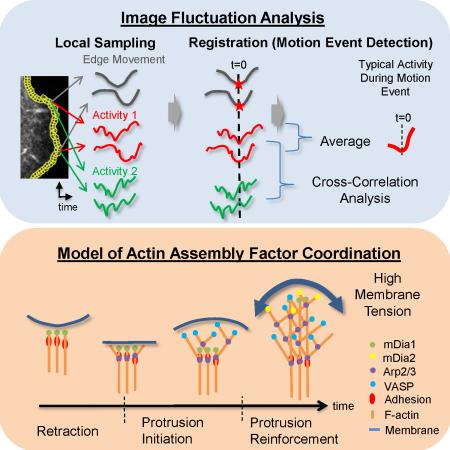
Summary
Highly redundant pathways often contain components whose functions are difficult to decipher from the responses induced by genetic or molecular perturbations. Here, we present a statistical approach that samples and registers events observed in images of intrinsic fluctuations in unperturbed cells to establish the functional hierarchy of events in systems with redundant pathways. We apply this approach to study the recruitment of actin assembly factors involved in the protrusion of the cell membrane. We find that the formin mDia1, along with nascent adhesion components, is recruited to the leading edge of the cell before protrusion onset, initiating linear growth of the lamellipodial network. Recruitment of Arp2/3, VASP, cofilin, and the formin mDia2 then promotes sustained exponential growth of the network. Experiments changing membrane tension suggest that Arp2/3 recruitment is mechano-responsive. These results indicate that cells adjust the overlapping contributions of multiple factors to actin filament assembly during protrusion on a ten-second timescale and in response to mechanical cues.
Graphical Abstract
Introduction
Many cellular processes rely on the integration of multiple pathways with overlapping yet differential functions (Kafri et al., 2009), and dissecting the contributions of these pathways is a fundamental challenge. Conventional perturbation approaches, in which pathways are inactivated by genetic or molecular methods or overactivated one at a time, are limited in their ability to probe the system, as they tend to lead to adaptation of the system to the perturbation, obscuring the bona fide function of the targeted component.
A prime example of a system with redundancy is actin-based cell protrusion (Krause and Gautreau, 2014). This protrusion machinery constantly adapts to ever-changing chemical and mechanical inputs. Two decades of work have established knowledge of most of the molecular players in the protrusion machinery. In particular, actin polymerization, the driving process behind cell protrusion, is promoted by an evolutionarily conserved multitude of actin assembly factors, all localized at the cell edge. However, it is still not clear whether these factors are functionally exchangeable or whether they are organized in a functional hierarchy.
Addressing this question by elimination of actin assembly factors, one at a time, offers limited answers, and, because the system often adapts to the absence of a single factor, may even cause considerable confusion. Indeed, the current discussion on what happens when functional Arp2/3 is absent from the protrusion machinery provides an illustrative example of the unexpected outcomes of perturbations in redundant molecular systems. For almost two decades the Arp2/3 complex has been described as the master regulator of actin assembly during cell protrusion (Pollard and Borisy, 2003). However, two independent studies recently have shown that cells in which the activity of Arp2/3 is inhibited still protrude efficiently (Suraneni et al., 2012; Wu et al., 2012), although both studies report severe defects or absence of lamellipodia.
This shows that filament growth mediated by other factors alone, such as members of the formin family of actin nucleators (Block et al., 2012; Yang et al., 2007; Zaoui et al., 2008) or actin modulators such as VASP or cofilin (Bear et al., 2002; Ghosh et al., 2004; Lacayo et al., 2007; Rottner et al., 1999), can drive cell protrusion. Yet, based on their observations of Arp2/3 inhibited systems, Suraneni et al. and Wu et al. disagree on what Arp2/3 does in the context of other actin assembly factors. Suraneni et al. report that fibroblasts missing Arp2/3 fail to sense chemotactic gradients, and therefore conclude that Arp2/3 is essential for amplifying the feedback between actin filament assembly and upstream signaling. Wu et al's data show that fibroblasts depleted of Arp2/3 are perfectly capable of responding to a chemotactic gradient but are deficient in following gradients of extracellular matrix ligands (haptotaxis). From this the authors conclude that Arp2/3 is not involved in the stimulation of actin assembly downstream of chemotactic signals, but that Arp2/3-driven lamellipodial networks are essential for the alignment of adhesions to influence the direction of migration in haptotactic gradients. We suspect that the differences between these data merely reflect the induction of differential adaptation processes. Specifically, it seems that in Suraneni et al.'s experiments the cells compensate for Arp2/3's contribution to actin-mediated protrusion, whereas in Wu et al. they compensate for Arp2/3's contribution to protrusion and to the putative feedback amplification of chemotactic signals. Thus, depending on the balance between compensating pathways, in one case the actin assembly factor system responds better to chemotaxis and in another case better to haptotaxis. The key issue is that neither data set permits inference of Arp2/3's function in the cell protrusion machinery unperturbed by genetic or molecular methods.
Because of functional overlap and feedback between actin assembly factors, the dissection of their hierarchy of operation requires an analysis of assembly factor dynamics in the unperturbed system. Thus far this has been accomplished by in vitro reconstitution of filopodia-like structures, where the stereotypical growth of actin filament bundles revealed temporal coordination and cooperation between Arp2/3 and mDia formins (Lee et al., 2010). Here we sought to develop a non-invasive experimental and computational approach that exploits spontaneous pathway fluctuations to achieve a similar analysis of the hierarchy of actin nucleation during cell protrusion in vivo. Our data reveal that cell protrusions are initiated by recruitment and activation of the formin mDia1. Recruitment of Arp2/3, previously thought of as the primary initiator of network assembly during protrusion events, follows this first step for the purpose of mechanical reinforcement of the growing actin filaments against increasing membrane tension (Ji et al., 2008) by autocatalytic nucleation of a dendritic network. Additional actin nucleators, such as mDia2, or actin modulators, such as VASP and cofilin, augment the growth of the Arp2/3-mediated network but play minor roles during the early protrusion steps.
Results
Fluctuations in membrane protrusion of the leading edge of migrating PtK1 epithelial cells
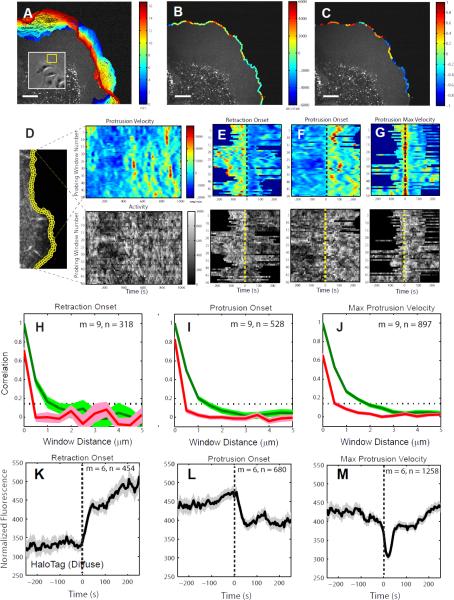
To investigate the spatiotemporal dynamics of cell protrusion, we imaged PtK1 cells stained with a membrane marker (CellMask Orange) at a 5 second frame interval. The state of motion at the leading edges of epithelial cells varies over short spatial and temporal scales (Figure 1A). This suggests that the underlying regulatory pathways, including the states of actin polymerization, are also spatiotemporally variable. To capture this morphogenic heterogeneity, we used automatic tracking of the cell edge to identify the state of protrusion and retraction locally in 500 nm-long sectors (Vilela et al., 2013). For each sector, we determined the protrusion or retraction velocity and the time since the onset of protrusion or retraction (Figure 1B-C and Data File1:Movie S1 and S2). Fast and slow protruding sectors, or protruding and retracting sectors, alternate over distances as short as 2 – 3 μm.
Figure 1. A local sampling and registration approach using intrinsic fluctuations at the leading edge of migrating PtK1 epithelial cells.
(A) Positions (blue, early; red, late time points) of the leading edge of a PtK1 cell stained with CellMask Orange membrane marker undergoing a wound healing response. Inset: Overview of epithelial cell sheet with protruding region highlighted by yellow box. (B) Protrusion (green to red colors) and retraction (green to blue colors) velocities tracked in edge sectors of 500 nm length. (C) Normalized times since protrusion (green to red colors) or retraction (green to blue colors) onset for each sector tracked in (B). A value 0 means the sector starts to protrude or retract. A value 1 or -1 means the sector terminates a protrusion or retraction segment, respectively. Scale bars: 10 μm. (D-G) Registration of spatially localized time series reporting molecular or cellular activities by alignment of motion events. (D) Left: Quantification of cell edge motion and molecular or cellular activities in 500 × 500 nm probing windows tracking the cell movement (see also Data File1:Movie S3). Right: Space-time map of instantaneous edge velocity (top) and activity (in this example, normalized fluorescence) (bottom); each row represents the velocity and activity time series, respectively, of one probing window. Registration of velocity map (top) and activity map (bottom) with respect to retraction onsets (t=0) (E), protrusion onsets (t=0) (F), and maximal protrusion velocity (t=0) (G). (H-J) Correlation of protrusion velocities between adjacent sampling windows; The red lines indicate Pearson's correlation coefficients between protrusion velocity time series acquired in sampling windows separated by the specified distances. The series are registered with respect to retraction onset (H), protrusion onset (I), and maximal protrusion velocity (J). The green lines indicate Pearson's correlation coefficients between sampling windows separated by the specified distances without considering time shifts in each case. The dotted lines indicate the 95% confidence level of the correlation values. For each condition the number n of time series sampled in m cells pooled from 4 independent experiments is indicated. (K-M) Normalized fluorescence intensity time series of HaloTag-TMR registered with respect to retraction onset, protrusion onset or maximal protrusion velocity. For each condition the number n of time series sampled in m cells pooled from 2 independent experiments is indicated. Solid lines indicate population averages. Shaded error bands about the population averages indicate 95% confidence intervals by bootstrap sampling.
Local sampling and temporal registration of spatially heterogeneous protrusion activities
To investigate the function of multiple actin nucleators and modulators in edge motion, we recorded time series of molecular and cellular events, such as the recruitment of a particular actin assembly factor or the local traction force level, in windows of about ~1 μm2, which tracked the cell edge (Figure 1D; Data File1:Movie S3). The size of these probing windows was chosen to be several-fold smaller than the 2 – 3 μm length scale of protrusion events, yet large enough to reduce by averaging the pixel-by-pixel noise of molecular events (Vilela et al., 2013). Because of the intrinsic variation of molecular events within an area of ~1 μm2, each time-series provided one instantiation of a stochastic process that relates these events to cell edge motion. Therefore, to answer questions like ‘when does a particular molecule arrive at the cell edge relative to the onset of a protrusion’, it was necessary to average over many time-series.
To this end, we registered (aligned) the time series of all windows of a cell using one of three edge motion events as a temporal fiduciary—retraction onset, protrusion onset, and instance of maximal protrusion velocity (Figure 1E-G). This registration required robust detection of the onsets of protrusion and retraction events (Extended Experimental Procedures, Figure S1). After registration, we averaged the intensity time series, first over all windows within an individual cell, and then across multiple cells. Of note, the accuracy of these final cell-population time series is highest closest to the event used for alignment and decreases with time before and after the event, primarily owing to the variation in the duration of individual protrusion and retraction segments.
Statistical independence of neighboring probing windows
For statistical analysis (mean, confidence internal, correlation) it is essential to determine the degree of interdependence between the time series obtained from neighboring probing windows. The information of neighboring time series is decoupled by two factors—(i) spatial heterogeneity (Figure 1B) and (ii) the time series were aligned relative to edge motion events detected independently for each probing window. To determine the degree of coupling, we calculated the correlation between the velocity time series registered with respect to retraction onset, protrusion onset, and maximal protrusion velocity. We also calculated spatial correlations of velocities between probing windows at the same time. For the latter we found significant correlations between probing windows over distances >1 μm (green lines in Figure 1H-J), suggesting that in our data sets neighboring time series were coupled indeed. However, after event alignment the length scale over which time series would be substantially correlated falls below 0.5 μm (red in Figure 1H-J). Thus, we concluded that each probing window generates a statistically independent sample of the stochastic protrusion process.
Characterization of cytoplasmic fluorescence dynamics using a diffuse fluorescent marker
To test the specificity of the signal of fluorescent actin assembly factors in reporting association of the protein with the lamellipodial actin network, we first characterized the temporal fluctuations of a diffuse fluorescent marker as a control signal. For this purpose, we used HaloTag, a fusion tag which forms a covalent bond to fluorescent ligands (Los et al., 2008). We expressed the HaloTag protein in PtK1 cells (Data File1:Movie S4) and labeled them with TMR (Tetramethylrhodamine) ligands. The analysis showed that the cytoplasmic fluorescence increases after retraction onset and decreases after protrusion onset (Figure 1K, L) because of cell volume changes during these motion events. The cytoplasmic fluorescence exhibits a sharp dip 20 sec after the protrusion velocity reaches its maximum (Figure 1M), suggesting that diffusion of HaloTag-TMR in the lamellipodia region is too slow to instantaneously follow cell edge movements.
Differential recruitment dynamics of actin assembly factors to lamellipodium
Building on these control experiments, we next characterized actin recruitment. We micro-injected trace amounts of Alexa 568 labeled actin into PtK1 cells and used live cell imaging to monitor fluctuations in fluorescence intensity as a proxy for actin recruitment to the lamellipodium (Data File1:Movie S5). As expected, the fluorescence of actin at the leading edge decreased after retraction onset and increased after protrusion onset (Figure 2A, B). Importantly, these dynamics were opposite to those observed for HaloTag-TMR (Figure 1K-M), indicating that the fluorescence signal from low amounts of Alexa 568–labeled actin predominantly reports the fraction of actin monomers incorporated in the filament network. The average fluorescence signal registered relative to maximal protrusion velocity displayed a sharp peak 10 sec after the reference event (Figure 2C), which is consistent with previous quantitative Fluorescent Speckle Microscopy analyses showing that the rate of actin assembly peaks after the time point of fastest protrusion (Ji et al., 2008).
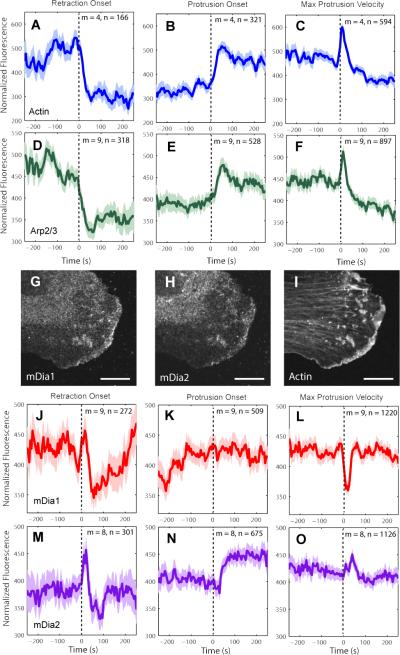
Figure 2. Recruitment of actin assembly factors to lamellipodium.
(A-F), (J-O) Normalized fluorescence intensity time series registered with respect to retraction onset, protrusion onset or maximal protrusion velocity. Time axis spans average duration of protrusion or retraction events (see Figure S6A and C). For each condition the number n of time series sampled in m cells pooled from multiple independent experiments (3 experiments for actin, 4 for Arp2/3, 4 for mDia1, and 4 for mDia2) is indicated. Solid lines indicate population averages. Shaded error bands about the population averages indicate 95% confidence intervals computed by bootstrap sampling. (G-I) Immuno-localization of formins mDia1 (G), mDia2 (H), and phalloidin-actin (I). Scale bars: 5 μm.
Next, we asked whether the temporal variation in actin filament assembly is related to temporal variations in recruitment and activation of actin assembly factors. We expressed HaloTag fusions to nucleator and modulator proteins and labeled them with TMR ligands. We first applied this approach to the Arp3 component of the Arp2/3 complex, and found an almost identical recruitment pattern to actin (Figure 2D-F, Data File1:Movie S6) with the exception that the actin signal increased just before protrusion onset, while the HaloTag-TMR-Arp3 signal began to increase only 10 – 20 s after protrusion onset (cf. Figure S2A vs. Figure S2B). This suggests that in spontaneous epithelial cell edge protrusions other factors may play a role in initiating lamellipodial actin filament assembly.
The involvement of Arp2/3 as a nucleator of lamellipodial actin filament assembly is well-established in PtK1 cells, but the roles of other nucleators such as formin family members have not been addressed systematically. During spontaneous protrusion events, the Rho family GTPase RhoA, an activating signal for members of the formin family including mDia1 (Watanabe et al., 1997), has been found active at the leading edge and in fact preceding the activation of the Rho family GTPase Rac1, which is upstream of Arp2/3 activation (Machacek et al., 2009; Machesky et al., 1999; Miki et al., 1998; Tkachenko et al., 2011). Thus, mDia1 was a possible candidate nucleator for the initiation of protrusions. In addition to mDia1, mDia2 also has been found at the leading edge of mouse melanoma B16F1 cells and by loss-of-function perturbation has been found to be required for lamellipodia formation (Yang et al., 2007). Therefore, we next investigated the recruitment dynamics of mDia1 and mDia2.
Although previous work in PtK1 cells suggested that formins were absent from the lamellipodium (Gupton et al., 2007), we found by reverse transcription PCR and Western blot that the formins mDia1 and mDia2 are both expressed in PtK1 cells (Figure S2C-D). Immunofluorescence imaging showed that both proteins are also localized to the leading edge (Figure 2G-I, Data File1:Movie S7, S8), along with Arp2/3 (Figure S2E-F). In contrast to Arp2/3, the fluorescence of HaloTag-TMR labeled mDia1 decreased after retraction onset but then consistently increased after 50 sec (Figure 2J) and plateaued before protrusion onset (Figure 2K). Hence mDia1 arrives at the leading edge much earlier than Arp2/3. Moreover, mDia1 recruitment underwent a transient dip ~15 sec after maximal protrusion velocity (Figure 2L), when Arp2/3 recruitment exhibited a sharp peak (Figure 2F), suggesting a hand-over from mDia1-based to Arp2/3-based actin nucleation during fast protrusion (Figure S3).
Similar to mDia1, mDia2 fluorescence showed a significant peak after retraction onset followed by a decrease (Figure 2M), but unlike mDia1, it significantly increased again only 30 sec after protrusion onset (Figure 2N), and had a distinct peak (rather than a dip) 40 sec after maximal protrusion velocity (Figure 2O). Hence mDia2 is recruited to the lamellipodium 30 sec after Arp2/3, similar to the findings in vitro (Lee et al., 2010) and consistent with the idea that formins can elongate Arp2/3-nucleated actin filaments (Block et al., 2012; Lee et al., 2010; Yang et al., 2007).
VASP and cofilin dynamics were similar to those of Arp2/3 (Figure S4A-F, Data File1:Movie S9, S10), with the exception that cofilin recruitment peaked 30 sec after maximal protrusion velocity, i.e. 20 sec later than actin, Arp2/3 and VASP (Figure S4F). Notably, none of the HaloTag-TMR labeled proteins exhibited the behaviors of diffuse HaloTag-TMR, implying that the level of labeling was low enough to specifically highlight the fraction of protein immobilized by association with the lamellipodial actin filament network. Taken together, our analyses revealed a differential recruitment of actin assembly factors during distinct phases of the protrusion-retraction cycle. Of all factors measured, mDia1 was the only one to arrive at the leading edge before protrusion. All other factors Arp2/3, VASP, cofilin, and mDia2 followed after protrusion onset.
Laboratory frame of reference reveals traction force and adhesion dynamics at the leading edge during protrusion
To convert actin assembly into edge protrusion, propulsive forces generated by growing filaments must be balanced by traction forces on the substrate at adhesions. Therefore, during protrusion, the coordinated action of actin assembly factors is expected to produce coupled variations in traction force and adhesion assembly. To test this coupling, we first applied our local sampling and registration approach to traction force microscopy data (Figure 3A, Data File1:Movie S11) and investigated how traction forces are coordinated with edge motion during protrusion-retraction cycles. We also monitored the assembly of adhesions by recruitment of HaloTag-TMR-labeled paxillin (Data File1:Movie S12) as a canonical marker for nascent adhesion formation (Nayal et al., 2006).
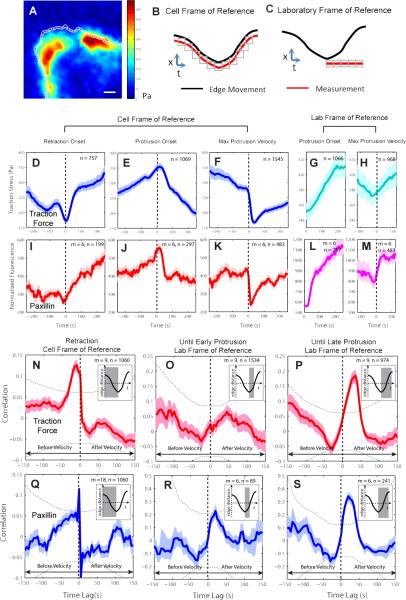
Figure 3. Traction force and adhesion dynamics in epithelial cell protrusion.
(A) Map of the magnitude of the traction force in a PtK1 cell with 1 × 1μm probing windows at the leading edge. Scale bar: 5 μm. (B) Definition of a cell frame of reference in which probing windows track the leading edge. (C) Definition of a laboratory frame of reference in which probing windows are placed at the sites of protrusion initiation and remain stationary to track activities associated with newly formed adhesions. (D–M) Time series of traction force (D–H) and normalized paxillin (adhesion marker) fluorescence intensity (I–M) in cell or laboratory frames of reference registered with respect to retraction onset, protrusion onset, or maximal protrusion velocity. (N–S) Pearson's cross-correlation of traction force and edge velocity (N–P) and of paxillin and edge velocity (Q–S) as a function of the time lag between the two variables using time series acquired during retraction phases in the cell frame of reference (N,Q), and time series acquired during early (from protrusion onset to 50 sec before maximal protrusion velocity) (O,R), and overall (from protrusion onset to 50 sec after maximal protrusion velocity) (P,S) protrusion phases in the laboratory frame of reference. For each condition the number n of time series sampled in m cells pooled from 3 independent experiments is indicated. Inset: The gray areas indicate the periods of the time series used for correlation analysis. Solid lines indicate population averages. Shaded error bands about the population averages indicate 95% confidence intervals computed by bootstrap sampling. Dotted lines (two arcs) indicate the significant correlation level with 95% confidence.
We quantified traction forces and paxillin intensity in two frames of reference. In a ‘cell frame of reference’, probing windows tracked the motion of the leading edge (Figure 3B, Data File1:Movie S3 and S13 (yellow windows)). In a ‘laboratory frame of reference’, probing windows were dynamically placed at the locations of protrusion onset but then left stationary to monitor the force evolution and protein recruitment at newly forming immobilized adhesions (Figure 3C and Data File1:Movie S13 (red windows)). Traction force levels at the cell edge had a basal level of ~350Pa and fluctuated ~50Pa about this value (Figure 3D–F). This is consistent with previous traction force measurements in the same model cell, which suggested that overall the cell cortex is pre-stressed by actomyosin contractility with superimposed smaller force variations that are associated with the dynamics of the actin protrusion machinery (Gardel et al., 2008).
In the cell frame of reference, traction forces at the leading edge started to increase at retraction onset (Figure 3D) until they dropped 25 sec after protrusion onset (Figure 3E). They displayed a significant dip when registered with respect to maximal protrusion velocity (Figure 3F), suggesting that adhesion sites are left behind from the advancing edge. The temporal patterns of traction forces were very similar to those of paxillin intensity (Figure 3I-K).
In the laboratory frame of reference, traction forces (Figure 3G, H) and paxillin intensity (Figure 3L, M) showed sustained increases as edge protrusion proceeded. We conclude from these data that adhesion formation is initiated before protrusion onset at the retracting edge. These nascent adhesions are then immobilized at protrusion initiation sites to counteract the propulsive forces generated at the advancing edge. Importantly, nascent adhesion formation happens only once at the onset of the protrusion cycle. The actin filament network formed after protrusion onset remains without substrate engagement until the onset of the next protrusion segment. Further support for this model was provided by analysis of the retrograde flow of actin filaments (Ponti et al., 2004) (Figure S4G-K).
Cross-correlation reveals timing between traction force and edge motion
We next investigated how traction forces are temporally coordinated with edge motions. First, we selected in the cell frame of reference all time series during retraction events, and correlated edge velocity and traction force as a function of the time lag between the time series (Figure 3N; Pearson's correlation coefficient). We found a positive correlation maximum at –10 seconds time lag indicating that the retraction velocity slows down 10 seconds after traction force begins to increase. We performed the same correlation analysis for protrusion events in the laboratory frame of reference. Using time series from protrusion onset to 50 seconds before maximal protrusion velocity, no significant correlation between edge velocity and traction force was observed (Figure 3O). However, when the time series were extended to 50 seconds after maximal protrusion velocity, a significant correlation maximum occurred at 40 seconds time lag (Figure 3P), indicating that traction force increases 40 seconds after the edge velocity increases. Thus, while the initial protrusion phase is nearly free of mechanical load on the growing actin filaments and coupled adhesions, beyond a critical point further edge advancement expands the plasma membrane under increasing membrane tension. The adhesion marker paxillin displayed similar correlations with edge motion (Figure 3Q-S). Taken together, our local image sampling and registration of traction forces and adhesion recruitment demonstrated a tight coordination of traction force, adhesion formation, and edge motion at length and time scales of single microns and tens of seconds.
Cross-correlation reveals coordination between actin nucleators
To further investigate the functional importance of mDia1 as an initiator of lamellipodial network assembly and cell protrusion, we performed dual-color live cell imaging of EGFP-mDia1 and SNAP-tag-TMR-actin, making it possible to directly correlate the activity of mDia1 and actin. By exploiting the spatiotemporal variation of the fluorescence signal associated with constitutive protrusion and retraction activity, we established direct statistical relationships between the two recruitment processes. In agreement with the recruitment sequence derived from single channel imaging of actin and actin nucleators, the dynamics of simultaneously imaged mDia1 and actin recruitment were similar prior to protrusion onset, but they diverged afterwards (Figure S5). Correlation analysis between EGFP-mDia1 and SNAP-tag-TMR-actin time series aligned relative to protrusion onset (Figure 4A) showed significant correlation values (0.15~0.25, p = 10−12 ~ 10−4, permutation t-test) in the upper triangle of the pairwise correlation matrix (Figure 4B-C; Extended Experimental Procedures), indicating that mDia1 systematically preceded actin polymerization and suggesting that mDia1 drives actin assembly before protrusion onset.
Figure 4. Temporal coordination between mDia1 and Arp2/3 in epithelial cell protrusion.
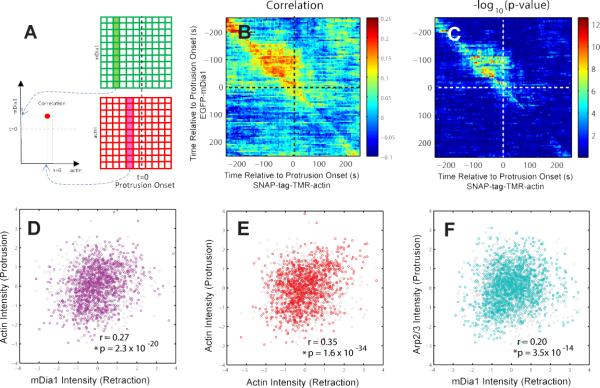
(A) Cross-correlation analysis between mDia1 and actin. (B) Pairwise Pearson's correlation coefficients of EGFP-mDia1- and SNAP-tag-TMR-actin- fluorescence intensity time series registered relative to protrusion onset (1,148 time series from 9 cells pooled from 2 independent experiments). (C) –log10(p-value) of the pairwise Pearson's correlation coefficients. (D-F) Scatter plots and Pearson's correlation coefficients of mean normalized intensities of mDia1 during retraction and of actin during protrusion (sample sizes as in Figure 4B) (D), actin during retraction and actin during protrusion (sample sizes as Figure 4B) (E), and mDia1 during retraction and Arp2/3 during protrusion (1,716 time series from 11 cells pooled from 2 independent experiments) (F). P-values are calculated using permutation t-test. The gray circles in Figure 4D-E indicate the distribution of the same data after randomizing the association between x- and y-component.
To test how the coupling of mDia1 and actin recruitment prior to protrusion influences lamellipodial and edge motion dynamics during protrusion, we correlated the mean EGFP-mDia1 intensity in a probing window during retraction with the mean SNAP-tag-TMR-actin intensity in the same window during protrusion. This analysis revealed that the level of mDia1 recruitment during retraction has a significant correlation (0.27, p= 2.3 × 10−20 , permutation t-test) with the amount of actin polymerization during protrusion (purple circles in Figure 4D), whereas there is no significant correlation (−0.02 p=0.56, permutation t-test) in the randomized decoupled data set (gray circles in Figure 4D). For comparison, we also did not find significant correlation between mDia1 during protrusion and actin during retraction (0.02, p = 0.5 , permutation t-test). On the other hand, we found a significant correlation (0.35, p= 1.6 × 10−34, permutation t-test) between actin recruitment during retraction and actin recruitment during protrusion (red circles in Figure 4E), whereas there is no significant correlation (−0.02, p=0.49, permutation t-test) in the randomized decoupled data set (gray circles in Figure 4E). Together, these analyses corroborate that mDia1drives actin assembly prior to protrusion and determines the amount of actin assembly during protrusion.
The recruitment sequence of actin nucleators derived from single channel imaging also indicated that actin polymerization after protrusion onset is dominated by Arp2/3. Thus, our correlation analysis between mDia1 and actin recruitment implies a connection between mDia1-mediated actin assembly prior to and Arp2/3 activation after protrusion onset. To test this, we performed dual-color imaging of EGFP-mDia1 and Arp3-HaloTag-TMR and correlated the mDia1 mean intensity during retraction and the Arp2/3 mean intensity during protrusion (Figure 4F). Indeed, compared to a much weaker correlation between mDia1 during protrusion and Arp2/3 during retraction (0.05, p= 0.04, permutation t-test), we found a significant correlation between the two variables (0.20, p= 3.5 × 10−14, permutation t-test) (cyan circles in Figure 4F), whereas there is no significant correlation (−0.02, p=0.52, permutation t-test) in the randomized decoupled data set (gray circles in Figure 4F).
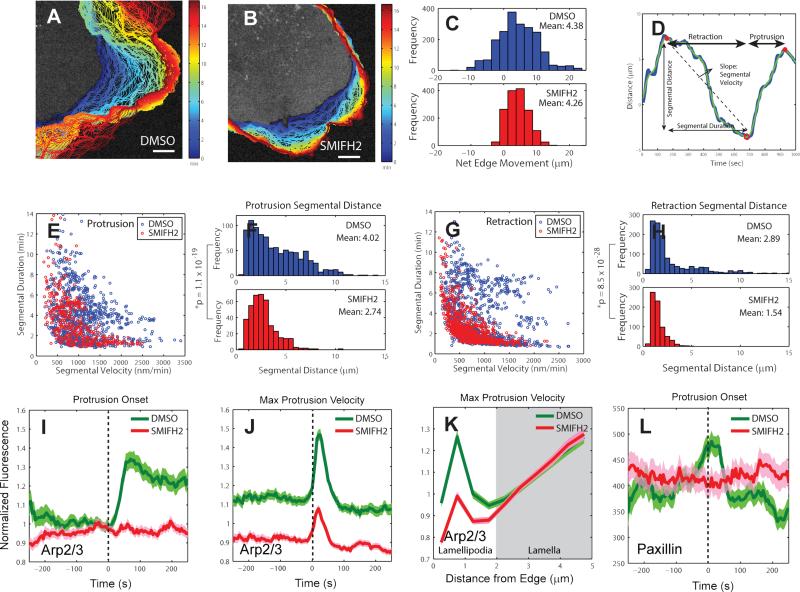
The formin inhibitor SMIFH2 reduced protrusion length and Arp2/3 activation at the leading edge
Our statistical analyses thus far suggest that early recruitment of mDia1 may nucleate the growth of linear actin structures, which are anchored to the substrate by nascent adhesions before protrusion starts and that this process plays an important role in coordinating the recruitment of Arp2/3, which enhances filament growth after protrusion onset. To confirm the coordination between mDia1 and Arp2/3 directly, we treated PtK1 cells with 20μM SMIFH2, a chemical inhibitor of formin FH2 domains (Rizvi et al., 2009). Formin-inhibited cells had less protrusive activity than cells under control conditions (Figure 5A-B), with a much tighter distribution of net distances travelled over the course of an entire movie (Figure 5C). This suggests that formin plays a role in controlling the distance range of protrusion-retraction cycles. To further investigate the role of formin, we identified protrusion and retraction segments for each probing window (Figure 5D) and calculated their duration and average velocity. The marginal distributions of the two parameters in control and formin-inhibited cells showed only weakly significant differences (Figure S6A-B). However, the joint distribution revealed that with formin-inhibition fast protrusion segments lasted for only a short time, while long-lasting protrusion segments tended to be slow. As a result, the distance traveled over one protrusion segment was significantly (p = 1.1 × 10−19, one tailed K-S test) reduced by formin inhibition (Figure 5F). In control cells the two parameters were less correlated (Figure 5E). Formin-inhibition also constrained the distance edge windows traveled over retraction segments (Figure 5G-H, Figure S6C-D). This explains why, when integrated over the course of an entire movie, the leading edges of formin-inhibited epithelial cells traveled the same net distances as those of control cells.
Figure 5. Effects of formin inhibition on protrusion length, Arp2/3 activation, and adhesion dynamics.
(A-B) Positions (blue, early; red, late time points) of leading edge of cells stained with CellMask Orange membrane marker and treated with DMSO (control) (A) and 20 μM SMIFH2 (formin inhibitor) (B). Scale bars: 5 μm. (C) Distribution of net edge movement in DMSO (n = 2,023 sectors from m = 9 cells) or SMIFH2 (n = 1498 sectors from m = 9 cells) treated cells, (D) Motion of a single 500nm-long edge sector over 16 minutes. Blue line: protrusion distance obtained by time-integration of protrusion velocity, Green line: spline-filtered protrusion distance, Red open circle (o): protrusion onset, Red solid circle (•): retraction onset. The time interval between a retraction onset and a protrusion onset is defined as a retraction segment. The time interval between a protrusion onset and a retraction onset is defined as a protrusion segment. (E) Velocity vs. duration of individual protrusion segments in DMSO- (n = 950 segments from m = 9 cells pooled from 3 independent experiments) and SMIFH2- (n = 399 segments from m = 9 cells pooled from 3 independent experiments) treated-cells. (F) Distribution of protrusion segmental distance in DMSO or SMIFH2-treated cells. n = 950 protrusion segments from m = 9 cells for DMSO and n = 399 protrusion segments from m = 9 cells for SMIFH2. The data were pooled from 3 independent experiments. P-value is calculated using one tailed K-S test. (G) Velocity vs. duration of individual retraction segments in DMSO (n = 1,180 segments from m = 9 cells) or SMIFH2 (n = 718 segments from m = 9 cells) treated cells. (H) Distribution of retraction segmental distance in DMSO or SMIFH2 treated-cells. n = 1,180 retraction segments from m = 9 cells for DMSO and n = 718 retraction segments from m = 9 cells for SMIFH2. The data were pooled from 3 independent experiments. P-value is calculated using one tailed K-S test. (I-J) Time series of Arp2/3 intensity in the lamellipodium, normalized to the Arp2/3 intensity in the region 1.5 – 2 μm from the cell edge, registered with respect to protrusion onset or maximal protrusion velocity in DMSO- or SMIFH2-treated-cells. (I) n = 556 time series sampled in m = 10 cells for DMSO and n = 602 time series sampled in m = 11 cells for SMIFH2, pooled from 3 independent experiments. (J) n = 1468 time series sampled in m = 10 cells for DMSO and n = 1467 time series sampled in m = 11 cells for SMIFH2, pooled from 3 independent experiments. (K) Spatial modulation of Arp2/3 fluorescence at time point of maximal protrusion velocity in DMSO- and SMIFH2-treated cells as a function of distance from the leading edge. (L) Normalized paxillin fluorescence intensity time series acquired in the cell frame of reference in DMSO- or SMIFH2-treated cells registered with respect to protrusion onset. n = 556 time series sampled in m = 8 cells for DMSO and n = 331 time series sampled in m = 11 cells for SMIFH2, pooled from 3 independent experiments. Solid lines indicate population averages. Shaded error bands about population averages indicate 95% confidence intervals computed by bootstrap sampling.
To test our prediction that the longer distances of protrusion segments in untreated cells originates in mDia1 modulating Arp2/3 recruitment, we compared Arp2/3 dynamics in cells without and with SMIFH2 treatment. Because the expression levels of Arp2/3-HaloTag-TMR varied between cells, we normalized the Arp2/3 intensity in the lamellipodium to the Arp2/3 intensity in the region 1.5 – 2 μm from the cell edge. Under formin inhibition, Arp2/3 levels did not significantly increase after protrusion onset and, compared to control cells, they exhibited only minor fluctuations after the time points of retraction onset and maximal protrusion velocity (Figure 5I-J; Figure S7A). Consistent with the dynamic analysis, the overall levels of Arp2/3 in the lamellipodium at maximal protrusion velocity were substantially reduced with SMIFH2 treatment, but they remained unchanged over the entire lamella region located behind the lamellipodium (>2 μm from the cell edge) (Figure 5K).
The finding that both mDia1 and paxillin were recruited to the leading edge before protrusion onset suggests that mDia1 could play an important role in adhesion dynamics. Moreover, a previous study showed that knock-down of mDia1 led to the disappearance of nascent adhesions (Zaoui et al., 2008). To probe the role of mDia1 in adhesion formation, we treated cells with 20 μM SMIFH2 and analyzed paxillin dynamics in the cell frame of reference. Recruitment of paxillin to the leading edge of SMIFH2 treated-cells at protrusion onset was highly variable with no detectable trends in the timing, in clear contrast to DMSO treated-cells, which had a peak in the paxillin recruitment at protrusion onset (Figure 5L). At maximal protrusion velocity, SMIFH2 treated-cells still showed a sharp decrease of paxillin fluorescence as the advancing cell edge rapidly moved away from stabilized nascent adhesions (Figure S7B). Hence, SMIFH2-treated cells were still capable of assembling adhesions at the leading edge, even though they lost the spatiotemporal coordination between protrusions and adhesions.
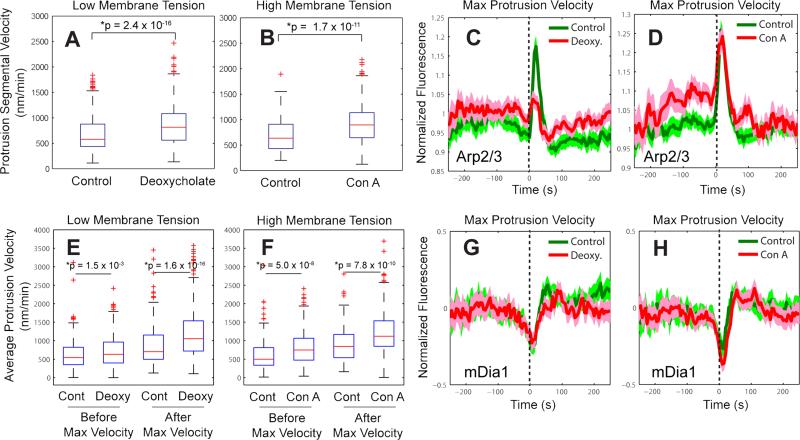
Changes in membrane tension revealed that Arp2/3 recruitment is mechanosensitive
Combining our observations of traction force fluctuations with the coordination of actin assembly factors led to the hypothesis that the Arp2/3 recruitment after protrusion onset could be controlled by the increasing force levels in nascent adhesions. Since these forces balance propulsive forces by actin polymerization at the cell edge, this would provide for a mechanism of mechano-responsive positive feedback, in which initial protrusion mediated by mDia1-driven filament assembly promotes the recruitment of Arp2/3 (and other assembly factors) in order to increase the rate of actin-based propulsion against increasing membrane tension and further reinforcement of Arp2/3 recruitment. Accordingly, the rate of Arp2/3 recruitment is predicted to depend on the rate of plasma membrane tension increase.
To test this prediction, we altered the membrane tension levels using deoxycholate (Raucher and Sheetz, 2000) and concanavalin A (Con A) (Supplemental Note 3) and used our statistical analysis to determine how the recruitment of Arp2/3 and mDia1 was affected. In previous studies variation in membrane tension was associated with increased (low tension) or decreased (high tension) efficiency of the load-dependent polymerization ratchet (Gauthier et al., 2011; Mogilner and Oster, 1996; Raucher and Sheetz, 2000) and Rac activation (Houk et al., 2012). Consistent with this inference, we found that reduced membrane tension using deoxycholate significantly (p=2.4 × 10−16, one tailed K-S test) increased protrusion segmental velocity (Figure 6A). However, increased membrane tension under Con A treatment also significantly increased (p = 1.7 × 10−11, one tailed K-S test) protrusion segmental velocity (Figure 6B). This would support a tension-induced positive feedback in the activation of actin assembly factors that compensates and even overcomes the load-dependent efficiency drop of the polymerization ratchet (Mogilner and Oster, 1996).
Figure 6. Mechanosensitivity of Arp2/3 dynamics during cell protrusion.
(A-B) Protrusion segmental velocity in control cells vs. cells treated with 400 μM deoxycholate to decrease membrane tension (A) (n = 1,950 time series sampled in m = 11 cells for control and n = 2,555 time series sampled in m = 12 cells for deoxycholate, pooled from 3 independent experiments) or cells treated with 1 μg/ml concanavalin A to increase membrane tension (B) (n = 1,796 time series sampled in m = 12 cells for control and n = 1,995 time series sampled in m = 10 cells for concanavalin A, pooled from 3 independent experiments). (C-D) Time series of Arp2/3 intensity in the lamellipodium, normalized to the Arp2/3 intensity in the region 1.5 – 2 μm from the cell edge registered with respect to maximal protrusion velocity in control vs. deoxycholate-treated cells (C) (n = 1,040 time series sampled in m = 6 cells for control and n = 1,448 time series sampled in m = 8 cells for deoxycholate, pooled from 2 independent experiments) or control vs. concanavalin A–treated cells (D) (n = 1,095 time series sampled in m = 8 cells for control and n = 1,054 time series sampled in m = 6 cells for concanavalin A, pooled from 2 independent experiments). (E) Average edge velocity between protrusion onset and maximal protrusion velocity vs. between maximal protrusion velocity and protrusion end in control vs. deoxycholate-treated cells. (F) Average edge velocity between protrusion onset and maximal protrusion velocity vs. between maximal protrusion velocity and protrusion end in control vs. concanavalin A–treated cells. (G-H) Normalized fluorescence intensity time series of mDia1 registered with respect to maximal protrusion velocity in control vs. deoxycholate- vs. concanavalin A–treated cells. (G) n = 1,182 time series sampled in m = 8 cells for control and n = 1,149 time series sampled in m = 7 cells for deoxycholate, pooled from 2 independent experiments. (H) n = 1,089 time series sampled in m = 8 cells for control and n = 1325 time series sampled in m = 8 cells for concanavalin A, pooled from 2 independent experiments. Solid lines indicate population averages. Shaded error bands about population averages indicate 95% confidence intervals computed by bootstrap sampling. P-values are calculated using one tailed K-S test.
To test this model, we investigated the changes in actin nucleator recruitment dynamics elicited by changes in membrane tension. Reducing membrane tension abolished the recruitment peak of Arp2/3 ~10 s after maximal protrusion velocity (Figure 6C). Together with the traction force measurements (Extended Experimental Procedures, Figure S7C), this suggests that during a protrusion cycle the membrane tension and the traction force at the leading edge increase, leading to Arp2/3 activation during reinforcement of cell protrusion. Increasing membrane tension did not alter the timing of the Arp2/3 peak after maximal protrusion velocity, but it substantially raised the rate of Arp2/3 recruitment prior to this time point (Figure 6D). Consistent with these recruitment patterns, the average edge velocity before maximal protrusion velocity was not greatly changed by lowering tension (Figure 6E), but was increased significantly by raising tension (Figure 6F). We also tested the mechanosensitivity of mDia1 formin. In contrast to Arp2/3, we did not find significant differences in mDia1 dynamics between control conditions and conditions of altered membrane tension (Figure 6G-H).
Discussion
The standard cell biological approach relying on molecular perturbations is limited when dealing with pathways with functional redundancy. Here we propose an approach to unraveling differential yet partially redundant functions in complex molecular systems by establishing the fine orchestration between molecular events in an unperturbed but dynamically fluctuating system. Implementing this approach required overcoming substantial technical challenges related to the heterogeneous and stochastic nature of the exploited fluctuations. To tackle this, we developed a local image sampling and registration approach that allowed us to extract hidden dynamic relations between intrinsically stochastic molecular activities. The main idea is that time series alignment relative to temporal fiduciaries enables us to characterize the dynamics of various molecular and cellular events, establishing the temporal hierarchy of molecular cascades.
Previously, correlation analysis between protrusion velocity and RhoGTPase activities has been used to establish the timing of RhoGTPase activation relative to protrusion events (Machacek et al., 2009). However, since the correlation coefficients were calculated in a temporally global manner, this approach could be limited when multiple temporal coordination mechanisms overlap within the same time series. By locating specific protrusion events such as protrusion and retraction onsets and maximum protrusion velocity, we were able to choose the intervals of time series to perform more specific correlation analyses.
To demonstrate this method, we mapped out the recruitment dynamics of actin assembly and related these data to edge movement, adhesion assembly, and force generation. Interestingly, each factor exhibited highly distinct dynamics (Figure 2 and Figure S4A-F). From this analysis, we built a precise event cascade that puts mDia1 recruitment at the top of a causal chain of molecular events driving cell protrusion. These experiments were strongly supported by statistical analyses of dual-color imaging of mDia1 and Arp2/3, which indicate dependence of Arp2/3 recruitment on mDia1, but not vice versa (Figure 4). To confirm this finding, we used small molecule inhibitors of formins to acutely disrupt mDia1 function and indeed found the prediction confirmed that without mDia1-initiated actin assembly significantly lower amounts of Arp2/3 were recruited to the leading edge and adhesion formation was disrupted (Figure 5I-K). This is consistent with previous studies that showed RhoA activity preceding Rac activity during spontaneous protrusion/retraction cycles (Machacek et al., 2009; Tkachenko et al., 2011) and knock-down of mDia1 abrogating nascent adhesions (Zaoui et al., 2008). Interestingly, recent studies also showed that mDia1 and Arp2/3 work together in actin organization in cell cortex (Bovellan et al., 2014) and Listeria comet tails (Fattouh et al., 2014).
Our work by no means questions the power of molecular biology and genetics as an experimental tool, especially for component discovery. It does, however, put a cautionary note on the interpretability of experimental outcomes in terms of the function of the targeted system component. The default interpretation of a molecular or genetic intervention experiment has to start with the conclusion that the data show how the system behaves when the targeted component is perturbed. This is fundamentally different from the conclusion what the targeted component contributes to the system behavior when it is present.
Our study also demonstrates the application of fluctuation analysis to reveal dynamic mechano-responses of cell protrusion. We show that Arp2/3 recruitment, but not mDia1 recruitment, is controlled, at least in part, by the level of membrane tension. This mechanism is elicited either globally by manipulations of tension homeostasis, or dynamically during spontaneous protrusion-retraction cycles where membrane tension changes transiently with the level of edge advancement. Hence, we suggest that Arp2/3's primary function in actin-mediated cell protrusion is the rapid upregulation of actin assembly under increasing force levels, regardless of whether the protrusion is initiated spontaneously, by chemotaxis, haptotaxis, or another guidance mechanism. We surmise that deducing this function of Arp2/3 required the direct observation of recruitment in the context of the dynamics of other system variables.
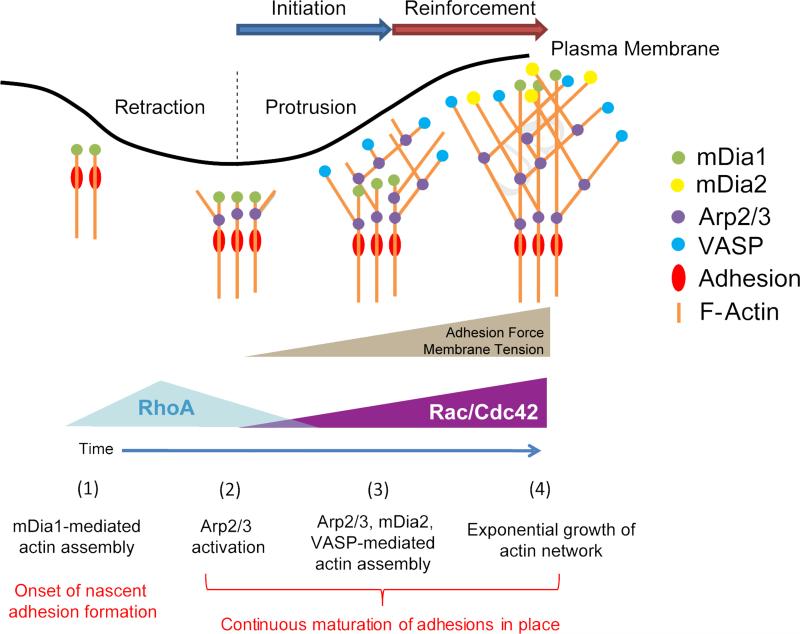
In summary, our data reveal a core component of an initiation-reinforcement cycle of actin filament assembly that drives persistent cell protrusion (Figure 7): mDia1 is the first nucleation factor recruited to the plasma membrane and activated during cell edge retraction. These early actin assembly events generate a filament scaffold for the recruitment of molecular components into nascent adhesions (Choi et al., 2008). At this point it is possible that other members of the formin family may be involved (Iskratsch et al., 2013), with the exception of mDia2 that has been characterized as a late factor. At protrusion onset, adhesions are spatially immobilized and Arp2/3 recruitment is activated. Arp2/3 binds to early-nucleated ‘mother filaments’ (Machesky et al., 1999) and promotes the exponential growth of a lamellipodial network in dendritic branches (Initiation). This accelerates membrane protrusion. As protrusion proceeds with adhesion maturation, increasing membrane tension slows down edge advancement while activating the mechano-responsive Arp2/3 recruitment to sustain actin filament assembly (reinforcement) (Ji et al., 2008). During this period, mDia1 spatially lags behind the Arp2/3-nucleated network (Figure 2L, Figure S3C). To corroborate heightened polymerization during reinforcement, additional factors such as VASP and mDia2, which compete with polymerization-blocking capping proteins for free filament ends, are recruited after the time point of maximal protrusion velocity. The transition from initiation to reinforcement happens over a time scale of 10 seconds. By inference, we suggest that it is accompanied by a switch from RhoA to Rac1 and Cdc42 signaling (Machacek et al., 2009). Owing to its mechanosensitivity, we expect that the protein components and duration of the initiation and reinforcement phases vary between different modes of cell migration and between different microenvironments. Our statistical framework for the analysis of dynamic image events will accelerate the determination of functional links between extracellular and intracellular mechananical and chemical guidance cues and the fine coordination of several redundant actin assembly pathways required for efficient cell protrusion during directed migration.
Figure 7. Model of initiation and reinforcement of actin assembly during cell protrusions mediated by multiple, functionally overlapping, assembly promoting pathways.
The formin mDia1 is recruited to the leading edge of the cell before protrusion onset, initiating linear growth of the lamellipodial network. Recruitment of Arp2/3 sensitive to membrane tension then promotes sustained exponential growth of the network, supported by additional assembly factors.
Experimental Procedures
Live cell imaging
PtK1 cells were imaged at 5 s interval for 1000 s using a 60x, 1.4 NA Plan Apochromat objective with 1.5x optovar, (for traction force microscopy and dual color imaging no optovar was used) for spinning disk confocal microscopy and using 100x, 1.49 NA Plan Apochromat objective for TIRF microscopy.
mDia formin inhibition
PtK1 cells were incubated with 20 μM SMIFH2 (Sigma-Aldrich) for 1 hour. For analysis of protrusion dynamics, cells were stained using CellMask Orange after incubation with SMIFH2. For HaloTag fusion proteins, HaloTag was labeled with TMR ligands after incubation with SMIFH2.
Membrane tension perturbation
PtK1 cells were incubated with either 400 μM dexoycholate (Sigma-Aldrich) for 30 minutes (decreasing membrane tension) or 1 μg/ml concanavalin A (Invitrogen) for 10 minutes (increasing membrane tension) and washed by the media before imaging. HaloTag labeling with TMR ligands was done before membrane tension perturbation.
Traction force microscopy
Preparation of soft substrates with far-red fluorescent beads for traction force microscopy is described elsewhere (Gutierrez et al., 2011). Cell traction forces were calculated by comparing images of tracer particles with and without cells on the substrate using Fourier transform traction cytometry method (Butler et al., 2002).
Image Analysis
Cell edges were segmented by intensity thresholding, Cell edge displacements were tracked, and probing windows were generated with a window size of 500 nm by 500 nm for fluorescence intensity/actin flow analysis and 1 μm by 1 μm for traction force analysis. Protrusion and retraction segments were identified on a per-window basis using a filtered edge displacement time series (Figure 5D and Figure S1A-B). Time-series of normalized fluorescence intensities, traction forces, actin retrograde flow sampled in individual windows were aligned relative to retraction/protrusion onsets and the instance of maximal protrusion velocity and averaged over all windows from multiple cells. The 95% confidence intervals about the average series were obtained by bootstrap resampling of the individual time series. All procedures are detailed in Extended Experimental Procedures.
Supplementary Material
Acknowledgements
K.L. was the recipient of Ruth L. Kirschstein NRSA Postdoctoral Fellowship from NIH. This work was funded by NIH grants GM071868 and GM098412. We thank Art Alberts for the gift of mDia2 antibody, Martin Schwartz for SNAP-tag-actin plasmids, J. Lim for helping with actin speckle imaging, A. Besser for providing the traction force microscopy software package, K. Jaqaman for the algorithm for segmentation of focal adhesions, and S. Besson for general software support. We thank J. Waters and Nikon Imaging Center at Harvard Medical School.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
K. L. initiated the project, designed the concept of event registration of the time series, wrote image analysis software for protrusion/correlation analysis, event registration, performed fluorescence live cell imaging, plasmid construction, immunofluorescence, reverse-transcription PCR, and wrote the final version of the manuscript and supplement; H. L. E. wrote the software for windowing; Y. O. performed traction force microscopy and western blotting; C. Z. prepared the soft-gels for traction force microscopy; A. G. and J. D. T. contributed to the writing of the manuscript. G. D. coordinated the study and wrote the final version of the manuscript and supplement. All authors discussed the results of the study.
References
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Block J, Breitsprecher D, Kuhn S, Winterhoff M, Kage F, Geffers R, Duwe P, Rohn JL, Baum B, Brakebusch C, et al. FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr. Biol. 2012;22:1005–1012. doi: 10.1016/j.cub.2012.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovellan M, Romeo Y, Biro M, Boden A, Chugh P, Yonis A, Vaghela M, Fritzsche M, Moulding D, Thorogate R, et al. Cellular control of cortical actin nucleation. Curr. Biol. 2014;24:1628–1635. doi: 10.1016/j.cub.2014.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ. Traction fields, moments, and strain energy that cells exert on their surroundings. Am. J. Physiol. Cell Physiol. 2002;282:C595–605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell. Biol. 2008;10:1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattouh R, Kwon H, Czuczman MA, Copeland JW, Pelletier L, Quinlan ME, Muise AM, Higgins DE, Brumell JH. The Diaphanous-Related Formins Promote Protrusion Formation and Cell-to-Cell Spread of Listeria monocytogenes. J. Infect. Dis. 2014;211:1185–95. doi: 10.1093/infdis/jiu546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardel ML, Sabass B, Ji L, Danuser G, Schwarz US, Waterman CM. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J. Cell Biol. 2008;183:999–1005. doi: 10.1083/jcb.200810060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier NC, Fardin MA, Roca-Cusachs P, Sheetz MP. Temporary increase in plasma membrane tension coordinates the activation of exocytosis and contraction during cell spreading. Proc. Natl. Acad. Sci. USA. 2011;108:14467–14472. doi: 10.1073/pnas.1105845108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- Gupton SL, Eisenmann K, Alberts AS, Waterman-Storer CM. mDia2 regulates actin and focal adhesion dynamics and organization in the lamella for efficient epithelial cell migration. J. Cell. Sci. 2007;120:3475–3487. doi: 10.1242/jcs.006049. [DOI] [PubMed] [Google Scholar]
- Gutierrez E, Tkachenko E, Besser A, Sundd P, Ley K, Danuser G, Ginsberg MH, Groisman A. High refractive index silicone gels for simultaneous total internal reflection fluorescence and traction force microscopy of adherent cells. PLoS One. 2011;6:e23807. doi: 10.1371/journal.pone.0023807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houk AR, Jilkine A, Mejean CO, Boltyanskiy R, Dufresne ER, Angenent SB, Altschuler SJ, Wu LF, Weiner OD. Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell. 2012;148:175–188. doi: 10.1016/j.cell.2011.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskratsch T, Yu CH, Mathur A, Liu S, Stevenin V, Dwyer J, Hone J, Ehler E, Sheetz M. FHOD1 is needed for directed forces and adhesion maturation during cell spreading and migration. Dev. Cell. 2013;27:545–559. doi: 10.1016/j.devcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L, Lim J, Danuser G. Fluctuations of intracellular forces during cell protrusion. Nat. Cell Biol. 2008;10:1393–1400. doi: 10.1038/ncb1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri R, Springer M, Pilpel Y. Genetic redundancy: new tricks for old genes. Cell. 2009;136:389–392. doi: 10.1016/j.cell.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 2014;15:577–590. doi: 10.1038/nrm3861. [DOI] [PubMed] [Google Scholar]
- Lacayo CI, Pincus Z, VanDuijn MM, Wilson CA, Fletcher DA, Gertler FB, Mogilner A, Theriot JA. Emergence of large-scale cell morphology and movement from local actin filament growth dynamics. PLoS Biol. 2007;5:e233. doi: 10.1371/journal.pbio.0050233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Gallop JL, Rambani K, Kirschner MW. Self-assembly of filopodia-like structures on supported lipid bilayers. Science. 2010;329:1341–1345. doi: 10.1126/science.1191710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. USA. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A, Oster G. Cell motility driven by actin polymerization. Biophys. J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayal A, Webb DJ, Brown CM, Schaefer EM, Vicente-Manzanares M, Horwitz AR. Paxillin phosphorylation at Ser273 localizes a GIT1-PIX-PAK complex and regulates adhesion and protrusion dynamics. J. Cell Biol. 2006;173:587–589. doi: 10.1083/jcb.200509075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- Raucher D, Sheetz MP. Cell spreading and lamellipodial extension rate is regulated by membrane tension. J. Cell Biol. 2000;148:127–136. doi: 10.1083/jcb.148.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi SA, Neidt EM, Cui J, Feiger Z, Skau CT, Gardel ML, Kozmin SA, Kovar DR. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 2009;16:1158–1168. doi: 10.1016/j.chembiol.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K, Behrendt B, Small JV, Wehland J. VASP dynamics during lamellipodia protrusion. Nat. Cell Biol. 1999;1:321–322. doi: 10.1038/13040. [DOI] [PubMed] [Google Scholar]
- Suraneni P, Rubinstein B, Unruh JR, Durnin M, Hanein D, Li R. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J. Cell Biol. 2012;197:239–251. doi: 10.1083/jcb.201112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachenko E, Sabouri-Ghomi M, Pertz O, Kim C, Gutierrez E, Machacek M, Groisman A, Danuser G, Ginsberg MH. Protein kinase A governs a RhoA-RhoGDI protrusion-retraction pacemaker in migrating cells. Nat Cell. Biol. 2011;13:660–667. doi: 10.1038/ncb2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela M, Halidi N, Besson S, Elliott H, Hahn K, Tytell J, Danuser G. Fluctuation analysis of activity biosensor images for the study of information flow in signaling pathways. Methods Enzymol. 2013;519:253–276. doi: 10.1016/B978-0-12-405539-1.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch BM, Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 2012;148:973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaoui K, Honore S, Isnardon D, Braguer D, Badache A. Memo-RhoA-mDia1 signaling controls microtubules, the actin network, and adhesion site formation in migrating cells. J. Cell Biol. 2008;183:401–408. doi: 10.1083/jcb.200805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.