Abstract
Background:
Pathology data contained within the electronic health record (EHR), and laboratory information system (LIS) of hospitals represents a potentially powerful resource to improve clinical care. However, existing reporting tools within commercial EHR and LIS software may not be able to efficiently and rapidly mine data for quality improvement and research applications.
Materials and Methods:
We present experience using a data warehouse produced collaboratively between an academic medical center and a private company. The data warehouse contains data from the EHR, LIS, admission/discharge/transfer system, and billing records and can be accessed using a self-service data access tool known as Starmaker. The Starmaker software allows users to use complex Boolean logic, include and exclude rules, unit conversion and reference scaling, and value aggregation using a straightforward visual interface. More complex queries can be achieved by users with experience with Structured Query Language. Queries can use biomedical ontologies such as Logical Observation Identifiers Names and Codes and Systematized Nomenclature of Medicine.
Result:
We present examples of successful searches using Starmaker, falling mostly in the realm of microbiology and clinical chemistry/toxicology. The searches were ones that were either very difficult or basically infeasible using reporting tools within the EHR and LIS used in the medical center. One of the main strengths of Starmaker searches is rapid results, with typical searches covering 5 years taking only 1–2 min. A “Run Count” feature quickly outputs the number of cases meeting criteria, allowing for refinement of searches before downloading patient-identifiable data. The Starmaker tool is available to pathology residents and fellows, with some using this tool for quality improvement and scholarly projects.
Conclusion:
A data warehouse has significant potential for improving utilization of clinical pathology testing. Software that can access data warehouse using a straightforward visual interface can be incorporated into pathology training programs.
Keywords: Clinical laboratory information system, data mining, electronic health records, immunoassays, medical informatics, systematized nomenclature of medicine
INTRODUCTION
Pathology testing is an integral part of modern medicine, with test results influencing diagnosis, prognosis, and management of the disease.[1,2] Hospital electronic health records (EHRs) contain a large repository of data that can potentially be used to improve utilization of laboratory testing and patient care.[3,4,5,6,7] However, a major challenge is having access to tools that can effectively and rapidly mine information from the EHR and other data sources. Complicated queries may traverse hospital flowsheet records, admission/discharge/transfer data, medical diagnoses, laboratory results, radiology reports, medication records, and other data sources. Laboratory data may be stored in discrete fields or buried within textual reports. While simple searches may be accomplished with reporting tools available within EHR software, complex queries may be much more challenging or even practically infeasible.
We report the use of a data warehouse for projects within a department of pathology at an academic medical center. There have been prior reports of the use of data warehouses for diverse biomedical applications such as clinical research investigations,[8,9] improving birth outcomes,[10,11] and microbiology/epidemiology analysis.[12] In this report, we provide a description of the data warehouse and also present application examples within pathology. The examples chosen were questions that were very difficult or even infeasible to answer using reporting tools from the EHR or laboratory information system (LIS) used by the institution.
Technical Background
Institutional details
The institution of this study is a 734 bed tertiary care academic medical center that includes an emergency room with level one trauma capability, pediatric and adult inpatient units, and multiple intensive care units (neonatal, pediatric, cardiovascular, medical, and surgical/neurologic), as well as primary and specialty outpatient services. This study was approved by the Institutional Review Board as a retrospective study in the time period from May 2, 2009, to July 22, 2014. During this time period, the EHR was Epic (Epic Systems, Inc., Madison, WI, USA), and the LIS was Cerner (Kansas City, MO, USA) “Classic” version 015. Both are managed by Health Care Information Systems (HCIS). The informatics architecture of the core laboratory has been described in detail elsewhere.[13]
Reporting tools are available in both the EHR and LIS. Outside of laboratory test results and basic demographics, searches using the LIS are not able to access many of the data elements found in the EHR such as patient medications, hospital flow sheet data, or demographic information other than name, gender, and age. The EHR has a large number of reporting tools available; however, the ability to do complex queries can be very challenging and time-consuming, especially when attempting to combine logic across different domains of data.
Data warehouse software
The data warehouse was created as a collaboration between the HCIS and Park Street Solutions (Naperville, IL, USA). Data are extracted weekly from the Clarity database associated with the medical center EHR system, including admission/data/transfer (ADT), laboratory orders and results, medication orders, and medication administration events, flow sheet entries, allergies, immunizations, and problems. Billing data such as admitting and discharge diagnoses, diagnosis-related groups, and procedure codes are extracted from the institution's financial data repository, also on a weekly basis. The database currently contains almost 2 billion clinical facts, representing six years of patient history from the two data sources.
The data model for the data warehouse is patient-centered and is driven by knowledge representation technology from Park Street Solutions. All patient facts are represented in terms of either standard clinical knowledge bases or ontologies such as ICD-9, Logical Observation Identifiers Names and Codes (LOINC), and Systematized Nomenclature of Medicine (SNOMED), or by means of HCIS’ own code systems. Core elements of the data warehouse are completely de-identified so that all queries and analytics can be carried out without exposing private health data, while users with sufficient privilege can re-identify data to support operations and quality improvement applications.
Starmaker is a self-service data access tool designed to query clinical data repositories and return tabular data for analysis and visualization. Starmaker allows data analysts and researchers to find patient cohorts and extract clinical data from the repository by specifying queries using simple click and drag techniques. Starmaker outputs data in comma-separated, tab-separated, and attribute-related file formats suitable for transfer to data analysis and visualization tools.
Starmaker users can express their data requirements in clinical and computational terms, rather than in terms of data structures and Structured Query Language (SQL). Complex Boolean logic include and exclude rules, date-based computing, unit conversion and reference scaling, and value aggregation are all implemented using a straightforward visual interface. Queries can use biomedical ontologies such as LOINC and SNOMED.
Starmaker allows its users to import lists of patients or to save the patients selected by Starmaker queries and supports the manual editing of saved lists. This provides a way to create a saved cohort that can be used on an ongoing basis for a variety of queries and analyses. Users can also build their own value sets, which are lists of codes or knowledge base concepts that form a clinically relevant group for analytic purposes. To store and organize queries, patient sets, value sets, and other user-developed objects, Starmaker provides a distributed object management system. Objects are stored by default in a private personal repository. Administrators can create shared repositories and folders, manage the access rights of individual users, and publish content to higher-level or public repositories. Queries perform well even in billion-row databases.
Starmaker generates queries that target an intermediate layer of stored procedures, functions, and other database objects. That layer provides a selection of powerful tools for clinical query expression that can be used by SQL query writers and application developers. Starmaker can be used to generate SQL that developers can copy and further modify, providing a rapid starting point for clinical query development.
Queries generated by Starmaker include a built-in Health Insurance Portability and Accountability Act (HIPAA) log-in step. The data access log captures which patients’ data have been interrogated, and the identity of the user to whom it has been disclosed. One feature of Starmaker is a “Run Count” that can rapidly return the number of cases meeting defined criteria without any output of HIPAA-sensitive data. The basic search query screen of Starmaker is shown in Figure 1.
Figure 1.
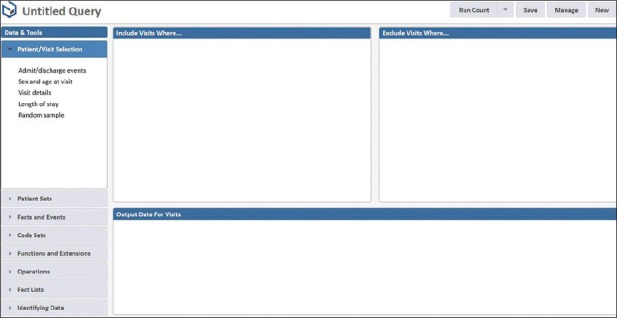
Basic screen for setting up searches using Starmaker
RESULTS
The following examples capture applications of Starmaker within the Department of Pathology.
Example Number 1: Are We Detecting the Expected Spectrum of Bloodstream Pathogens in Neutropenic Sepsis?
A data warehouse can be especially powerful for epidemiology questions.[12] One common clinical question is what pathogens are associated with neutropenic sepsis. This type of query involves the intersection of several areas of clinical and laboratory data (e.g., neutropenia, timeframe, laboratory diagnosis of pathogens). This type of search is very difficult using EHR reporting tools if going beyond a very limited time period (e.g., more than 1-month).
In Starmaker, the inclusion criteria included positive blood culture results, laboratory data for neutropenia (in this case using either automated or manual differential determinations of the neutrophil count), and inpatient admission status [Figure 2]. The exclusion criteria filter out positive blood cultures that are contaminants. The output data return the patient identifiers and the pathogen. This type of report can be readily modified to focus on specific patient populations (e.g., pediatric, dialysis) or to use other laboratory parameters (e.g., pancytopenia) as inclusion criteria.
Figure 2.
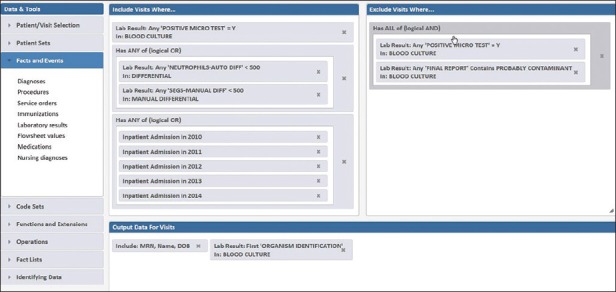
Starmaker search for positive blood cultures in neutropenic patients. The exclusion data filters out suspected contaminant organisms
Example Number 2: How Often are Angiotensin Converting Enzyme Levels Ordered in Patient on Angiotensin Converting Enzyme Inhibitors?
Patient medications can impact laboratory test results. One example occurs with the measurement of serum angiotensin converting enzyme (ACE) levels, a laboratory test frequently used in the workup of sarcoidosis. ACE levels are often elevated in patients with active sarcoidosis.[14] Previous studies have established that therapy with ACE inhibitors (e.g., captopril, enalapril, lisinopril, etc.) can reduce serum ACE levels; thus, this laboratory test should not be performed in patients taking ACE inhibitors.[15,16,17,18] However, ACE inhibitors are very commonly prescribed medications for hypertension and other indications,[19] leading to the potential for ordering of ACE levels for patients who are currently receiving ACE inhibitor therapy. How often does this occur?
This query could potentially be accomplished with reporting tools in the EHR. The search for patients who have had ACE levels performed is straightforward to set up and requires approximately four hours of computer run time per year of data retrieved, a strategy we have performed with utilization projects involving other laboratory tests.[20] The much bigger challenge is in retrieving patients who are on ACE inhibitors. Over the retrospective analysis period of slightly more than 5 years (May 02, 2009–July 22, 2014), the EHR contained 123 distinct medication order options involving nine different ACE inhibitors (benazepril, captopril, enalapril, fosinopril, lisinopril, perindopril, quinapril, ramipril, trandolapril), encompassing variations in formulations (e.g., generic versus branded name; combination products with other medications) and dosing patterns. Each of these orders is a separate search item in the EHR reporting tools system.
Utilizing Starmaker for this search was easier and much faster [Figure 3]. Pharmacy data could be searched using SNOMED terminology. A single search term could retrieve all ACE inhibitor formulations or focus on an individual ACE inhibitor (e.g. benazepril, lisinopril). Once constructed, a complete retrieval of all inpatient and outpatient encounters in a 5 year period that included an active prescription for an ACE inhibitor and a completed order for an ACE level involved less than one minute to retrieve results. This identified 108 patients who had an ACE level performed while on ACE inhibitor. A detailed analysis of this example of laboratory test misutilization is the subject of a separate study.
Figure 3.
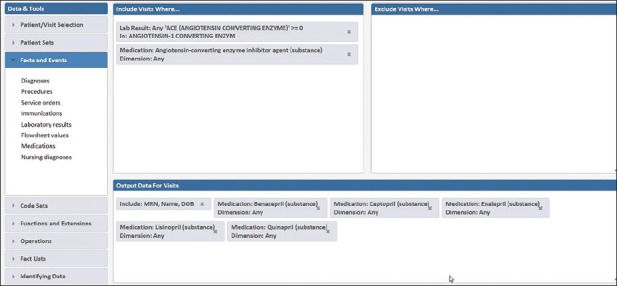
Starmaker search for patients receiving an angiotensin converting enzyme (ACE) inhibitor and an ACE level on same hospital inpatient or outpatient encounter. The output data includes which specific ACE inhibitor medication was active during the encounter
Example Number 3: What are the Potential Causes of False Positive Amphetamine Drug Screening Results?
Immunoassays (antibody-based tests) are commonly used for drug of abuse/toxicology screening on urine or other body fluids.[21,22] Immunoassays for drug screening may utilize monoclonal or polyclonal antibodies, with a specificity directed at one or more target compounds. In addition to immunoassays, mass spectrometry (MS)-based methods such as gas chromatography/MS or liquid chromatography-tandem mass spectrometry (LC/MS/MS) can provide specific and definitive identification of drugs and drug metabolites; such methods are often used for confirmation of positive immunoassay screening results or for detection of drugs known to be undetectable or not readily detected by immunoassays.[23,24] While an increasing number of clinical laboratories are using MS-based assays for drug of abuse testing, relatively few hospital-based clinical laboratories have the capability to do this testing with a fast turnaround time. Consequently, immunoassays continue to be used in many settings for rapid drug of abuse testing.
A limitation of the drug of abuse immunoassays is cross-reactivity with compounds structurally related to the target molecule(s) of the assay.[25,26] As an example, amphetamines are commonly included in the drug of abuse testing panels. Amphetamines immunoassays typically detect amphetamine and methamphetamine and, depending on assay antibody, may additionally detect “designer” amphetamines (e.g., 3,4-methylenedioxy-N-methylamphetamine, MDMA, “ecstasy”) or amphetamine-like compounds such as ephedrine, phentermine, or pseudoephedrine.[27,28]
At the institution, a search within the EHR revealed that over an approximately 5 year period of retrospective analysis (May 02, 2009–July 22, 2014), 6.0% of drug of abuse immunoassay screens for amphetamines were positive (1571 of 26,161 total tests). Of the positives, 101 failed to confirm with LC/MS/MS confirmatory testing by reference laboratory (6.4% of the 1,571 positives). Are there therapeutic medications that could be causing the false positives?
The package insert for the Roche Diagnostics (Indianapolis, IN, USA) Amphetamines II assay for cobas c501/c502 systems (the assay used at the institution) lists ten therapeutic medications (some of which such as amphetamine and methamphetamine are also common drugs of abuse) as having cross-reactivity of 0.14% or higher for the immunoassay (listed in decreasing order from most to least cross-reactive): amphetamine (either racemic or d-isomer), methamphetamine (d-greater than l-isomer in cross-reactivity), trazodone (via its metabolite m-chlorphenylpiperazine), labetalol (via its metabolite 1-methyl-3-phenylpropylamine), phentermine, pseudoephedrine, phendimetrazine, ephedrine, and phenylpropanolamine. Over the 5 year period of retrospective analysis, the EHR contained 207 distinct order options for these 10 medications, with the most options for pseudoephedrine (54), ephedrine (46), labetalol (27), and amphetamine (26). As with the ACE inhibitor described above, the sheer number of medication orders make searching within the EHR very time-consuming.
Utilizing Starmaker streamlined the search [Figure 4]. The population of interest (amphetamine screen-positive/confirmation-negative, n = 101) could be interrogated for how many occurred on patient encounters with active prescriptions for the ten medications described above. Of the ten medications, only labetalol was associated with more than one patient in the screen-positive/confirmation-negative category. Labetalol was a prescription medication in 19 of the 101 amphetamine screen-positive/confirmation-negative patients. Chart review of these 19 patients revealed that 10 of the 19 patients were women on labetalol for chronic management of hypertension during pregnancy. Six were trauma patients given labetalol prior to urine drug testing. These findings add to the literature (focused mainly on obstetric patients) that a metabolite of labetalol (1-methyl-3-phenylpropylamine) may be a cause of amphetamine-positive screens.[29,30,31] These findings led to changes in the laboratory handbook and interpretive comment on amphetamine drug testing results at the institution, along with education efforts directed at obstetrics/gynecology, the clinical service most impacted by this potential false positive.
Figure 4.
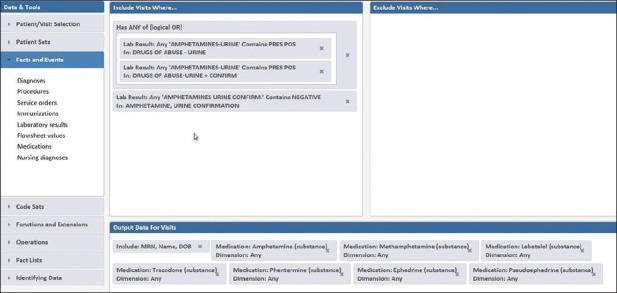
Starmaker search for patients with amphetamine-positive immunoassay screen but a negative confirmation. The output data includes whether specific drugs known to cross-react with the amphetamines immunoassay screen were active during the encounter
Example Number 4: What Restrictions Should be Placed on Automatic Reflex Confirmation of Drug Abuse Testing Results?
The scheme in Starmaker described above for the amphetamines can be readily adapted to similar queries for other drug of abuse testing questions such as estimating the number of positive drug screens caused by therapeutic medications. Querying larger numbers of individual medications or classes of medication is facilitated by SNOMED terminology. For commonly used medications (e.g., benzodiazepines, opiates), a high fraction of positive screening results for those drug classes may be caused by therapeutic use and not by nonmedical use. In setting policy for reflex confirmatory testing of positive screening results, can data analysis indicate how many positive results are likely explained by therapeutic medications?
As an example for the benzodiazepines, thirteen medications (alprazolam, chlordiazepoxide, clobazam, clonazepam, clorazepate, diazepam, estazolam, flurazepam, lorazepam, midazolam, oxazepam, temazepam, triazolam) with 207 distinct medication order options were in the EHR over the retrospective analysis period of 5 years (May 2, 2009–July 22, 2014). All of these benzodiazepines (either as the parent drug or metabolites or both) show greater than 50% cross-reactivity on the Roche Diagnostics Benzodiazepines Plus assay for cobas c501/c502 analyzers (the assay used at the institution).
A search was generated within Starmaker to capture the population of interest (presumptive positive benzodiazepine screens) and interrogate how many were associated with encounters in which prescription benzodiazepines were administered [Figure 5]. The results of this search showed that there were 4,991 positive benzodiazepine screens out of 26,192 total performed tests. Of these 4,991 positive screens, 3,308 (66.3%) occurred in patient encounters (mainly inpatient units and emergency department) associated with benzodiazepine prescriptions. These data suggest that reflex confirmation of all benzodiazepine positive screens will reveal a high percentage due to therapeutic medications. Therefore, unrestricted confirmation of all positive results is not likely to be cost-effective.
Figure 5.
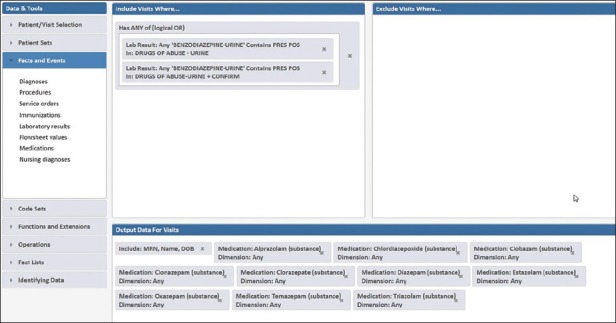
Starmaker search for patients with positive benzodiazepine immunoassay screen. The output data includes whether specific drugs known to cross-react with the benzodiazepines immunoassay screen were active during the encounter
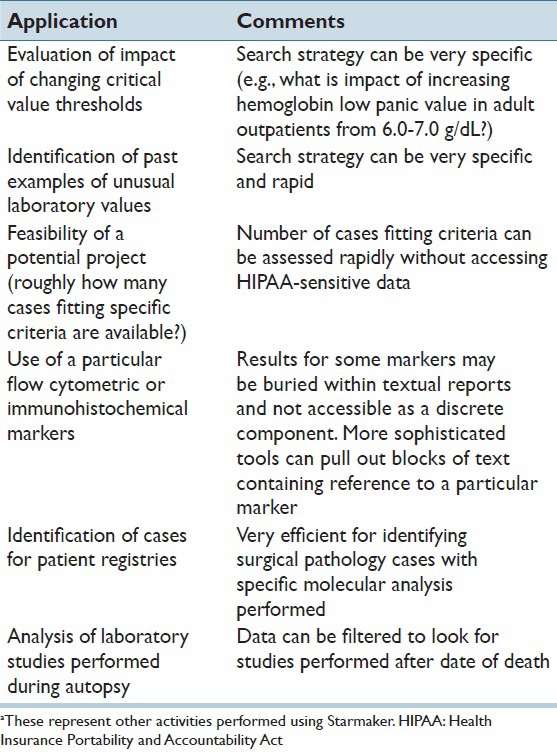
Table 1 lists additional applications of Starmaker used within the Department of Pathology. The ability of Starmaker to rapidly query laboratory data facilitates searches such as the impact of changing critical value thresholds or to identify how often extreme values occur (e.g., to establish auto-dilution protocols or to help determine which linearity calibration sets to use for certain assays). These types of searches may not need access of patient identifiers. In addition, we have found the application very useful in mining textual data out of reports, including for disease registries that increasingly seek very specific combinations of surgical pathology and molecular alterations. Finally, extensions within Starmaker can interrogate date of death to allow for a query of laboratory studies performed as part of autopsies.
Table 1.
Other applications of Starmakera
DISCUSSION
Hospitals contain vast repositories of pathology laboratory data that can potentially be used to improve utilization of laboratory testing and patient care.[3,4,5,6,7] However, efficient mining of this data can be quite challenging. EHR and LIS software typically have some data mining functions; however, these systems may not be able to interrogate data stored outside of their own systems. In addition, some hospitals may only allow data searches to be performed by specialized personnel. Data warehouses represent a powerful tool for complex data mining applications.[8,9,10,11,12] The warehouses can aggregate data from diverse sources (EHR, LIS, pharmacy, admission/discharge/transfer, billing records) in an architecture conducive to rapid searching without competition with production clinical environments.
In this report, we describe use and applications of Starmaker, a data warehouse tool, within an academic department of pathology. One of the goals within the department is to train pathology residents and fellows on data mining tools for scholarly or quality improvement projects. Tools like Starmaker make this goal feasible.
Financial Support and Sponsorship
Nil.
Conflicts of Interest
A.S. is CTO of Park Street Solutions which developed and markets the Starmaker software described in this report.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2015/6/1/45/161615
REFERENCES
- 1.Benson ES. The responsible use of the clinical laboratory. Clin Biochem. 1986;19:262–70. doi: 10.1016/s0009-9120(86)80038-8. [DOI] [PubMed] [Google Scholar]
- 2.Fang C, Otero HJ, Greenberg D, Neumann PJ. Cost-utility analyses of diagnostic laboratory tests: A systematic review. Value Health. 2011;14:1010–8. doi: 10.1016/j.jval.2011.05.044. [DOI] [PubMed] [Google Scholar]
- 3.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: Towards better research applications and clinical care. Nat Rev Genet. 2012;13:395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- 4.Jha AK, DesRoches CM, Campbell EG, Donelan K, Rao SR, Ferris TG, et al. Use of electronic health records in U.S. hospitals. N Engl J Med. 2009;360:1628–38. doi: 10.1056/NEJMsa0900592. [DOI] [PubMed] [Google Scholar]
- 5.Kudler NR, Pantanowitz L. Overview of laboratory data tools available in a single electronic medical record. J Pathol Inform. 2010;1:3. doi: 10.4103/2153-3539.63824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc. 2014;21:221–30. doi: 10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson BR. Managing laboratory test use: Principles and tools. Clin Lab Med. 2007;27:733–48, v. doi: 10.1016/j.cll.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Lyman JA, Scully K, Harrison JH., Jr The development of health care data warehouses to support data mining. Clin Lab Med. 2008;28:55–71. doi: 10.1016/j.cll.2007.10.003. vi. [DOI] [PubMed] [Google Scholar]
- 9.Mullins IM, Siadaty MS, Lyman J, Scully K, Garrett CT, Miller WG, et al. Data mining and clinical data repositories: Insights from a 667,000 patient data set. Comput Biol Med. 2006;36:1351–77. doi: 10.1016/j.compbiomed.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Goodwin LK, Iannacchione MA. Data mining methods for improving birth outcomes prediction. Outcomes Manag. 2002;6:80–5. [PubMed] [Google Scholar]
- 11.Prather JC, Lobach DF, Goodwin LK, Hales JW, Hage ML, Hammond WE. Medical data mining: Knowledge discovery in a clinical data warehouse. Proc AMIA Annu Fall Symp. 1997;2:101–5. [PMC free article] [PubMed] [Google Scholar]
- 12.Wisniewski MF, Kieszkowski P, Zagorski BM, Trick WE, Sommers M, Weinstein RA. Development of a clinical data warehouse for hospital infection control. J Am Med Inform Assoc. 2003;10:454–62. doi: 10.1197/jamia.M1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krasowski MD, Davis SR, Drees D, Morris C, Kulhavy J, Crone C, et al. Autoverification in a core clinical chemistry laboratory at an academic medical center. J Pathol Inform. 2014;5:13. doi: 10.4103/2153-3539.129450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieberman J. Elevation of serum angiotensin-converting-enzyme (ACE) level in sarcoidosis. Am J Med. 1975;59:365–72. doi: 10.1016/0002-9343(75)90395-2. [DOI] [PubMed] [Google Scholar]
- 15.Kamoun PP, Bardet JI, Di Giulio S, Grunfeld JP. Measurements of angiotensin converting enzyme in captopril-treated patients. Clin Chim Acta. 1982;118:333–6. doi: 10.1016/0009-8981(82)90021-3. [DOI] [PubMed] [Google Scholar]
- 16.Lieberman J. Effect of converting enzyme inhibitors on serum ACE test. Chest. 1990;98:1538. [PubMed] [Google Scholar]
- 17.Lieberman J, Zakria F. Effect of captopril and enalapril medication on the serum ACE test for sarcoidosis. Sarcoidosis. 1989;6:118–23. [PubMed] [Google Scholar]
- 18.Hamlin R, Pijpers FS, Mandigers PJ. Plasma ACE inhibition by five different ACE inhibitors. Vet Q. 1998;20 Suppl 1:S109. doi: 10.1080/01652176.1998.10807453. [DOI] [PubMed] [Google Scholar]
- 19.Piepho RW. Overview of the angiotensin-converting-enzyme inhibitors. Am J Health Syst Pharm. 2000;57 Suppl 1:S3–7. doi: 10.1093/ajhp/57.suppl_1.S3. [DOI] [PubMed] [Google Scholar]
- 20.Krasowski MD, Chudzik D, Dolezal A, Steussy B, Gailey MP, Koch B, et al. Promoting improved utilization of laboratory testing through changes in an electronic medical record: Experience at an academic medical center. BMC Med Inform Decis Mak. 2015;15:11. doi: 10.1186/s12911-015-0137-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melanson SE. The utility of immunoassays for urine drug testing. Clin Lab Med. 2012;32:429–47. doi: 10.1016/j.cll.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Moeller KE, Lee KC, Kissack JC. Urine drug screening: Practical guide for clinicians. Mayo Clin Proc. 2008;83:66–76. doi: 10.4065/83.1.66. [DOI] [PubMed] [Google Scholar]
- 23.Tenore PL. Advanced urine toxicology testing. J Addict Dis. 2010;29:436–48. doi: 10.1080/10550887.2010.509277. [DOI] [PubMed] [Google Scholar]
- 24.Wu AH, Gerona R, Armenian P, French D, Petrie M, Lynch KL. Role of liquid chromatography-high-resolution mass spectrometry (LC-HR/MS) in clinical toxicology. Clin Toxicol (Phila) 2012;50:733–42. doi: 10.3109/15563650.2012.713108. [DOI] [PubMed] [Google Scholar]
- 25.Krasowski MD, Pizon AF, Siam MG, Giannoutsos S, Iyer M, Ekins S. Using molecular similarity to highlight the challenges of routine immunoassay-based drug of abuse/toxicology screening in emergency medicine. BMC Emerg Med. 2009;9:5. doi: 10.1186/1471-227X-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kricka LJ. Principles of immunochemical techniques. In: Burtis CA, Ashwood ER, Bruns DE, editors. Tietz Textbook of Clinical Chemistry and Molecular Diagnostics. 4th ed. St. Louis, MO: Elsevier Saunders; 2006. pp. 219–43. [Google Scholar]
- 27.Krasowski MD, Ekins S. Using cheminformatics to predict cross reactivity of “designer drugs” to their currently available immunoassays. J Cheminform. 2014;6:22. doi: 10.1186/1758-2946-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrie M, Lynch KL, Ekins S, Chang JS, Goetz RJ, Wu AH, et al. Cross-reactivity studies and predictive modeling of “Bath Salts” and other amphetamine-type stimulants with amphetamine screening immunoassays. Clin Toxicol (Phila) 2013;51:83–91. doi: 10.3109/15563650.2013.768344. [DOI] [PubMed] [Google Scholar]
- 29.Dueñas-Garcia OF. False-positive amphetamine toxicology screen results in three pregnant women using labetalol. Obstet Gynecol. 2011;118:360–1. doi: 10.1097/AOG.0b013e3182263f1b. [DOI] [PubMed] [Google Scholar]
- 30.Gilbert RB, Peng PI, Wong D. A labetalol metabolite with analytical characteristics resembling amphetamines. J Anal Toxicol. 1995;19:84–6. doi: 10.1093/jat/19.2.84. [DOI] [PubMed] [Google Scholar]
- 31.Yee LM, Wu D. False-positive amphetamine toxicology screen results in three pregnant women using labetalol. Obstet Gynecol. 2011;117:503–6. doi: 10.1097/AOG.0b013e318206c07c. [DOI] [PubMed] [Google Scholar]


