Abstract
Honey bees are significant pollinators of agricultural crops and other important plant species. High annual losses of honey bee colonies in North America and in some parts of Europe have profound ecological and economic implications. Colony losses have been attributed to multiple factors including RNA viruses, thus understanding bee antiviral defense mechanisms may result in the development of strategies that mitigate colony losses. Honey bee antiviral defense mechanisms include RNA-interference, pathogen-associated molecular pattern (PAMP) triggered signal transduction cascades, and reactive oxygen species generation. However, the relative importance of these and other pathways is largely uncharacterized. Herein we review the current understanding of honey bee antiviral defense mechanisms and suggest important avenues for future investigation.
Introduction
Honey bees (Apis mellifera) are fascinating insects that play a critical role in agriculture as pollinators of crops (U.S. value over $15 billion/year) and plant species that enhance the biodiversity of both agricultural and non-agricultural landscapes [1]. Since 2006, honey bee populations in the U.S., Canada, and in some parts of Europe have experienced high annual losses [2–4]. An average of 33% of U.S. honey bee colonies die each year, and a fraction of these losses are attributed to Colony Collapse Disorder (CCD) [5–9]. Multiple biotic and abiotic factors contribute to colony health and survival (i.e., viruses, mites, microbes, bee genetics, weather, forage quality and availability, management practices, and agrochemical exposure) [9–12]. Understanding the most influential factors and potential synergistic effects on honey bee health is critical to developing pollinator management and conservation strategies that limit bee colony losses [13].
Several epidemiologic and temporal monitoring studies indicate the important role of pathogens in colony loss including viruses, bacteria, fungi, trypanosomatids, and mites [4,9,12,14–21]. The majority of honey bee infecting pathogens are RNA viruses, including Acute bee paralysis virus [22], Black queen cell virus [23], Israeli acute bee paralysis virus [24], Kashmir bee virus [25], Deformed wing virus [26], Kakugo virus [27], Varroa destructor virus-1 [28], Sacbrood virus [29], Slow bee paralysis virus [30], Cloudy wing virus [31], Big Sioux River virus [17,20], Aphid lethal virus (strain Brookings) [17,20], Chronic bee paralysis virus [32] (reviewed in [33,34]) and the Lake Sinai viruses (LSV1 and LSV2 [20], LSV3 [12], LSV4 [17], and LSV5 [35]. Honey bee virus infections may cause deformities, paralysis, death, or remain asymptomatic [33]. Bee viruses are transmitted via vertical and horizontal routes [36], including co-foraging with wild and managed bee populations [37–39]. The ectoparasitic mite Varroa destructor serves as a vector for several honey bee viruses [40–42] and causes colony loss by feeding on bee hemolymph and killing bee brood [43]. Several studies indicate that combinatorial effects of mites and viruses result in colony loss (reviewed in [34,44–46]). The relationship between colony health and pathogen prevalence and abundance is complex and dependent upon season, geographic location, pathogen strain, and both individual and colony level bee immune responses. Thus, temporal monitoring studies are key to understanding the relative impact of these variables on honey bee colony health.
The focus of the review is to summarize our current understanding of honey bee antiviral responses. Honey bees, like all other organisms, have evolved mechanisms to detect and limit virus infection. Knowledge of honey bee immune mechanisms is largely derived via comparison to the better-characterized immune responses in fruit-flies and mosquitoes. While comparative genomics is a useful approach for evaluating honey bee immune gene function, it is important to note that Western honey bees (Apis mellifera) are eusocial Hymenopteran insects, an order that diverged from the solitary Dipteran insects including fruit-flies and mosquitos approximately 300 million years ago [47–50]. General aspects of immunity, including detection of pathogen associated molecular patterns (PAMPs) and production of effector molecules are conserved in mammals, plants, and insects, and both plants and insects employ RNA interference (RNAi) as a major mechanism of antiviral defense [51–53]. These immune pathways provide a framework for understanding honey bee host – virus interactions.
Insect Immune Pathways
RNA interference (RNAi) is the major mechanism of antiviral defense in fruit-flies and mosquitos (reviewed in [53–58]). RNAi is a sequence specific, post-transcriptional gene and virus silencing mechanism that is triggered by double-stranded RNA (dsRNA). Direct evidence of the antiviral role of RNAi in insects has predominantly come from studies in Drosophila melanogaster, Aedes aegypti, and Anopheles gambiae, which involved experimental infections via injections with pure virus inocula, mutant-flies, or gene knock-down in mosquitos [59–63]. Likewise, field and laboratory based studies in Apis mellifera (Western honey bee) [64–69] and Apis cerana (Eastern honey bee) [70] indicate that RNAi-mediated antiviral immunity is important in honey bees (reviewed in [71]). In addition, dsRNA may serve as a non-sequence-specific virus associated molecular pattern (VAMP) that triggers innate antiviral immune pathways in fruit-flies [72] and honey bees [73,74], similar to the mammalian interferon response [75] (Figure 1, Tables 1 and S1).
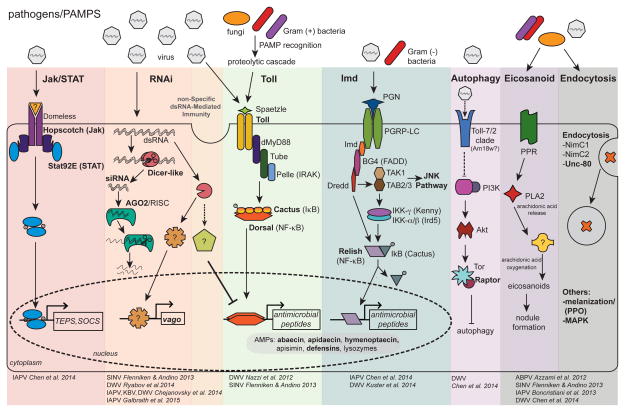
Figure 1. Honey Bee Immune Pathways - Highlighting Genes Implicated in Antiviral Immune Responses.
The honey bee genome encodes major members of insect immune pathways including: RNAi (RNA interference); Jak/STAT (Janus kinase/Signal Transducer and Activator of Transcription); Toll; NF-κB (Nuclear Factor κB); JNK (c-Jun N-terminal kinase); and MAPK (Mitogen-Activated Protein Kinases), as well as orthologs of genes involved in autophagy, eicosanoid biosynthesis, endocytosis, and melanization. Bold text indicates genes and proteins differentially expressed in virus-infected honey bees. Additional information including Apis mellifera (Am) gene accession numbers is provided in Tables 1 and S1. The first step in immune activation is host recognition of pathogen associated molecular patterns (PAMPs) including viral dsRNA, bacterial peptidoglycans, and fungal β-glucans. In general, the Toll pathway is involved in defense against Gram(+) bacteria and fungi and the Imd pathway is activated by Gram(−) bacteria, but specific host-pathogen interactions are unique. This is particularly true for host – virus interactions since data from fruit-flies, mosquitoes, and honey bees indicate differential activation of immune genes and pathways. The Jak/STAT pathway is activated via ligand binding to the Domeless receptor; while Drosophila melanogaster (Dm) express Domeless ligands (unpaired, upd, upd2, and upd3), a honey bee upd ortholog has not been identified. Following Domeless-ligand binding, Hopscotch Janus kinases are transphosphorylated, leading to phosphorylation and dimerization of STAT92E-like proteins. Activated STATs transcriptionally regulate antimicrobial effectors TEP7 (Thioester-containing protein 7), TEPA, TEPB, and the Jak/STAT inhibitor SOCS (Suppressor of Cytokine Signaling). The honey bee genome also encodes for D-PIAS (Protein Inhibitor of Activated STAT), another inhibitor of the Jak/STAT pathway. The RNAi-pathway is initiated by Dm Dicer-2 cleavage of viral dsRNA into 21–22 bp siRNAs; Am Dicer-like share ~30% aa identity with Dm Dicer-2. The siRNAs are then loaded into AGO2 (Argonaute-2), the catalytic component of the RISC (RNA Induced Silencing Complex). A single strand of the siRNA is retained in the RISC and used to specifically target cognate viral genome sequences for cleavage. In addition, Dm Dicer-2 serves as a dsRNA sensor that mediates a signal transduction cascade resulting in increased expression of Dm Vago and suppression of viral replication. Am Dicer-like may serve as a dsRNA sensor, and honey bees have a vago ortholog (Table S1), but the mechanism(s) of honey bee non-specific dsRNA-mediated antiviral responses require additional characterization. The Toll pathway is activated by a family of pathogen recognition receptors (PRRs) (e.g., peptidoglycan receptor proteins and Gram(−) binding proteins) that bind fungal and bacterial PAMPs. The Toll pathway is activated in some insect host-virus combinations, although the activation mechanism is unknown. Following PAMP binding, a serine protease cascade results in cleavage of pro-Spaetzle into mature Spaetzle. The honey bee genome encodes two putative spaetzle orthologs, which bind the membrane-anchored Toll receptor. Toll dimerization results in the recruitment of dMyD88, Tube, and Pelle. Pelle is likely involved in degradation of NF-κB inhibitors (e.g., Cactus-1, Cactus-2, Cactus-3), resulting in the release of transcription factors Dorsal-1A and Dorsal-1B. Nuclear translocation of Dorsal results in increased expression of antimicrobial peptides (AMPs). The Imd pathway is activated by Peptidoglycan recognition protein LC (PGRP-LC) binding to diaminopimelic-containing peptidoglycan of Gram(−) bacteria, followed by activation of the adaptor protein Immune deficiency (IMD), Relish phosphorylation by the IKK complex (IkB kinase), and cleavage of Relish by the caspase Dredd (Death-related ced-3/Nedd2-like). Relish transcriptionally regulates expression of AMPs and other genes involved in antimicrobial defense. The JNK pathway is also activated by TAB (Transforming growth factor-activated kinase 1) and TAK1 (Transforming growth factor-activated kinase 1 binding protein), resulting in AMP expression and/or apoptosis. In Drosophila, binding of vesicular stomatitis virus to the Toll-7 receptor promotes autophagy, likely by inhibiting the PI3/Akt/Tor (phosphatidylinositol 3-kinase/Protein kinase B/Target of rapamycin) pathway which suppresses autophagy. The honey bee genome encodes for one gene of the Toll-7/2 clade, 18-wheeler (am18w), which shares ~49% aa identity with Dm Toll-7 and ~45% aa identity with Dm Toll-2. The role of Am18w protein in antiviral defense and autophagy in honey bees is unknown. In insects, Eicosanoid biosynthesis begins with the induction of PLA2 (Phospholipase 2) from signal cascades downstream of viral, fungal, or bacterial PAMP recognition. Activated PLA2 hydrolyzes arachidonic acid (AA) from cellular phospholipids. Eicosonoid production likely occurs via oxidation of AA by an unidentified enzyme. Eicosanoids are critical for nodulation and aid in phagocytosis, micro-aggregation, adhesion, and release of prophenoloxidase (PPO) from hemocytes. Experimental evidence also suggests endocytosis, melanization, and MAPK pathways are involved in honey bee antiviral defense.
Table 1. Honey bee immune genes.
The Apis mellifera genome encodes major members of insect immune pathways including those depicted in Figure 1 and listed by gene name, pathway, and accession number in this table. Bold text indicates genes differentially expressed in virus-infected honey bees, and the specific virus and citation are provided for each. Transcript variants, the majority of which were predicted using Gnomon and the NCBI RefSeq Database, are listed although many have not been experimentally verified as expressed transcripts, nor been specifically implicated in antiviral defense. A list of additional honey bee immune related genes is provided in Supporting Table S1.
| Gene name | Pathway | Accession Number | Virus | Reference |
|---|---|---|---|---|
| abaecin | AMP | NM_001011617.1 | SINV | Flenniken and Andino 2013 |
| apidaecin 1 (apid 1) | AMP | NM_001011613.1 | SINV, DWV | Flenniken and Andino 2013, Kuster et al. 2014 |
| apidaecin 1 (apid73) | AMP | XM_006572699.1 | SINV, DWV | Flenniken and Andino 2013, Kuster et al. 2014 |
| apidaecin type 22 (apid22) | AMP | NM_001011642.1 | SINV, DWV | Flenniken and Andino 2013, Kuster et al. 2014 |
| hymenoptaecin | AMP | NM_001011615.1 | SINV, DWV | Flenniken and Andino 2013, Kuster et al. 2014 |
| defensin-2 | AMP | NM_001011638.1 | DWV | Kuster et el. 2014 |
| apisimin | AMP | NM_001011582.1 | ||
| defensin-1 | AMP | NM_001011616.2 | ||
| vago | antivir | XM_395092.4 | DWV | Ryabov et al. 2014 |
| nimrod c1 (nimc1) | EGF Family | XM_006561053.1 | SINV | Flenniken and Andino 2013 |
| phospholipase a2 (pla2) | Eicosanoid | NM_001011614.1 | ||
| unc-80/endocytosis | Endocytosis | XM_006558847.1 | SINV | Flenniken and Andino 2013 |
| dscam | IG superfamily | * see caption | SINV | Flenniken and Andino 2013 |
| relish (rel), var x1 | IMD | XM_006562219.1 | DWV | Kuster et el. 2014 |
| relish (rel), var x2 | IMD | XM_006562220.1 | DWV | Kuster et el. 2014 |
| relish (rel), var x3 | IMD | XM_006562221.1 | DWV | Kuster et el. 2014 |
| fadd | IMD | GB30399 | ||
| imd | IMD | NM_001163717.1 | ||
| ikkγ-kenny | IMD | XM_001120619.3 | ||
| ird5 | IMD | XM_623132.3 | ||
| pgrp-lc | IMD | XM_392452.5 | ||
| dredd | IMD | XM_001120830.1 | ||
| tab, var x1 | IMD | XM_001122664.3 | ||
| tab, var x2 | IMD | XM_006565777.1 | ||
| tak1, var x1 | IMD | XM_006572294.1 | ||
| tak1, var x2 | IMD | XM_397248.5 | ||
| d-pias, var x1 | Jak/STAT | XM_006561055.1 | IAPV | Chen et al. 2014 |
| d-pias, var x2 | Jak/STAT | XM_006561056.1 | IAPV | Chen et al. 2014 |
| d-pias, var x3 | Jak/STAT | XM_623568.4 | IAPV | Chen et al. 2014 |
| hopscotch (hop), var x1 | Jak/STAT | XM_001121783.3 | IAPV | Chen et al. 2014 |
| hopscotch (hop), var x2 | Jak/STAT | XM_006567688.1 | IAPV | Chen et al. 2014 |
| hopscotch (hop), var x3 | Jak/STAT | XM_006567689.1 | IAPV | Chen et al. 2014 |
| hopscotch (hop), var x4 | Jak/STAT | XM_006567690.1 | IAPV | Chen et al. 2014 |
| stat92e-like | Jak/STAT | XM_397181.5 | IAPV | Chen et al. 2014 |
| domeless | Jak/STAT | XM_003251652.2 | ||
| socs-5, var x1 | Jak/STAT | XM_006570603.1 | ||
| socs-5, var x2 | Jak/STAT | XM_624416.4 | ||
| tepb | Jak/STAT | XM_006570965.1 | ||
| tep7, var x1 | Jak/STAT | XM_006565440.1 | ||
| tep7, var x2 | Jak/STAT | XM_006565441.1 | ||
| tepa, var x1 | Jak/STAT | XM_006571765.1 | ||
| tepa, var x2 | Jak/STAT | XM_397416.4 | ||
| lysozyme 1 (lys) | Lysozyme | NC_007082.3 | ||
| lysozyme 2 (lys-2) | Lysozyme | NM_001120136.3 | ||
| lysozyme 3 (lys-3), var x1 | Lysozyme | XM_393161.5 | ||
| lysozyme 3 (lys-3), var x2 | Lysozyme | XM_006571783.1 | ||
| nimrod b (nimb) | Phagocytosis | GB12454 | ||
| nimrod a (nima) | Phagocytosis | XM_001120328.3 | ||
| nimrod c2 (nimc2), var x1 | Phagocytosis | XM_006561040.1 | ||
| nimrod c2 (nimc2), var x2 | Phagocytosis | XM_006561041.1 | ||
| nimrod c2 (nimc2), var x3 | Phagocytosis | XM_006561042.1 | ||
| nimrod c2 (nimc2), var x4 | Phagocytosis | XM_006561043.1 | ||
| pi3k, var x1 | PI3K-Akt-Tor | XM_006570469.1 | ||
| pi3k, var x2 | PI3K-Akt-Tor | XM_623894.3 | ||
| target of rapamycin (tor) | PI3K-Akt-Tor | XM_006566642.1 | ||
| akt-interacting protein-like | PI3K-Akt-Tor | XM_625206.4 | ||
| raptor | PI3K-Akt-Tor | XM_624057.4 | IAPV | Chen et al. 2014 |
| phenoloxidase subunit a3 (ppo) | PPO | NM_001011627.1 | ||
| argonaute 2 (ago2) | RNAi | XM_395048.5 | DWV | Galbraith et al. 2015 |
| dicer-like | RNAi | XM_006571316.1 | DWV | Galbraith et al. 2015 |
| lysyl oxidase-like 2 (lox2), var x1 | Scav. Receptor A | XM_006560641.1 | ||
| lysyl oxidase-like 2 (lox2), var x2 | Scav. Receptor A | XM_392090.4 | ||
| nf-κ-β inhibitor cactus 1 | Toll/TLR | NM_001163712.1 | DWV | Galbraith et al. 2015 |
| toll-6 | Toll/TLR | XM_393712.4 | DWV | Galbraith et al. 2015 |
| dorsal, var a | Toll/TLR | NM_001011577.1 | DWV | Nazzi et al 2012 |
| dorsal, var b | Toll/TLR | NM_001171006.1 | ||
| dorsal-2 (dl-2), var x1 | Toll/TLR | XM_006565455.1 | ||
| dorsal-2 (dl-2), var x2 | Toll/TLR | XM_395180.5 | ||
| ikappab kinase-like 2 (ik2) | Toll/TLR | XM_396937.5 | ||
| myd88, var x1 | Toll/TLR | NM_006560439.1 | ||
| myd88, var x2 | Toll/TLR | XM_006560440.1 | ||
| nf-kappa-β inhibitor cact1, var x1 | Toll/TLR | XM_006567107.1 | ||
| nf-kappa-β inhibitor cact1, var x2 | Toll/TLR | XM_006567108.1 | ||
| nf-kappa-β inhibitor cact2 | Toll/TLR | XM_394485.5 | ||
| nf-kappa-β inhibitor cact3, var 2 | Toll/TLR | XM_625153.4 | ||
| spaetzle-like, var x1 | Toll/TLR | XM_003250921.2 | ||
| spaetzle-like, var x2 | Toll/TLR | XM_006566961.1 | ||
| pelle, var x1 | Toll/TLR | XM_006565164.1 | ||
| pelle, var x2 | Toll/TLR | XM_623999.4 | ||
| traf6, var x1 | Toll/TLR | XM_006562507.1 | ||
| traf6, var x2 | Toll/TLR | XM_624204.4 | ||
| toll interacting protein (tollip) | Toll/TLR | XM_624414.4 | ||
| toll-1 | Toll/TLR | XM_006562720.1 | ||
| toll-10 | Toll/TLR | XM_006562853.1 | ||
| toll-8 | Toll/TLR | XM_393713.3 | ||
| tube protein (tub) | Toll/TLR | XM_001121229.3 | ||
| 18-wheeler (18-w)/toll like receptor | Toll/TLR | NM_001013361.1 |
Note dscam has 104 transcript variants: NM_001014991.1; XM_006567003.1-XM_006567105.1.
Other insect immune responses include melanization, encapsulation, reactive oxygen species production, and activation of signal transduction cascades that result in the production of antimicrobial peptides (AMPs) and other effector proteins (Figure 1, Tables 1 and S1). These pathways include the Toll, Imd (Immune Deficiency) and Jak/STAT (Janus kinase and Signal Transducer and Activator of Transcription) innate immune response pathways (Figure 1) (reviewed in [52,56,76–80]). There are numerous orthologous proteins utilized in plant, insect, and mammalian immune defense mechanisms (reviewed in [51,81]), and discovery of the Drosophila Toll pathway led to the identification of a repertoire of mammalian Toll-like receptors (TLRs) (reviewed in [81], [82]). The importance of the Toll, Imd, Jak/STAT, and other pathways in antiviral defense is variable and specific to individual virus-host interactions [76,80,83]. For example, the Toll pathway is involved in D. melanogaster and Aedes aegypti defense against Drosophila X virus [84] and Dengue [85], respectively, as dif loss of function mutants were more susceptible to virus infection. The Drosophila Imd pathway plays a larger role than the Toll pathway in limiting Sindbis virus [86] and Cricket paralysis virus (CrPV) [87], and the Jak-Stat pathway is critical to combating Drosophila C virus infection [88]. AMPs are small cationic peptides that penetrate microbial membranes, serve in innate immune signaling, and play additional uncharacterized functions (reviewed in [77,89]). While the role of AMPs in virus infection is not known, changes in AMP expression are used as indicators of immune pathway regulation. AMP induction in D. melanogaster varies, as some viruses induce expression (i.e., DXV and SINV) and others do not (i.e., CrPV and Rhabidovirus [90]). Numerous studies suggest the role of additional pathways in insect antiviral defense [72,80,88,90–92].
Honey Bee Antiviral Immune Responses
Bioinformatic analysis of the honey bee genome identified A. mellifera orthologs of insect immune genes and suggests that bees have fewer immune genes than D. melanogaster, Ae. aegypti, or An. gambiae [47,48,93]. The honey bee genome encodes the suite of genes required for RNAi including dicer-1, ago-2, r2d2, and dicer-like, which shares 30% nucleotide identity with Dm dicer-2 [47,94]. All the main components of the Toll, Imd, JNK, Tor, and Jak-STAT pathways have been identified (except upd), as well as immune effector proteins including AMPs (i.e., abaecin, hymenoptaecin, apidaecin, and defensin) and prophenoloxidases [48]. RNAi, Toll, Imd, endocytosis, MAPK, and non-specific dsRNA-mediated immune pathways have been implicated in honey bee antiviral defense (Figure 1, Tables 1 and S1).
A distinguishing feature of virus infection is the presence of long, double-stranded RNA molecules in the cytosol of the infected cell. Since long dsRNAs are not typical products of eukaryotic gene expression, these molecules are recognized as PAMPs in hosts including plants, arthropods, insects, and mammals [95]. Mammals have several receptors (e.g., TLR3, PKR, RIG-I, MDA-5) that upon binding dsRNA, activate signal transduction cascades, resulting in the transcriptional activation of genes involved in generating an “antiviral state” including thousands of interferon stimulated genes (reviewed in [96,97]). Importantly, long dsRNAs also serve as the substrate for RNAi-mediated antiviral responses. This first step of the antiviral small interfering RNA (siRNA) pathway is cleavage of cytosolic dsRNA by the Dicer enzyme (Figure 1). Initial studies implicating the role of RNAi in honey bee antiviral defense demonstrated that feeding sucrose solutions containing IAPV-specific dsRNA resulted in increased bee survival, lower levels of IAPV [64], larger colony size, and increased honey yields [67]. This also sparked commercial interest in dsRNA/RNAi-mediated antiviral treatments [67], and raised concerns regarding potential off-target effects and the use of RNAi-based insecticidal crops [98]. A subsequent laboratory-based study demonstrated that pre-treatment of larvae and adults with DWV-specific dsRNA prior to DWV-infection via feeding resulted in increased survival and decreased virus titers [65]. Likewise Apis cerana larvae pre-treated with virus-specific dsRNA had reduced levels of Chinese Sacbrood virus following infection via feeding [70].
One of the hallmarks of RNAi-mediated antiviral responses in insects is siRNA production. Small interfering RNAs produced by Dicer-2 cleavage are 21–22 bps in length, with an approximately 19 bp double-stranded RNA core, 5′-monophoshate ends, and two-nucleotide single-stranded overhangs at the 3′ hydroxyl ends; the single-strand siRNA retained in the holo-RNA Induced Silencing Complex (RISC) is modified (2′-O-methylated) at the 3′-end (reviewed in [99]). The first molecular evidence of virus-specific siRNAs in honey bee samples was obtained by Northern blot analysis [64,70]. Recently, Chejanovsky et al. evaluated siRNA populations isolated from bees obtained from either CCD-affected or unaffected colonies using high throughput sequencing and determined that there were more virus-specific (i.e., IAPV, KBV, and DWV) siRNA reads in CCD-affected samples [66,69]. These siRNAs were predominantly 22-nt long and distributed throughout the virus genome [66], indicating that the dsRNA replicative intermediate form of the IAPV genome was the Dicer substrate (reviewed in [100]). Further analysis of the IAPV-siRNAs from CCD-affected samples determined that most were negative-sense, and may thus serve as guide sequences that target the (+)ssRNA IAPV genome [66]. High throughput sequencing of small RNAs obtained from Varroa-infested, DWV-like, and VDV-1-infected bees identified a greater number of positive sense virus-specific siRNAs than negative sense siRNAs, and showed that DWV-like virus and siRNA abundance were proportional [69]. Interestingly, pupae with low virus levels that were exposed to few Varroa mites had 5-times more siRNAs than viral genomes, suggesting that when mite-pressure was low, the honey bee RNAi-mediated defense system was able to overcome virus replication [69].
Results to date indicate that honey bees utilize RNAi as an antiviral defense mechanism. Future studies that show increased virus copy number in response to experimental knock-down of dicer-like and/or argonaute-2 would provide additional evidence of an RNAi-mediated defense strategy in honey bees. Likewise, demonstrating siRNA incorporation into the RISC by sequencing only 2′-O-methylated siRNAs would provide additional experimental support for honey bee antiviral RNAi. The relative contribution of RNAi and other immune mechanisms requires further examination in the context of specific viruses, in different developmental stages and castes, and in a range of colony health (i.e., weak, healthy, CCD-affected). Genome integration of IAPV also requires further examination [64], since in D. melanogaster, both genome-integrated RNA viral sequences and RNAi are involved in limiting and maintaining persistent virus infections [63]. Together, these and other studies will reveal the relative role of RNAi in reducing or eliminating viruses in individual bees and colonies.
In D. melanogaster, Dicer-2 not only participates in RNAi, it also serves as a dsRNA sensor that upon binding results in the transcriptional activation of genes with antiviral function including vago [72] (Figure 1). Interestingly, Dicer-2 is a DEAD-box helicase motif containing protein, similar to the RIG-I-like family of mammalian cytosolic dsRNA sensors [72]. Recent evidence in honey bees suggests that dsRNA, regardless of its sequence-specificity, triggers an antiviral response that decreases viral burden [73]. Also, vago expression was increased in pupae that were orally infected with DWV [69]. Transcriptional profiling of Sindbis virus-infected and dsRNA-treated bees three days post-infection indicated that metabolic pathways were perturbed in both treatment groups. In addition, endocytosis and eicosanoid signaling pathways were differentially regulated in virus-infected bees, and dsRNA-treated bees differentially regulated genes involved in oxidative phosphorylation. The majority of differentially expressed genes were not involved in characterized innate immune pathways, albeit AMP expression was reduced (i.e., apidaecin and hymenoptaecin). Transcriptional changes in response to non-virus specific dsRNA (i.e., dsRNA-GFP) in developing honey bee workers were evaluated in a study aimed at investigating the off-target effects in RNAi-mediated gene knock-down experiments [74]. This study identified 1,400 differentially expressed genes, and gene ontology analyses determined that the affected genes included those involved in development, metabolism, immunity, stress response, and RNA processing and transport [74].
Several transcriptional level studies in honey bees implicate the involvement of uncharacterized genes/pathways in antiviral responses [18,69,73,101,102]. However, the roles of genes in the Toll, Imd, Jak-STAT, JNK, and RNAi pathways are the best characterized. Central players in honey bee immune signal transduction cascades include insect orthologs of a well-characterized mammalian transcription factor NF-κB, including Dorsal-1A, Dorsal-1B, and Relish (Figure 1, Tables 1 and S1). Nazzi et al. determined that dorsal-1A expression is key in limiting DWV infection [102]. Activation of NF-κB-family transcription factors results in the production of AMPs, which have unknown roles in antiviral immunity, and numerous other less well-characterized genes [19,48,103–105]. Symptomatic young bees experimentally infected with IAPV via feeding exhibited increased expression of Toll pathway members (i.e., toll-6, cactus, and hymenoptaecin) [101], whereas transcriptional profiling of IAPV positive bees from naturally infected colonies did not implicate either the Toll or Imd pathways in antiviral defense [18]. Young bees experimentally infected with Sindbis virus via injection and harboring very low levels of other bee pathogens expressed less apidaecin and hymenoptaecin than mock-infected controls [73]. Similarly, neither ABPV-challenge nor ABPV and E. coli co-challenge via injection resulted in AMP production (i.e., Defensin-1, Abaecin, and Hymenoptaecin) in adults or larvae, indicating that ABPV may suppress bee immune responses [106].
There are few general trends in the transcriptional response of honey bees to viruses due in large part to the relatively small number of studies performed to date and due to differences in virus-challenge methodologies (e.g., infection via injection, oral infection), experimental vs. natural infections, tissues examined, post-infection assay time, and developmental stage of the bee [107] (i.e., IAPV [18,101,108], DWV [44,69], SBV [70], CCD-affected [109], Sindbis virus [73]). Furthermore, variability between experimentally infected-bees may be attributed to differences in immune gene regulation between individuals within and between colonies, purity and strain of virus inoculum, varied microbiomes, and prevalence of pre-existing pathogens. In addition, there are relatively few predicted genes (~25%) that are involved in well-annotated pathways; 33% of the DEGs in naturally IAPV-infected adults had Drosophila orthogs and could be assigned putative function [18]. That said, differential expression of genes in immune, endocytic, and metabolic pathways are common to several data sets, but the directionality of regulation varies between studies and bee developmental stage [18,73,101]. Several investigations have focused on IAPV due to its association with colony health and the development of methods to produce IAPV-augmented infectious stocks via passaging bee viruses in pupae [108]. In adult bees, IAPV abundance is highest in the gut and hypopharyngeal gland and low in hemocytes (insect blood/immune cells) and the fat body, a tissue involved in metabolic activities (insect liver) [18,77,110,111]. Transcriptional profiling of IAPV-infected adults revealed differential expression of over 3,000 genes [18]. Functional analysis determined that genes involved in signal transduction and immune responses exhibited increased expression and that genes involved in metabolism and mitochondrial dysfunction had reduced expression [18]. In addition, IAPV-infection resulted in increased expression of genes involved in the TCA cycle II, protein ubiquitination, and eIF2 signaling, and that IAPV-infection reduced expression of genes in the γ-glutamyl cycle [18]. Chen et al. determined that IAPV-infection also perturbed expression of genes involved in insect immune pathways (i.e., oxidative phosphorylation, ABC transporter function, endocytosis, phagocytosis, TGF-beta signaling, Tor signaling, MAPK signaling, Jak-STAT signaling, and lysosomal degradation) [18]. Specific immune genes with increased expression in IAPV-infected adult honey bees include Jak/STAT pathway members (i.e., cbl, stat, pias, and hopscotch), Tor pathway members (i.e., gbl, mo25, dmel, and eIF4B), MAPK members (i.e., pointed, phi, and corkscrew), and genes involved in endocytosis (i.e., egfr, pastI, rabenosysn, and vacuolar protein sorting-associated protein 37B-like) [18] (Figure 1, Tables 1 and S1). It is noteworthy that IAPV-infected larvae had a different suite of DEGs with little overlap in the adult dataset [18]. Pupae infected with IAPV exhibited variable expression of ribosomal RNAs and increased expression of ribosomal protein S5a (RPS5), and glutathione S-transferase 1 [108]; bees from CCD-affected colonies also had increased rRNA expression [109]. The transcriptional profiles of the fat bodies from young, IAPV-infected worker bees [101] shared the most genes with IAPV-infected adult bees [18], and had little overlap with DEGs in bees infected with either E. coli bacteria [112] or microsporidia (Nosema spp.) [113], indicating that honey bee antiviral responses are distinct from immune responses mounted against other parasites. Increased expression of argonaute-2 and dicer-like in response to IAPV-infection also supports the role of a distinct antiviral response involving RNAi, Toll, and Jak-STAT pathways [101]. The research performed to date is informative, but additional studies are needed to better understand honey bee antiviral immune mechanisms at the transcriptional level (e.g., mechanisms of regulation of gene expression and the role of splice variants) and beyond.
Viruses and Other Stressors
The focus of this review is honey bee host – virus interactions, and honey bee antiviral responses, but honey bees live in a complex environment. The effects of viruses on bees, and the functionality of the bee immune responses, may be affected by the presence of other pathogens [12,19,20], the microbial context of infection (microbiome [114–117]), environmental factors including agrochemical exposure [104,118–121], and adequate nutrition [122–124]. Several studies indicate that bees infected with multiple pathogens have increased mortality and CCD-affected samples have a greater number of pathogens than control colonies [9,12,14]. While it is widely accepted that mite infestation is detrimental to honey bee colonies and that mites also serve as virus vectors [40–42], the mechanism(s) of synergistic detrimental interactions have not been fully elucidated [34,44,45,102,105].
Nazzi et al. investigated the combinatorial effects of mites and virus in both field and laboratory settings from the colony to the molecular level [102]. They determined that high mite infestation coupled with increasing levels of DWV from June to October resulted in increased colony mortality [102]. Transcriptome (RNASeq) analysis of adult bees in these colonies revealed lower expression of 19 immune genes including dorsal-1A, pathogen recognition receptors (AmSCR, B5 and B7 scavenger receptors, and C-type lectin 8), and immune signaling pathway members including hem, tak1, and socs [102] (Figure 1). Bees from colonies with both high mite and DWV levels exhibited increased expression of other immune genes including genes involved in pathogen recognition (PGRP-S2, nimC2, eater-like) and serine proteases [102]. Laboratory experiments confirmed that a combination of mites and DWV, but not mites alone, reduced dorsal-1A expression in adult bees [102]. Also, larvae in which dorsal-1A expression was reduced by RNAi-mediated knock-down harbored a greater number of DWV genome copies [102]. Recent studies by Kuster et al. demonstrated that DWV virus abundance increased up to 72 hours post experimental wounding or Varroa mite exposure [44]. Assessment of the transcriptional responses to wounding and mite exposure at times ranging from 24 – 240 hours post-capping demonstrated increased expression of immune genes (i.e., abaecin, apidaecin, defensin, hymenoptaecin, PGRPs, PPOact, and relish) and DWV infection (up to 72 hours) and reduction of mite numbers in conjunction with immune activation [44]. Cluster analysis suggested co-regulation of defensin and relish, and apidaecin and hymenoptaecin, whereas abaecin and PPOact were not associated with other immune gene regulation [44]. Interestingly, results to date indicate that mite pressure, independent of transmission, results in increased levels of DWV-like viruses with a VDV-1 CP coding region [69]. The interactions between the honey bee host, Varroa destructor, and viruses are not fully understood and require further investigation. Since honey bee colonies located in Newfoundland and Labrador, Canada [125], and several Hawaiian islands lack V. destructor [126], these populations provide unique opportunities to examine the effects of viruses on colony health and immune regulation.
Two sides to the story – Host vs. Virus Genetics
The genetic background of the host has implications on susceptibility to virus infection and disease severity. This is particularly relevant for honey bees as they live in colonies of ~ 30,000, the majority of which are sterile, genetic-half sisters, since queens typically mate with 12 drones [127]. Colony level diversity due to queen polyandry reduces the prevalence of honey bee diseases [128] and may result in varying transcriptional responses, variation between individual hemocyte populations, and differences in social immune mechanisms (e.g., grooming behavior, propolis production) [110,129]. Moreover, genetic diversity is not limited to the host, as the majority of honey bee viruses are RNA viruses with error prone polymerases that generate virus quasispecies over the course of infection [130]. Different virus variants within particular quasispecies populations may have greater or lesser pathogenicity in a particular host organism. In addition, different strains of honey bee viruses exhibit differential pathogenicity (i.e., DWV and IAPV) [18,69,126]. Recent studies determined that DWV strain prevalence was reduced in the presence of mites [126] and the recombinant strain of DWV, DWVv, is more virulent than other DWV-like viruses [69]. A greater appreciation of the existing virus genomic diversity across the globe is needed to better evaluate the effects of distinct virus strains on colony health. The development of infectious virus clones that are amenable to mutation (reverse genetic systems) are needed to verify strain-specific virulence and determine mechanism(s) of enhanced virulence or increased tolerance. Honey bees may vary in their degree of virus tolerance [79,131]. This should be explored at both the individual and colony levels, since the information gained may guide the use of virus susceptibility as an additional selectable trait in honey bee breeding programs [129,132,133]. In addition, further use and development of immortalized honey bee lines (i.e., AmE-711) [134], long-term cell cultures [135], and primary cell cultures [136,137], are required to further the field of honey bee virology. Future use of immortalized cell lines and infectious honey bee virus clones will serve to normalize future studies and lead to a better understanding of honey bee antiviral defense mechanisms.
Conclusion
Investigating virus-host interactions throughout all domains of life has led to a greater biological understanding of fundamental cellular processes and host-virus coevolution. Honey bee host – virus interactions likely depend upon bee age or developmental stage, additional biotic and abiotic variables, and genetics of both host and pathogen. Only with additional research in laboratory and field settings at both the individual bee and colony level, will the mechanisms of honey bee antiviral defense be understood. Undoubtedly, continued investigation of honey bee host-virus pairs will lead to the discovery of evolutionarily conserved immune defense strategies, as well as reveal numerous unique co-evolved relationships that are specific to each host-virus combination. It is a critical and exciting time to investigate honey bee antiviral response mechanisms.
Supplementary Material
The Apis mellifera genome encodes major members of insect immune pathways including those depicted in Figure 1 and listed by gene name, pathway, and accession number below. Bold text indicates genes differentially expressed in virus-infected honey bees, and the specific virus and citation are provided for each. A list of additional honey bee immune-related genes is included in worksheet 2.
Highlights.
Honey bee colony losses have been attributed to multiple factors including RNA viruses.
The RNAi, Toll, Jak/Stat, and non-specific dsRNA mediated immune pathways have been implicated in honey bee antiviral defense.
Transcriptional profiling of virus-infected bees illustrates differential expression of numerous uncharacterized honey bee genes.
Honey bee antiviral responses likely involve general response mechanisms, as well as unique co-evolved interactions; all require further investigation.
Acknowledgments
This work was supported in part by National Institutes of Health IDeA Program COBRE grant GM110732, National Science Foundation EPSCoR NSF-IIA-1443108, Project Apis m., and the Montana State University Agricultural Experiment Station. The Flenniken laboratory is also supported by the Montana Department of Agriculture, the Montana State Beekeepers Association, Montana State University, and the United States Department of Agriculture National Institute of Food and Agriculture, Agriculture and Food Research Initiative (USDA-NIFA-AFRI) program. Laura M. Brutscher is supported by the Project Apis m.-Costco Honey Bee Biology Fellowship. We would like to thank members of the Flenniken laboratory (Elisa Boyd, Ian Cavigli, Emma Garcia, and Madison Martin) for reviewing this manuscript prior to publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of interest, particularly those published within the last two years, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Calderone NW. Insect pollinated crops, insect pollinators and US agriculture: trend analysis of aggregate data for the period 1992–2009. PLoS ONE. 2012;7:e37235. doi: 10.1371/journal.pone.0037235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Zee R, Pisa L, Andonov S, Brodschneider R, Charriere J-D, Chlebo R, Coffey MF, Crailsheim K, Dahle B, Gajda A, et al. Managed honey bee colony losses in Canada, China, Europe, Israel and Turkey, for the winters of 2008–9 and 2009–10. J Apicult Res. 2012;51:91–114. [Google Scholar]
- 3.Clermont A, Eickermann M, Kraus F, Georges C, Hoffmann L, Beyer M. A survey on some factors potentially affecting losses of managed honey bee colonies in Luxembourg over the winters 2010/2011 and 2011/2012. J Apicult Res. 2014;53:43–56. [Google Scholar]
- **4.McMenamin AJ, Genersch E. ScienceDirect. Current Opinion in Insect Science. 2015 doi: 10.1016/j.cois.2015.01.015. This paper reviews recent large-scale honey bee colony losses, and the field and experimental data that supports the association of viruses with colony loss. [DOI] [PubMed] [Google Scholar]
- 5.vanEngelsdorp D, Hayes J, Underwood RM, Pettis J. A Survey of Honey Bee Colony Losses in the US, Fall 2007 to Spring 2008. PLoS ONE. 2008;3:e4071. doi: 10.1371/journal.pone.0004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *6.Steinhauer NA, Rennich K, Wilson ME, Caron DM, et al. A national survey of managed honey bee 2012–2013 annual colony losses in the USA: results from the Bee Informed Partnership. J Apicult Res. 2014;53:1–18. The Bee Informed Partnership provides data regarding the percentages of U.S. managed honey bee colonies lost each year; this data is essential to understanding high annual colony losses. [Google Scholar]
- 7.Spleen AM, Lengerich EJ, Rennich K, Caron D, Rose R, Pettis JS, Henson M, Wilkes JT, Wilson M, Stitzinger J, et al. A national survey of managed honey bee 2011–12 winter colony losses in the United States: results from the Bee Informed Partnership. 2013. [DOI] [Google Scholar]
- 8.vanEngelsdorp D, Caron D, Hayes J, Underwood R, Henson M, Rennich K, Spleen A, Andree M, Snyder R, Lee K, et al. A national survey of managed honey bee 2010–11 winter colony losses in the USA: results from the Bee Informed Partnership. J Apicult Res. 2012;51:115–124. [Google Scholar]
- **9.vanEngelsdorp D, Evans JD, Saegerman C, Mullin C, Haubruge E, Nguyen BK, Frazier M, Frazier J, Cox-Foster D, Chen Y, et al. Colony collapse disorder: a descriptive study. PLoS ONE. 2009;4:e6481. doi: 10.1371/journal.pone.0006481. This paper defines Colony Collapse Disorder (CCD), reviews other historically import large-scale bee losses, and provides a framework for the association of multiple parameters with colony loss. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis JD, Evans JD, Pettis J. Colony losses, managed colony population decline, and Colony Collapse Disorder in the United States. J Apicult Res. 2010;49:134–136. [Google Scholar]
- **11.Mullin CA, Frazier M, Frazier JL, Ashcraft S, Simonds R, vanEngelsdorp D, Pettis JS. High levels of miticides and agrochemicals in North American apiaries: implications for honey bee health. PLoS ONE. 2010;5:e9754. doi: 10.1371/journal.pone.0009754. This paper provides quantitative mass spectrometry data on over 100 pesticides and metabolites associated with honey bees and hive matrices; it is the most thorough investigation of honey bee agrochemical exposure to date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **12.Cornman RS, Tarpy DR, Chen Y, Jeffreys L, Lopez D, Pettis JS, vanEngelsdorp D, Evans JD. Pathogen webs in collapsing honey bee colonies. PLoS ONE. 2012;7:e43562. doi: 10.1371/journal.pone.0043562. This is an excellent review of honey bee pathogens and analyses of pathogen abundance and associations in CCD and non-CCD colonies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallant AL, Euliss NH, Browning Z. Mapping large-area landscape suitability for honey bees to assess the influence of land-use change on sustainability of national pollination services. PLoS ONE. 2014;9:e99268. doi: 10.1371/journal.pone.0099268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *14.Cox-Foster DL, Conlan S, Holmes EC, Palacios G, Evans JD, Moran NA, Quan P-L, Briese T, Hornig M, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. This paper describes the first metagenomic analysis of CCD-affected colonies. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen BK, Mignon J, Laget D, de Graaf DC, Jacobs FJ, vanEngelsdorp D, Brostaux Y, Saegerman C, Haubruge E. Honey bee colony losses in Belgium during the 2008–9 winter. J Apicult Res. 2010;49:337–339. [Google Scholar]
- *16.Genersch E, Ohe von der W, Kaatz H, Schroeder A, Otten C, Büchler R, Berg S, Ritter W, Mühlen W, Gisder S, et al. The German bee monitoring project: a long term study to understand periodically high winter losses of honey bee colonies. Apidologie. 2010;41:332–352. This paper described results from a four-year monitoring project and identified several factors associated with over-winter losses including mite infestation, DWV, ABPV, queen age, and weakness of colonies in autumn. [Google Scholar]
- 17.Ravoet J, Maharramov J, Meeus I, De Smet L, Wenseleers T, Smagghe G, de Graaf DC. Comprehensive Bee Pathogen Screening in Belgium Reveals Crithidia mellificae as a New Contributory Factor to Winter Mortality. PLoS ONE. 2013;8:e72443. doi: 10.1371/journal.pone.0072443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **18.Chen YP, Pettis JS, Corona M, Chen WP, Li CJ, Spivak M, Visscher PK, DeGrandi-Hoffman G, Boncristiani H, Zhao Y, et al. Israeli Acute Paralysis Virus: Epidemiology, Pathogenesis and Implications for Honey Bee Health. Plos Pathog. 2014;10:e1004261. doi: 10.1371/journal.ppat.1004261. This paper is an important contribution to better understanding the role of IAPV in colony loss and honey bee host-IAPV interactions (i.e., honey bee RNAi-mediated antiviral response, analysis of differentially expressed genes in IAPV-infected adults, and evidence for an IAPV encoded suppressor of RNAi). [DOI] [PMC free article] [PubMed] [Google Scholar]
- **19.Evans JD, Schwarz RS. Bees brought to their knees: microbes affecting honey bee health. Trends Microbiol. 2011;19:614–620. doi: 10.1016/j.tim.2011.09.003. This is an excellent review of honey bee associated microbes, including pathogens. [DOI] [PubMed] [Google Scholar]
- 20.Runckel* C, Flenniken* ML, Engel JC, Ruby JG, Ganem D, Andino R, DeRisi JL. Temporal analysis of the honey bee microbiome reveals four novel viruses and seasonal prevalence of known viruses, nosema, and crithidia. PLoS ONE. 2011;6:e20656. doi: 10.1371/journal.pone.0020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tentcheva D, Gauthier L, Zappulla N, Dainat B, Cousserans F, Colin ME, Bergoin M. Prevalence and seasonal variations of six bee viruses in Apis mellifera L and Varroa destructor mite populations in France. Appl Environ Microbiol. 2004;70:7185–7191. doi: 10.1128/AEM.70.12.7185-7191.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Govan VA, Leat N, Allsopp M, Davison S. Analysis of the complete genome sequence of acute bee paralysis virus shows that it belongs to the novel group of insect-infecting RNA viruses. Virology. 2000;277:457–463. doi: 10.1006/viro.2000.0616. [DOI] [PubMed] [Google Scholar]
- 23.Leat N, Ball B, Govan V, Davison S. Analysis of the complete genome sequence of black queen-cell virus, a picorna-like virus of honey bees. J Gen Virol. 2000;81:2111–2119. doi: 10.1099/0022-1317-81-8-2111. [DOI] [PubMed] [Google Scholar]
- 24.Maori E, Lavi S, Mozes-Koch R, Gantman Y, Peretz Y, Edelbaum O, et al. Isolation and characterization of Israeli acute paralysis virus, a dicistrovirus affecting honeybees in Israel: evidence for diversity due to intra- and inter-species recombination. J Gen Virol. 2007;88:3428–3438. doi: 10.1099/vir.0.83284-0. [DOI] [PubMed] [Google Scholar]
- 25.de Miranda JR, Drebot M, Tyler S, Shen M, Cameron CE, Stoltz DB, Camazine SM. Complete nucleotide sequence of Kashmir bee virus and comparison with acute bee paralysis virus. J Gen Virol. 2004;85:2263–2270. doi: 10.1099/vir.0.79990-0. [DOI] [PubMed] [Google Scholar]
- 26.Lanzi G, De Miranda J, Boniotti M, Cameron C, Lavazza A, Capucci L, Camazine S, Rossi C. Molecular and biological characterization of deformed wing virus of honeybees (Apis mellifera L. ) J Virol. 2006;80:4998–5009. doi: 10.1128/JVI.80.10.4998-5009.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiyuki T, Ohka S, Takeuchi H, Ono M, Nomoto A, Kubo T. Prevalence and phylogeny of Kakugo virus, a novel insect picorna-like virus that infects the honeybee (Apis mellifera L), under various colony conditions. 2006;80:11528–11538. doi: 10.1128/JVI.00754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ongus JR, Peters D, Bonmatin J-M, Bengsch E, Vlak JM, van Oers MM. Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor. J Gen Virol. 2004;85:3747–3755. doi: 10.1099/vir.0.80470-0. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh RC, Ball BV, Willcocks MM, Carter MJ. The nucleotide sequence of sacbrood virus of the honey bee: an insect picorna-like virus. J Gen Virol. 1999;80 ( Pt 6):1541–1549. doi: 10.1099/0022-1317-80-6-1541. [DOI] [PubMed] [Google Scholar]
- 30.de Miranda JR, Dainat B, Locke B, Cordoni G, Berthoud H, Gauthier L, Neumann P, Budge GE, Ball BV, Stoltz DB. Genetic characterization of slow bee paralysis virus of the honeybee (Apis mellifera L) J Gen Virol. 2010;91:2524–2530. doi: 10.1099/vir.0.022434-0. [DOI] [PubMed] [Google Scholar]
- 31.Carreck NL, Ball BV, Martin SJ. The epidemiology of cloudy wing virus infections in honey bee colonies in the UK. J Apicult Res. 2010;49:66–71. [Google Scholar]
- 32.Olivier V, Blanchard P, Chaouch S, Lallemand P, Schurr F, Celle O, Dubois E, Tordo N, Thiéry R, Houlgatte R, et al. Molecular characterisation and phylogenetic analysis of Chronic bee paralysis virus, a honey bee virus. Virus Res. 2008;132:59–68. doi: 10.1016/j.virusres.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 33.Chen YP, Siede R. Advances in Virus Research. Elsevier; 2007. Honey Bee Viruses; pp. 33–80. [DOI] [PubMed] [Google Scholar]
- 34.Genersch E, Aubert M. Emerging and re-emerging viruses of the honey bee (Apis mellifera L. ) Vet Res. 2010;41:54. doi: 10.1051/vetres/2010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Granberg F, Vicente-Rubiano M, Rubio-Guerri C, Karlsson OE, Kukielka D, Belák S, Sánchez-Vizcaíno JM. Metagenomic detection of viral pathogens in spanish honeybees: co-infection by aphid lethal paralysis, Israel acute paralysis and lake sinai viruses. PLoS ONE. 2013;8:e57459. doi: 10.1371/journal.pone.0057459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen YP, Pettis JS, Collins A, Feldlaufer MF. Prevalence and transmission of honeybee viruses. Appl Environ Microbiol. 2006;72:606–611. doi: 10.1128/AEM.72.1.606-611.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh R, Levitt AL, Rajotte EG, Holmes EC, Ostiguy N, vanEngelsdorp D, Lipkin WI, Depamphilis CW, Toth AL, Cox-Foster DL. RNA viruses in hymenopteran pollinators: evidence of inter-Taxa virus transmission via pollen and potential impact on non-Apis hymenopteran species. PLoS ONE. 2010;5:e14357. doi: 10.1371/journal.pone.0014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fürst MA, McMahon DP, Osborne JL, Paxton RJ, Brown MJF. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature. 2014;506:364–366. doi: 10.1038/nature12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazzei M, Carrozza ML, Luisi E, Forzan M, Giusti M, Sagona S, Tolari F, Felicioli A. Infectivity of DWV Associated to Flower Pollen: Experimental Evidence of a Horizontal Transmission Route. PLoS ONE. 2014;9:e113448. doi: 10.1371/journal.pone.0113448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boncristiani HF, Di Prisco G, Pettis JS, Hamilton M, Chen YP. Molecular approaches to the analysis of deformed wing virus replication and pathogenesis in the honey bee, Apis mellifera. Virol J. 2009;6:221. doi: 10.1186/1743-422X-6-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, Pettis J, Evans J, Kramer M. Transmission of Kashmir bee virus by the ectoparasitic mite Varroa destructor. Apidologie. 2004;35:441–448. [Google Scholar]
- 42.Shen M, Cui L, Ostiguy N, Cox-Foster D. Intricate transmission routes and interactions between picorna-like viruses (Kashmir bee virus and sacbrood virus) with the honeybee host and the parasitic varroa mite. J Gen Virol. 2005;86:2281–2289. doi: 10.1099/vir.0.80824-0. [DOI] [PubMed] [Google Scholar]
- 43.Rosenkranz P, Aumeier P, Ziegelmann B. Biology and control of Varroa destructor. J Invertebr Pathol. 2010;103 (Suppl 1):S96–119. doi: 10.1016/j.jip.2009.07.016. [DOI] [PubMed] [Google Scholar]
- **44.Kuster RD, Boncristiani HF, Rueppell O. Immunogene and viral transcript dynamics during parasitic Varroa destructor mite infection of developing honey bee (Apis mellifera) pupae. J Exp Biol. 2014;217:1710–1718. doi: 10.1242/jeb.097766. This paper provides quantitative transcriptional level data of immune gene regulation in mite-exposed pupae at multiple time points and shows little support for mite-mediated immune suppression; the introduction and discussion are well-researched and very informative. [DOI] [PubMed] [Google Scholar]
- **45.Nazzi F, Pennacchio F. Disentangling multiple interactions in the hive ecosystem. Trends Parasitol. 2014;30:556–561. doi: 10.1016/j.pt.2014.09.006. This short-opinion article reviews factors associated with colony loss and provides a framework for understanding the combinatorial effects of mites and viruses. [DOI] [PubMed] [Google Scholar]
- 46.Locke B, Forsgren E, de Miranda JR. Increased Tolerance and Resistance to Virus Infections: A Possible Factor in the Survival of Varroa destructor-Resistant Honey Bees (Apis mellifera) PLoS ONE. 2014;9:e99998. doi: 10.1371/journal.pone.0099998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **48.Evans JD, Aronstein K, Chen YP, Hetru C, Imler JL, Jiang H, Kanost M, Thompson GJ, Zou Z, Hultmark D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol Biol. 2006;15:645–656. doi: 10.1111/j.1365-2583.2006.00682.x. The honey bee immune genes and pathways described in this paper are the basis for honey bee immunology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grimaldi D, Engel MS. Evolution of the Insects. 2005. [Google Scholar]
- 50.Elsik CG, Worley KC, Bennett AK, Beye M, Camara F, Childers CP, de Graaf DC, Debyser G, Deng J, Devreese B, et al. Finding the missing honey bee genes: lessons learned from a genome upgrade. Bmc Genomics. 2014;15:86. doi: 10.1186/1471-2164-15-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ronald PC, Beutler B. Plant and animal sensors of conserved microbial signatures. Science. 2010;330:1061–1064. doi: 10.1126/science.1189468. [DOI] [PubMed] [Google Scholar]
- **52.Kingsolver MB, Huang Z, Hardy RW. Insect antiviral innate immunity: Pathways, effectors, and connections. J Mol Biol. 2013 doi: 10.1016/j.jmb.2013.10.006. This review article provides an excellent description of the immune mechanisms involved in insect innate antiviral immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *53.Ding S-W. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. doi: 10.1038/nri2824. This is an excellent review of RNAi in insects, nematodes, plants, and vertebrates; it summarizes key concepts and fundamental studies. [DOI] [PubMed] [Google Scholar]
- 54.van Rij RP, Berezikov E. Small RNAs and the control of transposons and viruses in Drosophila. Trends Microbiol. 2009;17:163–171. doi: 10.1016/j.tim.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 55.Flenniken ML, Kunitomi M, Tassetto M, Andino R. Insect Virology - The Antiviral Role of RNA Interference. Caister Academic Press. 2010 no volume. [Google Scholar]
- 56.Kingsolver MB, Hardy RW. Making connections in insect innate immunity. P Natl Acad Sci Usa. 2012;109:18639–18640. doi: 10.1073/pnas.1216736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *57.Nayak A, Tassetto M, Kunitomi M, Andino R. RNA interference-mediated intrinsic antiviral immunity in invertebrates. Curr Top Microbiol Immunol. 2013;371:183–200. doi: 10.1007/978-3-642-37765-5_7. This article provides and excellent review of RNAi-mediated insect antiviral immunity. [DOI] [PubMed] [Google Scholar]
- 58.Karlikow M, Goic B, Saleh M-C. RNAi and antiviral defense in Drosophila: Setting up a systemic immune response. Dev Comp Immunol. 2014;42:85–92. doi: 10.1016/j.dci.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 59.van Rij RP, Saleh M-C, Berry B, Foo C, Houk A, Antoniewski C, Andino R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X-H, Aliyari R, Li W-X, Li H-W, Kim K, Carthew R, Atkinson P, Ding S-W. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Campbell CL, Keene KM, Brackney DE, Olson KE, Blair CD, Wilusz J, Foy BD. Aedes aegypti uses RNA interference in defense against Sindbis virus infection. BMC Microbiol. 2008;8:47. doi: 10.1186/1471-2180-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keene KM, Foy BD, Sanchez-Vargas I, Beaty BJ, Blair CD, Olson KE. RNA interference acts as a natural antiviral response to O’nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. P Natl Acad Sci Usa. 2004;101:17240–17245. doi: 10.1073/pnas.0406983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saleh M-C, Tassetto M, van Rij RP, Goic B, Gausson V, Bassam B, Jacquier C, Antoniewski C, Andino R. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature. 2009;458:346–350. doi: 10.1038/nature07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *64.Maori E, Paldi N, Shafir S, Kalev H, Tsur E, Glick E, Sela I. IAPV, a bee-affecting virus associated with Colony Collapse Disorder can be silenced by dsRNA ingestion. Insect Mol Biol. 2009;18:55–60. doi: 10.1111/j.1365-2583.2009.00847.x. This paper provides initial evidence for the role of RNAi in honey bee antiviral defense. [DOI] [PubMed] [Google Scholar]
- **65.Desai SD, Eu Y-J, Whyard S, Currie RW. Reduction in deformed wing virus infection in larval and adult honey bees (Apis mellifera L) by double-stranded RNA ingestion. Insect Mol Biol. 2012;21:446–455. doi: 10.1111/j.1365-2583.2012.01150.x. This study demonstrated that pretreatment (via feeding) of both honey bee larvae and adults with dsRNA specific to DWV reduced viral burden, thus it provided additional evidence for the role of RNAi in antiviral defense. [DOI] [PubMed] [Google Scholar]
- **66.Chejanovsky N, Ophir R, Schwager MS, Slabezki Y, Grossman S, Cox-Foster D. Characterization of viral siRNA populations in honey bee colony collapse disorder. Virology. 2014;454–455:176–183. doi: 10.1016/j.virol.2014.02.012. Ultra-deep sequencing data was used to characterize siRNAs from bees obtained from CCD-affected and healthy colonies. Key findings from this study included that CCD-affected colonies had a greater number of 22-nt siRNAs than healthy colonies, siRNA sequences were distributed throughout the virus genomes, and that there were more negative-strand IAPV-siRNAs than positive-strand siRNAs. [DOI] [PubMed] [Google Scholar]
- 67.Hunter W, Ellis J, vanEngelsdorp D, Hayes J, Westervelt D, Glick E, Williams M, Sela I, Maori E, Pettis J, et al. Large-scale field application of RNAi technology reducing Israeli acute paralysis virus disease in honey bees (Apis mellifera, Hymenoptera: Apidae) Plos Pathog. 2010;6:e1001160. doi: 10.1371/journal.ppat.1001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang H, Xie J, Shreeve TG, Ma J, Pallett DW, King LA, Possee RD. Sequence recombination and conservation of Varroa destructor virus-1 and deformed wing virus in field collected honey bees (Apis mellifera) PLoS ONE. 2013;8:e74508. doi: 10.1371/journal.pone.0074508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **69.Ryabov EV, Wood GR, Fannon JM, Moore JD, Bull JC, Chandler D, Mead A, Burroughs N, Evans DJ. A Virulent Strain of Deformed Wing Virus (DWV) of Honeybees (Apis mellifera) Prevails after Varroa destructor-Mediated, or In Vitro, Transmission. Plos Pathog. 2014;10:e1004230. doi: 10.1371/journal.ppat.1004230. This is an interesting report of a DWV strain that has enhanced replication in bee pupae, which is not observed in mites. In addition, this study identified differentially expressed genes in mite-infested and virus-infected pupae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu X, Zhang Y, Yan X, Han R. Prevention of Chinese Sacbrood Virus Infection in Apis cerana using RNA Interference. Curr Microbiol. 2010;61:422–428. doi: 10.1007/s00284-010-9633-2. [DOI] [PubMed] [Google Scholar]
- **71.Niu J, Meeus I, Cappelle K, Piot N, Smagghe G. The immune response of the small interfering RNA pathway in the defense against bee viruses. Current Opinion in Insect Science. 2014;6:22–27. doi: 10.1016/j.cois.2014.09.014. This review describes the siRNA pathway, provides phylogenetic comparisons of Dicer-2 and Ago-2 from numerous bee species, reviews dsRNA-mediated gene silencing in both bumble bee and honey bees, and summarizes current experimental support for RNAi-mediated and non-specific dsRNA mediated antiviral responses in honey bees. [DOI] [PubMed] [Google Scholar]
- 72.Deddouche S, Matt N, Budd A, Mueller S, Kemp C, Galiana-Arnoux D, Dostert C, Antoniewski C, Hoffmann JA, Imler J-L. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat Immunol. 2008;9:1425–1432. doi: 10.1038/ni.1664. [DOI] [PubMed] [Google Scholar]
- *73.Flenniken ML, Andino R. Non-Specific dsRNA-Mediated Antiviral Response in the Honey Bee. PLoS ONE. 2013;8:e77263. doi: 10.1371/journal.pone.0077263. This study provides experimental evidence of a non-specific dsRNA-mediated antiviral response in bees infected with a model virus, and it describes differential gene expression in virus-infected and dsRNA-treated bees. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nunes F, Aleixo A, Barchuk A, Bomtorin A, Grozinger C, Simões Z. Non-Target Effects of Green Fluorescent Protein (GFP)-Derived Double-Stranded RNA (dsRNA-GFP) Used in Honey Bee RNA Interference (RNAi) Assays. Insects 2012, 3, 601–615. 2013;4:90–103. doi: 10.3390/insects4010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **76.Lamiable O, Imler J-L. Induced antiviral innate immunity in Drosophila. Curr Opin Microbiol. 2014;20:62–68. doi: 10.1016/j.mib.2014.05.006. This is an excellent review of fruit fly antiviral mechanisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 78.Ferrandon D, Imler J-L, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7:862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- **79.Merkling SH, van Rij RP. Beyond RNAi: Antiviral defense strategies in Drosophila and mosquito. J Insect Physiol. 2013;59:159–170. doi: 10.1016/j.jinsphys.2012.07.004. This is an outstanding review of fruit fly and mosquito antiviral mechanisms; it provides and comprehensive graphical representation of inducible immune pathways in Drosophila. [DOI] [PubMed] [Google Scholar]
- 80.Sabin LR, Hanna SL, Cherry S. Innate antiviral immunity in Drosophila. Curr Opin Immunol. 2010;22:4–9. doi: 10.1016/j.coi.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Valanne S, Wang J-H, Rämet M. The Drosophila Toll signaling pathway. J Immunol. 2011;186:649–656. doi: 10.4049/jimmunol.1002302. [DOI] [PubMed] [Google Scholar]
- 82.Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, Kopczynski C, Duyk G, Reichhart JM, Hoffmann JA. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev Cell. 2001;1:503–514. doi: 10.1016/s1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- 83.Asgari S. Insect Virology. Caister Academic Pr; 2010. [Google Scholar]
- 84.Zambon R, Nandakumar M, Vakharia V, Wu L. The Toll pathway is important for an antiviral response in Drosophila. P Natl Acad Sci Usa. 2005;102:7257–7262. doi: 10.1073/pnas.0409181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. Plos Pathog. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Avadhanula V, Weasner BP, Hardy GG, Kumar JP, Hardy RW. A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. Plos Pathog. 2009;5:e1000582. doi: 10.1371/journal.ppat.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Costa A, Jan E, Sarnow P, Schneider D. The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS ONE. 2009;4:e7436. doi: 10.1371/journal.pone.0007436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, Hoffmann JA, Imler J-L. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol. 2005;6:946–953. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- 89.Imler J-L, Bulet P. Antimicrobial peptides in Drosophila: structures, activities and gene regulation. Chem Immunol Allergy. 2005;86:1–21. doi: 10.1159/000086648. [DOI] [PubMed] [Google Scholar]
- 90.Carpenter J, Hutter S, Baines JF, Roller J, Saminadin-Peter SS, Parsch J, Jiggins FM. The transcriptional response of Drosophila melanogaster to infection with the sigma virus (Rhabdoviridae) PLoS ONE. 2009;4:e6838. doi: 10.1371/journal.pone.0006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.DiAngelo JR, Bland ML, Bambina S, Cherry S, Birnbaum MJ. The immune response attenuates growth and nutrient storage in Drosophila by reducing insulin signaling. P Natl Acad Sci Usa. 2009;106:20853–20858. doi: 10.1073/pnas.0906749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fischman BJ, Woodard SH, Robinson GE. Molecular evolutionary analyses of insect societies. P Natl Acad Sci Usa. 2011;108 (Suppl 2):10847–10854. doi: 10.1073/pnas.1100301108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 95.de Faria IJDS, Olmo RP, Silva EG, Marques JT. dsRNA Sensing During Viral Infection: Lessons from Plants, Worms, Insects, and Mammals. J Interferon Cytokine Res. 2013;33:239–253. doi: 10.1089/jir.2013.0026. [DOI] [PubMed] [Google Scholar]
- 96.Jensen S, Thomsen AR. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol. 2012;86:2900–2910. doi: 10.1128/JVI.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lundgren JG, Duan JJ. RNAi-Based Insecticidal Crops. Bioscience. 2013;63:657–665. [Google Scholar]
- 99.Wilson RC, Doudna JA. Molecular Mechanisms of RNA Interference. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. http://dx.doi.org/10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bronkhorst AW, van Rij RP. The long and short of antiviral defense: small RNA-based immunity in insects. Curr Opin Virol. 2014;7C:19–28. doi: 10.1016/j.coviro.2014.03.010. [DOI] [PubMed] [Google Scholar]
- **101.Galbraith DA, Yang X, Nino EL, Yi S, Grozinger C. Parallel Epigenomic and Transcriptomic Responses to Viral Infection in Honey Bees (Apis mellifera) PloS Pathogens. 2015;11(3):e1004713. doi: 10.1371/journal.ppat.1004713. This study assessed the transcriptional profile of IAPV-infected young bees with high throughput sequencing (RNA-Seq). They found that 753 genes were differentially expressed in IAPV-infected bees, including a few genes in the RNAi and Toll pathways. This study did an excellent job assessing pre-existing/confounding infections and therefore ensured that the gene regulation observed in their well-controlled experiments was most likely due to IAPV-infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **102.Nazzi F, Brown SP, Annoscia D, Del Piccolo F, Di Prisco G, Varricchio P, Vedova Della G, Cattonaro F, Caprio E, Pennacchio F. Synergistic parasite-pathogen interactions mediated by host immunity can drive the collapse of honeybee colonies. Plos Pathog. 2012;8:e1002735. doi: 10.1371/journal.ppat.1002735. This study provides experimental evidence of a central role for Am dorsal-1A in limiting DWV infection in the context of mite infestation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schlüns H, Crozier RH. Relish regulates expression of antimicrobial peptide genes in the honeybee, Apis mellifera, shown by RNA interference. Insect Mol Biol. 2007;16:753–759. doi: 10.1111/j.1365-2583.2007.00768.x. [DOI] [PubMed] [Google Scholar]
- 104.Boncristiani H, Underwood R, Schwarz R, Evans JD, Pettis J, vanEngelsdorp D. Direct effect of acaricides on pathogen loads and gene expression levels in honey bees Apis mellifera. J Insect Physiol. 2012;58:613–620. doi: 10.1016/j.jinsphys.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 105.Yang X, Cox-Foster DL. Impact of an ectoparasite on the immunity and pathology of an invertebrate: evidence for host immunosuppression and viral amplification. P Natl Acad Sci Usa. 2005;102:7470–7475. doi: 10.1073/pnas.0501860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *106.Azzami K, Ritter W, Tautz J, Beier H. Infection of honey bees with acute bee paralysis virus does not trigger humoral or cellular immune responses. Arch Virol. 2012;157:689–702. doi: 10.1007/s00705-012-1223-0. This study determined that ABPV-infected honey bee larvae did not produce AMPs, and suggested that ABPV may suppress bee immune responses, since AMPs were not produced by larvae co-challenged with ABPV and E. coli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *107.Bull JC, Ryabov EV, Prince G, Mead A, Zhang C, Baxter LA, Pell JK, Osborne JL, Chandler D. A strong immune response in young adult honeybees masks their increased susceptibility to infection compared to older bees. Plos Pathog. 2012;8:e1003083. doi: 10.1371/journal.ppat.1003083. This paper is an important step toward understanding age-dependent immunity in honey bees. They showed that older forager bees were more resistant to fungal infection due to age-related activation of immune pathways, where as younger house bees had a greater number of differentially expressed immune genes in response to fungal challenge. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Boncristiani HF, Evans JD, Chen Y, Pettis J, Murphy C. PLOS ONE: In Vitro Infection of Pupae with Israeli Acute Paralysis Virus Suggests Disturbance of Transcriptional Homeostasis in Honey Bees (Apis mellifera) PLoS ONE. 2013;8:e73429. doi: 10.1371/journal.pone.0073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Johnson RM, Evans JD, Robinson GE, Berenbaum MR. Changes in transcript abundance relating to colony collapse disorder in honey bees (Apis mellifera) P Natl Acad Sci Usa. 2009;106:14790–14795. doi: 10.1073/pnas.0906970106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Marringa WJ, Krueger MJ, Burritt NL, Burritt JB. Honey bee hemocyte profiling by flow cytometry. PLoS ONE. 2014;9:e108486. doi: 10.1371/journal.pone.0108486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hillyer JF, Strand MR. Mosquito hemocyte-mediated immune responses. Current Opinion in Insect Science. 2014;3:14–21. doi: 10.1016/j.cois.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Richard F-J, Holt HL, Grozinger CM. Effects of immunostimulation on social behavior, chemical communication and genome-wide gene expression in honey bee workers (Apis mellifera) Bmc Genomics. no date:13. doi: 10.1186/1471-2164-13-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Holt HL, Aronstein KA, Grozinger CM. Chronic parasitization by Nosema microsporidia causes global expression changes in core nutritional, metabolic and behavioral pathways in honey bee workers (Apis mellifera) Bmc Genomics. 2013:14. doi: 10.1186/1471-2164-14-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *114.Moran NA, Hansen AK, Powell JE, Sabree ZL. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS ONE. 2012;7:e36393. doi: 10.1371/journal.pone.0036393. This study furthers our understanding of the honey bee microbiome, an important factor to consider in bee health and immune defense. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *115.Mattila HR, Rios D, Walker-Sperling VE, Roeselers G, Newton I. Characterization of the active microbiotas associated with honey bees reveals healthier and broader communities when colonies are genetically diverse. PLoS ONE. 2012;7(3):e32962. doi: 10.1371/journal.pone.0032962. This study furthers our understanding of the honey bee microbiome, an important factor to consider in bee health and immune defense. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rainey SM, Shah P, Kohl A, Dietrich I. Understanding the Wolbachia-mediated inhibition of arboviruses in mosquitoes: progress and challenges. Journal of General Virology. 2014;95:517–530. doi: 10.1099/vir.0.057422-0. [DOI] [PubMed] [Google Scholar]
- 117.Hamilton PT, Perlman SJ. Host defense via symbiosis in Drosophila. Plos Pathog. 2013;9:e1003808. doi: 10.1371/journal.ppat.1003808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Doublet V, Labarussias M, de Miranda JR, Moritz RFA, Paxton RJ. Bees under stress: sublethal doses of a neonicotinoid pesticide and pathogens interact to elevate honey bee mortality across the life cycle. Environ Microbiol. 2014 doi: 10.1111/1462-2920.12426. [DOI] [PubMed] [Google Scholar]
- *119.Di Prisco G, Cavaliere V, Annoscia D, Varricchio P, Caprio E, Nazzi F, Gargiulo G, Pennacchio F. Neonicotinoid clothianidin adversely affects insect immunity and promotes replication of a viral pathogen in honey bees. P Natl Acad Sci Usa. 2013 doi: 10.1073/pnas.1314923110. This work examined synergistic effects of agrochemical exposure and virus-infection in honey bees and determined that abidaecin expression was reduced and DWV virus levels were greater in clothianidin treated bees. Overall this study indicated that the Toll pathway is important for antiviral defense, and that Toll signaling is reduced by clothianidin exposure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Locke B, Forsgren E, Fries I, de Miranda JR. Acaricide treatment affects viral dynamics in Varroa destructor-infested honey bee colonies via both host physiology and mite control. 2012 doi: 10.1128/AEM.06094-11. no volume. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *121.Pettis JS, vanEngelsdorp D, Johnson J, Dively G. Pesticide exposure in honey bees results in increased levels of the gut pathogen Nosema. Naturwissenschaften. 2012;99:153–158. doi: 10.1007/s00114-011-0881-1. This study provides evidence of the interaction between agrochemical exposure and pathogen load; honey bees from colonies exposed to sublethal amounts of imidacloprid had higher levels of Nosema than bees from control colonies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *122.Wheeler MM, Robinson GE. Diet-dependent gene expression in honey bees: honey vs. sucrose or high fructose corn syrup. Sci Rep. 2014;4:5726. doi: 10.1038/srep05726. This study furthers our understanding of the honey bee nutrition, an important factor to consider in bee health and immune defense. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *123.DeGrandi-Hoffman G, Chen Y, Huang E, Huang MH. The effect of diet on protein concentration, hypopharyngeal gland development and virus load in worker honey bees (Apis mellifera L) J Insect Physiol. 2010;56:1184–1191. doi: 10.1016/j.jinsphys.2010.03.017. This study demonstrated that bees fed either protein supplement or pollen, in addition to high fructose corn syrup (HFCS) containing water, had lower DWV loads, than bees fed only HFCS-containing water. This controlled study is an important contribution toward understanding the role of nutrition in honey bee health immune defense. [DOI] [PubMed] [Google Scholar]
- 124.Di Pasquale G, Salignon M, Le Conte Y, Belzunces LP, Decourtye A, Kretzschmar A, Suchail S, Brunet J-L, Alaux C. Influence of pollen nutrition on honey bee health: do pollen quality and diversity matter? PLoS ONE. 2013;8:e72016. doi: 10.1371/journal.pone.0072016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shutler D, Head K, Burgher-MacLellan KL, Colwell MJ, Levitt AL, Ostiguy N, Williams GR. Honey Bee Apis mellifera Parasites in the Absence of Nosema ceranae Fungi and Varroa destructor Mites. PLoS ONE. 2014;9:e98599. doi: 10.1371/journal.pone.0098599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Martin SJ, Highfield AC, Brettell L, Villalobos EM, Budge GE, Powell M, Nikaido S, Schroeder DC. Global honey bee viral landscape altered by a parasitic mite. Science. 2012;336:1304–1306. doi: 10.1126/science.1220941. [DOI] [PubMed] [Google Scholar]
- 127.Tarpy DR, vanEngelsdorp D, Pettis JS. Genetic diversity affects colony survivorship in commercial honey bee colonies. Naturwissenschaften. 2013;100:723–728. doi: 10.1007/s00114-013-1065-y. [DOI] [PubMed] [Google Scholar]
- 128.Tarpy DR, Seeley TD. Lower disease infections in honeybee (Apis mellifera) colonies headed by polyandrous vs monandrous queens. Naturwissenschaften. 2006;93:195–199. doi: 10.1007/s00114-006-0091-4. [DOI] [PubMed] [Google Scholar]
- 129.Evans JD, Spivak M. Socialized medicine: individual and communal disease barriers in honey bees. J Invertebr Pathol. 2010;103 (Suppl 1):S62–72. doi: 10.1016/j.jip.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 130.Lauring AS, Andino R. Quasispecies theory and the behavior of RNA viruses. Plos Pathog. 2010;6:e1001005. doi: 10.1371/journal.ppat.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ayres JS, Schneider DS. Tolerance of infections. Annu Rev Immunol. 2012;30:271–294. doi: 10.1146/annurev-immunol-020711-075030. [DOI] [PubMed] [Google Scholar]
- 132.Cobey S, Sheppard WS. Status of breeding practices and genetic diversity in domestic US honey bees. Honey Bee Colony. 2012 [Google Scholar]
- 133.Buechler R, Andonov S, Bienefeld K, Costa C, Hatjina F, Kezic N, Kryger P, Spivak M, Uzunov A, Wilde J. Standard methods for rearing and selection of Apis mellifera queens. J Apicult Res. 2013:52. [Google Scholar]
- **134.Goblirsch MJ, Spivak MS, Kurtti TJ. A cell line resource derived from honey bee (Apis mellifera) embryonic tissues. PLoS ONE. 2013;8:e69831. doi: 10.1371/journal.pone.0069831. This paper describes a honey bee cell line (AmE-711) derived from fragmented honey bee embryonic tissue. Immortalized cell lines are important for investigating bee cell biology and honey bee host - pathogen interactions. Future use of these cells lines for honey bee virus-propagation will provide more uniform inoculum for virus-host interaction studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bergem M, Norberg K, Aamodt RM. Long-term maintenance of in vitro cultured honeybee (Apis mellifera) embryonic cells. BMC Dev Biol. 2006;6:17. doi: 10.1186/1471-213X-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Barbara GS, Grünewald B, Paute S, Gauthier M, Raymond-Delpech V. Study of nicotinic acetylcholine receptors on cultured antennal lobe neurones from adult honeybee brains. Invert Neurosci. 2008;8:19–29. doi: 10.1007/s10158-007-0062-2. [DOI] [PubMed] [Google Scholar]
- 137.Kreissl S, Bicker G. Dissociated neurons of the pupal honeybee brain in cell culture. J Neurocytol. 1992;21:545–556. doi: 10.1007/BF01187116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Apis mellifera genome encodes major members of insect immune pathways including those depicted in Figure 1 and listed by gene name, pathway, and accession number below. Bold text indicates genes differentially expressed in virus-infected honey bees, and the specific virus and citation are provided for each. A list of additional honey bee immune-related genes is included in worksheet 2.