Abstract
Although HLA-A3 and A11 have been reported to be ligands for KIR3DL2, evidences for in vivo relevance of this interaction is still missing. To explore the functional importance of KIR3DL2 allelic variation, we analyzed the autoimmune disease pemphigus foliaceus, known to be negatively associated with activating KIR genes. The frequency of KIR3DL2*001 was increased in patients (OR=2.04, p=0.007). The risk was higher for the presence of both KIR3DL2*001 and HLA-A3 or A11 (OR=3.76, p=0.013), providing the first evidence that HLA-A3 and A11 may interact with KIR3DL2 in vivo. The non-synonymous single nucleotide polymorphism 1190T (rs3745902) was associated with protection (OR=0.52, p=0.018). This SNP results in a threonine to methionine substitution. Individuals who have methionine in this position exhibit a lower percentage of KIR3DL2 positive cells and also lower intensity of KIR3DL2 on expressing cells; additionally, we show that the expression of KIR3DL2 is independent of other killer cell immunoglobulin-like receptors. Pemphigus foliaceus is a very unique complex disease strongly associated with immune-related genes. It is the only autoimmune disease known to be endemic, showing a strong correlation with environmental factors. Our data demonstrate that this relatively unknown autoimmune disease may facilitate understanding of the molecular mechanisms of KIR3DL2 ligand recognition.
Keywords: KIR, autoimmunity, allele polymorphism, ligands, expression, pemphigus foliaceus, MHC, natural killer cells
1. Introduction
Killer cell immunoglobulin-like receptors (KIR) are expressed on the surface of natural killer (NK) cells and T cells, regulating the balance of activating and inhibitory signals [1]. These receptors are important for immune defense [2] and influence placentation during pregnancy [3]. KIR genes are located on chromosome 19q13.42 within the leukocyte receptor complex [4],[5]. They vary in number (presence/absence polymorphism) and in allelic polymorphism. Two gene content haplogroups have been reported: haplogroup A is characterized by many inhibitory genes and only one short tailed activating gene (KIR2DS4) and is the most frequent in worldwide populations; haplogroup B shows high diversity of combinations of activating genes [6].
The framework KIR3DL2 is the longest KIR gene and spans 16,256bp. It is also one of the most polymorphic KIR, with 86 described alleles [7]. The KIR3DL2 receptor comprises a 140-kDa dimeric molecule with three extracellular domains and a long intracellular tail, carrying two immune receptor tyrosine-based inhibitory motifs (ITIM) [8]. A long cytoplasmic tail is normally a hallmark of an inhibitory KIR; in contrast, activating KIR generally exhibit a short tail and associate with the immune receptor tyrosine-based activating motif (ITAM)-containing signaling chain DAP12 [9],[10] . Thus, the nomenclature of this gene family is based on the number of extracellular domains and the size of the cytoplasmic tail; i.e. KIR3DL, three extracellular domains, long cytoplasmic tail [11].
In vitro studies have suggested that HLA-A3 and HLA-A11 are ligands of KIR3DL2 [12] but these interactions appear to be weak, and highly peptide dependent. Indeed, to date only one peptide (and variants thereof) has been reported to support A3 and A11 recognition by KIR3DL2. Moreover, the in vivo significance of this interaction is unclear, especially in light of the finding that this interaction does not lead to fully functional NK cells, in contrast to other inhibitory KIR/ligand pairs [13].
Although KIR polymorphism has been studied in many Brazilian populations [14]-[16], KIR3DL2 allele diversity has not been well characterized. In addition, there is little information about the role of KIR3DL2 allele polymorphism in diseases. Here, we analyzed the influence of KIR3DL2 alleles in an autoimmune disease cohort from Brazil. Pemphigus foliaceus (PF) is an autoimmune blistering disease of skin characterized by autoantibodies against desmoglein-1, a molecule important for cell adhesion [17]. Many genes, including HLA class II, have been reported to associate with differential susceptibility to PF [18]-[20]. Activating KIR genes are often associated with protection in infectious diseases and susceptibility to autoimmunity [21]. However, we recently showed that the presence of higher numbers and ratios of activating KIR genes protect from PF [22]. Pemphigus is endemic not only in Brazil, but also in Tunisia and Colombia and the disorder is sporadically seen around the world [23],[24]. PF is strongly related to environmental factors, possibly due to substances contained in the saliva of hematophagous insects or to infectious microorganisms that trigger the disease in susceptible individuals [25],[26]. This particularity of PF may explain why activating KIR has been associated with protection against the development of this disease.
Allelic polymorphism of inhibitory KIR may result in functional differences, shifting the balance of inhibitory and activating signals in NK cells. KIR3DL2 is highly polymorphic and present in virtually all haplotypes [6],[27]. In addition to the fact that all individuals carry this gene, KIR3DL2 is highly expressed on NK cells [28]. KIR3DL2 is, therefore, a strong candidate for disease association studies as some KIR3DL2 allotypes could present differential inhibitory effects and affect susceptibility to diseases. Moreover, we previously demonstrated that activating KIR protect against PF [22]. Therefore, we hypothesized that stronger inhibitory KIR3DL2 allotypes could confer risk to PF.
Here, we show that the allele KIR3DL2*001 and the single nucleotide polymorphism 1190T (rs3745902) are associated with differential susceptibility to pemphigus foliaceus. We present genetic epidemiological support for an in vivo interaction between KIR3DL2 and HLA-A3 and A11. Moreover we also show that the protective SNP 1190T marks KIR3DL2 differential expression levels suggesting the necessity for a threshold of inhibition for the development of PF.
2. Results and Discussion
2.1. KIR3DL2*001 increases the susceptibility to PF
KIR allelic polymorphism and its effect on disease outcome are not well characterized. KIR presence/absence polymorphism as well as combinations of KIR-HLA have been associated with several infectious and autoimmune diseases [21],[29]. In contrast to other autoimmune diseases in which KIR polymorphism has been associated, we have reported that activating KIR genes are protective against PF [22]. Here, we hypothesized that different KIR3DL2 allotypes could be greater inhibitory than others, what could contribute to shift the balance of activating and inhibitory signals on the NK cell surface. Based on previous results, allotypes that show greater inhibition could potentially confer risk to PF.
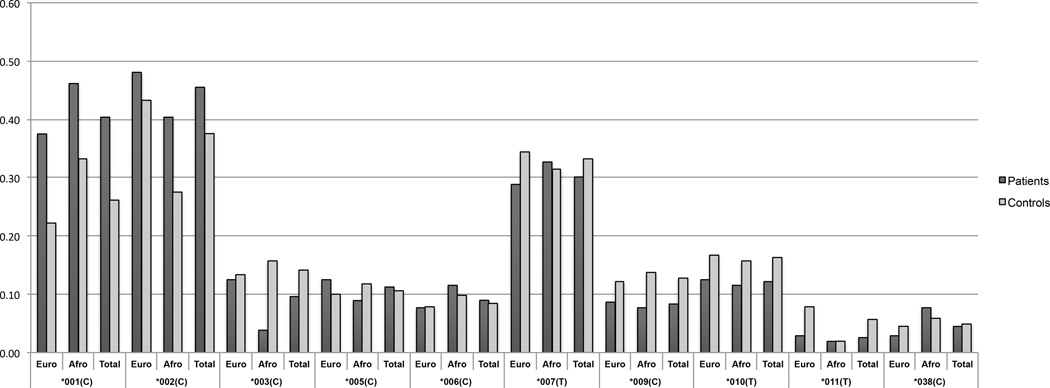
To test this hypothesis we sequenced KIR3DL2 in patients and controls. The frequencies of all alleles are shown in Supplementary Table S1 and frequencies of the most common alleles are shown in Figure 1. The allele KIR3DL2*001 was associated with increased susceptibility to PF for both carrier and allele frequencies in Euro-descendants (OR=2.1, p=0.015; OR=2.04, p=0.007 respectively) (Table 1). A statistically non-significant increase of KIR3DL2*001 was seen in the Afro-descendants. The risk was increased for homozygotes KIR3DL2*001/001 (OR=3.83; p=0.025), showing an additive effect. The HLA ligand specificity of KIR3DL2 remains unclear, although HLA-A*03 and A*11 tetramers have been shown to bind to KIR3DL2 when folded with specific EBV peptides [12]. The fact that the susceptibility was increased when we analyzed carriage of KIR3DL2*001 together with the presence of at least one copy of HLA-A3 or HLA-A11 (OR=3.76, p=0.013 – Table 2) suggests that these HLA-A molecules interact with KIR3DL2 in vivo. Furthermore, the presence of ligands without the receptor was not associated with PF (Table 2). Together with previous data showing that activating KIR genes protect from PF, these results suggest that KIR3DL2*001 may bind HLA-A3 and A11 in vivo and that this interaction stronger inhibitory receptor than other alleles. In addition, we tested the combination of KIR3DL2*001 with HLA-Bw6 (previously associated with increased susceptibility to PF [22]) or other common class I alleles. No additive effect was seen for KIR3DL2*001 + Bw6 (OR=1.89 p=0.02) or KIR3DL2*001 + other common HLA-A, B or C alleles (data not shown), corroborating the hypothesis that A3 and A11 may be functional ligands of KIR3DL2.
Figure 1. Carrier frequencies for each KIR3DL2 allele in patients and controls.
Only the most frequent alleles are shown. Euro= Euro-descendants; Afro= Afro-descendants; Total=total sample. (C) or (T)= presence of the variant 1190C or 1190T (rs3745902).
Table 1.
3DL2*001 and the SNP 1190T are associated to pemphigus foliaceus
| Patients | Controls | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P | A | F (%) | P | A | F(%) | p | OR | 95% CI | ||
| Allele frequencies | ||||||||||
| 001 | Euro | 46 | 160 | 22.3 | 22 | 156 | 12.4 | 0.007 | 2.04 | 1.17 – 3.54 |
| Afro | 29 | 75 | 27.9 | 18 | 81 | 18.2 | 0.070 | 1.74 | 0.89 – 3.39 | |
| Total | 75 | 235 | 24.2 | 40 | 237 | 14.4 | 0.002 | 1.89 | 1.24 – 2.89 | |
| Carrier frequencies | ||||||||||
| 001 | Euro | 39 | 65 | 37.5 | 20 | 70 | 22.2 | 0.015 | 2.10 | 1.11 – 3.97 |
| Afro | 24 | 28 | 46.2 | 17 | 34 | 33.3 | 0.129 | 1.71 | 0.77 – 3.81 | |
| Total | 63 | 93 | 40.4 | 37 | 104 | 26.2 | 0.010 | 1.90 | 1.16 – 3.11 | |
| Genotypes | ||||||||||
| 001/001 | Total | 12 | 144 | 7.7 | 3 | 138 | 2.1 | 0.025 | 3.83 | 1.06 – 13.87 |
| 001/other | Total | 51 | 105 | 32.7 | 34 | 107 | 24.1 | 0.087 | 1.55 | 0.93 – 2.60 |
| SNPs | ||||||||||
| 322A | Euro | 40 | 64 | 38.5 | 45 | 45 | 50.0 | 0.071 | 0.63 | 0.35 – 1.11 |
| Afro | 27 | 25 | 51.9 | 24 | 27 | 47.1 | 0.308 | 0.70 | 0.35 – 1.38 | |
| 337G | Euro | 29 | 75 | 27.9 | 24 | 66 | 26.7 | 0.490 | 1.06 | 0.56 – 2.00 |
| Afro | 9 | 43 | 17.3 | 15 | 36 | 29.4 | 0.146 | 0.50 | 0.20 – 1.28 | |
| 1190T | Euro | 40 | 64 | 38.5 | 49 | 44 | 54.4 | 0.018 | 0.52 | 0.29 – 0.93 |
| Afro | 23 | 29 | 44.2 | 21 | 30 | 41.2 | 0.752 | 1.13 | 0.52 – 2.47 | |
P=presence; A=absent; F=frequency, p=p-value; OR=odds ratio, CI=confidence interval. For simplification, KIR3DL2*001 is represented as 001. Bold highlights the significant associations.
Table 2.
Association of KIR3DL2 variants and HLA-A ligands with pemphigus foliaceus
| Patients | Controls | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P | A | F (%) | P | A | F(%) | p | OR | 95% CI | ||
| Carrier frequencies | ||||||||||
| HLA-A ligand | Euro | 47 | 59 | 44.3 | 23 | 46 | 33.3 | 0.146 | 1.59 | 0.85 – 2.99 |
| Afro | 23 | 35 | 39.7 | 13 | 21 | 38.2 | 0.887 | 1.06 | 0.44 – 2.53 | |
| Total | 70 | 94 | 42.7 | 36 | 67 | 35.0 | 0.210 | 1.36 | 0.83 – 2.31 | |
|
3DL2*001+ HLA-A ligand |
Euro | 18 | 61 | 22.8 | 4 | 51 | 7.3 | 0.017 | 3.76 | 1.20 – 11.82 |
| Afro | 6 | 29 | 17.1 | 3 | 25 | 10.7 | 0.360 | 1.72 | 0.39 – 7.61 | |
| Total | 24 | 90 | 21.1 | 7 | 76 | 8.4 | 0.016 | 2.89 | 1.18 – 7.09 | |
|
1190T+HLA-A ligand |
Euro | 10 | 69 | 12.7 | 8 | 48 | 14.3 | 0.488 | 0.87 | 0.32 – 2.36 |
| Afro | 5 | 30 | 14.3 | 3 | 25 | 10.7 | 0.569 | 1.19 | 0.26 – 5.44 | |
| Total | 15 | 99 | 13.2 | 11 | 73 | 13.1 | 0.582 | 1.00 | 0.44 – 2.32 | |
HLA-A ligand: presence of A3 and/or A11. P=presence; A=absent; F=frequency, p=p-value; OR=odds ratio, CI=confidence interval. For simplification, KIR3DL2*001 is represented as 001. Bold highlights the significant association
In addition to A3 and A11, KIR3DL2 has also been shown to recognize B27, a family of HLA alleles closely associated with ankylosing spondylitis and B27-associated-arthritides [30]-[32]. In our cohort, the frequency of HLA-B*27 is very low (f≤0.03 in patients and controls; Supplementary Table S2), and therefore, not informative. HLA-A*03, in contrast, is one of the most common HLA-A alleles (Supplementary Table S2) and combined with HLA-A*11, represents a ligand frequency higher than B*27 in the majority of worldwide populations, making these alleles the most likely primary KIR3DL2 in vivo ligands.
2.2. Cytoplasmic variant 1190T protects from PF
We next examined if individual SNPs, rather than alleles, were related to the susceptibility to PF. We excluded those SNPs present in low frequency or those that could be explained predominantly by a single allele. Three remaining SNPs were tested (Table 1), two in exon 3 (322G>A, rs654686; 337C>G, rs3188286) and one in exon 9 (1190C>T, rs3745902). The variant 1190T was negatively associated with PF in Euro-descendants (OR=0.52, p=0.018). No significant association for this variant was detected in Afro-descendants. As we reported before [22], the KIR relative effect in Euro-descendants is possibly higher than in Afro-descendants as a consequence of differences between ethnicities and the complexity of this disease.
The SNP 1190C>T causes an amino acid change (Thr376Met) in the long cytoplasmic tail. Although no major changes in mature protein are predicted for this replacement, the physicochemical properties of these two amino acids differ. Methionine is hydrophobic while threonine is polar due to the presence of a hydroxyl group. In the Grantham scale, which measures the physicochemical distance between all amino acid pairs and ranges from 5 to 215, the value for threonine and methionine is 81 [33]. Low values indicate conservative and high values indicate radical replacements. Even though it is an intermediate value, the frequency of amino acid replacements that show such difference is relatively low in mammalian proteins. Zhang [34], studying mammalian nuclear genes, estimated that only 17% of the transitions cause non-synonymous substitutions that alter the polarity of the amino acids.
Another more important characteristic that differs between these two amino acids is that threonine may be phosphorylated by protein kinases in eukaryotic cells. Although tyrosine phosphorylation is critical for both ITIM and ITAM function by facilitating the recruitment of the protein tyrosine phosphatases Src-homology domain containing phosphatase (SHP)-1 and -2, serine/threonine phosphorylation of KIR cytoplasmic domains can also play an important role in receptor expression and cycling, as has been reported for KIR3DL1 [35]. Importantly, Thr376 lies at position -1 relative to the tyrosine residue predicted to be critical within the KIR3DL2 ITIM. Moreover, experimental evidence supports a role for amino acids neighboring the ITIM tyrosine in controlling the ability of the receptor to interact with downstream molecules such as SHP-1. In particular, substitution at position -2 relative to the tyrosine in the ITIM can prevent its interaction with the protein phosphatases SHP-1 and SHP-2 [36]. Although to date position -1 has not been directly implicated in ITIM function, the Thr376Met it is certainly conceivable that this substitution may alter the receptor’s inhibitory function.
2.3. KIR3DL2 exhibits differential expression levels that correlate with 1190T
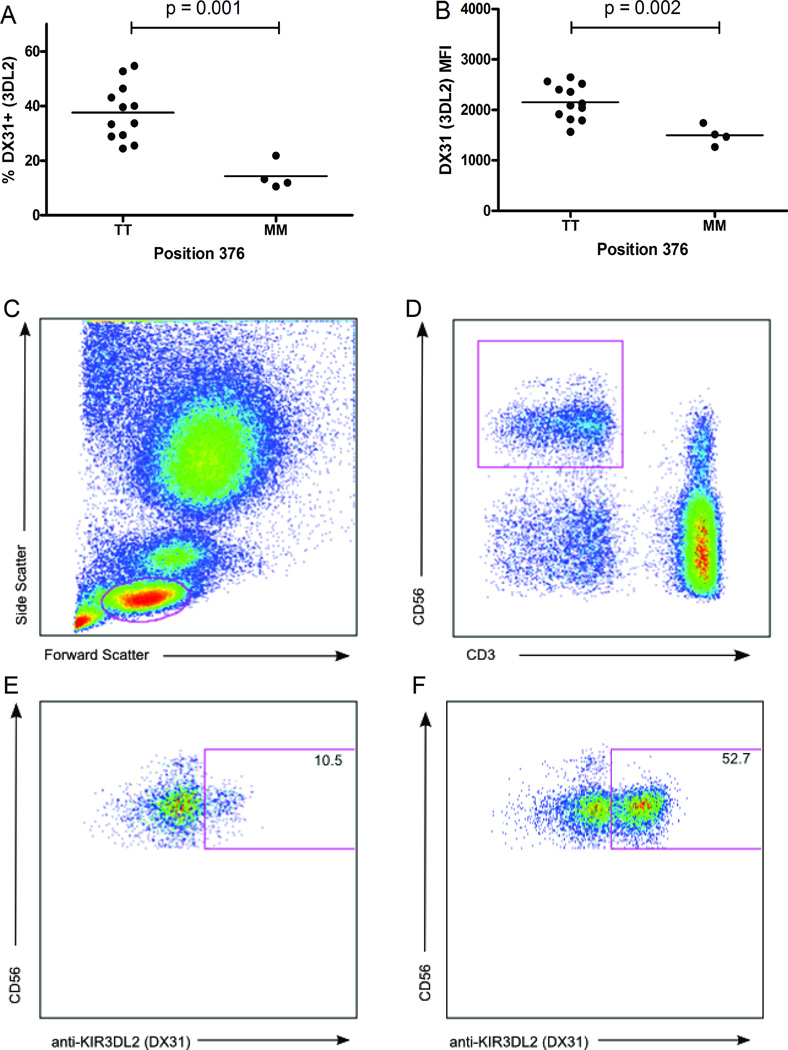
In addition to the inhibitory capacity of the ITIM, this motif has also been recently implicated in the endocytosis of KIR in its unphosphorylated form due to interactions with the adaptor protein AP-2 [37]. Thus, modulation of this motif may also influence KIR3DL2 function via control of receptor internalization and/or cell surface expression levels. We tested this possibility by examining if the 1190C>T SNP might correlate with KIR3DL2 expression. Direct examination of KIR3DL2 expression levels on the NK cells of healthy donors revealed that the percentage of KIR3DL2+ NK cells was 2.6 fold higher in 1190C homozygotes as compared donors homozygous for 1190T (Figure 2). In addition to the percentage of positive NK cells, the 1190C>T SNP also correlated with differential expression levels of KIR3DL2; MFIs of KIR3DL2+ NK cells in 1190T homozygotes was 1.4 fold lower than in 1190C homozygotes. Our data are the first to correlate differential expression of KIR3DL2 with a single nucleotide polymorphism and they reaffirm the functional significance of this SNP and corroborate our hypothesis. The lower expression frequency of Met376 allotypes reduces the size of the NK cell subset that is inhibited by KIR3DL2, and the lower level of receptor expression on positive cells would be predicted to reduce their inhibitory potential. Thus the NK cell population in 1190T individuals is biased towards lower inhibition in two ways, offering an attractive hypothesis as to why this SNP is negatively associated with PF.
Figure 2. Position 376 (1190C>T) marks KIR3DL2 expression.
Each dot represents an individual. Position 376 is codified by the SNP rs3745902; 1190T=methionine (M); 1190C=threonine (T). A) Percentage of KIR3DL2 (DX31) positive NK cells versus TT and MM genotypes; B) KIR3DL2 (DX31) median fluorescence intensity versus genotypes TT and MM. C) Forward and side scatter plot of peripheral blood mononuclear cells (PBMCs). The lymphocyte population is selected. D) Natural Killer (NK) cells are identified using CD56 and CD3 antibodies. NK positive cells are selected as CD56 positive and CD3 negative cells. Anti-KIR3DL2 (DX31) positive cells are identified with NK positive cells in a donor with E) low expression and F) high expression.
2.4. 1190T association is apparently independent of gene content haplotypes
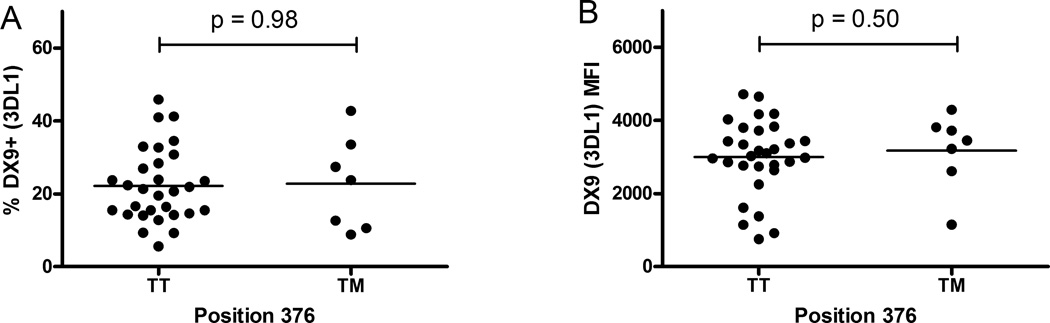
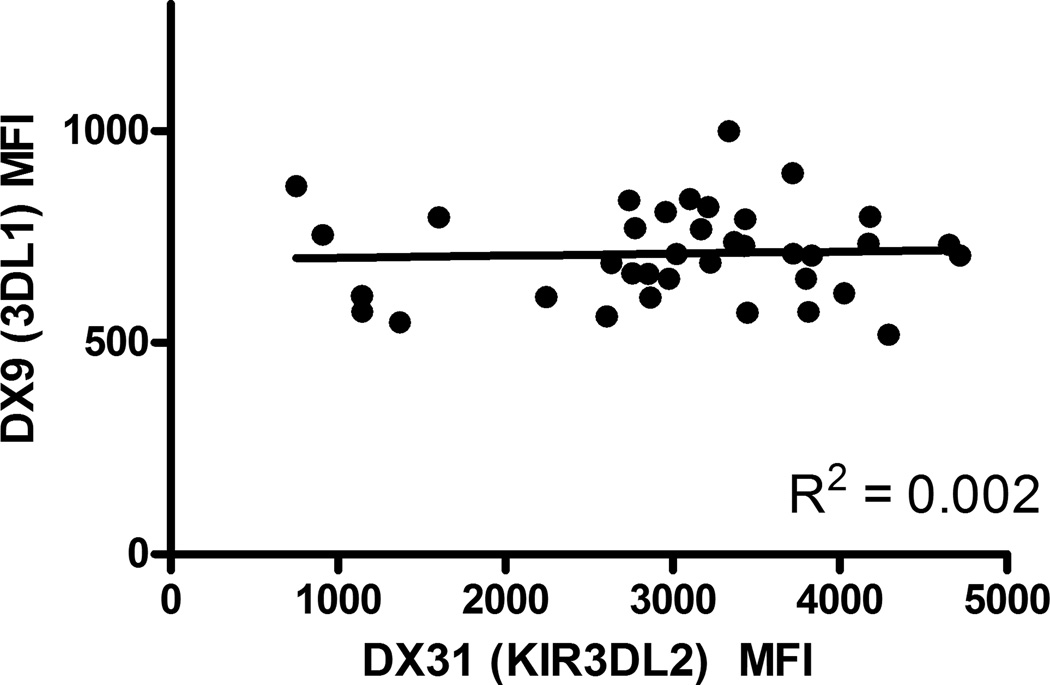
There is strong linkage disequilibrium (LD) between KIR3DL2 alleles and other genes [38],[39]. Presence of haplotype A has been shown to be associated with increased susceptibility to PF [22]. Thus we wondered whether haplotype A might exhibit overall higher KIR expression suggesting the LD between KIR3DL2 and other genes in the haplotype might be, in fact, responsible for the correlation between 1190T and differential expression levels. To answer this question, we analyzed KIR3DL1 expression in a subset of individuals in which KIR3DL2 expression was also measured. When we separated them based on amino acid position 376 caused by SNP 1190C>T genotypes, TT and TM had a very similar percentage of KIR3DL1+ cells (p=0.98) with similar levels of KIR3DL1 expression per cell (p=0.50) (Figure 3). We also did not see any correlation between KIR3DL1 and KIR3DL2 expression among these individuals (Figure 4). These results suggest the differential expression levels of KIR3DL2 are in fact independent of KIR3DL1. Therefore, we conclude that KIR haplotype gene content is not responsible for the differential expressions levels seen in KIR3DL2.
Figure 3. Position 376 (1190C>T) does not correlate with KIR3DL1 expression.
Each dot represents an individual. Position 376 is codified by the SNP rs3745902; 1190T=methionine (M); 1190C=threonine (T). Due the linkage disequilibrium between KIR genes and alleles, we could not find a representative number of MM/3DL1+ individuals. A) Percentage of KIR3DL1 (DX9) positive NK cells versus TT and TM genotypes; B) KIR3DL1 (DX9) median fluorescence intensity versus genotypes TT and TM.
Figure 4. KIR3DL2 expression does not correlate with KIR3DL1 expression.
KIR3DL1 (DX9) versus KIR3DL2 (DX31) median fluorescence intensity. Each dot represents an individual. KIR3DL1 and KIR3DL2 expression were measured in the same individuals.
2.5. KIR3DL2*001 and 1190T possibly have distinct effects on PF susceptibility
Interestingly, the concomitant presence of the variant 1190T and the ligands HLA-A3 and A11 has no effect on PF disease susceptibility (Table 2). Given the cytoplasmic location of this variant we would not expect it to affect binding affinities, rather our data show these KIR3DL2 alleles have lower expression levels perhaps reducing KIR3DL2 expression to levels too low to effectively function as an inhibitory receptor. In contrast, the odds ratio of KIR3DL2*001 combined with presence of A3/A11 (OR=3.76) is much stronger than the odds ratio of the presence of KIR3DL2*001 alone (OR=2.04). Therefore, we see a stronger (additive) susceptibility effect when we combined KIR3DL2*001+A3/A11. KIR3DL2*002 is the most frequent allele in our cohort and like KIR3DL2*001 does not carry the variant 1190T. The fact the highly expressed allele KIR3DL2*002 is not associated with PF (OR=1.21; p=0.507) tells us that there are likely two distinct factors contributing to susceptibility to PF: a) differential inhibition conferred by KIR3DL2*001 compared to other highly expressed alleles, and b) the differential levels of KIR3DL2 expression associated with 1190T.
To test this hypothesis, we performed regression analysis with stepwise selection. In our model, we included the presence of variants KIR3DL2*001, 1190T, the concomitant presence of KIR3DL2*001+ligand (A3 and/or A11) and the presence of homozygosity for haplotype A. The logistic regression analysis showed that both the presence of the variant 1190T (OR=0.46; p=0.02; log likelihood=182.268) and concomitant presence of KIR3DL2*001+ligand (OR=3.2; p=0.04 log likelihood=177.833) retained in the model and explained the result. Similar result was found when we tested a model with all KIR3DL2 alleles and variants (not shown). In total our data lead us to predict a model where PF associated peptides presented in the context of A3 or A11 are recognized well by KIR3DL2*001 promoting disease. KIR3DL2*002, in contrast, may only recognize these ligands weakly, while the poorly expressed 1190T alleles fail to effectively inhibit NK cells regardless of their binding affinities. The reason why KIR3DL2*001 and KIR3DL2*002 would exhibit degrees of function is unknown. However, it is worth noting that these two molecules differ by only one amino acid (Glu137Asp) and that nearby residues (138 and 140) have been shown to be HLA contact residues in the related receptor KIR3DL1 [40]. Considering the similarity of KIR3DL2 and KIR3DL1, and assuming these receptors bind similarly to their respective ligands, Glu137Asp would be expected to be very close to HLA binding region of KIR3DL2. Although Glu137Asp is a relatively conservative change between two negatively charged amino acids, its proximity to residues that may contact HLA could substantially change the ability of these KIR3DL2 allotypes to bind ligand. Alternatively, other characteristics of these allotypes may differ, such as alterations in receptor stability or folding. Formally testing these hypotheses will require identification of peptides recognized by KIR3DL2 in pemphigus foliaceus patients and functional assays to verify the impact of the substitution Glu137Asp on KIR3DL2 binding.
3. Conclusion
KIR3DL2*001 is associated with increased susceptibility to PF in a gene dose and ligand-dependent manner, suggesting that it may be a potent inhibitor as compared to the other KIR3DL2 alleles. To our knowledge, no other studies have revealed apparent interactions between KIR3DL2 and HLA-A3 and A11 in vivo. Moreover, we find that KIR3DL2 exhibits differential expression levels that correlate with the SNP 1190C>T. Lower expression of KIR3DL2 protects against PF possibly due to an overall decrease in inhibitory signals within NK cells. This effect is independent of KIR3DL1 and apparently independent of gene content haplotypes. This is the first study showing that allele specific KIR3DL2 differential expression levels are associated with disease. Unfortunately, allelic information is still lacking for the majority of KIR studies and these data are crucial in comprehending the role that these genes play in other diseases. Our data demonstrate that even complex diseases such as pemphigus can yield invaluable knowledge regarding KIR-dependent mechanisms that regulate immune responses.
4. Material and Methods
4.1. Samples
A total of 156 patients and 141 controls without history of the disease were analyzed in this present study. Patients were contacted mainly at Hospital Adventista do Pênfigo, Campo Grande, MS, Brazil, a specialized hospital located at the endemic area. All individuals voluntarily agreed to participate and written informed consent was obtained from all participants. In accordance with Brazilian Federal laws, this study was approved by the Human Research Ethics Committee of the Federal University of Parana and the CONEP (Comissão Nacional de Ética em Pesquisa). Because different populations may differ in allele frequencies, the individuals were separated according to their predominant ancestry: Euro-descendants (Euro n=104 patients and n=90 controls) and Afro-descendants (Afro, n=52 patients and n=51 controls). This approach has been validated by previous population genetic studies from our group, which showed that the distribution of alleles known to be restricted to populations autochthonous from one continent follows a gradient among the population strata, as expected if the classification discriminated the strata according to relative contributions of the ancestral populations [41],[42]. Euro-descendants and Afro-descendants were also analyzed as a single population sample when the frequencies between them did not differ statistically; total sample may give us a better representation of the whole population.
4.2. KIR3DL2 and HLA genotyping
All individuals were genotyped for presence of KIR3DL2 in a former study [22]. Here, we amplified exons 3, 4, 5, 7, 8 and 9 and also intron 7 using gene specific primers and the products were sequenced using the Big Dye terminator kit (Applied Biosystems). Specific PCR-SSP primers were designed to solve the 002/010 or 010/015, 001/007 or 006/010 ambiguities. All primer sequences are available on request. The HLA genotyping was performed using the LABType® SSO reagent kits (One Lambda, USA).
4.3. Statistical analyzes
Tests of population differentiation were performed by analyzes of 2×2 contingency tables, calculating the exact p-value by the metropolis algorithm. The p-value of 0.05 was adopted as the significance limit. The Mantel-Haenszel method [43] was applied for calculating the odds ratio (OR) and the 95% confidence intervals. Logistic regression analyzes with stepwise selection were carried out by IBM SPSS Statistics software.
4.4. Flow cytometry
KIR3DL2 and KIR3DL1 expression was assessed on a cohort of healthy, predominantly Caucasian, donors. Whole blood was stained with anti-CD3, anti-CD56 and anti-KIR antibodies (anti-3DL2 (DX31; L. Lanier, UCSF) and anti-3DL1 (DX9, BD Biosciences)). After red blood cell lysis, the cells were washed, fixed and analyzed using a BD LSRII flow cytometer using Diva software. Data was analyzed using FlowJo analysis software.
Supplementary Material
Acknowledgments
To CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) for a research fellowship. We thank all staff of Hospital Adventista do Penfigo for allowing us to work in this institution and all patients and controls who voluntarily agreed to participate of this work. This project was funded by CNPq, PRONEX, Institutos do Milênio, Fundação Araucária, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Parham P. Killer cell immunoglobulin-like receptor diversity: balancing signals in the natural killer cell response. Immunology Letters. 2004;92:11–13. doi: 10.1016/j.imlet.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 2.Parham P. Influence of KIR diversity on human immunity. Adv. Exp. Med. Biol. 2005;560:47–50. doi: 10.1007/0-387-24180-9_6. [DOI] [PubMed] [Google Scholar]
- 3.Trowsdale J, Moffett A. NK receptor interactions with MHC class I molecules in pregnancy. Semin. Immunol. 2008;20:317–320. doi: 10.1016/j.smim.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Wende H, Colonna M, Ziegler A, Volz A. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm. Genome. 1999;10:154–160. doi: 10.1007/s003359900961. [DOI] [PubMed] [Google Scholar]
- 5.Liu WR, Kim J, Nwankwo C, Ashworth LK, Arm JP. Genomic organization of the human leukocyte immunoglobulin-like receptors within the leukocyte receptor complex on chromosome 19q13.4. Immunogenetics. 2000;51:659–669. doi: 10.1007/s002510000183. [DOI] [PubMed] [Google Scholar]
- 6.Hsu KC, Liu X-R, Selvakumar A, Mickelson E, O'Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. The Journal of Immunology. 2002;169:5118–5129. doi: 10.4049/jimmunol.169.9.5118. [DOI] [PubMed] [Google Scholar]
- 7.Robinson J, Marsh SGE, Mistry K, McWilliam H, Lopez R. IPD--the Immuno Polymorphism Database. Nucleic Acids Research. 2010;38:D863–D869. doi: 10.1093/nar/gkp879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu. Rev. Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 9.Isakov N. ITAMs: immunoregulatory scaffolds that link immunoreceptors to their intracellular signaling pathways. Recept. Channels. 1998;5:243–253. [PubMed] [Google Scholar]
- 10.Long EO. Regulation of immune responses through inhibitory receptors. Annu. Rev. Immunol. 1999;17:875–904. doi: 10.1146/annurev.immunol.17.1.875. [DOI] [PubMed] [Google Scholar]
- 11.Long EO, Colonna M, Lanier LL. Inhibitory MHC class I receptors on NK and T cells: a standard nomenclature. Immunol. Today. 1996;17:100. doi: 10.1016/0167-5699(96)80590-1. [DOI] [PubMed] [Google Scholar]
- 12.Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, Rowland-Jones S, et al. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur. J. Immunol. 2004;34:1673–1679. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- 13.Fauriat C, Andersson S, Björklund AT, Carlsten M, Schaffer M, Björkström NK, Baumann BC, et al. Estimation of the size of the alloreactive NK cell repertoire: studies in individuals homozygous for the group A KIR haplotype. J. Immunol. 2008;181:6010–6019. doi: 10.4049/jimmunol.181.9.6010. [DOI] [PubMed] [Google Scholar]
- 14.Augusto DG, Piovezan BZ, Tsuneto LT, Callegari-Jacques SM, Petzl-Erler ML. KIR gene content in amerindians indicates influence of demographic factors. Barbour JD, ed. PLoS ONE. 2013;8:e56755. doi: 10.1371/journal.pone.0056755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Augusto DG, Zehnder-Alves L, Pincerati MR, Martin MP, Carrington M, Petzl-Erler ML. Diversity of the KIR gene cluster in an urban Brazilian population. Immunogenetics. 2012;64:143–152. doi: 10.1007/s00251-011-0565-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollenbach JA, Augusto DG, Alaez C, Bubnova L, Fae I, Fischer G, Gonzalez-Galarza FF, et al. 16 thIHIW: Population Global Distribution of Killer Immunoglobulin-like Receptor (KIR) and Ligands. Int J Immunogenet. 2012 doi: 10.1111/iji.12028. n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chidgey M. Desmosomes and disease: an update. Histol. Histopathol. 2002;17:1179–1192. doi: 10.14670/HH-17.1179. [DOI] [PubMed] [Google Scholar]
- 18.Petzl-Erler ML, Santamaria J. Are HLA class II genes controlling susceptibility and resistance to Brazilian pemphigus foliaceus (fogo selvagem)? Tissue Antigens. 1989;33:408–414. doi: 10.1111/j.1399-0039.1989.tb01684.x. [DOI] [PubMed] [Google Scholar]
- 19.Pavoni DP, Petzl-Erler ML, Roxo VMMS, Marquart Filho A. Dissecting the associations of endemic pemphigus foliaceus (Fogo Selvagem) with HLA-DRB1 alleles and genotypes. Genes Immun. 2003;4:110–116. doi: 10.1038/sj.gene.6363939. [DOI] [PubMed] [Google Scholar]
- 20.Malheiros D, Petzl-Erler ML. Individual and epistatic effects of genetic polymorphisms of B-cell co-stimulatory molecules on susceptibility to pemphigus foliaceus. Genes Immun. 2009;10:547–558. doi: 10.1038/gene.2009.36. [DOI] [PubMed] [Google Scholar]
- 21.Kulkarni S, Martin MP, Carrington M. The Yin and Yang of HLA and KIR in human disease. Semin. Immunol. 2008;20:343–352. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Augusto DG, Lobo-Alves SC, Melo MF, Pereira NF, Petzl-Erler ML. Activating KIR and HLA Bw4 ligands are associated to decreased susceptibility to pemphigus foliaceus, an autoimmune blistering skin disease. Zimmer J, ed. PLoS ONE. 2012;7:e39991. doi: 10.1371/journal.pone.0039991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robledo MA, Prada S, Jaramillo D, Leon W. South American pemphigus foliaceus: study of an epidemic in El Bagre and Nechi, Colombia 1982 to 1986. Br. J. Dermatol. 1988;118:737–744. doi: 10.1111/j.1365-2133.1988.tb02590.x. [DOI] [PubMed] [Google Scholar]
- 24.Hans-Filho G, Aoki V, Rivitti E, Eaton DP, Lin MS, Diaz LA. Endemic pemphigus foliaceus (fogo selvagem)--1998. The Cooperative Group on Fogo Selvagem Research. Clin. Dermatol. 1999;17:225–235. doi: 10.1016/s0738-081x(99)00014-0. discussion 105-6. [DOI] [PubMed] [Google Scholar]
- 25.Hans-Filho G, Santos dos V, Katayama JH, Aoki V, Rivitti EA, Sampaio SA, Friedman H, et al. An active focus of high prevalence of fogo selvagem on an Amerindian reservation in Brazil. Cooperative Group on Fogo Selvagem Research. J Investig Dermatol. 1996;107:68–75. doi: 10.1111/1523-1747.ep12298213. [DOI] [PubMed] [Google Scholar]
- 26.Aoki V, Millikan RC, Warren SJP, Hans-Filho G, Eaton DP, Rivitti EA, Hilario-Vargas J, et al. Environmental risk factors in endemic pemphigus foliaceus (fogo selvagem) J Invest Dermat SP. 2004;9:34–40. doi: 10.1111/j.1087-0024.2004.00833.x. [DOI] [PubMed] [Google Scholar]
- 27.Martin AM, Freitas EM, Witt CS, Christiansen FT. The genomic organization and evolution of the natural killer immunoglobulin-like receptor (KIR) gene cluster. Immunogenetics. 2000;51:268–280. doi: 10.1007/s002510050620. [DOI] [PubMed] [Google Scholar]
- 28.Björkström NK, Fauriat C, Bryceson YT, Sandberg JK, Ljunggren H-G, Malmberg K-J. Analysis of the KIR repertoire in human NK cells by flow cytometry. Methods Mol. Biol. 2010;612:353–364. doi: 10.1007/978-1-60761-362-6_24. [DOI] [PubMed] [Google Scholar]
- 29.Khakoo SI, Carrington M. KIR and disease: a model system or system of models? Immunol. Rev. 2006;214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- 30.Giles J, Shaw J, Piper C, Wong-Baeza I, McHugh K, Ridley A, Li D, et al. HLA-B27 homodimers and free H chains are stronger ligands for leukocyte Ig-like receptor B2 than classical HLA class I. J. Immunol. 2012;188:6184–6193. doi: 10.4049/jimmunol.1102711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong-Baeza I, Ridley A, Shaw J, Hatano H, Rysnik O, McHugh K, Piper C, et al. KIR3DL2 binds to HLA-B27 dimers and free H chains more strongly than other HLA class I and promotes the expansion of T cells in ankylosing spondylitis. J. Immunol. 2013;190:3216–3224. doi: 10.4049/jimmunol.1202926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowness P, Ridley A, Shaw J, Chan AT, Wong-Baeza I, Fleming M, Cummings F, et al. Th17 cells expressing KIR3DL2+ and responsive to HLA-B27 homodimers are increased in ankylosing spondylitis. J. Immunol. 2011;186:2672–2680. doi: 10.4049/jimmunol.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J. Rates of conservative and radical nonsynonymous nucleotide substitutions in mammalian nuclear genes. J. Mol. Evol. 2000;50:56–68. doi: 10.1007/s002399910007. [DOI] [PubMed] [Google Scholar]
- 35.Alvarez-Arias DA, Campbell KS. Protein kinase C regulates expression and function of inhibitory killer cell Ig-like receptors in NK cells. The Journal of Immunology. 2007;179:5281–5290. doi: 10.4049/jimmunol.179.8.5281. [DOI] [PubMed] [Google Scholar]
- 36.Vély F, Vivier E. Conservation of structural features reveals the existence of a large family of inhibitory cell surface receptors and noninhibitory/activatory counterparts. The Journal of Immunology. 1997;159:2075–2077. [PubMed] [Google Scholar]
- 37.Boll W, Ohno H, Songyang Z, Rapoport I, Cantley LC, Bonifacino JS, Kirchhausen T. Sequence requirements for the recognition of tyrosine-based endocytic signals by clathrin AP-2 complexes. EMBO J. 1996;15:5789–5795. [PMC free article] [PubMed] [Google Scholar]
- 38.Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, Parham P. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. The Journal of Immunology. 2002;168:2307–2315. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- 39.Gourraud P-A, Meenagh A, Cambon-Thomsen A, Middleton D. Linkage disequilibrium organization of the human KIR superlocus: implications for KIR data analyses. Immunogenetics. 2010;62:729–740. doi: 10.1007/s00251-010-0478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vivian JP, Duncan RC, Berry R, O'Connor GM, Reid HH, Beddoe T, Gras S, et al. Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature. 2011;479:401–405. doi: 10.1038/nature10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braun-Prado K, Vieira Mion AL, Petzl-Erler ML, Farah Pereira N, Culpi L. HLA class I polymorphism, as characterised by PCR-SSOP, in a Brazilian exogamic population. Tissue Antigens. 2000;56:417–427. doi: 10.1034/j.1399-0039.2000.560504.x. [DOI] [PubMed] [Google Scholar]
- 42.Probst CM, Tsuneto LT, Bompeixe EP, Petzl-Erler ML, Pereira NF, de O Dalalio MM, Visentainer JE. HLA polymorphism and evaluation of European, African, and Amerindian contribution to the white and mulatto populations from Paraná, Brazil. Hum. Biol. 2000;72:597–617. [PubMed] [Google Scholar]
- 43.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.