Abstract
This review discusses polymicrobial interactions with the host in both health and disease. As our ability improves to identify specific bacterial clonal types both with respect to abundance and location in the oral biofilm we will learn more concerning their contribution to both oral health and disease. Recent studies examining host-bacterial interactions have revealed that commensal bacteria not only protect the host simply by niche occupation, but that bacterial interactions with host tissue can promote the development of proper tissue structure and function. These data indicate that our host-associated polymicrobial communities, such as those found in the oral cavity, co-evolved with us and have become an integral part of who we are. Understanding the microbial community factors that underpin associations with host tissue that contribute to periodontal health may also reveal how dysbiotic periodontopathic oral communities disrupt normal periodontal tissue functions in disease. A disruption of the oral microbial community creates dysbiosis, either by overgrowth of specific or nonspecific microorganisms or changes in the local host response where the community can now support a disease state. Dysbiosis provides the link between systemic changes (e.g., diabetes), exogenous risk factors (e.g., smoking), and the dysbiotic community and can drive the periodontal tissue destruction. Many other risk factors associated with periodontal disease such as stress, aging, and genetics also likely affect the microbial community, and more research is needed utilizing sophisticated bacterial taxonomic techniques to better elucidate these effects on the microbiome and to develop strategies to target the dysbiotic mechanisms and improve periodontal health.
The etiology of periodontitis has been elusive. Hypotheses have ranged from microbial etiologies to those that employ trauma or disuse as possible reasons for the localized tissue destruction and alveolar bone loss characteristic of the disease [54]. The observation that careful removal of dental plaque, the microbial biofilm that coats the tooth and tooth root surface was an effective approach to reduce both gingivitis and periodontitis led to the clear recognition that oral microbes somehow contributed to the disease process [38]. Subsequently the importance of the host component to the disease process was recognized [47]. Periodontitis was proposed to be a dysfunctional host response to the oral biofilm that was altered in both the amounts and types of bacteria. However, the contribution of oral bacteria to the disease process remains unclear. Hypotheses that purport specific periodontopathic bacteria to those that claim a nonspecific bacterial etiology are both applicable today as well as the idea that it is the host that drives the microbial changes with contribution of microbes to the disease process as secondary [4]. These positions are not mutually exclusive, and it may be that all are correct in different situations.
However, at least one component of periodontitis is clear; the composition of the oral microbial biofilm is significantly different in healthy sites when compared to clinically defined disease sites [12, 43]. This observation, combined with advances in characterizing the microbial composition of host associated polymicrobial communities, and the recognition that these microbial communities have well evolved and intricate communication systems between themselves as well as the host has led to the notion that novel therapeutic interventions may arise from a better understanding of the oral microbial community. Clinical advances have already been made in the treatment of certain forms of inflammatory bowel disease where polymicrobial intestinal transplants have shown promise in restoring a healthy relationship between the intestine and its microbial inhabitants [30]. Furthermore, new therapeutic regimens involving selective bacterial killing [1, 18, 58], probiotic [42, 48, 60], and prebiotic [70] approaches all require a more intimate knowledge of the polymicrobial dependencies for survival in the oral biofilm [49, 50, 73].
Therefore, this review emphasizes polymicrobial interactions with the host in both health and disease. It is hoped that by gaining a better understanding of how bacterial communities contribute to our oral health and the effects of altering these communities on disease, more accurate and useful diagnostic and therapeutic interventions can be elucidated.
Polymicrobial analysis
The analysis of polymicrobial communities is severely hampered by the application of the Linnaeus system for the definition of a species. Species is defined for most forms of animal and plant life when two individuals, male and female, can produce fertile offspring [16]. With the discovery of “animalcules” as van Leeuwenhoek called bacteria it was only natural that they would be classified according to the system employed for all other forms of life. However, the main mechanisms of bacterial genetic recombination do not involve sexual reproduction, and, therefore, it has been recognized for quite some time that the use of genus and species names for bacteria is inadequate. We have survived this inadequacy by employing a variety of genotypic and phenotypic typing systems to better understand those bacterial species that are significantly associated with different disease states.
A different approach for bacterial taxonomy employing the sequence of 16S rRNA, a highly conserved bacterial gene involved in building the scaffolding for protein synthesis has been found to be extremely useful for describing the bacterial members of polymicrobial communities. Employing the 16S rRNA sequence was first championed by Dr. Carl Woese [67] and now is routinely being used to describe the microbial diversity found in polymicrobial communities including dental plaque [11]. There are a number of different 16S rRNA sequencing techniques which either allow a more definitive description of the species composition by determining the entire 16S rRNA sequence or deep sequencing applications that reveal low abundance members of the community albeit at a less definitive species resolution. One feature that all 16S rRNA analyses of polymicrobial communities have revealed is that there is much more microbial species diversity in dental plaque than previously thought. The main reason for this is that the 16S sequence approach does not rely on culturing methods, and hence those bacterial species that are not yet cultivatable can now be detected. The actual amount of microbial diversity reported by different sequence analysis studies varies due to mostly technical reasons [11]. However, the presence of a large number of bacterial species not yet cultivated presents new opportunities to understand polymicrobial dental plaque. For example, understanding the metabolic and structural co-dependent interactions which preclude cultivation will most certainly reveal novel bacterial-bacterial and bacterial–host interactions that help shape either periodontal health or disease.
However, characterization of the oral polymicrobial community even by the most discriminating and complete 16S rRNA approach is insufficient to adequately describe the microbial composition. The reason for this is the presence of bacterial genes that differ among nearly identical 16S rRNA species. This is a result of the genetic recombination mechanisms employed by bacteria such as transformation where select genes can be transferred into the bacteria genome to render that specific bacterial clone a novel phenotype. If the phenotype confers a selective advantage, such as an improved ability to use an available nutrient source, in the niche where the gene is acquired, the clone can multiply and become a new member of the polymicrobial community. These events are common and contribute to the dynamic nature of the oral microbial community [64]. In fact, the acquisition of new genes is so common within the same bacterial species that the terms pan and core bacterial genomes has entered our lexicon [8]. A pan genome represents all the genes found in a specific species and includes the core genome, which are highly conserved within the species, as well as the unique genes that are only found in specific strains or clones of the species [44]. Identification of specific clones within a particular species will reveal the true diversity of polymicrobial communities. For example, it has been found in the nasopharyngeal cavity that different clones of Streptococcus mitis, Streptococcus oralis and Streptococcus infantis can appear and disappear as well as fluctuate in their relative amounts over time in the same individual [5]. More relevant to periodontitis, the highly leukotoxic JP2 clone of Aggregatibacter actinomycetemcomitans has been reported to be an etiologic agent of localized aggressive (juvenile) periodontitis [26].
As our ability improves to identify specific bacterial clonal types both with respect to abundance and location in the oral biofilm we will learn more concerning their contribution to oral health and disease. For example, identifying bacterial functions which confer a selective advantage to bacterial clones will reveal those functions of community structure that contribute both to our periodontal health, as well as bacterial functions that may directly relate to disease. Accordingly, novel probiotic, prebiotic, and selective bacterial killing approaches can then be initiated.
The healthy microbiome
The oral cavity is highly populated with numerous polymicrobial communities each occupying highly specific niches that differ in both anatomical location and as well as nutrient availability [43]. Oral host colonization is a reflection of the proficient ability of bacteria to adapt to a variety of different niches through high rates of genetic recombination as discussed above. One consequence of robust commensal bacteria colonization is to prevent pathogenic bacterial colonization through a process termed colonization resistance. Colonization resistance has long been recognized as an important function of commensal bacteria, and the detrimental effects of commensal depletion by broad spectrum antibiotic application have been documented [7, 28]. However, more recently studies examining host-bacterial interactions have revealed that commensal bacteria not only protect the host simply by niche occupation, but that bacterial interactions with host tissue promote the development of proper tissue structure and function. These data indicate that our host associated polymicrobial communities, such as those found in the oral cavity, co-evolved with us and have become an integral part of who we are [39]. Therefore a more transcendent view of human biology where we begin to understand how this microbial component is integrated into human physiology (considering the microbiome as a human organ) [3] may be helpful to understanding how these polymicrobial communities can become dysfunctional in their interactions with host tissues and then impair proper host tissue structure and function.
The most significant and clear studies with respect to the ability of polymicrobial commensal communities to actively mature proper tissue structure and function are found in the intestine [22, 31, 41, 66, 68]. These studies have heavily relied upon comparisons in tissue structure and function between normal or specific-pathogen-free and germ-free mice. Germ-free mice are completely devoid of bacteria and are generated by sterile Caesarean section, raised aseptically in an isolator with sterile filtered air, and are housed using sterile food, water, and bedding. Specific-pathogen-free mice are only devoid of known mouse pathogens and contain a normal repertoire of oral and intestinal bacteria [41]. Therefore, comparisons between these two mouse groups can reveal those attributes of host tissue that are formed in response to bacterial colonization. For example, early studies in germ-free mice demonstrated that commensal bacteria have a direct impact on the morphology of the intestine [17, 20]. Bacteria are responsible for degradation of mucus glycoproteins, and their absence results in an enlargement of the cecum. The villi of the small intestine are longer and the crypts are shorter and contain fewer cells in germ-free animals. In addition, commensal bacteria have effects on intestinal motility. Effects of bacteria can also be seen on the development and function of the intestinal immune system. Commensal bacteria are required for the complete development of Peyer’s patches, the lamina propria, and the intraepithelial spaces, all three of the main immune elements found in the intestine [17, 20]. More recently, studies in germ-free mice have revealed that the commensal bacteria induce angiogenesis, contributing to the development of the complex vascular beds found just underneath the mucosal surface [57]. Furthermore, it has been found that constitutive intercellular adhesion marker-1 expression in these vessels is also regulated by the presence of the commensal microbiota [35]. In fact the state of “controlled” inflammation that normally exists in the intestine has been attributed to both the quality and quantity of intestinal commensal microorganisms [9, 10].
The human oral cavity is similar to the intestinal system in that both contain highly evolved polymicrobial communities, and both have state of “controlled” inflammation [9, 36, 46]. However significantly less is understood concerning the contribution of the oral polymicrobial community on oral tissue structure and function. This is somewhat surprising since one well described manifestation of the inflammatory surveillance that occurs in healthy periodontal tissue is the constant translocation of neutrophils from the highly vascularized periodontal tissue into the gingival sulcus [63]. Neutrophil migration out of the vasculature and into the surrounding tissue is a key protective attribute of periodontal tissue and is highly regulated such that too much or too little migration can both result in disease [19, 53]. Since neutrophil migration is well known to respond to bacterial inflammatory signals in other tissues, it would be expected to be regulated either directly or indirectly by the oral bacterial polymicrobial community. Indeed studies have revealed that healthy human junctional epithelium, the tissue in closest contact with the polymicrobial oral community, express an interleukin-8 gradient to guide neutrophils through periodontal tissue toward the sulcus, and it was postulated that oral commensal colonization was the stimulus [62]. However, initial studies in germ-free rats demonstrated that neutrophils were found transiting in junctional epithelial tissue, indicating that commensal bacterial colonization was evidently not required for this key protective mechanism [51]. Furthermore, the expression of carcinoembryonic antigen-related cell adhesion molecule-1, a cell adhesion molecule postulated to contribute the structural integrity of this loosely organized tissue, and secretory leukocyte protease inhibitor, a molecule that protects the host from host-mediated protease-induced tissue damage, is specifically expressed in the junctional epithelium, yet studies in germ-free mice demonstrated that bacterial colonization was not required for their expression [27, 29]. These observations demonstrate that certain structural and functional aspects of periodontal tissue, specifically the junctional epithelium, the tissue in closest contact with the oral polymicrobial community, are host developmentally driven even in the absence of bacterial signals.
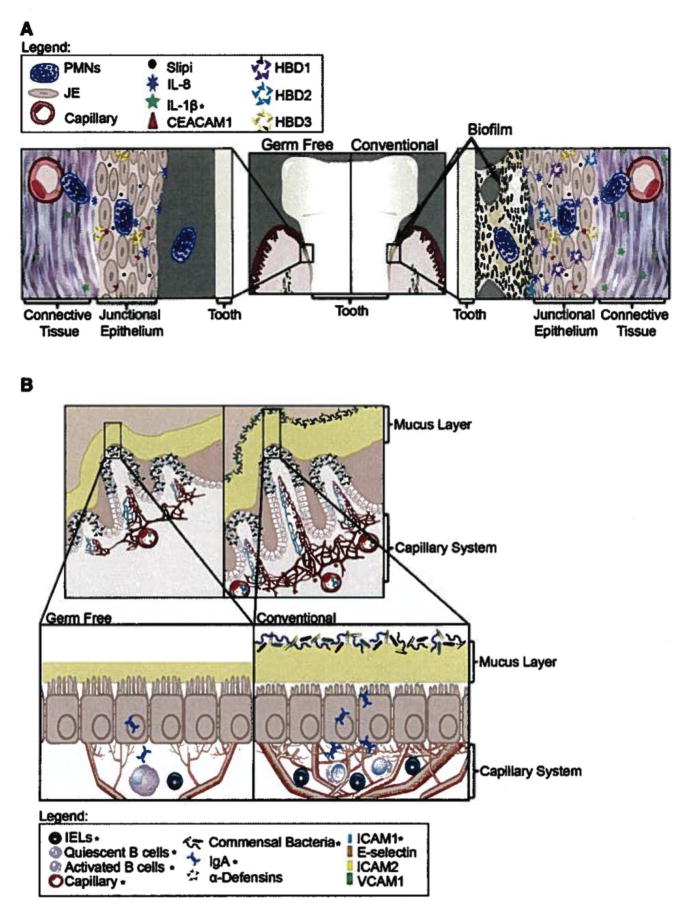
More recent studies have shown that the effect of bacterial colonization of the periodontium is more subtle than that observed in intestinal tissue (Fig. 1) [11]. Although neutrophils are present in the junctional epithelial tissue of germ-free mice, two independent studies have found that commensal colonization significantly increases the number of neutrophils found in gingival tissue [65, 71]. Therefore, a key protective mechanism found in periodontal tissue, the transit of neutrophils into the gingival space [24](45), is orchestrated by both host developmental and bacterial induced programs. Furthermore, it has been shown that the chemokine ligands that attract and direct neutrophil migration in mouse periodontal tissue are also programmed by both host- and bacterially-induced programs [71]. However, in this case the host developmental program is associated with chemokine (C-X-C motif) ligand 1 (CXCL1) whereas commensal colonization specifically induces the expression of the closely related neutrophil chemokine ligand CXCL2 through the myeloid differentiation primary response gene (88) toll-like receptor (MYD88-TLR) activation pathway [71]. Therefore, clearly, the oral polymicrobial community significantly contributes to the periodontal innate host defense status in clinically healthy tissue; however, their contribution is superimposed on host developmental programs.
Fig. 1.
(A) Current knowledge of microbial influence on the junctional epithelium based on cumulative data from human and mouse studies. The architecture of junctional epithelium and the presence of neutrophils are similar between germ-free and conventional mice. Several molecules appear to change dramatically with the addition of bacteria but many are unchanged [14]. (B) Overview of current knowledge of microbial influence on the intestinal epithelium. The architecture of the intestinal tissue is changed markedly with the addition of bacteria: the crypts are deeper, the capillary network is more extensive, the mucus layer is reduced, cilia are shorter, and many differences are seen with immune cells and molecules as indicated [32]. The figure indicates the relative location and abundance of innate immune cells/molecules (*indicates changes due to microbial interactions confirmed in germ-free studies.) Reprinted with permission from Cell Host & Microbe 10, October, 2011 ©2011 Elsevier Inc.
Periodontal tissue is similar to intestinal tissue with respect to polymicrobial communities significantly contributing to creating a proper functioning tissue. Understanding the microbial community factors that underpin associations with host tissue that contribute to periodontal health may also reveal how dysbiotic periopathogenic oral communities disrupt normal periodontal tissue functions in disease. It appears that bacterial interactions contributing to periodontal health may be just as complicated as those seen in disease.
Dysbiosis: change in composition and numbers of oral microbiome; the unhealthy microbiome
Periodontitis is a dysbiotic disease [6]. This is not a new concept, rather it is a better name to describe what was previously described as a shift from mostly gram positive bacteria found in healthy sites to mostly gram negative bacteria found in clinically diseased sites [43]. Dysbiosis, as the term implies, is a symbiosis that has gone awry. It is the concept that some diseases are due to a decrease in the number of beneficial symbionts and/or an increase in the number of pathogens [6]. Instead of contributing to healthy tissue function as described above, the oral polymicrobial community interferes with normal periodontal tissue function. This is the definition of disease, when normal tissue function is disturbed. There are numerous cases of dysbiotic diseases including inflammatory bowel disease, otitis media, and bacterial vaginosis [6]. A key contribution of describing periodontitis as a dysbiotic disease is its emphasis on bacterial community and the ramifications thereof in understanding periodontitis etiology. For example, one ramification of bacterial community thinking is that not all bacteria in the community need to have the same function. This notion was recently demonstrated where it was shown that Porphyromonas gingivalis, a designated periodontopathogen [55], required other members of the oral microbial community to elicit periodontitis in a mouse model of disease [23]. In this study, germ-free mice failed to develop periodontitis when administered P. gingivalis alone, in contrast specific-pathogen-free mice that contained a normal repertoire of oral commensal bacteria did develop disease. Furthermore, it was shown that although P. gingivalis was a minor component of the oral microbial community after administration, its presence was associated with a significant increase in the quantity of the rest of the oral polymicrobial community. Therefore, the presence of even small amounts of P. gingivalis in this mouse model of disease created a dysbiotic oral polymicrobial community as evidenced by the increase in bacterial numbers. It was found that the ability of P. gingivalis to sabotage the mouse complement system was likely responsible for the increase in the oral microbial community resulting in alveolar bone loss. Therefore, the function of P. gingivalis in this community setting was to interfere with host protective mechanisms facilitating the overgrowth of the entire polymicrobial community.
The idea that P. gingivalis inhibited innate host functions as a service to the community by allowing it to gain access to the needed protein rich substrates in gingival crevicular fluid was originally proposed when it became apparent that effects such as P. gingivalis lipopolysaccharide inhibition of host toll-like receptor-4 responses had the potential to affect the entire oral polymicrobial community [13]. Based upon these data P. gingivalis was proposed to be a keystone species in the oral polymicrobial community and provided another scientific rationale for its high association with disease. Interference with numerous different host component protection mechanisms are likely to alter the homeostatic balance that evolution has produced for proper functioning periodontal tissue [13, 15]. This is becoming evident when examining periodontitis that originates in select knockout mice that have reduced innate defense functions that are critical to neutrophil migration such as CXCR2 [23, 69] and double E-and P-selectin knockout mice [45]. In both of these models of disease a dysbiotic oral polymicrobial community has been described. The observation that impairment of host protective mechanisms creates dysbiotic oral polymicrobial communities, which are altered in both their composition and numbers, lends credence to the nonspecific hypothesis [40, 61] for the etiology of periodontitis. Numerous different types of changes in the oral polymicrobial community can result in disease and no one bacterium is directly responsible for the observed bone destruction.
However, a challenge to the nonspecific hypothesis was recently described in a different mouse model of disease [33]. In this model silk ligatures are tied around the base of select teeth to induce disease, whereas in the study described above disease is initiated by overwhelming repeated inoculations with P. gingivalis by oral gavage. In the ligature model wild-type mice developed disease as determined by alveolar bone loss, yet disease did not occur in nucleotide-binding oligomerization domain-containing protein-1 (Nod1) knockout mice [33]. Nod1 is a pattern recognition receptor able to detect the presence of bacterial peptidoglycan. Although this could be thought of as another example of how innate defense functions are required for proper maintenance of the oral polymicrobial community, the authors demonstrated that in fact it was a single member of the oral polymicrobial commensal micobiome, designated NI1060, which accumulated in the community and could initiate the disease when administered alone. This member of the community, with characteristics similar to A. actinomycetemcomitans, was able to induce Nod1 responses, and these were sufficient to result in alveolar bone loss. This study lends support to the notion that indeed there are specific periopathogens, and that creating dysbiotic situations (such as ligature placement) facilitates their outgrowth and subsequent detrimental effects on periodontal function. Local factors in the oral cavity such as heavy calculus, restoration overhangs, areas of food impaction, or even active caries might produce similar dysbiotic effects in humans.
Finally, the fact that the inflammatory response alters the periodontal environment precludes definitive conclusions concerning if the dysbiotic community created the inflammation or if the inflammation created the dysbiotic community [4]. It has been long recognized that periodontitis clearly does not fit into Koch’s postulates, and the inflammatory environment of disease could alter the polymicrobial community. Therefore, conclusions concerning the periodontopathic potential of bacteria routinely found in diseased sites have usually been interpreted with caution and the word “association” being employed as opposed to causal. However, more recent observations that a new class of compounds that actively resolve inflammation, appropriately termed resolvins, can restore the dysbiotic community to an almost normal healthy composition [25], has prompted a reevaluation of the contribution of the oral polymicrobial community to periodontitis etiology [4]. It is suggested that host changes to the periodontal environment are sufficient to create periodontal polymicrobial dysbiosis, and simply reducing the host inflammatory response nearly restores healthy homeostasis. This is the most likely explanation for periodontitis observed in select innate host deficient knockout mice such as CXCR2 [69] and P- and E-selectin mice [45] where the oral polymicrobial community is dysbiotic and “natural” periodontitis is observed. In addition, it is also likely that in certain individuals dysregulated inflammation due to undefined host factors may be the cause of the dysbiotic community and, therefore, resolution of the inflammation should be sufficient to treat both the dysbiosis and periodontitis.
Therefore, the non-specific, specific, and host mediated etiologies for the development of periodontitis can all be observed in different animal models of disease. However, these etiologies must be taken with some caution since it is not clear how each of these animal models will translate to humans. For example, in humans, there is fairly good evidence that A. actinomycetemcomitans JP2 is responsible for many cases of localized aggressive periodontitis, which affects adolescents of African descent [26]. However more recent studies have revealed that A. actinomycetemcomitans may be necessary but insufficient by itself to cause disease in humans but rather a consortium of this bacterium with Streptococcus parasanguinis and Filifactor alocis may be more predictive of disease [34]. This data is consistent with the notion that even in situations where a clear periopathogen was purportedly identified, other members of the dysbiotic community may significantly contribute to disease. For example if a community member functions to provide a key metabolite to the pathogen, then drugs that inhibit production of the metabolite by the community member could decrease pathogenicity indirectly. This reinforces our need to understand the dysbiotic community and suggests that novel therapeutic intervention strategies may arise from learning more about bacterial-bacterial interactions [49, 50, 73] in the context of host responses.
What causes dysbiosis?
In animal models of periodontal disease we know that both environmental and genetic factors can create dysbiotic oral polymicrobial communities. For example, both antibiotic pre-treatment and gavage [2, 23] as well as tying ligatures around the base of teeth [25, 33, 72] both create environmental conditions which alter the amount and composition of the oral microbial community and result in loss of alveolar bone. Likewise, CXCR2 [23, 69], E- and P-selectin [45], and lymphocyte function-associated antigen-1 [23] knockout mice all naturally develop a periodontitis that is associated with a change in the amount and composition of the oral polymicrobial community.
In humans, periodontitis risk factors also can be categorized as environmental or genetic although less is known about how these may create dysbiotic communities. Environmental factors include smoking and obesity [21], and both have been shown to be associated with sub gingival and intestinal dysbiotic communities, respectively. In fact, it has recently been found that smoking creates a sub gingival microbiome in healthy sites that more closely resembles diseased sites, suggesting that smoking creates “an at-risk-for-harm environment” to create periodontitis [37]. Diabetes is also associated with periodontitis and as it has been postulated [21] a more complete analysis of the periodontal microbiome is necessary to determine if this disease is associated with a dysbiotic periodontal community. Human genetic risk factors have been and remain the subject of numerous studies, which have been reviewed elsewhere [56, 59]. It will be interesting to see in the future if any of the putative periopathogenic single nucleotide polymorphisms in host response and other genes [52] are associated with dysbiotic oral communities.
Conclusion
The oral microbial community has evolved in symbiosis with the human periodontium to usually create health. A disruption of this community creates a dysbiosis, either by overgrowth of specific or nonspecific microorganisms or changes in the local host response where the community can now support a disease state (Fig. 2). Dysbiosis provides the link between systemic changes (diabetes), exogenous risk factors (smoking), and the dysbiotic community and drives periodontal tissue destruction. Many of the other risk factors associated with periodontal disease such as stress, aging, and genetics also likely affect the microbial community, and more research is needed utilizing sophisticated bacterial taxonomic techniques to better elucidate these effects on the microbiome and to develop strategies to target the dysbiotic mechanisms and improve periodontal health.
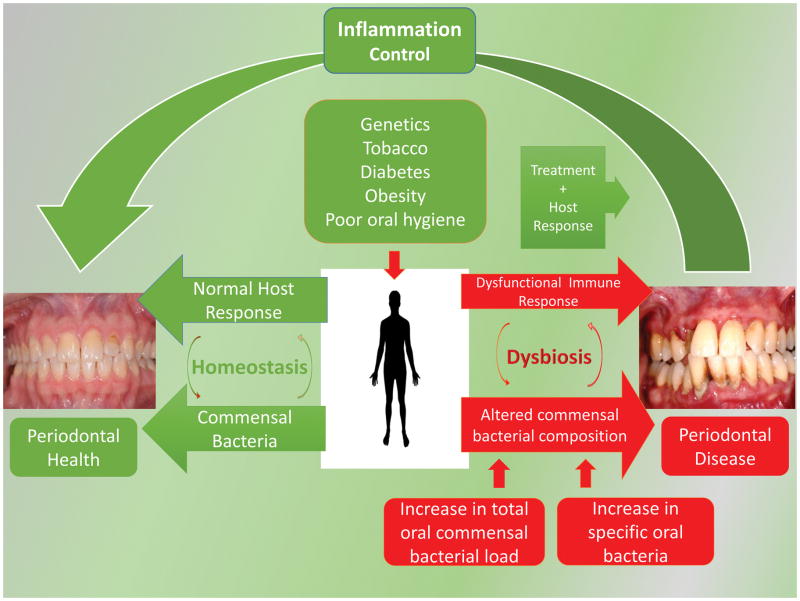
Fig. 2.
Periodontal Health and Disease as a function of the periodontal oral community and the host response. Periodontal health results from a homeostatic balance between the oral microbial community amount and composition and the corresponding host response. Risk factors can alter this balance and create a dysbiotic oral microbial community with an altered oral commensal bacterial composition. Bacterial community changes which result in a dysfunctional host response include an increase the total number of bacteria (22) and/or the outgrowth of specific bacteria (31). Periodontal treatment regimens can restore homeostasis.
Acknowledgments
The authors would like to thank Dr. Margaret Collins for her assistance with the manuscript, the Winter Quarter 2014 session of Periodontics 574 at the University of Washington for helpful discussions, and Dr. Amir Emam for designing Fig. 2. RPD also thanks current and past members of his laboratory for useful discussions and is supported by NIDCR DE012768 and DE18274
Contributor Information
Frank A. Roberts, Email: froberts@u.washington.edu, University of Washington, School of Dentistry, Department of Periodontics, 1959 NE Pacific Street, Box 357444, Seattle Washington 98195-7444, Phone: 206-685-9046, Fax: 206-616-7478
Richard P. Darveau, Email: rdarveau@u.washington.edu, University of Washington, School of Dentistry, Department of Periodontics, 1959 NE Pacific Street, Box 357444, Seattle Washington 98195-7444, Phone: 206-543-9514, Fax: 206-616-7478
References
- 1.Allaker RP, Douglas CW. Novel anti-microbial therapies for dental plaque-related diseases. International journal of antimicrobial agents. 2009;33:8–13. doi: 10.1016/j.ijantimicag.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 2.Baker PJ. Genetic control of the immune response in pathogenesis. Journal of periodontology. 2005;76:2042–2046. doi: 10.1902/jop.2005.76.11-S.2042. [DOI] [PubMed] [Google Scholar]
- 3.Baquero F, Nombela C. The microbiome as a human organ. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2012;18(Suppl 4):2–4. doi: 10.1111/j.1469-0691.2012.03916.x. [DOI] [PubMed] [Google Scholar]
- 4.Bartold PM, Van Dyke TE. Periodontitis: a host-mediated disruption of microbial homeostasis. Unlearning learned concepts. Periodontol 2000. 2013;62:203–217. doi: 10.1111/j.1600-0757.2012.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bek-Thomsen M, Tettelin H, Hance I, Nelson KE, Kilian M. Population diversity and dynamics of Streptococcus mitis, Streptococcus oralis, and Streptococcus infantis in the upper respiratory tracts of adults, determined by a nonculture strategy. Infection and immunity. 2008;76:1889–1896. doi: 10.1128/IAI.01511-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berezow AB, Darveau RP. Microbial shift and periodontitis. Periodontol 2000. 2011;55:36–47. doi: 10.1111/j.1600-0757.2010.00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brook I. Bacterial interference. Critical reviews in microbiology. 1999;25:155–172. doi: 10.1080/10408419991299211. [DOI] [PubMed] [Google Scholar]
- 8.Brunner J, Wittink FR, Jonker MJ, de Jong M, Breit TM, Laine ML, de Soet JJ, Crielaard W. The core genome of the anaerobic oral pathogenic bacterium Porphyromonas gingivalis. BMC microbiology. 2010;10:252. doi: 10.1186/1471-2180-10-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cebra JJ. Influences of microbiota on intestinal immune system development. The American journal of clinical nutrition. 1999;69:1046S–1051S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- 10.Chadwick VS, Anderson RP. Microorganisms and their products in inflammatory bowel disease. Elsevier Science; Amsterdam: 1992. [Google Scholar]
- 11.Curtis MA, Zenobia C, Darveau RP. The relationship of the oral microbiotia to periodontal health and disease. Cell host & microbe. 2011;10:302–306. doi: 10.1016/j.chom.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darveau RP, Tanner A, Page RC. The microbial challenge in periodontitis. Periodontology 2000. 1997;14:12–32. doi: 10.1111/j.1600-0757.1997.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 13.Darveau RP. The oral microbial consortium’s interaction with the periodontal innate defense system. DNA and cell biology. 2009;28:389–395. doi: 10.1089/dna.2009.0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nature reviews Microbiology. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 15.Darveau RP, Hajishengallis G, Curtis MA. Porphyromonas gingivalis as a potential community activist for disease. Journal of dental research. 2012;91:816–820. doi: 10.1177/0022034512453589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Queiroz K. Species concepts and species delimitation. Systematic biology. 2007;56:879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- 17.Duncan HE, Edberg SC. Host-microbe interaction in the gastrointestinal tract. Critical reviews in microbiology. 1995;21:85–100. doi: 10.3109/10408419509113535. [DOI] [PubMed] [Google Scholar]
- 18.Eckert R, Sullivan R, Shi W. Targeted antimicrobial treatment to re-establish a healthy microbial flora for long-term protection. Advances in dental research. 2012;24:94–97. doi: 10.1177/0022034512453725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eskan MA, Jotwani R, Abe T, Chmelar J, Lim JH, Liang S, Ciero PA, Krauss JL, Li F, Rauner M, Hofbauer LC, Choi EY, Chung KJ, Hashim A, Curtis MA, Chavakis T, Hajishengallis G. The leukocyte integrin antagonist Del-1 inhibits IL-17-mediated inflammatory bone loss. Nature immunology. 2012;13:465–473. doi: 10.1038/ni.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiology and molecular biology reviews : MMBR. 1998;62:1157–1170. doi: 10.1128/mmbr.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genco RJ, Borgnakke WS. Risk factors for periodontal disease. Periodontol 2000. 2013;62:59–94. doi: 10.1111/j.1600-0757.2012.00457.x. [DOI] [PubMed] [Google Scholar]
- 22.Gordon HA, Pesti L. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol Rev. 1971;35:390–429. doi: 10.1128/br.35.4.390-429.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, Darveau RP, Curtis MA. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell host & microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hart TC, Shapira L, Van Dyke TE. Neutrophil defects as risk factors for periodontal diseases. Journal of periodontology. 1994;65:521–529. doi: 10.1902/jop.1994.65.5s.521. [DOI] [PubMed] [Google Scholar]
- 25.Hasturk H, Kantarci A, Goguet-Surmenian E, Blackwood A, Andry C, Serhan CN, Van Dyke TE. Resolvin E1 regulates inflammation at the cellular and tissue level and restores tissue homeostasis in vivo. Journal of immunology. 2007;179:7021–7029. doi: 10.4049/jimmunol.179.10.7021. [DOI] [PubMed] [Google Scholar]
- 26.Haubek D, Ennibi OK, Poulsen K, Vaeth M, Poulsen S, Kilian M. Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: a prospective longitudinal cohort study. Lancet. 2008;371:237–242. doi: 10.1016/S0140-6736(08)60135-X. [DOI] [PubMed] [Google Scholar]
- 27.Hayashi Y, Matsunaga T, Yamamoto G, Nishii K, Usui M, Yamamoto M, Tachikawa T. Comprehensive analysis of gene expression in the junctional epithelium by laser microdissection and microarray analysis. Journal of periodontal research. 2010;45:618–625. doi: 10.1111/j.1600-0765.2010.01276.x. [DOI] [PubMed] [Google Scholar]
- 28.He X, McLean JS, Guo L, Lux R, Shi W. The social structure of microbial community involved in colonization resistance. The ISME journal. 2014;8:564–574. doi: 10.1038/ismej.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heymann R, Wroblewski J, Terling C, Midtvedt T, Obrink B. The characteristic cellular organization and CEACAM1 expression in the junctional epithelium of rats and mice are genetically programmed and not influenced by the bacterial microflora. Journal of periodontology. 2001;72:454–460. doi: 10.1902/jop.2001.72.4.454. [DOI] [PubMed] [Google Scholar]
- 30.Hold GL, Smith M, Grange C, Watt ER, El-Omar EM, Mukhopadhya I. Role of the gut microbiota in inflammatory bowel disease pathogenesis: What have we learnt in the past 10 years? World journal of gastroenterology : WJG. 2014;20:1192–1210. doi: 10.3748/wjg.v20.i5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 32.Hooper LV. Bacterial contributions to mammalian gut development. Trends in microbiology. 2004;12:129–134. doi: 10.1016/j.tim.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Jiao Y, Darzi Y, Tawaratsumida K, Marchesan JT, Hasegawa M, Moon H, Chen GY, Nunez G, Giannobile WV, Raes J, Inohara N. Induction of bone loss by pathobiont-mediated Nod1 signaling in the oral cavity. Cell host & microbe. 2013;13:595–601. doi: 10.1016/j.chom.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaplan JB, Schreiner HC, Furgang D, Fine DH. Population structure and genetic diversity of Actinobacillus actinomycetemcomitans strains isolated from localized juvenile periodontitis patients. Journal of clinical microbiology. 2002;40:1181–1187. doi: 10.1128/JCM.40.4.1181-1187.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komatsu S, Berg RD, Russell JM, Nimura Y, Granger DN. Enteric microflora contribute to constitutive ICAM-1 expression on vascular endothelial cells. American journal of physiology Gastrointestinal and liver physiology. 2000;279:G186–191. doi: 10.1152/ajpgi.2000.279.1.G186. [DOI] [PubMed] [Google Scholar]
- 36.Kornman KS, Page RC, Tonetti MS. The host response to the microbial challenge in periodontitis: assembling the players. Periodontol 2000. 1997;14:33–53. doi: 10.1111/j.1600-0757.1997.tb00191.x. [DOI] [PubMed] [Google Scholar]
- 37.Kumar PS. Personal Communication.
- 38.Lang NP. Commentary: bacteria play a critical role in the etiology of periodontal disease. Journal of periodontology. 2014;85:211–213. doi: 10.1902/jop.2013.130699. [DOI] [PubMed] [Google Scholar]
- 39.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loesche WJ. Chemotherapy of dental plaque infections. Oral sciences reviews. 1976;9:65–107. [PubMed] [Google Scholar]
- 41.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nature reviews Immunology. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 42.Maekawa T, Hajishengallis G. Topical treatment with probiotic Lactobacillus brevis CD2 inhibits experimental periodontal inflammation and bone loss. Journal of periodontal research. 2014 doi: 10.1111/jre.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Advances in dental research. 1994;8:263–271. doi: 10.1177/08959374940080022001. [DOI] [PubMed] [Google Scholar]
- 44.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Current opinion in genetics & development. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 45.Niederman R, Westernoff T, Lee C, Mark LL, Kawashima N, Ullman-Culler M, Dewhirst FE, Paster BJ, Wagner DD, Mayadas T, Hynes RO, Stashenko P. Infection-mediated early-onset periodontal disease in P/E-selectin-deficient mice. Journal of clinical periodontology. 2001;28:569–575. doi: 10.1034/j.1600-051x.2001.028006569.x. [DOI] [PubMed] [Google Scholar]
- 46.Page RC, Schroeder HE. Pathogenesis of inflammatory periodontal disease. A summary of current work. Laboratory investigation; a journal of technical methods and pathology. 1976;34:235–249. [PubMed] [Google Scholar]
- 47.Page RC, Kornman KS. The pathogenesis of human periodontitis: an introduction. Periodontology 2000. 1997;14:9–11. doi: 10.1111/j.1600-0757.1997.tb00189.x. [DOI] [PubMed] [Google Scholar]
- 48.Raff A, Hunt LC. Probiotics for periodontal health: a review of the literature. Journal of dental hygiene. 2012;86:71–81. [PubMed] [Google Scholar]
- 49.Ramsey MM, Whiteley M. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1578–1583. doi: 10.1073/pnas.0809533106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramsey MM, Rumbaugh KP, Whiteley M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS pathogens. 2011;7:e1002012. doi: 10.1371/journal.ppat.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rovin S, Costich ER, Gordon HA. The influence of bacteria and irritation in the initiation of periodontal disease in germfree and conventional rats. Journal of periodontal research. 1966;1:193–204. doi: 10.1111/j.1600-0765.1966.tb01860.x. [DOI] [PubMed] [Google Scholar]
- 52.Shaffer JR, Polk DE, Wang X, Feingold E, Weeks DE, Lee MK, Cuenco KT, Weyant RJ, Crout RJ, McNeil DW, Marazita ML. Genome-Wide Association Study of Periodontal Health Measured by Probing Depth in Adults Ages 18–49 years. G3. 2014;4:307–314. doi: 10.1534/g3.113.008755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shin J, Hosur KB, Pyaram K, Jotwani R, Liang S, Chavakis T, Hajishengallis G. Expression and function of the homeostatic molecule Del-1 in endothelial cells and the periodontal tissue. Clinical & developmental immunology. 2013;2013:617809. doi: 10.1155/2013/617809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Socransky SS, Haffajee AD. Evidence of bacterial etiology: a historical perspective. Periodontology 2000. 1994;5:7–25. doi: 10.1111/j.1600-0757.1994.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 55.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. Journal of clinical periodontology. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 56.Stabholz A, Soskolne WA, Shapira L. Genetic and environmental risk factors for chronic periodontitis and aggressive periodontitis. Periodontology 2000. 2010;53:138–153. doi: 10.1111/j.1600-0757.2010.00340.x. [DOI] [PubMed] [Google Scholar]
- 57.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suci P, Young M. Selective killing of Aggregatibacter actinomycetemcomitans by ciprofloxacin during development of a dual species biofilm with Streptococcus sanguinis. Archives of oral biology. 2011;56:1055–1063. doi: 10.1016/j.archoralbio.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 59.Taba M, Jr, Souza SL, Mariguela VC. Periodontal disease: a genetic perspective. Brazilian oral research. 2012;26(Suppl 1):32–38. doi: 10.1590/s1806-83242012000700006. [DOI] [PubMed] [Google Scholar]
- 60.Teughels W, Durukan A, Ozcelik O, Pauwels M, Quirynen M, Haytac MC. Clinical and microbiological effects of Lactobacillus reuteri probiotics in the treatment of chronic periodontitis: a randomized placebo-controlled study. Journal of clinical periodontology. 2013;40:1025–1035. doi: 10.1111/jcpe.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Theilade E. The non-specific theory in microbial etiology of inflammatory periodontal diseases. Journal of clinical periodontology. 1986;13:905–911. doi: 10.1111/j.1600-051x.1986.tb01425.x. [DOI] [PubMed] [Google Scholar]
- 62.Tonetti MS, Imboden MA, Gerber L, Lang NP, Laissue J, Mueller C. Localized expression of mRNA for phagocyte-specific chemotactic cytokines in human periodontal infections. Infection and immunity. 1994;62:4005–4014. doi: 10.1128/iai.62.9.4005-4014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tonetti MS, Imboden MA, Lang NP. Neutrophil migration into the gingival sulcus is associated with transepithelial gradients of interleukin-8 and ICAM-1. Journal of periodontology. 1998;69:1139–1147. doi: 10.1902/jop.1998.69.10.1139. [DOI] [PubMed] [Google Scholar]
- 64.Tribble GD, Rigney TW, Dao DH, Wong CT, Kerr JE, Taylor BE, Pacha S, Kaplan HB. Natural competence is a major mechanism for horizontal DNA transfer in the oral pathogen Porphyromonas gingivalis. mBio. 2012;3 doi: 10.1128/mBio.00231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsukamoto Y, Usui M, Yamamoto G, Takagi Y, Tachikawa T, Yamamoto M, Nakamura M. Role of the junctional epithelium in periodontal innate defense and homeostasis. Journal of periodontal research. 2012;47:750–757. doi: 10.1111/j.1600-0765.2012.01490.x. [DOI] [PubMed] [Google Scholar]
- 66.Umesaki Y, Setoyama H. Structure of the intestinal flora responsible for development of the gut immune system in a rodent model. Microbes Infect. 2000;2:1343–1351. doi: 10.1016/s1286-4579(00)01288-0. [DOI] [PubMed] [Google Scholar]
- 67.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proceedings of the National Academy of Sciences of the United States of America. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu J, Gordon JI. Inaugural Article: Honor thy symbionts. Proc Natl Acad Sci U S A. 2003;100:10452–10459. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu JJ, Ruddy MJ, Wong GC, Sfintescu C, Baker PJ, Smith JB, Evans RT, Gaffen SL. An essential role for IL-17 in preventing pathogen-initiated bone destruction: recruitment of neutrophils to inflamed bone requires IL-17 receptor-dependent signals. Blood. 2007;109:3794–3802. doi: 10.1182/blood-2005-09-010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zarco MF, Vess TJ, Ginsburg GS. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral diseases. 2012;18:109–120. doi: 10.1111/j.1601-0825.2011.01851.x. [DOI] [PubMed] [Google Scholar]
- 71.Zenobia C, Luo XL, Hashim A, Abe T, Jin L, Chang Y, Jin ZC, Sun JX, Hajishengallis G, Curtis MA, Darveau RP. Commensal bacteria-dependent select expression of CXCL2 contributes to periodontal tissue homeostasis. Cellular microbiology. 2013;15:1419–1426. doi: 10.1111/cmi.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zenobia C, Hasturk H, Nguyen D, Van Dyke TE, Kantarci A, Darveau RP. Porphyromonas gingivalis Lipid A Phosphatase Activity Is Critical for Colonization and Increasing the Commensal Load in the Rabbit Ligature Model. Infection and immunity. 2014;82:650–659. doi: 10.1128/IAI.01136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu Y, Dashper SG, Chen YY, Crawford S, Slakeski N, Reynolds EC. Porphyromonas gingivalis and Treponema denticola synergistic polymicrobial biofilm development. PloS one. 2013;8:e71727. doi: 10.1371/journal.pone.0071727. [DOI] [PMC free article] [PubMed] [Google Scholar]