Abstract
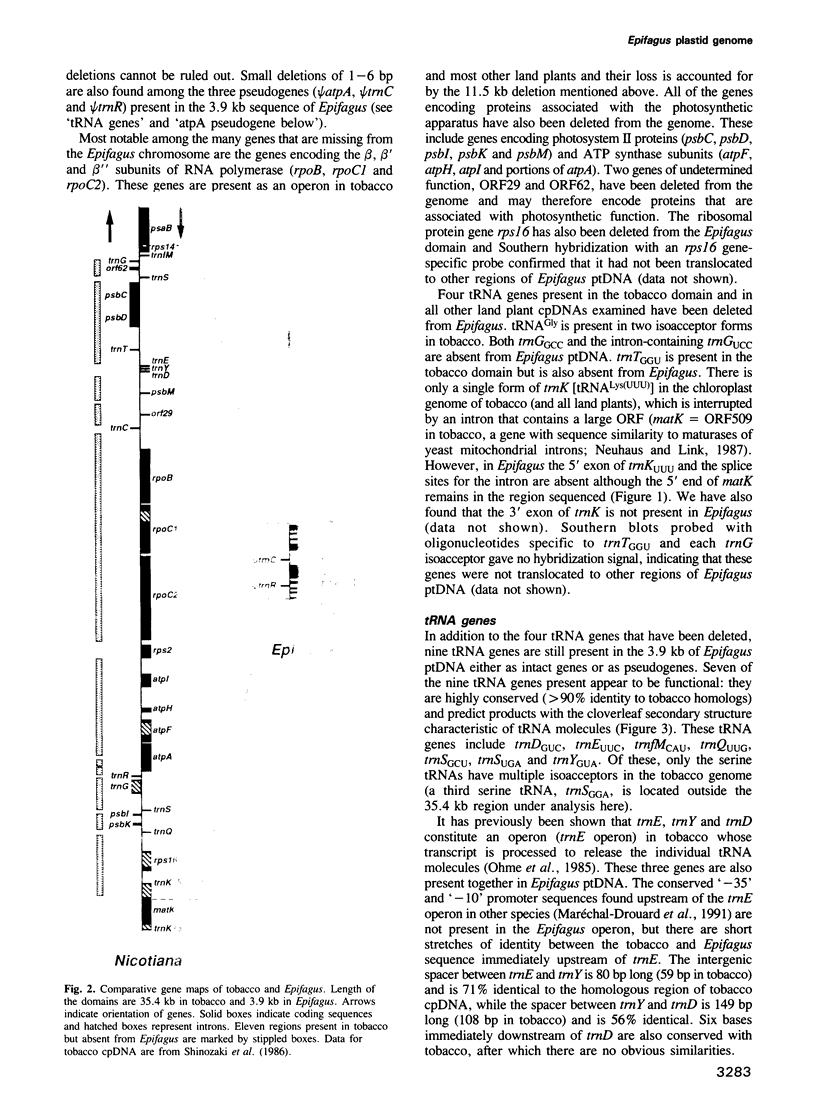
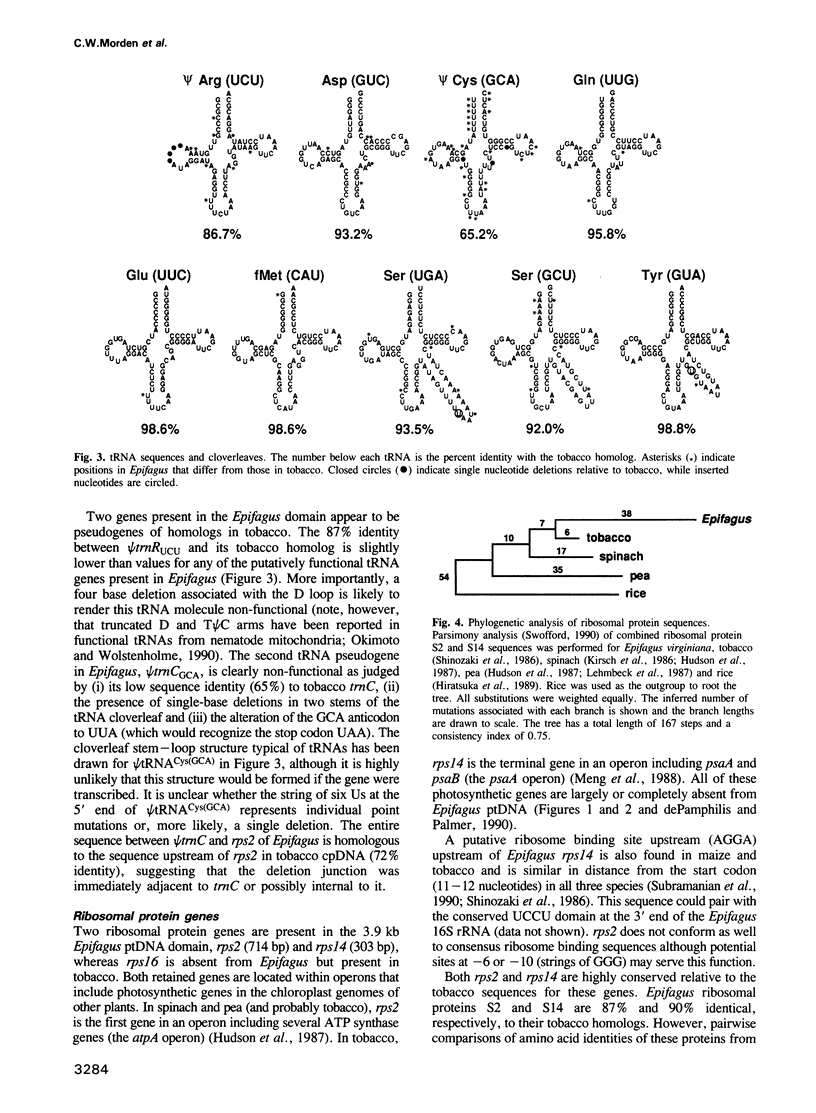
The non-photosynthetic, parasitic flowering plant Epifagus virginiana has recently been shown to contain a grossly reduced plastid genome that has lost many photosynthetic and chloro-respiratory genes. We have cloned and sequenced a 3.9 kb domain of plastid DNA from Epifagus to investigate the patterns of evolutionary change in such a reduced genome and to determine which genes are still present and likely to be functional. This 3.9 kb domain is colinear with a 35.4 kb region of tobacco chloroplast DNA, differing from it by a minimum of 11 large deletions varying in length from 354 bp to 11.5 kb, as well as by a number of small deletions and insertions. The nine genes retained in Epifagus encode seven tRNAs and two ribosomal proteins and are coextensive and highly conserved in sequence with homologs in photosynthetic plants. This suggests that these genes are functional in Epifagus and, together with evidence that the Epifagus plastid genome is transcribed, implies that plastid gene products play a role in processes other than photosynthesis and gene expression. Genes that are completely absent include not only photosynthetic genes, but surprisingly, genes encoding three subunits of RNA polymerase, four tRNAs and one ribosomal protein. In addition, only pseudogenes are found for two other tRNAs. Despite these defunct tRNA genes, codon and amino acid usage in Epifagus protein genes is normal. We therefore hypothesize that the expression of plastid genes in Epifagus relies on the import of nuclear encoded tRNAs and RNA polymerase from the cytoplasm.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldauf S. L., Palmer J. D. Evolutionary transfer of the chloroplast tufA gene to the nucleus. Nature. 1990 Mar 15;344(6263):262–265. doi: 10.1038/344262a0. [DOI] [PubMed] [Google Scholar]
- Basu M. K., Wilson H. J. Mercury risk from teeth. Nature. 1991 Jan 10;349(6305):109–109. doi: 10.1038/349109a0. [DOI] [PubMed] [Google Scholar]
- Chen H. C., Wintz H., Weil J. H., Pillay D. T. Three mitochondrial tRNA genes from Arabidopsis thaliana: evidence for the conversion of a tRNAPhe gene into a tRNATyr gene. Nucleic Acids Res. 1989 Apr 11;17(7):2613–2621. doi: 10.1093/nar/17.7.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello P. S., Gray M. W. RNA editing in plant mitochondria. Nature. 1989 Oct 19;341(6243):662–666. doi: 10.1038/341662a0. [DOI] [PubMed] [Google Scholar]
- Diamond A. M., Montero-Puerner Y., Lee B. J., Hatfield D. Selenocysteine inserting tRNAs are likely generated by tRNA editing. Nucleic Acids Res. 1990 Nov 25;18(22):6727–6727. doi: 10.1093/nar/18.22.6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driesel A. J., Crouse E. J., Gordon K., Bohnert H. J., Herrmann R. G., Steinmetz A., Mubumbila M., Keller M., Burkard G., Weil J. H. Fractionation and identification of spinach chloroplast transfer RNAs and mapping of their genes on the restriction map of chloroplast DNA. Gene. 1979 Aug;6(4):285–306. doi: 10.1016/0378-1119(79)90070-2. [DOI] [PubMed] [Google Scholar]
- Gantt J. S., Baldauf S. L., Calie P. J., Weeden N. F., Palmer J. D. Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 1991 Oct;10(10):3073–3078. doi: 10.1002/j.1460-2075.1991.tb07859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Gray M. W., Boer P. H. Organization and expression of algal (Chlamydomonas reinhardtii) mitochondrial DNA. Philos Trans R Soc Lond B Biol Sci. 1988 May 31;319(1193):135–147. doi: 10.1098/rstb.1988.0038. [DOI] [PubMed] [Google Scholar]
- Gruissem W., Elsner-Menzel C., Latshaw S., Narita J. O., Schaffer M. A., Zurawski G. A subpopulation of spinach chloroplast tRNA genes does not require upstream promoter elements for transcription. Nucleic Acids Res. 1986 Oct 10;14(19):7541–7556. doi: 10.1093/nar/14.19.7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberto J. M., Lamattina L., Bonnard G., Weil J. H., Grienenberger J. M. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature. 1989 Oct 19;341(6243):660–662. doi: 10.1038/341660a0. [DOI] [PubMed] [Google Scholar]
- Hancock K., Hajduk S. L. The mitochondrial tRNAs of Trypanosoma brucei are nuclear encoded. J Biol Chem. 1990 Nov 5;265(31):19208–19215. [PubMed] [Google Scholar]
- Heckman J. E., Sarnoff J., Alzner-DeWeerd B., Yin S., RajBhandary U. L. Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3159–3163. doi: 10.1073/pnas.77.6.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka J., Shimada H., Whittier R., Ishibashi T., Sakamoto M., Mori M., Kondo C., Honji Y., Sun C. R., Meng B. Y. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 1989 Jun;217(2-3):185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- Hu J., Bogorad L. Maize chloroplast RNA polymerase: the 180-, 120-, and 38-kilodalton polypeptides are encoded in chloroplast genes. Proc Natl Acad Sci U S A. 1990 Feb;87(4):1531–1535. doi: 10.1073/pnas.87.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Troxler R. F., Bogorad L. Maize chloroplast RNA polymerase: the 78-kilodalton polypeptide is encoded by the plastid rpoC1 gene. Nucleic Acids Res. 1991 Jun 25;19(12):3431–3434. doi: 10.1093/nar/19.12.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G. S., Mason J. G., Holton T. A., Koller B., Cox G. B., Whitfeld P. R., Bottomley W. A gene cluster in the spinach and pea chloroplast genomes encoding one CF1 and three CF0 subunits of the H+-ATP synthase complex and the ribosomal protein S2. J Mol Biol. 1987 Jul 20;196(2):283–298. doi: 10.1016/0022-2836(87)90690-5. [DOI] [PubMed] [Google Scholar]
- Izuchi S., Terachi T., Sakamoto M., Mikami T., Sugita M. Structure and expression of tomato mitochondrial genes coding for tRNA(Cys) (GCA), tRNA(Asn) (GUU) and tRNA(Tyr) (GUA): a native tRNA(Cys) gene is present in dicot plants but absent in monocot plants. Curr Genet. 1990 Oct;18(3):239–243. doi: 10.1007/BF00318387. [DOI] [PubMed] [Google Scholar]
- Joyce P. B., Gray M. W. Chloroplast-like transfer RNA genes expressed in wheat mitochondria. Nucleic Acids Res. 1989 Jul 25;17(14):5461–5476. doi: 10.1093/nar/17.14.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmbeck J., Stummann B. M., Henningsen K. W. Sequence of two regions of pea chloroplast DNA, one with the genes rps14, trnfM and trnG-GCC, and one with the genes trnP-UGG and trnW-CCA. Nucleic Acids Res. 1987 Apr 24;15(8):3630–3630. doi: 10.1093/nar/15.8.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerbs S., Bräutigam E., Parthier B. Polypeptides of DNA-dependent RNA polymerase of spinach chloroplasts: characterization by antibody-linked polymerase assay and determination of sites of synthesis. EMBO J. 1985 Jul;4(7):1661–1666. doi: 10.1002/j.1460-2075.1985.tb03834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G., Gutman G. A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987 May;4(3):203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Maréchal-Drouard L., Neuburger M., Guillemaut P., Douce R., Weil J. H., Dietrich A. A nuclear-encoded potato (Solanum tuberosum) mitochondrial tRNA(Leu) and its cytosolic counterpart have identical nucleotide sequences. FEBS Lett. 1990 Mar 26;262(2):170–172. doi: 10.1016/0014-5793(90)80181-h. [DOI] [PubMed] [Google Scholar]
- Maréchal-Drouard L., Weil J. H., Guillemaut P. Import of several tRNAs from the cytoplasm into the mitochondria in bean Phaseolus vulgaris. Nucleic Acids Res. 1988 Jun 10;16(11):4777–4788. doi: 10.1093/nar/16.11.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng B. Y., Tanaka M., Wakasugi T., Ohme M., Shinozaki K., Sugiura M. Cotranscription of the genes encoding two P700 chlorophyll a apoproteins with the gene for ribosomal protein CS14: determination of the transcriptional initiation site by in vitro capping. Curr Genet. 1988 Oct;14(4):395–400. doi: 10.1007/BF00419998. [DOI] [PubMed] [Google Scholar]
- Morden C. W., Golden S. S. Sequence analysis and phylogenetic reconstruction of the genes encoding the large and small subunits of ribulose-1,5-bisphosphate carboxylase/oxygenase from the chlorophyll b-containing prokaryote Prochlorothrix hollandica. J Mol Evol. 1991 May;32(5):379–395. doi: 10.1007/BF02101278. [DOI] [PubMed] [Google Scholar]
- Narita J. O., Rushlow K. E., Hallick R. B. Characterization of a Euglena gracilis chloroplast RNA polymerase specific for ribosomal RNA genes. J Biol Chem. 1985 Sep 15;260(20):11194–11199. [PubMed] [Google Scholar]
- Neuhaus H., Link G. The chloroplast tRNALys(UUU) gene from mustard (Sinapis alba) contains a class II intron potentially coding for a maturase-related polypeptide. Curr Genet. 1987;11(4):251–257. doi: 10.1007/BF00355398. [DOI] [PubMed] [Google Scholar]
- Ohme M., Kamogashira T., Shinozaki K., Sugiura M. Structure and cotranscription of tobacco chloroplast genes for tRNAGlu(UUC), tRNATyr(GUA) and tRNAAsp(GUC). Nucleic Acids Res. 1985 Feb 25;13(4):1045–1056. doi: 10.1093/nar/13.4.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K., Fukuzawa H., Kohchi T., Sano T., Sano S., Shirai H., Umesono K., Shiki Y., Takeuchi M., Chang Z. Structure and organization of Marchantia polymorpha chloroplast genome. I. Cloning and gene identification. J Mol Biol. 1988 Sep 20;203(2):281–298. doi: 10.1016/0022-2836(88)90001-0. [DOI] [PubMed] [Google Scholar]
- Okimoto R., Wolstenholme D. R. A set of tRNAs that lack either the T psi C arm or the dihydrouridine arm: towards a minimal tRNA adaptor. EMBO J. 1990 Oct;9(10):3405–3411. doi: 10.1002/j.1460-2075.1990.tb07542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. D. Contrasting modes and tempos of genome evolution in land plant organelles. Trends Genet. 1990 Apr;6(4):115–120. doi: 10.1016/0168-9525(90)90125-p. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfitzinger H., Guillemaut P., Weil J. H., Pillay D. T. Adjustment of the tRNA population to the codon usage in chloroplasts. Nucleic Acids Res. 1987 Feb 25;15(4):1377–1386. doi: 10.1093/nar/15.4.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfitzinger H., Weil J. H., Pillay D. T., Guillemaut P. Codon recognition mechanisms in plant chloroplasts. Plant Mol Biol. 1990 May;14(5):805–814. doi: 10.1007/BF00016513. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. An anticodon change switches the identity of E. coli tRNA(mMet) from methionine to threonine. Nucleic Acids Res. 1990 Jan 25;18(2):285–289. doi: 10.1093/nar/18.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science. 1988 Nov 4;242(4879):765–768. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986 Sep;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L., Shaw J. RNA editing and the mitochondrial cryptogenes of kinetoplastid protozoa. Cell. 1989 May 5;57(3):355–366. doi: 10.1016/0092-8674(89)90911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlin D., Hu J., Bogorad L. Binding and transcription of relaxed DNA templates by fractions of maize chloroplast extracts. Proc Natl Acad Sci U S A. 1989 Feb;86(3):876–880. doi: 10.1073/pnas.86.3.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dePamphilis C. W., Palmer J. D. Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature. 1990 Nov 22;348(6299):337–339. doi: 10.1038/348337a0. [DOI] [PubMed] [Google Scholar]




