2009;35:171. doi: 10.1016/j.burns.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim W.S. Park B.S. Sung J.H. Yang J.M. Park S.B. Kwak S.J. Wound healing effect of adipose-derived stem cells: a critical role of secretory factors on human dermal fibroblasts. J Dermatol Sci. 2007;48:15. doi: 10.1016/j.jdermsci.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 11.Park B.S. Jang K.A. Sung J.H. Park J.S. Kwon Y.H. Kim K.J. Adipose-derived stem cells and their secretory factors as a promising therapy for skin aging. Dermatol Surg. 2008;34:1323. doi: 10.1111/j.1524-4725.2008.34283.x. [DOI] [PubMed] [Google Scholar]
- 12.Rajashekhar G. Traktuev D.O. Roell W.C. Johnstone B.H. Merfeld-Clauss S. Van Natta B. IFATS collection: Adipose stromal cell differentiation is reduced by endothelial cell contact and paracrine communication: role of canonical Wnt signaling. Stem Cells. 2008;26:2674. doi: 10.1634/stemcells.2008-0277. [DOI] [PubMed] [Google Scholar]
- 13.Altman A.M. Yan Y. Matthias N. Bai X. Rios C. Mathur A.B. IFATS collection: Human adipose-derived stem cells seeded on a silk fibroin-chitosan scaffold enhance wound repair in a murine soft tissue injury model. Stem Cells. 2009;27:250. doi: 10.1634/stemcells.2008-0178. [DOI] [PubMed] [Google Scholar]
- 14.Tapp H. Deepe R. Ingram J. Kuremsky M. Hanley E.N. Gruber H.E. Adipose-derived mesenchymal stem cells from the sand rat: transforming growth factor beta and 3D co-culture with human disc cells stimulate proteoglycan and collagen type I rich extracellular matrix. Arthritis Res Ther. 2008;10:R89. doi: 10.1186/ar2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuji W. Inamoto T. Yamashiro H. Ueno T. Kato H. Kimura Y. Adipogenesis induced by human adipose tissue-derived stem cells. Tissue Eng Part A. 2009;15:83. doi: 10.1089/ten.tea.2007.0297. [DOI] [PubMed] [Google Scholar]
- 16.Wang H.J. Pieper J. Schotel R. van Blitterswijk C. Lamme E.N. Stimulation of skin repair is dependent on fibroblast source and presence of extracellular matrix. Tissue Eng. 2004;10:1054. doi: 10.1089/ten.2004.10.1054. [DOI] [PubMed] [Google Scholar]
- 17.Yang X. Shah J.D. Wang H. Nanofiber enabled layer-by-layer approach toward three-dimensional tissue formation. Tissue Eng Part A. 2009;15:945. doi: 10.1089/ten.tea.2007.0280. [DOI] [PubMed] [Google Scholar]
- 18.Ayres C.E. Jha B.S. Sell S.A. Bowlin G.L. Simpson D.G. Nanotechnology in the design of soft tissue scaffolds: innovations in structure and function. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2010;2:20. doi: 10.1002/wnan.55. [DOI] [PubMed] [Google Scholar]
- 19.Sill T.J. von Recum H.A. Electrospinning: applications in drug delivery and tissue engineering. Biomaterials. 2008;29:1989. doi: 10.1016/j.biomaterials.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Andrews K.D. Hunt J.A. Upregulation of matrix and adhesion molecules induced by controlled topography. J Mater Sci Mater Med. 2008;19:1601. doi: 10.1007/s10856-008-3377-6. [DOI] [PubMed] [Google Scholar]
- 21.Rossi A. Pasqui D. Barbucci R. Gerli R. Weber E. The topography of microstructured surfaces differently sffects fibrillin deposition by blood and lymphatic endothelial cells in culture. Tissue Eng Part A. 2009;15:525. doi: 10.1089/ten.tea.2007.0421. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y. Ji Y. Ghosh K. Clark A.F. Huang L. Rafailovich M.H. Effects of fiber orientation and diameter on the behavior of human dermal fibroblasts on electrospun PMMA scaffolds. J Biomed Mater Res Part A. 2009;90:1092. doi: 10.1002/jbm.a.32165. [DOI] [PubMed] [Google Scholar]
- 23.Guelcher S.A. Goldstein A.S. Effect of Fiber diameter and alignment of electrospun polyurethane meshes on mesenchymal progenitor cells. Tissue Eng Part A. 2009;15:2435. doi: 10.1089/ten.tea.2008.0295. [DOI] [PubMed] [Google Scholar]
- 24.Jones L.J. Gray M. Yue S.T. Haugland R.P. Singer V.L. Sensitive determination of cell number using the CyQUANT cell proliferation assay. J Immunol Methods. 2001;254:85. doi: 10.1016/s0022-1759(01)00404-5. [DOI] [PubMed] [Google Scholar]
- 25.Agarwal S. Wendorff J.H. Greiner A. Progress in the field of electrospinning for tissue engineering applications. Adv Mater. 2009;21:3343. doi: 10.1002/adma.200803092. [DOI] [PubMed] [Google Scholar]
- 26.Aviss K.J. Gough J.E. Downes S. Aligned electrospun polymer fibres for skeletal muscle regeneration. Eur Cell Mater. 2010;19:193. doi: 10.22203/ecm.v019a19. [DOI] [PubMed] [Google Scholar]
- 27.Cai Z.X. Mo X.M. Zhang K.H. Fan L.P. Yin A.L. He C.L. Fabrication of chitosan/silk fibroin composite nanofibers for wound-dressing applications. Int J Mol Sci. 2010;11:3529. doi: 10.3390/ijms11093529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nandakumar A. Fernandes H. de Boer J. Moroni L. Habibovic P. van Blitterswijk C.A. Fabrication of bioactive composite scaffolds by electrospinning for bone regeneration. Macromol Biosci. 2010;10:1365. doi: 10.1002/mabi.201000145. [DOI] [PubMed] [Google Scholar]
- 29.Wu H. Fan J. Chu C.C. Wu J. Electrospinning of small diameter 3-D nanofibrous tubular scaffolds with controllable nanofiber orientations for vascular grafts. J Mater Sci Mater Med. 2010;21:3207. doi: 10.1007/s10856-010-4164-8. [DOI] [PubMed] [Google Scholar]
- 30.Xu C. Aligned biodegradable nanofibrous structure: a potential scaffold for blood vessel engineering. Biomaterials. 2004;25:877. doi: 10.1016/s0142-9612(03)00593-3. [DOI] [PubMed] [Google Scholar]
- 31.Yoshimoto H. A biodegradable nanofiber scaffold by electrospinning and its potential for bone tissue engineering. Biomaterials. 2003;24:2077. doi: 10.1016/s0142-9612(02)00635-x. [DOI] [PubMed] [Google Scholar]
- 32.Rho K.S. Jeong L. Lee G. Seo B.M. Park Y.J. Hong S.D. Electrospinning of collagen nanofibers: effects on the behavior of normal human keratinocytes and early-stage wound healing. Biomaterials. 2006;27:1452. doi: 10.1016/j.biomaterials.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Zong X. Bien H. Chung C.Y. Yin L. Fang D. Hsiao B.S. Electrospun fine-textured scaffolds for heart tissue constructs. Biomaterials. 2005;26:5330. doi: 10.1016/j.biomaterials.2005.01.052. [DOI] [PubMed] [Google Scholar]
- 34.Moffat K.L. Kwei A.S.P. Spalazzi J.P. Doty S.B. Levine W.N. Lu H.H. Novel nanofiberbased scaffold for rotator cuff repair and augmentation. Tissue Eng Part A. 2009;15:115. doi: 10.1089/ten.tea.2008.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santoro S.A. Walsh J.J. Staatz W.D. Baranski K.J. Integrin-mediated platelet adhesion and platelet activation C-NH2. Cell. 1991;2:905. doi: 10.1091/mbc.2.11.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vandenberg P. Ries A. Luckenbill-edds L. Characterization of a Type IV Collagen Major Cell Binding Site with Affinity to the cd/ l and the a2 1 Integrins. Cell. 2000;113:1475. doi: 10.1083/jcb.113.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boudreau N.J. Jones P.L. Extracellular matrix and integrin signalling: the shape of things to come. Biochem J. 1999;339:481. [PMC free article] [PubMed] [Google Scholar]
- 38.Baker B.M. Nathan A.S. Gee A.O. Mauck R.L. The influence of an aligned nanofibrous topography on human mesenchymal stem cell fibrochondrogenesis. Biomaterials. 2010;31:6190. doi: 10.1016/j.biomaterials.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozturk N. Girotti A. Kose G.T. Rodríguez-Cabello J.C. Hasirci V. Dynamic cell culturing and its application to micropatterned, elastin-like protein-modified poly(N-isopropylacrylamide) scaffolds. Biomaterials. 2009;30:5417. doi: 10.1016/j.biomaterials.2009.06.044. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y. Franco A. Huang L. Gersappe D. Clark R.F. Rafailovich M.H. Control of cell migration in two and three dimensions using substrate morphology. Exp Cell Res. 2009;315:2544. doi: 10.1016/j.yexcr.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 41.Clark P. Connolly P. Curtis A.S.G. Dow J.A.T. Wilkinson C.D.W. Cell guidance by ultrafine topography in vitro. J cell Sci. 1991;99:73. doi: 10.1242/jcs.99.1.73. [DOI] [PubMed] [Google Scholar]
- 42.Wang J.H.C. Jia F. Gilbert T.W. Woo S.L.Y. Cell orientation determines the alignment of cell-produced collagenous matrix. J Biomech. 2003;36:97. doi: 10.1016/s0021-9290(02)00233-6. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt S. Friedl P. Interstitial cell migration: integrin-dependent and alternative adhesion mechanisms. Cell Tissue Res. 2010;339:83. doi: 10.1007/s00441-009-0892-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garchía A.J. Get a grip: integrins in cell-biomaterial interactions. Biomaterials. 2005;26:7525. doi: 10.1016/j.biomaterials.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 45.Davey G. Buzzai M. Assoian R.K. Reduced expression of (alpha)5(beta)1 integrin prevents spreading-dependent cell proliferation. J Cell Sci. 1999;112:4663. doi: 10.1242/jcs.112.24.4663. [DOI] [PubMed] [Google Scholar]
- 46.Postlethwaite A.E. keski- Oja J. Moses H.L. Kang A.H. Stimulation of the chemotactic migration of human fibroblasts by transforming growth factor beta. J Exp Med. 1994;165:251. doi: 10.1084/jem.165.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ellis I.R. Schor S.L. Differential effects of TGF-β1 on hyaluronan synthesis by fetal and adult skin fibroblasts: implications for cell migration and wound healing. Exp Cell Res. 1996;228:326. doi: 10.1006/excr.1996.0332. [DOI] [PubMed] [Google Scholar]
- 48.Evans R.A. Tian Y.C. Robert Steadman and Aled Owain Phillips, TGF-β1- mediated fibroblast-myofibroblast terminal differentiation- the role of smad proteins. Exp Cell Res. 2003;282:90. doi: 10.1016/s0014-4827(02)00015-0. [DOI] [PubMed] [Google Scholar]
- 49.Vaughan M.B. Howard E.W. Tmasek J.J. Transforming growth factor-β1 promotes the morphological and functional differentiation of the myofibroblast. Exp Cell Res. 2001;1:180. doi: 10.1006/excr.2000.4869. [DOI] [PubMed] [Google Scholar]
- 50.Gailit J. Welch M.P. Clark R.A.F. TGF-β1 stimulates expression of keratinocyte integrins during re-epithelialization of cutaneous wounds. J Invest Dermatol. 1994;103:221. doi: 10.1111/1523-1747.ep12393176. [DOI] [PubMed] [Google Scholar]
- 51.Hynes R.O. The extracellular matrix: not just pretty fibrils. Science. 2009;326:1216. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schultz G.S. Wysocki A. Interactions between extracellular matrix and growth factors in wound healing. Wound Repair Regen. 2009;17:153. doi: 10.1111/j.1524-475X.2009.00466.x. [DOI] [PubMed] [Google Scholar]
- 53.Annes J.P. Chen Y. Munger J.S. Rifkin D.B. Integrin alphaVbeta6-mediated activation of latent TGF-beta requires the latent TGF-beta binding protein-1. J Cell Bio. 2004;165:723. doi: 10.1083/jcb.200312172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jenkins R.G. Su X. Su G. Scotton C.J. Camerer E. Laurent G.J. Davis G.E. Chambers R.C. Matthay M.A. Sheppard D. Ligation of protease-activated receptor 1 enhances αvβ6 integrin – dependent TGF-β activation and promotes acute lung injury. J Clin Invest. 2006;116:1606. doi: 10.1172/JCI27183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keski-oja J. Koli K. Melchner H.V. TGF-b activation by traction? Trends Cell Biol. 2004;14:657. doi: 10.1016/j.tcb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 56.Boyd D.D. Regulation of matrix metalloproteinase gene expression. J Cell Physiol. 2007;211:19. doi: 10.1002/jcp.20948. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.