Abstract
Trichothiodystrophy (TTD) is a rare multisystem disorder, characterized by sulfur deficient hair with alternating dark and light “tiger tail” banding on polarized light microscopy. TTD is caused by mutations in DNA repair/transcription genes XPD, XPB or TTDA, and in TTDN1, a gene of unknown function. While most TTD patients are photosensitive, patients with TTDN1 mutations were reported to be non-photosensitive. We followed a cohort of 36 TTD patients from 2001 to 2013. We describe 5 patients from 4 families with defects in the TTDN1 gene: 4 had no photosensitivity while 1 patient exhibited cutaneous burning. Deep phenotyping of our cohort revealed differences between the patients with and without TTDN1 mutations. Delayed bone age and seizure disorders were overrepresented in the TTDN1 group (p=0.009 and p=0.024, respectively), while some characteristic TTD clinical, laboratory, and imaging findings were absent. The 3 oldest TTDN1 patients displayed autistic behaviors in contrast to the characteristic friendly, socially interactive personality in the other patients. DNA sequencing revealed deletion mutations in TTDN1 ranging in size from a single base pair to over 120kb. These data identify a distinct phenotype relationship in TTD caused by TTDN1 mutations and suggest a different mechanism of disease.
Introduction
Trichothiodystrophy (TTD) is a rare, autosomal recessive disorder associated with defects in DNA repair/transcription genes. Diagnostic features include short, brittle, sulfur-deficient hair with a pattern of alternating light and dark “tiger-tail” bands under polarized light microscopy (Baden et al. 1976; Price et al. 1980; Liang et al. 2005; Liang et al. 2006). Additional features include photosensitivity in approximately half of these patients, ichthyosis, developmental delay, short stature, congenital dysmorphism, maternal pregnancy complications, collodian membrane at birth, and hematologic, ophthalmologic, and skeletal abnormalities. TTD is also associated with early mortality; affected patients have a 20-fold increased risk of death before age 10 (Faghri et al. 2008).
Causative mutations are found in the DNA repair/ transcription genes, XPD, XPB or TTDA (DiGiovanna and Kraemer 2012). TTDN1 (Trichothiodystrophy nonphotosensitive 1; C7ORF11, MPLKIP), a gene of unknown function, is a cause of the non-photosensitive TTD phenotype (Nakabayashi et al. 2005; Stefanini et al. 2010). Previous reports of TTDN1-associated TTD have identified several heterogeneous deletions in the gene, ranging from one base pair to 11-31 kb in size (Nakabayashi et al. 2005; Botta et al. 2007). Patients reported in the literature have been uniformly non-photosensitive, however no genotype-phenotype correlations have been found beyond these observations (Nakabayashi et al. 2005; Botta et al. 2007).
Our study employed deep phenotyping, a comprehensive analysis of phenotypic abnormalities using a detailed database (Boland et al. 2013), to more precisely describe a subgroup of patients with this rare genetic disorder. We sought a genotype-phenotype correlation in order to gain better insight both into the function of the underlying gene and the overall pathogenicity of the disorder. This may also serve as a model for future phenotyping studies given burgeoning interest in this methodology [reviewed in (Robinson 2012)]. In the following study, we report 5 additional TTD patients with confirmed defects in TTDN1 from our cohort and suggest that there is a distinct phenotype in TTDN1 patients.
Results and Discussion
Clinical features and mutations in 5 TTDN1 patients
We evaluated 36 patients with features consistent with TTD at the NIH (Liang et al. 2005; Liang et al. 2006; Schlucker et al. 2006; Boyle et al. 2008; Moslehi et al. 2010; Zhou et al. 2010; Brooks et al. 2011; Tamura et al. 2011; Tamura et al. 2012; Zhou et al. 2013; Atkinson et al. 2014). Of the 36 patients enrolled in our protocol, 20 were identified with mutations in XPD, 3 with TTDA mutations, 8 with unknown mutations, and 5 with mutations in TTDN1. The TTDN1 patients ranged in age from 1 to 14 years at last NIH evaluation, and came from 4 different families (Fig. 1a-f). Two patients (TTD487BE and TTD488BE) are affected siblings (Figure 1 c and d).
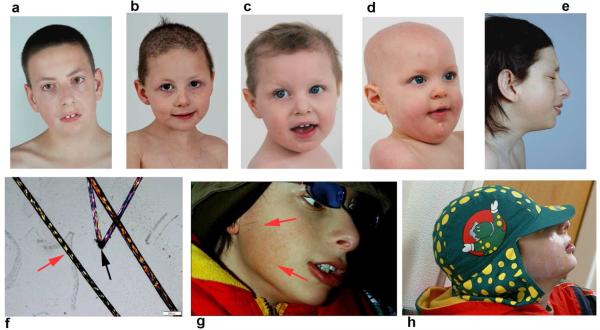
Fig. 1. Characteristics of 5 TTDN1 patients.
All patients have coarse hair with characteristic “tiger-tail” banding and hair shaft abnormalities under polarized light microscopy. a) Patient TTD402BE, 14 year old boy. b) Patient TTD480BE, 4 year old girl. c) Patient TTD487BE, 2 year old girl. d) Patient TTD488BE, age 1(brother of patient TTD487BE). e) Patient TTD343BE, 14 year old boy. f) Alternating light and dark (“tiger-tail”) banding (red arrow), and hair shaft abnormalities (black arrow) in TTD402BE under polarized light microscopy. g and h) Patient TTD343BE is exquisitely sensitive to UV, g)burning (red arrows) on brief exposure to an uncovered fluorescent light fixture in an area of the face that was not completely shielded by a hat with brim and sideshield (h) compared to his appearance before exposure (e).. The other TTDN1 patients are non-photosensitive. Parents of these 5 patients gave permission to publish these images of their children and their clinical information.
Patient Cases and TTDN1 Mutations
Patient TTD402BE, a Caucasian male (Fig 1 a), was born at 36 weeks gestation following a pregnancy complicated by preterm labor. He was hypotonic at birth, but neither collodian membrane nor cataracts were present. In early childhood his scalp hair was sparse and brittle, but improved in texture and length by age 6. He was mildly photophobic, but his skin did not burn on exposure to UV. He developed recurrent otitis media, requiring 2 sets of PE (pressure equalizing) tubes. All developmental milestones were delayed; he walked at 19 months and continued to display poor expressive speech and articulation as well as intellectual impairment. He was diagnosed with autism spectrum disorder at age 10 based on repetitive and obsessive behaviors, poor expressive speech, and difficulty in socialization. At age 14, he was 50th percentile for height and 75th for weight. He exhibited hair findings typical of TTD, including coarse, sulfur-deficient hair (sulfur content 2.9%, normal = 5.0%) with tiger-tail banding on polarized light microscopy and sparse lateral eyebrows. Minimal facial freckling, beaked nails, and xerosis (but not ichthyosis) were noted on skin exam. Other features included high-arched palate, retrognathia, mild pectus excavatum, mild genu valgus, pes planus, scoliosis, myopia, nystagmus, and sialorrhea. Skeletal imaging at age 10 years revealed generalized osteopenia without central osteosclerosis, spina bifida occulta at C7/T1, and normal bone age. CT bone densitometry found his average bone mineral density to be 128.7 mg/cc which is 2.72 standard deviations below age matched control (z-score). A unilateral left kidney was found on abdominal ultrasound. There was no evidence of dysmyelination on brain MRI at age 10 years despite his developmental delay. Blood results including hemoglobin, hematocrit, MCV, and RBC, neutrophil and lymphocyte counts, as well as hemoglobin electrophoresis, were normal. DNA sequencing of TTD402BE's cells revealed two different heterozygous mutations in the initiation codon of TTDN1. The first allele contained the maternally inherited mutation c.2T>G; a paternally inherited mutation c.2T>C was identified in the second allele (Table 1, Figs. 2 and S1). This likely results in the loss of translation initiation and thus no protein production.
Table 1.
Mutations of all known TTD patients with defects in TTDN1
| ALLELE 1 (maternal) | ALLELE 2 (paternal) | ||||||
|---|---|---|---|---|---|---|---|
| Patient | Photo-sensitive | Age/Sex | Genomic mutation | Protein change | Genomic mutation | Protein change | References |
| TTD5PV | No | 8/F | Deletion of 11-31kb | No protein | Homozygous | Botta et al. 2007, Fois et al. 1988 | |
| TTD10RO | No | 3/M | Deletion of at least 150 kb | No protein | Homozygous | Botta et al. 2007 | |
| TTD16PV | No | 4/F | c.148_152delCACAC | p.His50Alafs*8 | Homozygous | Botta et al. 2007 | |
| TTD31PV | No | 3/F | c.148_152delCACAC | p.His50Alafs*8 | c.277delT | p.Ser93Profs*60 | Botta et al. 2007 |
| TTD1MA | No | 6/F | c.229delC | p.Arg77Glyfs*76 | Homozygous | Botta et al. 2007 | |
| TTD11RO | No | 3/F | c.277delT | p.Ser93Profs*60 | Homozygous | Botta et al. 2007 | |
| Moroccan Siblings | No | 16/M 4/F 13m/F |
c.187_188delGG | 57 residue truncated protein | Homozygous | Nakabayashi et al. 2005, Przedborski et al. 1990 | |
| Amish brittle-hair brain syndrome | No | 20 Amish patients | c.480A>G | p.Met144Val | Homozygous | Nakabayashi et al. 2005, Jackson et al. 1974 | |
| TTD9PV | No | 3/F | Partial deletion of exon 1 and entire exon 2 | No protein (probable) | Homozygous | Nakabayashi et al. 2005, Rizzo et al. 1992 | |
| TTD402BE | No | 14/M | c.2T>C (initiation codon) | No protein | c.2T>G (initiation codon) | No protein | This paper |
| TTD480BE | No | 4/F | Deletion of ~120kb | No protein | c.227delG | p.Gly76Alafs*77 | This paper |
| TTD487BE TTD488BE |
No | 2/F 1/M |
c. 277delT | p.Ser93Profs*60 | Deletion of ~92 kb | No protein | This paper |
| TTD343BE | Yes | 14/M | 4 bp insertion & deletion of ~5kb starting at c.279 | No protein | Homozygous | This paper | |
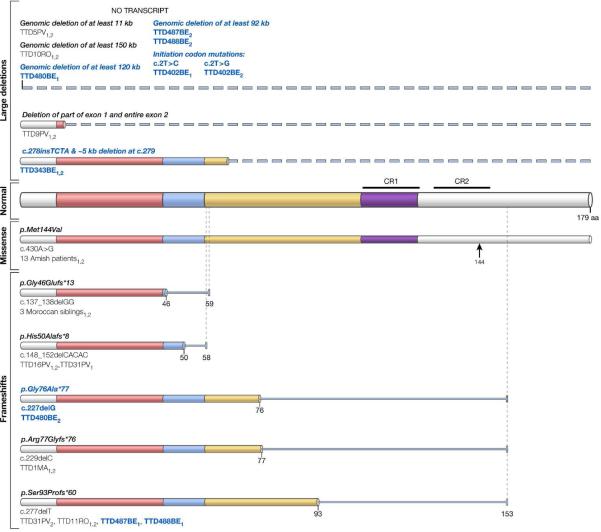
Figure 2. Mutations in the TTDN1 protein.
The 179 amino acid TTDN1 protein is shown. (Normal protein) Glycine/proline rich regions are colored, with low complexity regions shown in blue and purple and highly conserved C-terminal regions (CR1 and CR2) marked. Mutations reported in Nakabayashi et al. (2005) and reported in Botta et al. (2005) are shown with standard black lettering and mutations reported in this paper are in bold blue lettering (Table 1). The mutations above the normal protein are large deletions (indicated by dashed lines). Mutations below the normal protein are missense (indicated by an arrow) and one or two base frameshifts followed by an abnormal amino acid sequence (indicated by narrow lines ending at the site of the premature termination codon). Alleles are denoted by a 1 and/or 2 following the patient identification numbers. Mutation nomenclature and cDNA numbering follow standard formatting (Botta et al., 2007).
Patient TTD480BE, a Caucasian female (Figure 1b), was born at term after an uncomplicated pregnancy. The neonatal period was unremarkable and the patient did not have low birth weight, collodian membrane or congenital cataracts; feeding difficulties were noted in infancy. She was not photosensitive. She experienced recurrent otitis media necessitating 3 sets of PE tubes. All developmental milestones were delayed. She walked at age 3 and at her last NIH visit at age 4, she spoke few rudimentary words. At this visit, her height and weight were in the 10th and 5th percentiles respectively. She displayed sparse, brittle, sulfur-deficient hair (sulfur content 2.4%, normal = 5.0%) with tiger-tail banding under polarized light microscopy (Fig. 1f), and sparse eyebrows with broken eyelashes. Skin exam revealed generalized xerosis (not ichthyosis) with multiple freckle-like macules over the nose and cheeks, a periumbilical café au lait spot, and dystrophic, peeling fingernails and toenails. Other features included diffuse hypotonia, sialorrhea, strabismus, hyperopia, epicanthal folds with mild telecanthus, high nasal bridge, micrognathia, genu valgus, pes planus, and clinodactyly of the right hand. Imaging showed delayed bone age (estimated age 2 years, 3 months) with mild osteopenia, and right sided aortic arch with aberrant left subclavian artery. There was no evidence of dysmyelination on brain MRI. Hemoglobin, hematocrit, MCV, hemoglobin A2, and RBC, neutrophil, and lymphocyte counts were all normal. Three years later, she was undergoing evaluation for autism spectrum disorder due to new onset repetitive behaviors and verbal delay. Array comparative genome hybridization (aCGH) revealed a greater than 120 kb deletion in the maternally inherited allele, resulting in hemizygosity in TTDN1. A single base pair deletion was found at c.227 in the paternally inherited TTDN1 allele, leading to a frame shift and stop codon formation 77 residues downstream (Table 1, Figs. 2 and S2). It is unlikely that a functional protein would result from these mutations.
Patients TTD487BE and TTD488BE are Caucasian siblings, a sister and brother respectively (Figure 1 c and d). They were each born at term following uncomplicated pregnancies. Both siblings were non-photosensitive, mildly hypotonic, and experienced recurrent upper respiratory infections and otitis media. They were diagnosed with developmental delay at age 1 for TTD487BE, and 8 months for TTD488BE. Cardiac structural defects were found in both siblings - atrial septal defect in TTD487BE and pulmonic stenosis in TTD488BE. Atonic seizure disorder was diagnosed in TTD487BE at 2 years of age and in TTD488BE at 18 months. At NIH, TTD487BE was 10th percentile for height and 25th for weight at age 2, while TTD488BE was 97th percentile for height and 50th for weight at age 1. Both siblings exhibited characteristic sparse, brittle, and sulfur-deficient hair (sulfur content 2.6% for TTD487BE and 2.35% for TTD488BE, normal = 5.0%) with tiger-tail banding under polarized light. Eyebrows and eyelashes were also sparse. Additional features for TTD487BE included epicanthal folds with mild telecanthus, micrognathia, simplified ears, protruding umbilicus, bilateral 4th-5th digit clinodactyly with low placed thumbs, genu valgus, pes planus, and sialorrhea. Follicular papules along the extremities and hypoplastic toenails were noted on her skin exam. TTD488BE displayed left ear crease, and cyanosis of the bilateral feet in addition to epicanthal folds, protuberant umbilicus, simplified ears, low placed thumbs and sialorrhea. His skin exam revealed keratosis pilaris and peeling nails, but not ichthyosis. MRI of the brain did not show evidence of dysmyelination in either patient. Radiographic studies revealed delayed bone age for both (estimated age 1 year, 6 months for TTD487BE and newborn for TTD488BE), but no osteosclerosis. Blood work was normal in both patients, with the exception of a low neutrophil count in TTD487BE. DNA sequence analysis revealed a single base deletion at c.277 on the maternally inherited allele for both siblings, leading to a frame shift and stop codon formation 60 residues downstream. An aCGH revealed an approximately 92 kb deletion in the paternally inherited allele, resulting in hemizygosity in TTDN1 for both siblings (Table 1, Figs. 2 and S3).
Patient TTD343BE, a Caucasian male (Figure 1e, 1g-h), was born at term following an uncomplicated pregnancy. However, he developed fetal distress during labor and was delivered via emergency Caesarian-section. His birth weight was low (2450 grams). His infancy was remarkable for hypotonia, difficulty feeding, and failure to thrive. He had a history of severe photosensitivity, with burning and blistering after brief exposure to the sun. His mother described that, as an infant, if his bassinette was placed near a window “he would scream and his skin would become raw”. Consequently, he wore sunscreen and a hat with visor, neck and ear shields and sunscreen even when indoors (Figure 1h). We observed this exquisite photosensitivity when he began to complain of discomfort during a brief exposure to an unshielded fluorescent light while waiting in a hallway at NIH. Shortly thereafter, he developed a burn in an uncovered area of his face (Fig 1g). He developed recurrent upper respiratory and GI infections, necessitating multiple hospitalizations. He did not meet his developmental milestones appropriately, walking independently at age 4 and first speaking at age 8. At age 10, he was diagnosed with autism based on his language delay, repetitive behaviors and difficulty in socialization. He consistently grew at or below the 5th percentile for height and weight; at the NIH, he was <3rd percentile for both height and weight at age 14 years. He displayed typical hair findings for TTD, including coarse, sulfur-deficient hair (sulfur content 2.4%, normal = 5.0%) with tiger-tail banding under polarized light with few shaft abnormalities. His hair grew long and thick, an unusual finding for TTD patients. His eyebrows and eyelashes were sparse and broken. Generalized xerosis and spooned, peeling nails were noted on skin exam. Additional features included epicanthal folds, prominent nose tip and high nasal bridge, smooth philtrum, mild micrognathia, retrognathia, pectus excavatum, and genu valgus, as well as pes planus, bilateral 2nd-3rd toe syndactyly and sialorrhea. Imaging revealed delayed bone age (estimated age 12 years, 6 months), but no evidence of dysmyelination on brain MRI. At age 24 he developed epilepsy with grand mal seizures. DNA sequencing of TTD343BE revealed homozygosity for a 4 bp insertion in the TTDN1 gene, followed by a 5 kb deletion starting at c.279 (Table 1, Fig. 2 and S4). Both parents were found to be heterozygous for this mutation. Because the deletion spans both exons of TTDN1, PCR amplification of genomic DNA did not yield a band for TTD343BE (Fig. S5). Bands can be seen for the 4 remaining TTDN1 patients, all of whom have at least one copy of the gene.
No increased sensitivity to UVC in TTDN1 cells
Defects in TTDN1 were believed to be a cause of the non-photosensitive TTD phenotype, as no reported cases in the literature were described to develop cutaneous burning on sun exposure (Nakabayashi et al. 2005; Botta et al. 2007). Similarly, 4 out of 5 TTDN1 patients in our cohort were non-photosensitive; however, 1 patient (TTD343BE) did exhibit exquisite photosensitivity. Of the 30 reported cases of TTDN1 [Table 1], no other patients were noted to be photosensitive.
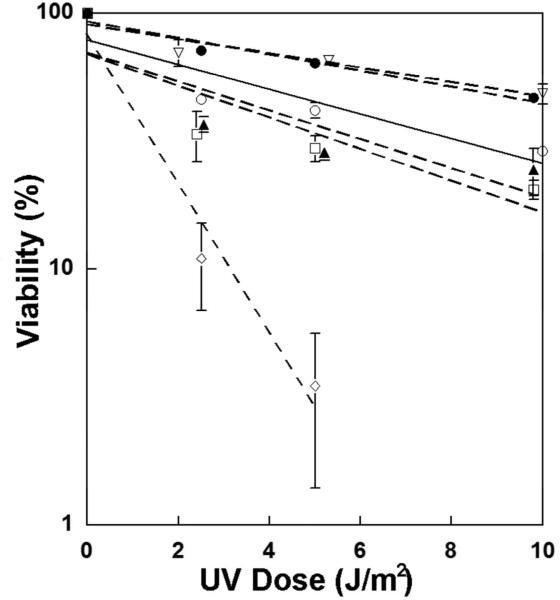
We performed UVC hypersensitivity studies on fibroblasts from each of the 4 TTDN1 families to measure their cellular sensitivity to UVC (Fig. 3). Fibroblasts from the photosensitive patient, TTD343BE, had a cellular response profile similar to those of the other, non-photosensitive, TTDN1 patients and the normal control. In contrast, fibroblasts from photosensitive XP and TTD patients with mutations in XPD showed increased sensitivity to UV (Figure 3 and (Boyle et al. 2008)). By dissociating cell death from clinical photosensitivity, this result suggests that cell death is not required to stimulate inflammatory mediators which result in cutaneous burning. These findings also provide further evidence that TTDN1 does not play a role in transcription coupled - NER, unlike photosensitive TTD-associated genes XPD, TTDA and XPB (Nakabayashi et al. 2005; Stefanini et al. 2010). The TTDN1 protein has been reported to interact with Polo-like kinase 1 to help regulate cell cycle progression (Zhang et al. 2007), however this has not been confirmed.
Figure 3.
Post-UV survival of TTDN1, XP and normal cell lines. Cell cultures were exposed to the indicated dose of UVC and post-UVC viability was assessed after 4 days using an MTS assay. One normal (FXP0223 [open circle]), four TTDN1 (TTD402BE [closed triangle], TTD480BE [closed circle], TTD487BE [open triangle] and TTD343BE [open square]), and one XP (XP29BE, open diamond) cells were used. Overlapping points were displaced for clarity. Interestingly, fibroblasts from photosensitive and non-photosensitive TTDN1 patients were not hypersensitive to UVC in contrast to cells from an XP patient with mutations in the XPD gene.
Deep phenotyping reveals a distinct TTDN1 genotype-phenotype relationship
Our detailed clinical analysis (Table S1) of a cohort of 36 patients exhibiting features of TTD uncovered a distinct genotype-phenotype relationship in patients with defects in TTDN1. As compared to patients with mutations in XPD, TTDA or unidentified genes, delayed bone age and seizure disorders were overrepresented in the TTDN1 group. There were 3 patients from 2 families with TTDN1 mutations (TTD343BE, and siblings TTD487BE and TTD488BE) diagnosed with seizure disorders, but only 3 out of the 31 other TTD patients (p=0.024). Additionally, 4 patients from 3 families with TTDN1 defects had delayed bone age (TTD343BE, TTD480BE, and siblings TTD487BE and TTD488BE), as compared to 4 out of 23 TTD patients with other mutations (p=0.009). These findings are consistent with earlier case reports (Table 1): seizure disorders were reported in two separate families with confirmed TTDN1 defects (Jackson et al. 1974; Przedborski et al. 1990), as was delayed bone age (Fois et al. 1988; Przedborski et al. 1990).
Our TTDN1 patients had characteristic hair findings of tiger-tail banding and brittle, sulfur-deficient hair, recurrent infections, intellectual impairment, hypotonia, and osteopenia, as seen in TTD. However, other features consistent with TTD, including pregnancy complications, low birth weight, collodian membrane at birth, reduced height and weight, cataracts, thalassemia-like changes on blood work, and imaging findings of brain dysmyelination (Viprakasit et al. 2001; Kraemer et al. 2007; Faghri et al. 2008; Tamura et al. 2011; Brooks et al. 2011; DiGiovanna and Kraemer 2012) were underrepresented or absent. TTD is also associated with a strikingly sociable, outgoing, and friendly personality. In contrast, the 3 oldest TTDN1 patients within our cohort (TTD402BE, TTD343BE, and TTD480BE) displayed autistic features, including repetitive behaviors, obsessive interests, and difficulty socializing. Similar autistic features in TTD patients with confirmed TTDN1 defects have been previously reported in case studies, reinforcing this genotype-phenotype correlation (Przedborski et al. 1990; Rizzo et al. 1992). Taken together, the overrepresentation of seizure disorders, delayed bone age, and autistic features in this subgroup, as well as the absence of other characteristic TTD features, represents a distinct phenotype and suggests a different mechanism of disease for patients with TTDN1 defects.
Eight TTDN1 mutations were associated with a TTD phenotype in the literature (Table 1 and Figure 2). Seven of these mutations were deletions which involved a one or two base pairs, leading to a frame shift followed by a premature stop codon, or large deletions, preventing protein production. Most mutations were found in a single patient or family, however one cohort of 20 affected individuals of Amish background was reported with a common homozygous TTDN1 missense mutation (Jackson et al. 1974). DNA analysis of our cohort revealed large deletions in TTDN1 in 3 families leading to hemizygosity, a finding consistent with previous reports (Table 1 and Figure 2). These deletions would be expected to involve the adjacent SUGCT (succinylCoA:glutarate-CoA transferase) gene on chromosome 7. Mutations in SUGCT have been reported to be associated with autosomal recessive glutaric aciduria type 3 (Sherman et al. 2008; Marlaire et al. 2014). However, our TTDN1 patients had deletions of only one SUGCT allele and the two patients tested had normal urinary organic acids. A single base pair deletion discovered in 1 family was also reported in two unrelated patients in the literature (Botta et al. 2007), indicating perhaps the location of a mutational hotspot. Other mutations in TTDN1 were found in 2 families; we report compound heterozygous missense mutations in the initiation codon in one family, and in the second, a single base pair deletion occurring near a previously reported mutation. The mutations uncovered in our cohort are expected to result in a null protein, which suggests that TTDN1 is not essential for life (Nakabayashi et al. 2005; Botta et al. 2007), in contrast to other TTD causing genes that are part of the basal transcription factor, TFIIH (XPB, XPD, and TTDA) (DiGiovanna and Kraemer 2012)..
The identification of genotype-phenotype correlations in complex genetic diseases like TTD will allow clinicians to better predict the underlying genetic defect, providing more timely diagnosis and more specific prognostic information. This may be especially useful in patients for whom mutations could not be identified. For example, one previously reported TTD patient described as having autism, mental retardation and seizures, was unable to be genotyped (Schepis et al. 1997). Based on additional findings of delayed bone age and normal laboratory blood values, our findings suggest that this patient may have had a defect in TTDN1. In the future, we seek to further clarify genotype-phenotype relationships for TTD patients with XPD and TTDA mutations using the deep phenotyping methodology outlined in this study. With the information we collect, we also hope to identify features that may predict early mortality in these patients, identify causative mutations in patients with unknown defects, and further our understanding of disease pathogenesis in TTD.
Materials and Methods
Patients with features consistent with TTD were enrolled in an NCI Institutional Review Board approved DNA repair disease natural history protocol 99-C-0099, which adhered to the Declaration of Helsinki Protocols. Informed consent was obtained, following which patients underwent thorough skin exam, skin biopsy for fibroblast culture and blood draw for both diagnostic and research purposes. Diagnostic blood tests for each patient included Chem 20, hepatic, lipid, mineral, and iron panels, complete blood count with differential, immunoglobulin A, E, G, and M levels, hemoglobin electrophoresis, thyroid function tests, vitamin D and vitamin B12 levels, and additional tests according to medical need. Hair samples were also collected from each patient for examination under polarized light microscopy (Liang et al. 2005) and amino acid analysis (Liang et al. 2006). Further assessments included ophthalmology (Brooks et al. 2011), audiology, neurology, radiology, and others as medically indicated (Atkinson et al. 2014).
Clinical documentation, imaging and laboratory results were collected and reviewed for all TTD patients utilizing the NIH Clinical Center Biomedical Translational Research Information System (BTRIS), in addition to records obtained from outside facilities. We extracted more than 160 features to build a large clinical database using Microsoft Excel (Table S1). These features were used to compare patients with defects in TTDN1 to those with other mutations within our cohort. Statistical analysis included Fisher's exact tests. Two-sided p-values were used with a cutoff of p<0.05.
Clinical protocol, cells, and culture conditions
Skin biopsies were obtained and sent to the Human Genetic Mutant Cell Repository (Camden, NJ) or Fisher BioServices, NCI Frederick Central Repository (Frederick, MD) for establishment of fibroblast cell cultures. Fibroblast cultures from five TTD (TTD402BE, FXP0109; TTD480BE, FXP0416; TTD487BE, FXP0390; TTD488BE, FXP0388; TTD343BE, JA1451) and 1 XP-D (XP29BE, GM11613) patients, and normal control (FXP0223) were grown in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) containing 4mM glutamine and 10% fetal calf serum (FCS) (Atlanta Biologicals) in an 8% CO2 humidified incubator at 37°C as described previously (Zhou et al. 2013).
Post-UV Cell survival
The measurement of cell viability 4 days after UVC exposure was carried out as described previously using a 3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-4 (-sulfophenyl)-2H-tetrazolium (MTS) (Promega, Madison, WI) (Imoto et al. 2002).
PCR Amplification and Nucleotide Sequence Analysis
To identify the genetic changes, entire coding region of the TTDN1 (C7orf11) gene including splice donor and acceptor sites of exon 1 and exon 2 were polymerase chain reaction (PCR) amplified. The primers were designed on the basis of GenBank reference sequences (accession nos. NC_000007 for genomic sequence and NM_138701 for mRNA sequence). TTDN1 cDNA sequences were followed with numbering based on base 1 as the A of the ATG initiation codon. The following forward and reverse primer sequences were used to amplify entire exon 1 and exon 2 regions of the TTDN1 gene: exon 1- 5’-CGAGGTTTTCGGCTTTGGCTC-3’, 5’-TCTCTAAGCCTCAGTTCGCTC-3’ (PCR fragment: 510 bp); exon 2 – 5’-ACTTTTAAGCAGCAATGTGATTCC-3’, 5’-GTATTGTTGGCTTATATCCACTAC-3’ (PCR fragment: 252 bp). The PCR amplification was achieved using primer pairs, Advantage GC genomic PCR Kit (Clontech Laboratories, Inc.) and patients’ DNA separated either from cells or blood. The PCR steps were conducted as follows: 94°C for 1 min, then 35 cycles of amplification (94°C for 20 s and 62°C for 2 min), ending with 62°C for 3 min. The PCR products were resolved on a 2% agarose gel. The PCR bands in the gel were visualized with EZ-Vision (Amresco) and UV light and photographed. After agarose gel purification the PCR product was subcloned into pCR2.1-TOPO vector (TOPO TA cloning kit; Invitrogen) as per the vendor's protocol in order to separate both mutant alleles. Mutations were confirmed by nucleotide sequencing. PCR was used to map the homozygous large deletion in genomic DNA from patient TTD343BE. The forward primer (5’- TTTTGGTAAAGGACTA - 3’) in C7orf11 gene and the reverse primer (5’- ATTCACCAAAACTAAT - 3’) in GRCh38 Primary Assembly Sequence ID: NC_000007.14 that flanks the deletion gives a 441 bp PCR product using Advantage GC genomic PCR Kit. The PCR steps were 94°C for 1 min, then 35 cycles of amplification (94°C for 20 s and 52°C for 2 min), ending with 52°C for 3 min. Sequencing of direct PCR or cloned PCR products was performed by cycle sequencing employing dideoxy termination chemistry and a Prism Model 3700 Capillary Array Sequencer using PCR primers as described (Zhou et al. 2013). An aCGH was performed by the National Cancer Institute Clinical Molecular Profiling Core using a custom designed high density DNA repair gene array as previously described (Tan et al. 2013).
Supplementary Material
Acknowledgements
This research was made possible through the National Institutes of Health (NIH) Medical Research Scholars Program, a public-private partnership supported jointly by the NIH and generous contributions to the Foundation for the NIH from Pfizer Inc., The Doris Duke Charitable Foundation, The Alexandria Real Estate Equities, Inc. and Mr. and Mrs. Joel S. Marcus, and the Howard Hughes Medical Institute, as well as other private donors. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. We would like to thank Carine Nadem and Jared Jagdeo for their help with DNA sequencing, Daniel Edelman, Holly Stevenson and Yonghong Wang of the NCI Clinical Molecular Profiling Core for assistance with aCGH studies, and John Crawford and Mary King, photographers, and Alan Hoofring, lead medical illustrator, of the NIH Medical Arts and Photography group for assistance with Figures 1 and 2. We are indebted to our patients and families for their gracious participation.
Abbreviations
- TTD
trichothiodystrophy
- XP
xeroderma pigmentosum
- MCV
mean corpuscular volume
- RBC
red blood cell
- WBC
white blood cell
- NER
nucleotide excision repair
- NIH
National Institutes of Health
- PE
pressure equalizer
- aCGH
array comparative genome hybridization
Footnotes
An abstract of this study was presented at the 2014 meeting of the Society for Investigative Dermatology in Albuquerque, NM (Heller ER, Khan SG, Tamura D, DiGiovanna JJ, Kraemer KH. Mutations in TTDN1 are associated with a unique trichothiodystrophy phenotype. J Invest Dermatol. 2014; 134(S1): S75.)
Conflict of Interest: The authors state no conflict of interest.
References
- Atkinson EC, Thuara D, Tamura D, et al. Growth and nutrition in children with trichothiodystrophy. Journal of Ped Gastroenterology and Nutrition. 2014 doi: 10.1097/MPG.0000000000000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden HP, Jackson CE, Weiss L, et al. The physicochemical properties of hair in the BIDS syndrome. Am.J Hum.Genet. 1976;28(5):514–521. [PMC free article] [PubMed] [Google Scholar]
- Boland MR, Hripcsak G, Shen Y, et al. Defining a comprehensive verotype using electronic health records for personalized medicine. J Am.Med.Inform.Assoc. 2013;20(e2):e232–e238. doi: 10.1136/amiajnl-2013-001932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta E, Offman J, Nardo T, et al. Mutations in the C7orf11 (TTDN1) gene in six nonphotosensitive trichothiodystrophy patients: no obvious genotype phenotype relationships. Hum.Mutat. 2007;28(1):92–96. doi: 10.1002/humu.20419. [DOI] [PubMed] [Google Scholar]
- Boyle J, Ueda T, Oh KS, et al. Persistence of repair proteins at unrepaired DNA damage distinguishes diseases with ERCC2 (XPD) mutations: cancer-prone xeroderma pigmentosum vs. non-cancer-prone trichothiodystrophy. Hum.Mutat. 2008;29(10):1194–1208. doi: 10.1002/humu.20768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BP, Thompson AH, Clayton JA, et al. Ocular manifestations of trichothiodystrophy. Ophthalmology. 2011;118(12):2335–2342. doi: 10.1016/j.ophtha.2011.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiovanna JJ, Kraemer KH. Shining a light on xeroderma pigmentosum. J Invest Dermatol. 2012;132(3 Pt 2):785–796. doi: 10.1038/jid.2011.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghri S, Tamura D, Kraemer KH, et al. Trichothiodystrophy: a systematic review of 112 published cases characterises a wide spectrum of clinical manifestations. J Med.Genet. 2008;45(10):609–621. doi: 10.1136/jmg.2008.058743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fois A, Balestri P, Calvieri S, et al. Trichothiodystrophy without photosensitivity. Biochemical, ultrastructural and DNA repair studies. Eur.J.Pediatr. 1988;147:439–441. doi: 10.1007/BF00496431. [DOI] [PubMed] [Google Scholar]
- Imoto K, Kobayashi N, Katsumi S, et al. The total amount of DNA damage determines ultraviolet-radiation-induced cytotoxicity after uniformor localized irradiation of human cells. J.Invest Dermatol. 2002;119(5):1177–1182. doi: 10.1046/j.1523-1747.2002.19514.x. [DOI] [PubMed] [Google Scholar]
- Jackson CE, Weiss L, Watson JH. “Brittle” hair with short stature, intellectual impairment and decreased fertility: an autosomal recessive syndrome in an Amish kindred. Pediatrics. 1974;54(2):201–207. [PubMed] [Google Scholar]
- Kraemer KH, Patronas NJ, Schiffmann R, et al. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: A complex genotype-phenotype relationship. Neuroscience. 2007;145(4):1388–1396. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Kraemer KH, Morris A, et al. Characterization of tiger-tail banding and hair shaft abnormalities in trichothiodystrophy. J Am.Acad.Dermatol. 2005;52(2):224–232. doi: 10.1016/j.jaad.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Liang C, Morris A, Schlucker S, et al. Structural and molecular hair abnormalities in trichothiodystrophy. J.Invest Dermatol. 2006;126(10):2210–2216. doi: 10.1038/sj.jid.5700384. [DOI] [PubMed] [Google Scholar]
- Marlaire S, Van SE, Veiga-da-Cunha M. C7orf10 encodes succinate hydroxymethylglutarate CoA-transferase, the enzyme that converts glutarate to glutaryl-CoA. J Inherit.Metab Dis. 2014;37(1):13–19. doi: 10.1007/s10545-013-9632-0. [DOI] [PubMed] [Google Scholar]
- Moslehi R, Signore C, Tamura D, et al. Adverse effects of trichothiodystrophy DNA repair and transcription gene disorder on human fetal development. Clin.Genet. 2010;77(4):365–373. doi: 10.1111/j.1399-0004.2009.01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K, Amann D, Ren Y, et al. Identification of C7orf11 (TTDN1) gene mutations and genetic heterogeneity in nonphotosensitive trichothiodystrophy. Am.J Hum.Genet. 2005;76(3):510–516. doi: 10.1086/428141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price VH, Odom RB, Ward WH, et al. Trichothiodystrophy: sulfur-deficient brittle hair as a marker for a neuroectodermal symptom complex. Arch.Dermatol. 1980;116(12):1375–1384. doi: 10.1001/archderm.116.12.1375. [DOI] [PubMed] [Google Scholar]
- Przedborski S, Ferster A, Goldman S, et al. Trichothiodystrophy, mental retardation, short stature, ataxia, and gonadal dysfunction in three Moroccan siblings [see comments]. Am.J.Med.Genet. 1990;35:566–573. doi: 10.1002/ajmg.1320350424. [DOI] [PubMed] [Google Scholar]
- Rizzo R, Pavone L, Micali G, et al. Trichothiodystrophy: report of a new case with severe nervous system impairment. J.Child Neurol. 1992;7:300–303. doi: 10.1177/088307389200700311. [DOI] [PubMed] [Google Scholar]
- Robinson PN. Deep phenotyping for precision medicine. Hum.Mutat. 2012;33(5):777–780. doi: 10.1002/humu.22080. [DOI] [PubMed] [Google Scholar]
- Schepis C, Elia M, Siragusa M, et al. A new case of trichothiodystrophy associated with autism, seizures, and mental retardation. Pediatr.Dermatol. 1997;14:125–128. doi: 10.1111/j.1525-1470.1997.tb00219.x. [DOI] [PubMed] [Google Scholar]
- Schlucker S, Liang C, Strehle KR, et al. Conformational differences in protein disulfide linkages between normal hair and hair from subjects with trichothiodystrophy: a quantitative analysis by Raman microspectroscopy. Biopolymers. 2006;82(6):615–622. doi: 10.1002/bip.20515. [DOI] [PubMed] [Google Scholar]
- Sherman EA, Strauss KA, Tortorelli S, et al. Genetic mapping of glutaric aciduria, type 3, to chromosome 7 and identification of mutations in c7orf10. Am.J Hum.Genet. 2008;83(5):604–609. doi: 10.1016/j.ajhg.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanini M, Botta E, Lanzafame M, et al. Trichothiodystrophy: from basic mechanisms to clinical implications. DNA Repair (Amst) 2010;9(1):2–10. doi: 10.1016/j.dnarep.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Tamura D, Khan SG, Merideth M, et al. Effect of mutations in XPD(ERCC2) on pregnancy and prenatal development in mothers of patients with trichothiodystrophy or xeroderma pigmentosum. Eur.J.Hum.Genet. 2012;20(12):1308–1310. doi: 10.1038/ejhg.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura D, Merideth M, DiGiovanna JJ, et al. High-risk pregnancy and neonatal complications in the DNA repair and transcription disorder trichothiodystrophy: report of 27 affected pregnancies. Prenat Diagn. 2011;31(11):1046–1053. doi: 10.1002/pd.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Anzick SL, Khan SG, et al. Chimeric negative regulation of p14ARF and TBX1 by a t(9;22) translocation associated with melanoma, deafness, and DNA repair deficiency. Hum.Mutat. 2013;34(9):1250–1259. doi: 10.1002/humu.22354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viprakasit V, Gibbons RJ, Broughton BC, et al. Mutations in the general transcription factor TFIIH result in beta-thalassaemia in individuals with trichothiodystrophy. Hum.Mol.Genet. 2001;10(24):2797–2802. doi: 10.1093/hmg/10.24.2797. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Tian Y, Chen Q, et al. TTDN1 is a Plk1-interacting protein involved in maintenance of cell cycle integrity. Cell Mol.Life Sci. 2007;64(5):632–640. doi: 10.1007/s00018-007-6501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Khan SG, Tamura D, et al. Brittle hair, developmental delay, neurologic abnormalities, and photosensitivity in a 4-year-old girl. J Am.Acad.Dermatol. 2010;63(2):323–328. doi: 10.1016/j.jaad.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Khan SG, Tamura D, et al. Abnormal XPD-induced nuclear receptor transactivation in DNA repair disorders: trichothiodystrophy and xeroderma pigmentosum. Eur.J.Hum.Genet. 2013;21(8):831–837. doi: 10.1038/ejhg.2012.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.