Abstract
Alzheimer’s disease (AD) is a severe1 age-related neurodegenerative disorder characterized by accumulation of amyloid-β (Aβ) plaques and neurofibrillary tangles, synaptic and neuronal loss, and cognitive decline. Several genes have been implicated in AD, but chromatin state alterations during neurodegeneration remain uncharacterized. Here, we profile transcriptional and chromatin state dynamics across early and late pathology in the hippocampus of an inducible mouse model of AD-like neurodegeneration. We find a coordinated downregulation of synaptic plasticity genes and regulatory regions, and upregulation of immune response genes and regulatory regions, which are targeted by factors that belong to the ETS family of transcriptional regulators, including PU.1. Human regions orthologous to increasing-level enhancers show immune cell-specific enhancer signatures as well as immune cell expression quantitative trait loci (eQTL), while decreasing-level enhancer orthologs show fetal-brain-specific enhancer activity. Notably, AD-associated genetic variants are specifically enriched in increasing-level enhancer orthologs implicating immune processes in AD predisposition. Indeed, increasing enhancers overlap known AD loci lacking protein-altering variants and implicate additional loci that do not reach genome-wide significance. Our results reveal new insights into the mechanisms of neurodegeneration and establish the mouse as a useful model for functional studies of AD regulatory regions.
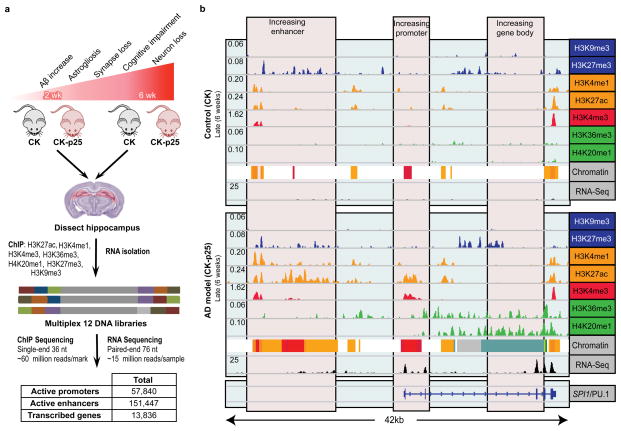
Gene expression2,3 and genetic variation4 studies suggest gene-regulatory changes may underlie AD, but regulatory epigenetic alterations during neurodegeneration remain uncharacterized, given the inaccessible nature of human brain samples. To address this need, we profiled transcriptional and epigenomic changes during neurodegeneration in the hippocampus of the CK-p25 mouse model of AD5–7 and CK littermate controls at both early and late stages of neurodegeneration (2 weeks and 6 weeks after p25 induction). CK-p25 mice, in which accumulation of the Cdk5 activator p25 is inducible, exhibit DNA damage, aberrant gene expression and elevated amyloid-β levels at early stages7, followed by neuronal and synaptic loss and cognitive impairment at late stages5,6.
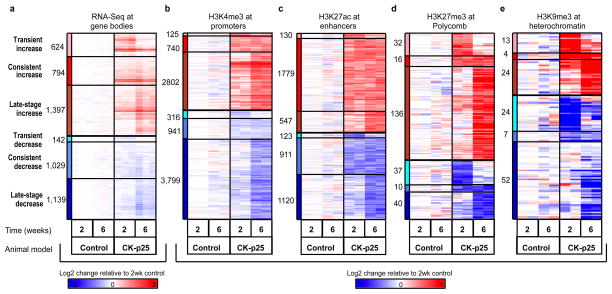
For transcriptome analysis, we used RNA sequencing to quantify gene expression changes for 13,836 ENSEMBL genes (see Methods; Extended Data Fig. 1a; Supplementary Table 1). We found 2,815 upregulated genes and 2,310 downregulated genes in the CK-p25 AD mouse model as compared to CK littermate controls (at q < 0.01; Supplementary Table 1), which we classified into transient (2 weeks only), late-onset (6 weeks only), and consistent (both) expression classes (Fig. 1a; Extended Data Fig. 4a, Supplementary Table 1). These showed distinct functional enrichments (Fig. 1a; Supplementary Table 2), with transient-increase genes enriched in cell cycle functions (P < 10−92), consistent-increase genes enriched in immune (P < 10−10) and stimulus response functions (P < 10−4), and consistent- and late-decrease genes enriched in synaptic and learning functions (P < 10−12).
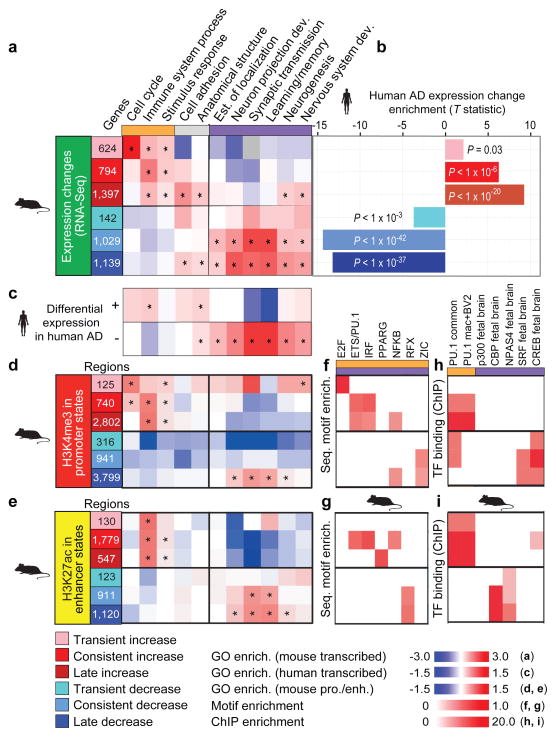
Figure 1. Conserved gene expression changes between mouse and human AD are associated with immune and neuronal functions.
a, Six distinct temporal classes of differentially expressed genes are denoted, transient (early) increase (pink) or decrease (light blue), consistent increase (red) or decrease (blue), and late (6 wk) increase (dark red) or decrease (navy blue). Expression is shown relative to the mean of three replicates at 2 week control (CK) mice. Shown are the most significant distinct biological process Gene Ontology categories in each class of differentially regulated genes (asterisk denotes enrichment of hypergeometric P < 0.01). Grey boxes indicate no overlapping genes. b, T-statistic identifying the bias of each differentially regulated class of genes in AD cases relative to controls; negative t denotes lower expression in AD, positive t denotes higher expression in AD. c, Enrichment of Gene Ontology categories for differentially expressed genes between AD cases and controls in human2. Enrichment of each Gene Ontology category examined in the gene expression analysis was calculated for d, H3K4me3 promoters (red) and e, H3K27ac enhancers (yellow). Asterisk denotes categories with a binomial P < 0.01. Enrichment of regulatory motifs within changing f, promoters (top) and g, enhancers (bottom) in the mouse AD model. Overlap of changing h, promoters (top) and i, enhancers (bottom) with regions shown to be bound by immune (orange) and neuronal (purple) transcriptional factors (TF) and co-factors profiled using ChIP-seq in mouse immune and neuronal tissues15–19.
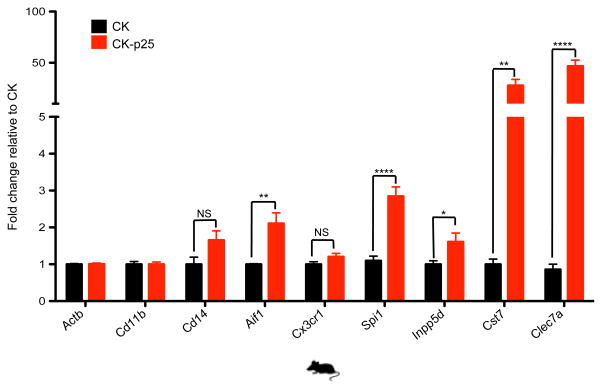
These coordinated neuronal and immune changes are consistent with the pathophysiology of AD2 and probably reflect both cell-type-specific expression changes as well as changes in cell composition. Indeed, comparison with expression in microglia8 (the resident immune cells of the brain) shows that both, cell type composition (P = 2.7 × 10−4) and microglia-specific activation (P = 2.9 × 10−6) significantly contribute to the gene expression changes (see Methods). Additionally, reverse transcription followed by quantitative PCR (RT-qPCR) of increased-level genes in purified CD11b+ CD45low microglia populations confirms cell-type-specific activation for five of the seven microglia-specific genes tested (Extended Data Fig. 2).
Confirming the biological relevance of our mouse model for human AD, the observed changes in gene expression in mouse, especially for the consistent and late classes, agreed with gene expression differences between 22 AD patients and 9 controls in human post-mortem laser capture microdissected hippocampal gray matter2 (Fig. 1b). The enriched Gene Ontology classes also agreed between mouse and human, with higher immune gene expression and lower neuronal gene expression in AD patients (Fig. 1c).
For epigenome analysis, we used chromatin immunoprecipitation sequencing (ChIP-seq) to profile seven chromatin marks9: H3K4me3 (associated primarily with active promoters); H3K4me1 (enhancers); H3K27ac (enhancer/promoter activation); H3K27me3 (Polycomb repression); H3K36me3 and H4K20me1 (transcription); and H3K9me3 (heterochromatin) (Extended Data Fig. 1a). We used ChromHMM (http://compbio.mit.edu/ChromHMM/) to learn a chromatin state model (see Methods, Extended Data Fig. 3a) defined by recurrent combinations of histone modifications, consisting of promoters, enhancers, transcribed, bivalent, repressed, heterochromatin and low-signal states (Extended Data Fig. 3a). We defined 57,840 active promoters using H3K4me3 peaks within promoter chromatin states, and 151,447 active enhancer regions using H3K27ac peaks within enhancer chromatin states (Extended Fig. 1a; Supplementary Table 3; see Methods).
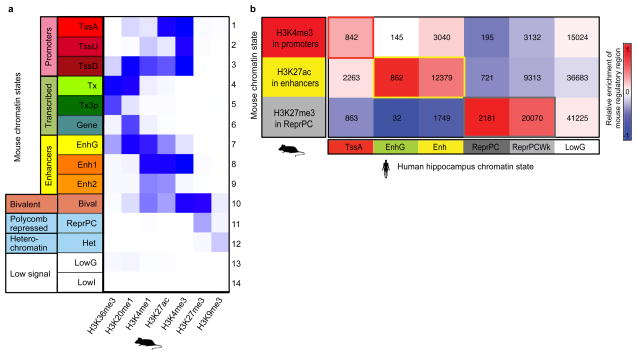
We mapped orthologous genes between mouse and human using ENSEMBL one-to-one orthologs (see Methods). We also mapped orthologous noncoding regions using multiple mammalian sequence alignments, mapping each mouse peak to its best human match (see Methods). We found matches for 90% of promoter regions, 84% of enhancers, 74% of Polycomb-repressed regions, and 33% of heterochromatin regions (Supplementary Table 3). Comparing our mouse chromatin states to human hippocampus chromatin states10, we found significant epigenomic conservation at orthologous noncoding regions (Extended Data Fig. 3b), consistent with recent results11.
We quantified epigenomic changes in promoter regions using relative differences in H3K4me3 levels resulting in 3,667 increased-level and 5,056 decreased-level peaks (q < 0.01, Extended Data Fig. 4b and Supplementary Table 3), which we classified into transient, consistent, and late-stage, as for gene expression changes. For enhancer regions, we used relative levels of H3K27ac, resulting in 2,456 increased-level and 2,154 decreased-level peaks (Extended Data Fig. 4c; Supplementary Table 3). Only a very small number of peaks showed differences in Polycomb-repressed and heterochromatin regions, leading us to focus on enhancer and promoter changes for the remaining analyses (Extended Data Fig. 4d, e; Supplementary Table 3).
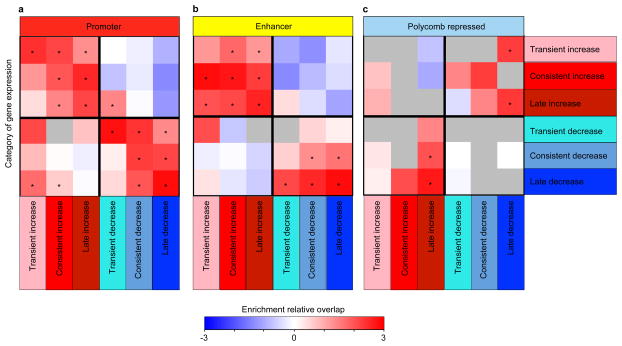
Genes flanking increased- and decreased-level regulatory regions (see Methods) showed consistent gene expression changes for both promoter and enhancers regions (Extended Data Fig. 5), and were consistently enriched in immune and stimulus-response functions for increased-level enhancers and promoters, and in synapse and learning-associated functions for deceased-level enhancers and promoters (Fig. 1d, e), consistent with our Gene Ontology results of changing gene expression levels.
Increased- and decreased-level regulatory regions showed distinct regulatory motif enrichments (Fig. 1f, g). Increased-level peaks were enriched in NFκB, E2F, PPARG, IRF and PU.1 (ref. 12) transcription factor motifs for both enhancers and promoters, consistent with immune regulator targeting. Decreased-level peaks in enhancers were enriched for DNA-binding RFX motifs, and peaks in promoters were enriched for zinc-finger ZIC motifs, two known neurodevelopmental regulators13,14.
Consistent with the observed motif enrichments, increased-level enhancers and promoters showed in vivo binding of PU.1 in mouse embryos15,16 (Fig. 1h, i). Only increased-level promoters were bound in macrophages and BV-2 microglial-like cells17–19 that are both implicated in AD20, while both increased- and decreased-level promoters were bound in several immune cell lineages (Fig. 1h). The PU.1 regulator itself (encoded by the SPI1 gene), showed increased expression and enhancer levels (Extended Data Fig. 1b), possibly contributing to immune enhancer and promoter upregulation, consistent with roles for PU.1, ETS-1, and other ETS family members in microglia activation and proliferation during neurodegeneration21,22. By contrast, neuronal function regulators were not enriched in increased-level enhancers (except for a weak enrichment of fetal brain CREB, Fig. 1i), consistent with primarily immune and inflammatory function of these regions.
Decreased-level enhancers and promoters were targeted by different regulators, suggesting distinct regulatory programs. Decreased-level promoters were preferentially bound by CREB and SRF (P < 10−21 and P < 10−16), two known regulators of neuronal activity in cortical neurons23, and decreased-level enhancers were preferentially bound by CBP (Phypergeometric = 5.4 × 10−20), a known co-activator for neuronal activity16 (Fig. 1h, i). Surprisingly, p300-bound regions15 did not show any enrichment, suggesting distinct roles for CBP and p300, despite a general association with enhancers for both. The distinct neuronal and immune targeting of decreased-level and increased-level regulatory regions provides a mechanistic basis for the expression differences observed for neuronal and immune genes, and suggests potential therapeutic targets for reversing observed alterations during neurodegeneration.
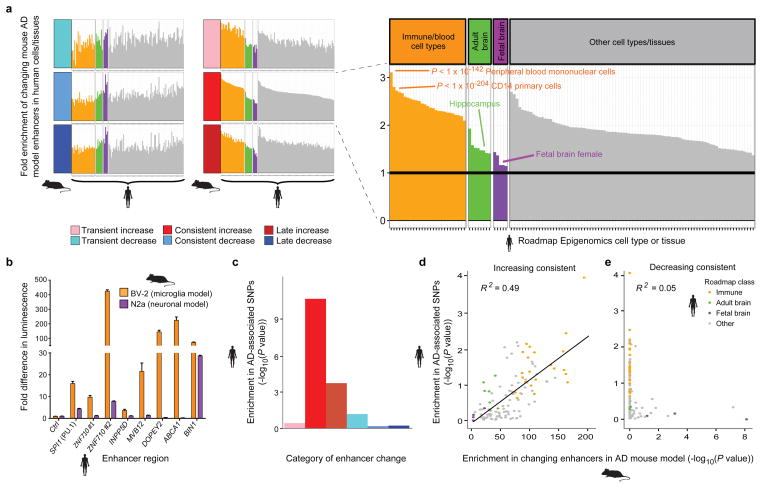
On the basis of chromatin state annotations in 127 human cell types and tissues10 (Fig. 3a and Supplementary Table 4) regions orthologous to increased-level enhancers in mouse showed immune cell enhancer activity in human (P < 10−100), while orthologs of decreased-level enhancers in mouse showed fetal brain tissue enhancer activity in human (P < 10−8 consistent; P < 10−17 late-stage; Fig. 2a; Supplementary Table 4). Adult brain tissues (including hippocampus) were not as strongly enriched, suggesting changes are biased towards neuronal plasticity. These results are consistent with decreased neuronal plasticity, and increased microglial activation and proliferation during AD progression24.
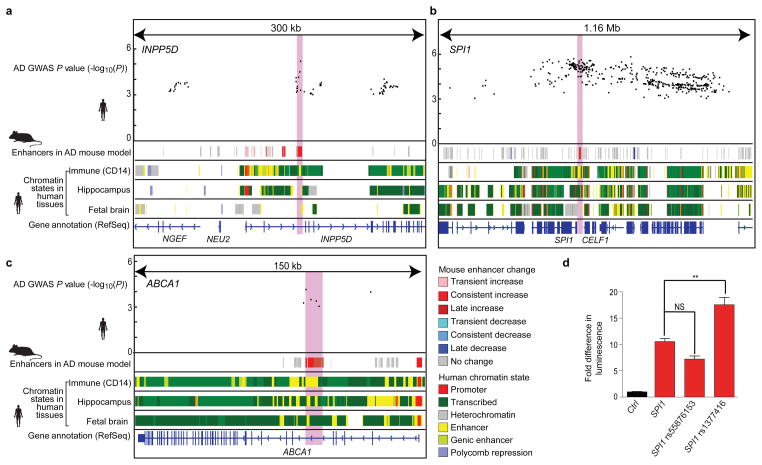
Figure 3. Increasing enhancer orthologs help interpret AD-associated noncoding loci.
Overlap of disease-associated SNPs (top) with increasing enhancers (2nd row, red) and immune enhancers in human (CD14+ primary cells) is shown for genome-wide significant (INPP5D and CELF1 (containing the SPI1 gene); a and b) and below-significance (ABCA1; c) AD GWAS loci. Roadmap chromatin state annotations for immune cells (CD14+ primary; E029), hippocampus (E071), and fetal brain (E81), with colors as shown in the key. Light red highlight denotes increasing enhancer regions tested in luciferase assay. kb, kilobases; Mb, megabases. d, AD associated SNP rs1377416 amplifies in vitro luciferase activity of putative enhancer region 38,313–37,359 base pairs (bp) upstream of SPI1 (PU.1) gene in BV-2 cells. n=3, P < 0.0001, one-way analysis of variance (ANOVA); **P < 0.01, Tukey’s multiple comparison post-hoc test. NS, nonsignificant.
Figure 2. AD GWAS loci are preferentially enriched in increasing enhancer orthologs with immune function.
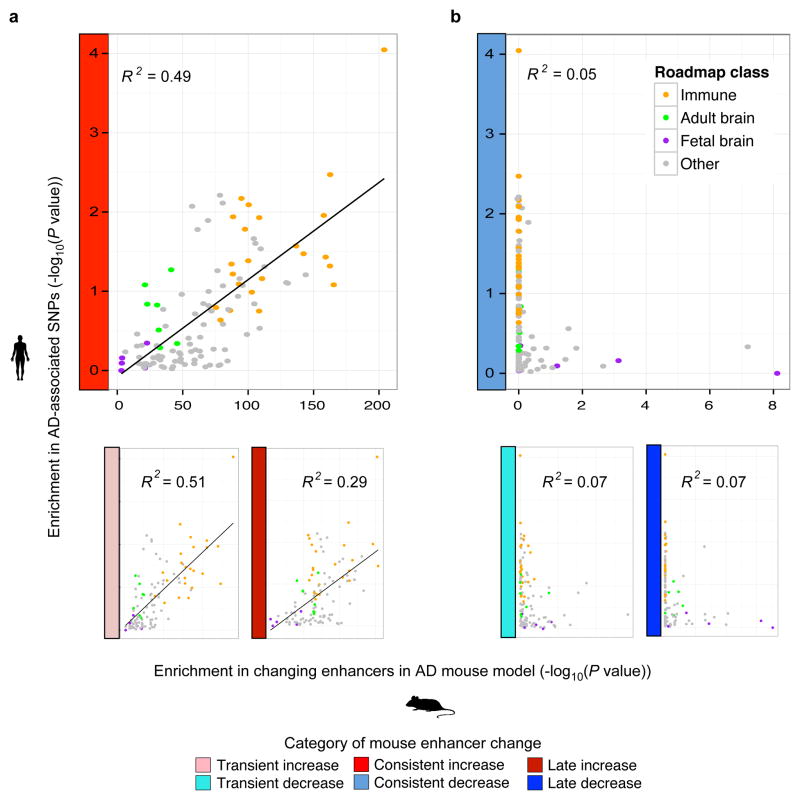
a, Enrichment (y-axis) of changing mouse AD enhancer orthologs, with a focus on consistently increasing (red) category of enhancers, in 127 cell and tissue types profiled by the Roadmap Epigenomics Consortium10 (columns). Roadmap samples are grouped into fetal brain (purple), adult brain (green), immune/blood cell types (orange), and all other (grey). b, Cell-type-specific fold luciferase reporter expression change relative to control (ctrl) for selected increasing enhancer regions in BV-2 microglia (orange) vs N2A neurons (purple) (n=3, *P < 0.05, two-tailed t-test). c, Enrichment of AD-associated SNPs (y-axis, binomial P value) in human regions orthologous to the mouse enhancers. d,e, Enrichment of AD-associated SNPs (y-axis, permutation P value) in tissue-specific enhancer annotations from the Roadmap Epigenomics Consortium (points), relative to their enrichment for d, consistently increasing and e, consistently decreasing orthologous enhancer regions in the mouse AD model (x-axis, hypergeometric P value). Linear regression trend line and R2, based on Pearson correlation, is shown.
To verify whether the increased-level putative enhancer regions were indeed functional, we used a luciferase reporter assay to evaluate their ability to drive in vitro gene expression in immortalized murine microglial (BV-2) and neuroblastoma (N2a) cell lines. Eight of the nine increased-level human orthologs tested were indeed able to drive in vitro reporter expression. Two of these, BIN1 and ZNF710, were active in both cell types, while the remaining six showed a BV-2 cell-specific increase in luciferase expression (Fig. 2b and Supplementary Table 5) confirming both, functional conservation and tissue specificity of increased-level enhancer regions implicated by our mouse model of AD.
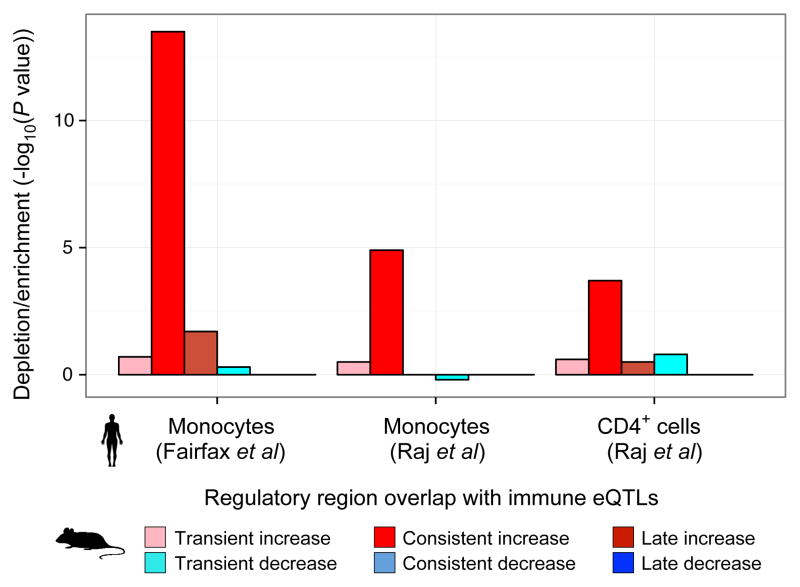
Human orthologs of increased-level enhancers were also enriched for expression quantitative trait loci (eQTLs) in CD4+ T-cells and CD14+ monocytes25,26 (Extended Data Fig. 6, Supplementary Table 6), indicating they harbor driver mutations controlling immune cell regulatory programs. The enrichment was strongest for CD14+ monocytes (Extended Data Fig. 6), which also showed the highest enhancer enrichment and is consistent with the observed inflammatory response Gene Ontology category.
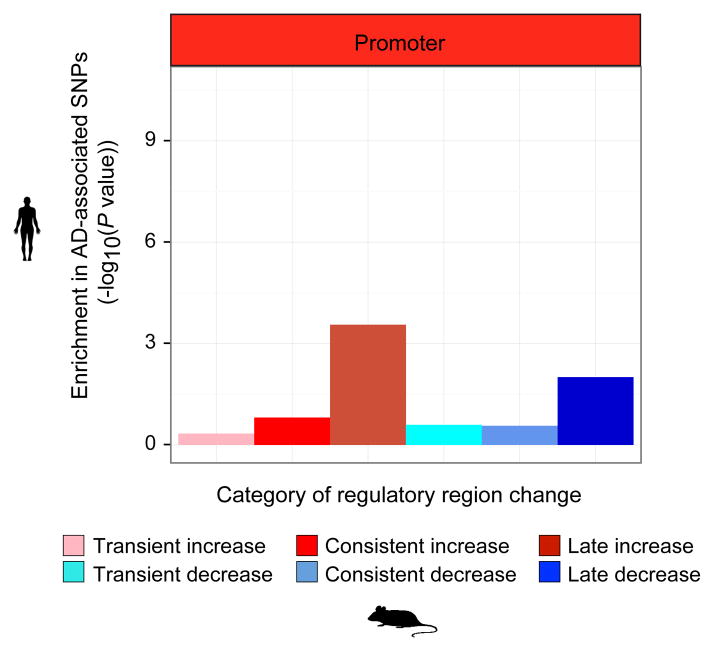
To test whether the implicated regulatory regions are causal, we examined their enrichment for AD-associated variants from genome-wide association studies (GWAS). Genetic variants associated with AD in a meta-analysis of ~74,000 individuals4, were enriched in increased-level enhancer orthologs (Fig. 2c), (4.4-fold enrichment, binomial P = 1.2 × 10−10 at GWAS cutoff P < 0.001; 9.7-fold enrichment, binomial P < 3.7 × 10−6 at GWAS cutoff P < 10−5). By contrast, decreased-level enhancer orthologs were surprisingly not enriched (0.61-fold), suggesting a causal role specifically for immune-related processes. Promoter regions were only weakly enriched, strongly implicating distal enhancers in mediating AD predisposition (Extended Data Fig. 7).
Across diverse cell types and tissues, we found concordance between the enrichment for AD GWAS single nucleotide polymorphisms (SNPs) and the enrichment for increased-level enhancer orthologs (R2 = 0.49; Fig. 2d, Extended Data Fig. 8a, left and Supplementary Table 4), with CD14+ immune cells being the most enriched in both, followed by other immune cell types, and with fetal brain enhancers showing the smallest enrichment in both. By contrast, decreasing enhancers orthologs showed a very weak correlation (R2 < 0.08) (Fig. 2e, Extended Data Fig. 8b, right and Supplementary Table 4). The increased-level enhancer ortholog enrichment for AD GWAS SNPs persisted both within CD14+ enhancers (3.0-fold enrichment, binomial P = 1.3 × 10−5) and outside CD14+ enhancers (3.4-fold, P = 0.005) suggesting it is not solely a feature of CD14+ cell type enrichment (see Methods).
These results are consistent with enhanced microglial expression of CD14 in brains of animal models of AD, and a regulatory role of the CD14 receptor in microglial inflammatory response, which modulates amyloid-β deposition24. Thus, the enrichment of AD-associated variants in CD14+ primary immune cells, but not neuronal cells, indicates that AD genetic predisposition is primarily associated with immune function, while decrease in neuronal plasticity may be affected primarily by non-genetic effects, such as diet, education, physical activity and age, which are thought to lead to epigenetic changes related to cognitive reserve27.
We next utilized the epigenomic annotations of increased-level enhancer orthologs to gain insights into AD-associated loci (Supplementary Table 7). Among the 20 genome-wide significant AD-associated loci4, 11 contain no protein-altering SNPs in linkage disequilibrium (LD), indicating they may have noncoding roles. Of these, 5 localize within increased-level enhancer orthologs, including two well-established GWAS loci (PICALM, BIN1), and three loci (INPP5D, CELF1 (also containing the SPI1 gene), PTK2B) only recently recognized as significant by combining all AD cohorts.
For INPP5D (Fig. 3a), a known regulator of inflammation28, the most significant variants localize within an increased-level enhancer ortholog, which also shows CD14+ enhancer activity. In the CELF1 locus (Fig. 3b) a large region of association spans several genes, but the strongest genetic signal (P = 2 × 10−6) localizes upstream of SPI1 (PU.1), and specifically within an increased-level enhancer ortholog that is also active in immune cells. We confirmed that the AD-associated C-T substitution, rs1377416, in the SPI1 enhancer leads to increased in vitro enhancer activity in murine BV-2 microglia cells using a luciferase reporter assay (Fig. 3d). In addition, the AD-associated SNP rs55876153 near SPI1, which overlaps an increased-level mouse enhancer ortholog, is in strong linkage disequilibrium (LD = 0.89, see Methods) with a known SPI1 eQTL, rs10838698 (ref. 25), even though it did not significantly alter enhancer activity in the luciferase assay.
Outside known GWAS loci, an additional 22 weakly-associated regions (3.9-fold, P < 4.9 × 10−7) contain variants within increased-level enhancer orthologs (Supplementary Table 7), of which 17 lack protein-altering variants in LD (R2 < 0.4), providing strong candidates for directed experiments. One such example includes ABCA1 (P = 6.9 × 10−5, Fig. 3c), a paralog of AD-associated ABCA7 and encoding a glial-expressed transporter that influences APOE metabolism in the central nervous system29. The region lacks protein-altering variants and all five SNPs in the cluster of association lie specifically within an increased-enhancer ortholog, which is also active in CD14+ immune cells and, to a lesser extent, in human hippocampus and fetal brain.
Overall, our study revealed contrasting changes in immune and neuronal genes and regulatory regions during AD-like neurodegeneration in mouse, strong human-mouse conservation of gene expression and epigenomic signatures, and enrichment of AD-associated loci in increased-level enhancer orthologs in human. While immune genes are known to be among the most significant genetic loci associated with AD, the depletion of neuronal promoters and enhancers is particularly striking for a cognitive disorder with well-established environmental and experiential factors that include diet, exercise, education and age. These results are consistent with a model in which increased immune susceptibility to environmental factors during aging and cognitive decline is mediated by interactions between genetically driven immune cell dysregulation and environmentally driven epigenomic alteration in neuronal cells.
Our study also illustrates the power of model organisms for the study of human disease progression, especially for disorders affecting inaccessible tissues for which only post-mortem samples are available in human. We find that molecular changes in both genes and regulatory regions are highly conserved between human AD and CK-p25 neurodegeneration, enabling detailed studies of the molecular signatures associated with disease progression across diverse environmental conditions, in a variety of brain regions and cell types, and in response to therapeutic agents before or after disease onset.
Lastly, our results suggest specific therapeutic targets for AD, including putative causal nucleotides lying in increased-level enhancer orthologs that may be targeted by CRISPR/Cas9 genome editing30, and trans-acting regulators. In particular, the transcription factor PU.1 is implicated as a therapeutic target by its genetic association with AD, as well as the enrichment of the PU.1 motif and the PU.1 in vivo binding sites at increased-level regulatory regions during mouse neurodegeneration. The conservation of neuronal and immune regulatory circuitry between mouse and human suggests that CK-p25 mice may offer a powerful model for studying the gene-regulatory and cognitive effects of such interventions.
Materials and Methods
Animals
All mouse work was approved by the Committee on Animal Care of the Division of Comparative Medicine at MIT. Adult (3-months-old) female double-transgenic CK-p25 (ref. 5) mice and their respective control littermates were used for the experiments. Brain tissue was collected at either 2 or 6 weeks following p25 induction. Upon dissection tissue was flash-frozen in liquid nitrogen. No animals were excluded from the study and no randomization or blinding was required.
Chromatin Immunoprecipitation (ChIP)
Mouse hippocampus was harvested immediately after euthanasia. Chromatin immunoprecipitation was then performed as described in Broad ChIP protocol (http://www.roadmapepigenomics.org/protocols/type/experimental/). Briefly, tissues were minced and crosslinked in 1% formaldehyde (Thermo Scientific) for 15 min at room temperature and quenched with glycine for 5 min (Sigma). The samples were homogenized in cell lysis buffer containing proteinase inhibitors (complete, Roche) and chromatin was then fragmented to a size range of ~200–500 bp using a Branson 250 digital sonifier. Solubilized chromatin was then diluted and incubated with ~1 μg antibody at 4°C overnight. Immune complexes were captured with Protein A-sepharose beads, washed and eluted. Enriched chromatin was then subjected to crosslink reversal and proteinase K digestion at 65°C, phenol-chloroform extraction and ethanol precipitation. Isolated ChIP DNA was resuspended and quantified using the Qubit assay (Invitrogen). H3K4me1 (Abcam, ab8895), H3K4me3 (Millipore, #07-473), H3K9me3 (Abcam, ab8898), H3K27me3 (Millipore, #07-449), H3K27ac (Abcam, ab4729), H3K36me3 (Abcam, ab9050) and H4K20me1 (Abcam, ab9051) were used to immunoprecipitate endogenous proteins.
ChIP-seq high-throughput sequencing, read mapping and quality control
Sequencing libraries were prepared from ~1–5 ng ChIP (or input) DNA as described previously31. Gel electrophoresis was used to retain library fragments between 300 and 600 bp. Prior to sequencing, libraries were quantified using Qubit (Invitrogen) and quality-controlled using Agilent’s Bioanalyzer. 36 bp single end sequencing was performed using the Illumina HiSeq 2000 platform according to standard operating procedures. For each histone modification, five biological replicate datasets were produced with corresponding whole cell extract controls, except for H3K4me3, H4K20me1and H3K27me3 in the 2 wk control (CK) sample where optimal amount of reads for sufficient coverage was obtained from four biological replicates. Reads were mapped to the mm9 reference mouse genome using MAQ v0.7.1-9 using default parameters32. Reads mapping to multiple locations were discarded. Duplicates were marked and filtered using PICARD (http://picard.sourceforge.net/). After filtering, roughly 55–60 million unique reads were obtained for each histone modification in each condition (~9–12 million reads per replicate) and ~110–145 million reads in total for the whole-cell extract controls in each condition. All replicate datasets passed quality control based on ENCODE ChIP-seq data standards based on read quality, read mapping statistics, library complexity and strand cross-correlation analysis (to measure signal-to-noise ratios)33.
RNA sequencing
Mouse brains were homogenized and total RNA was extracted using Trizol reagent (Ambion). Total RNA was quality-controlled using Agilent’s Bioanalyzer and prepared for sequencing using Illumina’s TruSeq Stranded Total RNA Sample Preparation Kit with Ribo-Zero. High-throughput sequencing was performed on an Illumina HiSeq 2000 platform. Roughly 15 million 76 pair-end reads were generated for each dataset. Sequence reads were aligned to mouse mm9 genome with Bowtie. Based on the reproducibility of the results (Fig. 2a), three replicate biological datasets were produced for each condition. A small number of replicates suffice for RNA-seq studies34 and we were able to detect large-scale changes in read counts in coherent Gene Ontology categories, with similarities to human AD (Fig. 2c, d). Therefore, we decided that additional replicates were not necessary.
Peak calling and signal coverage tracks for ChIP-seq data
For each histone modification in each condition, mapped reads were pooled across ChIP-seq replicates and regions of enrichment (peaks) were identified for the pooled ChIP-seq dataset relative to the pooled control using the MACS2 peak caller (version 2.0.10.20130712)35 (https://github.com/taoliu/MACS/) using a relaxed P value of 0.01. For each histone modification, overlapping peaks (at least 1 bp overlap) were merged across all conditions to obtain a non-redundant master list of regions of enrichment. Master lists of broad domains of enrichment for the diffused marks H3K27me3, H3K9me3, H3K36me3 and H4K20me1 were obtained by allowing merging peaks across conditions that were within 1kb of each other. Genome-wide signal coverage tracks representing per-base fold enrichment and the likelihood ratio of ChIP relative to control were also computed using MACS2.
Learning combinatorial chromatin states
We used ChromHMM to learn combinatorial chromatin states jointly across all four conditions36. ChromHMM was trained using all seven chromatin marks in virtual concatenation mode across all conditions. Reads from replicate datasets were pooled before learning states. The ChromHMM parameters used are as follows: reads were shifted in the 5′ to 3′ direction by 100 bp; for each ChIP-seq dataset, read counts were computed in non-overlapping 200 bp bins across the entire genome; each bin was discretized into two levels, 1 indicating enrichment and 0 indicating no enrichment. The binarization was performed by comparing ChIP-seq read counts to corresponding whole-cell extract control read counts within each bin and using a Poisson P value threshold of 1 × 10−4 (the default discretization threshold in ChromHMM). We trained several models with the number of states ranging from 12 states to 23 states. We decided to use a 14 state model for all further analyses since it captured all the key interactions between the chromatin marks and larger number of states did not capture significantly new interactions. To assign to assign biologically meaningful mnemonics to the states, we used the ChromHMM package to compute the overlap and neighborhood enrichments of each state relative to coordinates of known gene annotations. The trained model was then used to compute the posterior probability of each state for each genomic bin in each condition. The regions were labeled using the state with the maximum posterior probability. The chromatin state models and browser tracks can be downloaded from http://www.broadinstitute.org/~anshul/projects/liz/segmentation/results/S14/webpage_14.html
Differential analysis and visualization
We used the DEseq2 method that models read count statistics from replicates across multiple conditions to identify differentially expressed genes and regions of enrichment of histone marks37. Our procedures are consistent with the standards for ChIP-seq and RNA-seq analysis determined by rigorous benchmarking as a part of the ENCODE project33. The minimal recommended depth for sufficient sensitivity of peak detection for histone marks for the human or mouse genome is ~20 million mapped reads33. However, due to limited amount of starting material obtained from a single mouse, we obtained ~10 million unique mapped reads from each biological replicate. Directly, using read counts from the original replicates would result in significant loss of power to detect differential events. To improve to improve sensitivity, for each histone mark in each condition, we pooled mapped reads from all replicates and created a pair of pseudo-replicates with equal number of reads (~30M) by randomly subsampling (without replacement) from the pool. Reads were then extended to the predominant fragment length. Extended-read counts were computed within all regions in the master peak list of a histone mark for all pseudo-replicates in all conditions and the table of counts was used as input to DEseq2. The raw data is available online (NCBI GEO GSE65159).
For RNA-seq data, the number of reads overlapping ENSEMBL gene models38 were determined by HT-Seq (http://www-huber.embl.de/users/anders/HTSeq/). The raw data is available online (NCBI GEO GSE65159). To ensure that the genes we chose were sufficiently quantifiable, we remove every gene where fewer than 20 reads were found across all samples. The resulting set of genes is found in Supplementary Table 1.
IGV39 is used to visualize the histone marks, gene expression, chromatin state, and AD GWAS data relative to the RefSeq gene model. Gene expression levels shown are raw read density. Levels of histone marks plotted are the log-likelihood ratio of ChIP signal relative to whole-cell extract control.
Within the DEseq2 framework of generalized linear models, we used a combination of different models to determine the significantly regulated genes and significantly regulated histone mark levels. We compared the set of all 2 week and 6 week controls to the three following groups: (1) the 2 week CK-p25 samples; (2) the 6 week CK-p25 samples; (3) a group containing both the 2 week and 6 week samples. The first two tests identified changes that might be 2-week or 6-week specific. The third test identified changes that might be too subtle to detect at any one time point alone. In each case, the most basic equation (count ~ CKp25 status) was used, but for a subset of samples. A stringent threshold of q < 0.01 (Benjamini Hochberg) was used to determine significantly changing genes expression levels and histone mark levels. Next, to determine the temporal bias of genes expression levels and histone marks we built another model (count ~ time), which compared the 2 week and 6 week CK-p25 samples. Levels considered likely to change (q<0.5) were categorizes as transient (2 week bias) or late-stage (6 week bias). The results of the RNA-seq analysis are found in Supplementary Table 1, while the results of the histone mark analysis are in Supplementary Table 2.
For the histone modifications, we defined promoters using H3K4me3 peaks labeled with the promoter state annotation under any of the conditions (CK-p25/control and 2 weeks/6 weeks). We define enhancers based on peaks of H3K27ac labeled by the enhancer chromatin state. We define Polycomb-repressed regions based on peaks H3K27me3 labeled by the Polycomb-repressed chromatin state. Our definitions are consistent with known roles of these histone modifications40. Defining the boundaries of the regulatory regions using the peaks of the relevant histone modifications, and not the chromatin states, maximizes our power to detect changes in histone mark levels.
Pathway and Gene Ontology analysis for the gene expression data were then generated through the use of DAVID41,42. We present the most significant biological process Gene Ontology category result as well as a subset of non-redundant less significant categories that still pass our threshold significant (q < 0.01). For the regulatory regions, GREAT (with default parameters) was used to find the fold enrichment in the same Gene Ontology categories43.
Statistical framework for comparing CK-p25 changing genes and regulatory regions to other data sets
A common theme throughout the analysis is the characterization of regulatory regions that change in the CK-p25 mouse model. The most stringent control for this characterization are genes or regions of the same type that do not change in CK-p25. Due to the six categories of direction (increasing, decreasing) and temporal pattern (transient, consistent, late-stage), we chose a discrete statistical framework as opposed to trying to define a ranking across these different conditions. To measure the overlap between these discrete categories and other discrete data sets, we could use either a hypergeometric P value or a binomial P value. For every test in the material described below, we computed both significance values and obtained consistent results, with only minor differences in exact P value. In general, we chose the hypergeometric test, which is the most direct to look at overlap of annotated regions. As opposed to the overlap of the CK-p25 mouse categories with other ChIP-seq peaks, the overlap with transcription factor binding site motifs or SNPs can be thought of as sampling with replacement, which lends itself to the binomial P value.
Comparison of histone marks and gene expression
As described above, DESeq2 was used determine the log fold change in expression at 2 and 6 weeks in CK-p25 mice relative to control. Each enhancer and promoter was mapped to the closest ENSEMBL gene model based on distance to transcription start. For each category of histone mark direction and temporal pattern, we examined the enrichment of each category of CK-p25 gene expression change relative to unchanging genes. The significance of the enrichment is calculated using a hypergeometric test.
Identification of orthologous human regions
The promoter (H3K4me3 peaks annotated as transcription start site by chromatin state), enhancer (H3K27ac peaks annotated as enhancer by chromatin state), and Polycomb-repressed regions (H3K27me3 peaks annotated as Polycomb-repressed by chromatin state) were mapped to the human genome. BED files representing the coordinates of these peaks in mm9 were mapped to mm10 using liftover44. Those peaks were mapped compared to the human genome the UCSC multiple alignment chain files (http://hgdownload.soe.ucsc.edu/goldenPath/mm10/multiz60way/)45. More specifically, the alignments that overlap the mouse peak and include hg19 were extracted. We calculated the human mouse pairwise alignment for each multiple alignment using the “globalms” function of biopython (http://biopython.org/, version 1.59; python version 2.71). The highest scoring pairwise alignment formed base of the orthologous region in human. This region was extended on either side using lower scoring multiple alignments. The orthologous region in hg19 was required to be greater than 30 bp and no more than twice the length of the region in mouse. The mean conservation was examined using the PHASTCons score across placental mammals46 based on the same 60-way multiple sequence alignment. The mapped enhancer regions were annotated with their chromatin state in human hippocampus, and across all 127 cell types and tissues, using BEDTools47. The information from human tissues was collected according to protocols described in more detail in the companion publication as a part of the Roadmap Epigenomics project10 (http://www.roadmapepigenomics.org/). The protocols are approved by the NIH and no sequence information from identifiable subjects is provided.
Computational analysis of cell type proportion
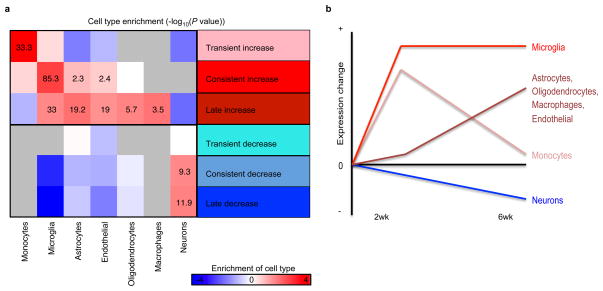
To computationally estimate the relative composition of the neural and immune cell types we compared the changing expression patterns in our dataset to a set of established cell-type specific markers48–50. This analysis shows that indeed it is likely that cell type composition is changing in the CK-p25 mouse model, consistent with a known decrease in number of neurons and astrogliosis at six weeks5. In summary, a transient enrichment of monocyte specific transcripts was observed at 2 weeks, a consistent enrichment of microglial-specific transcripts was enriched at 2 weeks and 6 weeks, while astrocyte, oligodendrocyte, and endothelial-specific markers were primarily increased at 6 weeks (Extended Data Fig. 9a, b). We could also detect a signature of neuronal loss, primarily at 6 weeks as well (Extended Data Fig. 9a, b). Based on these results alone, it is possible that changes in cell type composition are contributing to some of the differences we observe in our mouse model.
We also compared our data to a published study of microglial activation in another mouse model of AD8, to computationally dissect out changes likely due to cell type proportion versus changes due to activation within cells. If the changes in our mouse model were primarily due to cell type proportion, then the increase we observed in the CK-p25 mice should be proportional to the expression level of those genes microglia. If the changes we observed were primarily due to activation, then the changes we observe in the CK-p25 mouse should be proportional to the amount of activation found in during neurodegeneration8. Using the genes with published gene expression changes during activation8, we modeled these two possibilities as a linear regression problem and examined the relative significance of both hypotheses in the R programming language:
CK-p25 log fold change ~ Microlglial Expression + Microglial Activation log fold change
We found that the changes in the CK-p25 mice were significantly related to the changes in cell activation (P = 2.9 × 10−6) as well as the changes in cell type proportion (P = 2.7 × 10−4), suggesting that both cell activation and composition changes occur.
Comparison of gene expression in mouse model and human AD
To examine the relationship between AD in the mouse model and human, we mapped each 1-1 orthologous gene from mouse to human in ENSEMBL (http://www.biomart.org/) 51. For each category of expression change in mouse, we examined how that set of genes behaved in human AD cases relative to controls in whole hippocampus52 as well as laser capture microdissected hippocampal grey matter2. To make this comparison we first downloaded both datasets from GEO (GSE1297, GSE28146), applied a variance stabilization normalization, and then used limma53 to find the log fold change in expression of all cases relative to controls. For each category of mouse gene expression, we calculated a P value based on a t-test for the bias of genes to increase or decrease in human AD relative to control. Because the original study52 had more confounders due to changes in grey/white matter proportion, we focused our analysis on the 22 cases and 9 controls from the laser capture samples2.
Enrichment of cofactors and transcription factors
Peaks representing both neural15,16 and immune17–19 enhancers or transcription factor binding were used to annotate the H3K27ac enhancers and H3K4me3 promoters. We used a hypergeometric test to evaluate whether or not these external annotations were enriched in the set of increased-level or decreased-level enhancers relative to the enhancers whose levels do not change. This same procedure was used to look at the enrichment of the CK-p25 enhancer orthologs in Roadmap Epigenome data. In this case, only enhancers that map to human are taken to be the background.
The putative binding sites based on transcription factor binding site motifs were identified independent of conservation and have been previously published54. The transcription factor binding sites were further clustered based on similarity55. The least significant of two statistical test was used as a stringent measure of binding site enrichment. (1) The real transcription factor binding site motifs in the category of interest were compared to shuffled control motifs that preserved nucleotide content. (2) The real transcription factor binding site motifs in the category of interest were compared to the real motifs in enhancers that are stable in the CK-p25 mice. To estimate the significance for test (1), we use a binomial P value because the length distribution is different for changing regulator regions compared to unchanging. Then we estimate the probability of finding a site per base pair. To estimate the significance for test (2), we use hypergeometric test. After identifying significant transcription factor binding sites in categories or regulatory regions, we collapsed the results into clusters of almost identical motifs, representing families. The group members can be found in a companion manuscript10 as well as online (http://www.broadinstitute.org/~pouyak/motifs-table/).
Luciferase reporter assay
14 oligonucleotide gBlocks (IDT), ranging in 500–1000 nt in length, and corresponding to 10 enhancer regions were synthesized. Each gBlock contained a constant 5′ GCTAGCCTCGAGGAT and 3′ ATCAAGATCTGGCCT region, for direct cloning into an EcoRV (NEB) linearized minimal promoter firefly luciferase vector pGL4.23[luc2/minP] (Promega). The resulting reporter constructs were verified by DNA sequencing. BV-2 cells were kindly provided by Dr. Bruce Yankner. N2a cells were purchased from the American Type Culture Collection and maintained following their protocols. Briefly, cells were grown in RPMI-1640 and DMEM respectively, supplemented with 10% fetal bovine serum (FBS) and 1% pen/strep, and split 1:10 every 3 days. Cells were seeded into 24-well plates 1 day before transfection. Transfections into BV-2 and N2a cells were performed with 1 μg of a pGL4.23 plasmid and 200 ng of Renilla luciferase construct pGL4.74[Rluc/TK] (Promega). Luciferase activities were measured 24 h post-transfection using the Dual-Glo Luciferase Assay (Promega) and an EnVision 2103 Multilabel Plate Reader (PerkinElmer) and normalized to Renilla luciferase activity. All assays were performed in triplicate.
Microglia isolation
2 week induced CK-p25 mice and age-matched controls were perfused with 50 mls PBS to wash away blood and minimize macrophage contamination in the brains. Hippocampal tissue was harvested immediately after perfusion and a single-cell suspension was prepared as described previously56. Fluorescence-activated cell sorting (FACS) was then used to purify CD11b+ CD45low microglia cells using APC conjugated CD11b mouse clone M1/70.15.11.5 (Miltenyi Biotec, #130-098-088) and PE conjugated CD45 antibody (BD Pharmingen, #553081). Cells were collected directly into RNA lysis buffer (Qiagen, #74104).
cDNA synthesis and qPCR
Total RNA was extracted using the RNeasy Mini kit (Qiagen, #74104) according to manufacturer’s instructions. RNA concentration and purity was determined using Agilent’s Bioanalyzer and reverse transcribed using iScript cDNA Synthesis Kit (Biorad, #170-8891). For gene expression analysis cDNA from three biological replicates was quantitatively amplified on a thermal cycler (BioRad) using SYBR green (Biorad) and gene-specific primers (Supplementary Table 8). The comparative Ct method57 was used to examine differences in gene expression. Values were normalized to expression levels of Cd11b (also known as Itgam). Three technical replicates were used for each gene.
eQTL Analysis
The human orthologous regions to mouse enhancers that change in the CK-p25 mouse were compared to control for the their enrichment to overlap regulatory SNPs from published eQTL studies in immune cell types under a variety of conditions25,26. Because the eQTLs were processed separately, we applied our own threshold (P < 1 × 10−4). We then calculated enrichment of human orthologs of different categories CK-p25 enhancers relative to stable regions and used a binomial P value to estimate the significance.
Enrichment of AD GWAS SNPs in Roadmap enhancers
The enrichment of AD GWAS SNPs that map to Roadmap enhancer regions is calculated based on permutations of SNPs. Briefly, SNPs were permuted 1,000,000 times preserving distance to gene, minor allele frequency, and a number of SNPs in LD. The thousand genomes projects database was used as the reference for this information.
Comparison of regulatory regions to AD meta-analysis
The enrichment of CK-p25 human enhancer orthologs in AD was calculated by comparing the number changing regions that overlap SNPs4 to unchanging regions that overlap SNPs. We calculate the significance using a binomial P value, where the probability of success in the changing enhancers is based on the frequency in the unchanging enhancers. The results for the consistently increasing enhancers were slightly more significant when using a hypergeometric test instead of the binomial. To test whether the enrichment of increasing enhancer orthologous regions was due to the overlap with CD14+ cell enhancers, we repeated the above enrichment procedure within the set of CK-p25 enhancer orthologs that also overlap CD14+ cell enhancers. The enrichment using this control was still significant (3.0-fold enrichment, binomial P = 1.3 × 10−5). AD GWAS SNPs that were in a mouse enhancer orthologs were expanded using an LD of 0.8 and then tested for potential coding SNPs58 or eQTLs (Supplementary Table 7).
Extended Data
Extended Data Figure 1. Epigenomic and transcriptomic profiling of a mouse model of AD.
a, Experimental design and progression pathology in the CK-p25 mice. b, Gene expression and histone modification levels at the SPI1 locus at 6 weeks of inducible p25 overexpression. Profiled are histone marks associated with repression (blue); histone marks associated with enhancers (orange); histone marks associated with promoters (red); histone marks associated with gene bodies (green); RNA-seq (black).
Extended Data Figure 2. Differential microglia-specific gene expression changes in the CK-p25 mice.
RT-qPCR of selected microglia markers and immune response genes shows upregulation of gene expression in FAC-sorted CD11b+ CD45Low microglia from 2 week induced CK-p25 mice (red bars) relative to respective controls (black bars). Actb (b-actin) was used as a negative control. Values were normalized to Cd11b expression (n=3, *P < 0.05, two-tailed t-test); NS, non-significant.
Extended Data Figure 3. Chromatin state conservation.
a, Combinatorial patterns of seven histone modifications profiled were used to define promoter (1-3; A, active; D, downstream; U, upstream), gene body (4-6; tx, transcribed, 3P, 3 prime), enhancer (7-9; G, genic, 1 = strong, 2 = weak), bivalent (10), repressed Polycomb (11), heterochromatin (12), and low signal (13-14) chromatin states. Darker blue indicates a higher enrichment of the measured histone mark (x axis) to be found in a particular state (y-axis). b, Promoter, enhancer, and repressed chromatin states in mouse hippocampus (rows), as profiled in this study, align to matching chromatin states in human (columns), as profiled by the Roadmap Epigenomics Consortium10. Shading indicates enrichment relative to human chromatin state abundance (columns). The number of regions overlapping is shown in each cell of the heatmap.
Extended Data Figure 4. Differential gene expression and histone mark levels at regulatory regions in CK-p25 mice.
a–e, Shown are six distinct classes of differentially modified regions: transient (early) increase (pink) or decrease (light blue), consistent increase (red) or decrease (blue), and late (6 wk) increase (dark red) or decrease (navy blue). The heatmap shows the log fold change relative to 2 week controls for a, gene expression; b, H3K4me3 peaks at “TSS (transcription start site)” chromatin state; c, H3K27ac peaks at enhancer chromatin state; d, H3K27me3 peaks overlapping the Polycomb repressed chromatin state; e, H3K9me3 peaks overlapping the heterochromatin chromatin state. Numbers denote peaks falling into each category.
Extended Data Figure 5. Relationship between changes of gene expression and regulatory regions in CK-p25 mice.
For each class of gene expression change in the CK-p25 model (x axis), overlap with different histone modifications is shown (y axis) for a, H3K4me3 at promoters; b, H3K27ac at enhancers; c, H3K27me3 at Polycomb repressed regions. Histone modifications were mapped to the nearest transcription start site (Supplementary Table 3) to show the enrichment of the changing regulatory regions relative to those that are stable in CK-p25. The significance is calculated based on the hypergeometric P value of the overlap.
Extended Data Figure 6. Enrichment of immune cell eQTLs in increasing mouse enhancers.
Enrichment of eQTL SNP (y axis; −log10(binomial P < 10−4)) in monocytes and CD4+ (ref. 25,26) is compared to the orthologous regions of CK-p25 affected enhancers relative to enhancers whose levels do not change.
Extended Data Figure 7. Weak enrichment of AD GWAS SNPs at differential CK-p25 promoters.
Enrichment of AD-associated SNPs (y-axis, binomial P value) in human regions orthologous to different classes of mouse promoters
Extended Data Figure 8. Enrichment of tissue-specific enhancer annotations from the Roadmap Epigenomics Consortium for AD-associated SNPs and mouse enhancers.
Enrichment of AD-associated SNPs (y-axis, permutation P value) in tissue-specific enhancer annotations from the Roadmap Epigenomics Consortium (points), relative to their enrichment for a, increased-level and b, decreased-level (colors of different classes along y-axis) of orthologous enhancer regions in the mouse AD model (x-axis, hypergeometric P value). Linear regression trend line and R2, based on Pearson correlation is shown.
Extended Data Figure 9. Cell type composition.
a, For each class of gene expression change (x-axis), shown is the enrichment of cell-type-specific gene markers from published datasets48–50. The macrophage and monocyte categories are computed relative to microglia49,50. The enrichment is calculated relative to the genes that do not change in expression level in the CK-p25 mice. Cells in the heatmap are labeled based on the −log10(P value) (hypergeometric t-test). Cases where no genes overlapped are shown in grey. b, Summary of a, showing the inferred change in cell type composition across time.
Supplementary Material
For each gene with at least 20 reads in one sample, provided is the basic gene annotations used and which category of expression change the gene falls in. Also provided is the log fold change and false discovery rate adjusted P value (when applicable) for the statistical tests used to identify significant genes and their profile. These include all CK-p25 samples compared to control, two week CK-p25 compared to control, six week CK-p25 compared to control, and two week CK-p25 compared to six week CK-p25.
For each category of gene expression change, shown is the significant gene biological process gene ontology categories (FDR < 0.01) and the relevant statistics for the enrichment, including fold enrichment, P value, and false discovery rate.
Annotation information is provided for each H3K4me3 peak at an enhancer (H3K4me3_tss), each H3K27ac peak at an enhancer (H3K27ac_enh), and each H3K27me3 peak at a Polycomb repressed regions (H3K27me3_narrow_pcm). First, mouse annotation is shown, which includes location (inter=intergenic) as well as the the ensembl id, symbol, and distances of the closest transcription start site. Also shown is the log fold change (lfc) and adjusted P value (adjp), for the two week CK-p25 mice relative to control (2Wk), the six week CK-p25 mice relative to control (6Wk), both groups of CK-p25 together (CKp25), and the six week CK-p25 mice compared to the two week CK-p25 mice. The corresponding coordinates in human with the percentage of the mouse region that was able to be mapped to human and mean placental mammal conservation score is also shown. Cases where the region could not be mapped to human are indicated by “NA”.
Every row contains a roadmap cell type or tissue, which is annotated by its enrichment for AD GWAS SNPs based on a permutation test70 or based on its enrichment in the human regions orthologous to mouse enhancers whose level does not change in CK-p25. “inc” denoted increasing CK-p25 mouse enhancers while “dec” denotes a decreasing enhancer category.
Listed are chromosome (Chr), starting and ending location coordinates, length, and overlap with DNase hypersensitivity sites (DHS), for each increasing putative enhancer region fragment tested. Upregulation (+) and no change (−) in enhancer activity relative to control (minimal promoter only) construct is denoted in BV-2 microglia. The effect of each (rsid designated) SNP on the activity of the respective enhancer fragment is denoted by Up, Down or No change, relative to the activity of the reference enhancer fragment sequence. ns, non-significant.
Every row contains a SNP/gene pair along with P value that was annotated as an eQTL in the literature40,41 and overlaps the human region orthologous to a mouse enhancer that increases consistently in the CK-p25 model. SNPs that implicated in the AD GWAS meta analysis6 are also highlighted.
List of all AD GWAS SNPs6 that overlap the orthologs of regulatory regions that increase in the CK-p25 mouse model. Certain features of the mouse regulatory regions including the category of change temporal dynamics (category), the ensembl ID of gene with the closest transcription start site (closest tss), and its symbol (symbol), are annotated. Also provided are basic features of the mapping of this feature to human, including the genomic coordinates and the percentage of the region that mapped from mouse to human. For the human SNP, annotated are the position, the distance to the nearest protein coding gene, the gene’s ENSEMBL ID, and a description of where the SNP lies (location). Haploreg71 was used to find SNPs in LD that lie in protein coding regions and is presented in the following format: linkage disequilibrium, gene symbol, type of mutation. Missense variants are highlighted in red. Each SNP is annotated based on Roadmap enhancers in CD14 immune cells, the adult hippocampus, and male fetal brain.
Acknowledgments
We thank Alison Mungenast for critical reading and editing of the manuscript and invaluable discussions about the project, Mali Taylor for mouse colony maintenance, and Xiaolan Zhang, Robbyn Issner, Holly Whitton and Charles Epstein, for technical assistance with ChIP-seq library preparation. We thank Pouya Kheradpour for the transcription factor binding site motif scan of the mouse genome. This work was partially supported by the Belfer Neurodegeneration Consortium funding and NIH/NINDS/NIA (RO1NS078839) to L-HT, Early Postdoc Mobility fellowship from the Swiss National Science Foundation (P2BSP3_151885) to HM, and NIH/NHGRI (R01HG004037-07 and RC1HG005334) to MK.
Footnotes
Author Contributions
This study was designed by EG, ARP, AK, MK and L-HT, and directed and coordinated by MK and L-HT. EG initiated, planned and performed the experimental work. ARP performed computational analysis to characterize differential gene expression and histone mark levels, identify orthologous human regions and enriched transcription factor binding sites, and compare regulatory regions to human AD meta-analysis data. AK contributed to the computational analysis by generating mouse chromatin states and the quantification and control of ChIP datasets. HM helped with isolation and gene expression analysis of specific cell type populations. GQ performed permutation test comparing human Roadmap enhancers to AD GWAS SNPs. The manuscript was written by EG, ARP, L-HT and MK, and commented on by all authors.
All data are available from the NCBI GEO database under accession number GSE65159, the NIH Roadmap (http://www.roadmapepigenomics.org/data) and NCBI Epigenomics portal (http://www.ncbi.nlm.nih.gov/epigenomics).
The authors declare no competing financial interests.
References
- 1.Alzheimer’s_Association. 2014 Alzheimer’s disease facts figures. Alzheimers Dement. 2014;10 (2):e47–92. doi: 10.1016/j.jalz.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Blalock EM, Buechel HM, Popovic J, Geddes JW, Landfield PW. Microarray analyses of laser-captured hippocampus reveal distinct gray and white matter signatures associated with incipient Alzheimer’s disease. J Chem Neuroanat. 2011;42 (2):118–126. doi: 10.1016/j.jchemneu.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang B, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153 (3):707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013 doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruz JC, Tseng HC, Goldman JA, Shih H, Tsai LH. Aberrant Cdk5 activation by p25 triggers pathological events leading to neurodegeneration and neurofibrillary tangles. Neuron. 2003;40 (3):471–483. doi: 10.1016/s0896-6273(03)00627-5. [DOI] [PubMed] [Google Scholar]
- 6.Fischer A, Sananbenesi F, Pang PT, Lu B, Tsai LH. Opposing roles of transient and prolonged expression of p25 in synaptic plasticity and hippocampus-dependent memory. Neuron. 2005;48 (5):825–838. doi: 10.1016/j.neuron.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 7.Cruz JC, et al. p25/cyclin-dependent kinase 5 induces production and intraneuronal accumulation of amyloid beta in vivo. Journal of Neuroscience. 2006;26 (41):10536–10541. doi: 10.1523/JNEUROSCI.3133-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orre M, et al. Isolation of glia from Alzheimer’s mice reveals inflammation and dysfunction. Neurobiol Aging. 2014;35 (12):2746–2760. doi: 10.1016/j.neurobiolaging.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489 (7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng Y, et al. Principles of regulatory information conservation between mouse and human. Nature. 2014;515 (7527):371–375. doi: 10.1038/nature13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallant S, Gilkeson G. ETS transcription factors and regulation of immunity. Arch Immunol Ther Exp (Warsz) 2006;54 (3):149–163. doi: 10.1007/s00005-006-0017-z. [DOI] [PubMed] [Google Scholar]
- 13.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107 (50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aruga J. The role of Zic genes in neural development. Mol Cell Neurosci. 2004;26 (2):205–221. doi: 10.1016/j.mcn.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Visel A, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152 (4):895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465 (7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.May G, et al. Dynamic analysis of gene expression and genome-wide transcription factor binding during lineage specification of multipotent progenitors. Cell Stem Cell. 2013;13 (6):754–768. doi: 10.1016/j.stem.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38 (4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crotti A, et al. Mutant Huntingtin promotes autonomous microglia activation via myeloid lineage-determining factors. Nat Neurosci. 2014;17 (4):513–521. doi: 10.1038/nn.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prinz M, Priller J. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci. 2014;15 (5):300–312. doi: 10.1038/nrn3722. [DOI] [PubMed] [Google Scholar]
- 21.Gomez-Nicola D, Fransen NL, Suzzi S, Perry VH. Regulation of microglial proliferation during chronic neurodegeneration. J Neurosci. 2013;33 (6):2481–2493. doi: 10.1523/JNEUROSCI.4440-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jantaratnotai N, et al. Upregulation and expression patterns of the angiogenic transcription factor ets-1 in Alzheimer’s disease brain. J Alzheimers Dis. 2013;37 (2):367–377. doi: 10.3233/JAD-122191. [DOI] [PubMed] [Google Scholar]
- 23.Lyons MR, West AE. Mechanisms of specificity in neuronal activity-regulated gene transcription. Prog Neurobiol. 2011;94 (3):259–295. doi: 10.1016/j.pneurobio.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed-Geaghan EG, Reed QW, Cramer PE, Landreth GE. Deletion of CD14 attenuates Alzheimer’s disease pathology by influencing the brain’s inflammatory milieu. J Neurosci. 2010;30 (46):15369–15373. doi: 10.1523/JNEUROSCI.2637-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fairfax BP, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343 (6175):1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raj T, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344 (6183):519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stern Y. Cognitive reserve in ageing and Alzheimer’s disease. Lancet Neurol. 2012;11 (11):1006–1012. doi: 10.1016/S1474-4422(12)70191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam PY, Yoo SK, Green JM, Huttenlocher A. The SH2-domain-containing inositol 5-phosphatase (SHIP) limits the motility of neutrophils and their recruitment to wounds in zebrafish. J Cell Sci. 2012;125 (Pt 21):4973–4978. doi: 10.1242/jcs.106625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krimbou L, et al. Molecular interactions between apoE and ABCA1: impact on apoE lipidation. J Lipid Res. 2004;45 (5):839–848. doi: 10.1194/jlr.M300418-JLR200. [DOI] [PubMed] [Google Scholar]
- 30.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8 (11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473 (7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18 (11):1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22 (9):1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8 (9):1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9 (9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9 (3):215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11 (10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flicek P, et al. Ensembl 2013. Nucleic Acids Res. 2013;41 (Database issue):D48–55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14 (2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffman MM, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41 (2):827–841. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4 (1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 42.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37 (1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28 (5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hinrichs AS, et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34 (Database issue):D590–598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanchette M, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14 (4):708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15 (8):1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26 (6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34 (36):11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hickman SE, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16 (12):1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Butovsky O, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2013;17 (1):131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vilella AJ, et al. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19 (2):327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blalock EM, et al. Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A. 2004;101 (7):2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21 (9):2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 54.Lindblad-Toh K, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478 (7370):476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kheradpour P, Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014;42 (5):2976–2987. doi: 10.1093/nar/gkt1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guez-Barber D, et al. FACS purification of immunolabeled cell types from adult rat brain. J Neurosci Methods. 2012;203 (1):10–18. doi: 10.1016/j.jneumeth.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25 (4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 58.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40 (Database issue):D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For each gene with at least 20 reads in one sample, provided is the basic gene annotations used and which category of expression change the gene falls in. Also provided is the log fold change and false discovery rate adjusted P value (when applicable) for the statistical tests used to identify significant genes and their profile. These include all CK-p25 samples compared to control, two week CK-p25 compared to control, six week CK-p25 compared to control, and two week CK-p25 compared to six week CK-p25.
For each category of gene expression change, shown is the significant gene biological process gene ontology categories (FDR < 0.01) and the relevant statistics for the enrichment, including fold enrichment, P value, and false discovery rate.
Annotation information is provided for each H3K4me3 peak at an enhancer (H3K4me3_tss), each H3K27ac peak at an enhancer (H3K27ac_enh), and each H3K27me3 peak at a Polycomb repressed regions (H3K27me3_narrow_pcm). First, mouse annotation is shown, which includes location (inter=intergenic) as well as the the ensembl id, symbol, and distances of the closest transcription start site. Also shown is the log fold change (lfc) and adjusted P value (adjp), for the two week CK-p25 mice relative to control (2Wk), the six week CK-p25 mice relative to control (6Wk), both groups of CK-p25 together (CKp25), and the six week CK-p25 mice compared to the two week CK-p25 mice. The corresponding coordinates in human with the percentage of the mouse region that was able to be mapped to human and mean placental mammal conservation score is also shown. Cases where the region could not be mapped to human are indicated by “NA”.
Every row contains a roadmap cell type or tissue, which is annotated by its enrichment for AD GWAS SNPs based on a permutation test70 or based on its enrichment in the human regions orthologous to mouse enhancers whose level does not change in CK-p25. “inc” denoted increasing CK-p25 mouse enhancers while “dec” denotes a decreasing enhancer category.
Listed are chromosome (Chr), starting and ending location coordinates, length, and overlap with DNase hypersensitivity sites (DHS), for each increasing putative enhancer region fragment tested. Upregulation (+) and no change (−) in enhancer activity relative to control (minimal promoter only) construct is denoted in BV-2 microglia. The effect of each (rsid designated) SNP on the activity of the respective enhancer fragment is denoted by Up, Down or No change, relative to the activity of the reference enhancer fragment sequence. ns, non-significant.
Every row contains a SNP/gene pair along with P value that was annotated as an eQTL in the literature40,41 and overlaps the human region orthologous to a mouse enhancer that increases consistently in the CK-p25 model. SNPs that implicated in the AD GWAS meta analysis6 are also highlighted.
List of all AD GWAS SNPs6 that overlap the orthologs of regulatory regions that increase in the CK-p25 mouse model. Certain features of the mouse regulatory regions including the category of change temporal dynamics (category), the ensembl ID of gene with the closest transcription start site (closest tss), and its symbol (symbol), are annotated. Also provided are basic features of the mapping of this feature to human, including the genomic coordinates and the percentage of the region that mapped from mouse to human. For the human SNP, annotated are the position, the distance to the nearest protein coding gene, the gene’s ENSEMBL ID, and a description of where the SNP lies (location). Haploreg71 was used to find SNPs in LD that lie in protein coding regions and is presented in the following format: linkage disequilibrium, gene symbol, type of mutation. Missense variants are highlighted in red. Each SNP is annotated based on Roadmap enhancers in CD14 immune cells, the adult hippocampus, and male fetal brain.