Abstract
Inactivation of human drug-metabolizing cytochrome P450 3A4 (CYP3A4) could lead to serious adverse events such as drug-drug interactions and toxicity. However, when properly controlled, CYP3A4 inhibition may be beneficial as it can improve clinical efficacy of co-administered therapeutics that otherwise are quickly metabolized by CYP3A4. Currently, the CYP3A4 inhibitor ritonavir and its derivative cobicistat are prescribed to HIV patients as pharmacoenhancers. Both drugs were designed based on the chemical structure/activity relationships rather than the CYP3A4 crystal structure. To unravel the structural basis of CYP3A4 inhibition, we compared the binding modes of ritonavir and ten analogues using biochemical, mutagenesis and x-ray crystallography techniques. This review summarizes our findings on the relative contribution of the heme-ligating moiety, side chains and the terminal group of ritonavir-like molecules to the ligand binding process, and highlights strategies for a structure-guided design of CYP3A4 inactivators.
1. Introduction
Humans have 57 isoforms of cytochrome P450 that catalyze the oxidation of a variety of endogenous molecules and xenobiotics. Cytochrome P450 3A4 (CYP3A4) is one of the most abandant and important drug-metabolizing isoforms that clears over a half of prescribed pharmaceuticals [1]. It has been demonstrated both biochemically and structurally that CYP3A4 has a large and highly malleable active site cavity which can accommodate a wide range of chemically diverse compounds [2]. Moreover, CYP3A4 has the ability to simultaneously bind two or more molecules of the same or different nature, one of which could serve as an effector and modulate the rate or/and the site of metabolism of the other molecule. Drug oxidation and clearance may also be affected by compounds that inhibit CYP3A4. Some pharmaceuticals can act both as substrates and inactivators of CYP3A4. Inactivation of CYP3A4 then could result in drug plasma levels rising to toxic levels.
A carefully controlled CYP3A4 inactivation, however, may be beneficial. One area where CYP3A4 inhibitors are currently exploited is in the treatment of HIV infection, during which sufficient plasma levels of HIV protease inhibitors (PIs), the key components of highly active antiretroviral therapy, must be maintained in order to prevent the emergence of drug-resistant variants. Co-administration of CYP3A4 inactivators, such as ritonavir and cobicistat (Fig 1), enhances pharmacokinetics and improves clinical efficacy of PIs that otherwise are quickly metabolized by CYP3A4 [3, 4]. Both ritonavir and its derivative cobicistat were developed based on the chemical structure/activity relationships rather than the CYP3A4 crystal structure and, therefore, the precise mechanism of their action remained unclear. To better understand the structural basis of CYP3A4 inactivation, we undertook a series of studies on ritonavir and its analogues. The results, as well as strategies for a rational CYP3A4 inhibitor design, are summarized in this review.
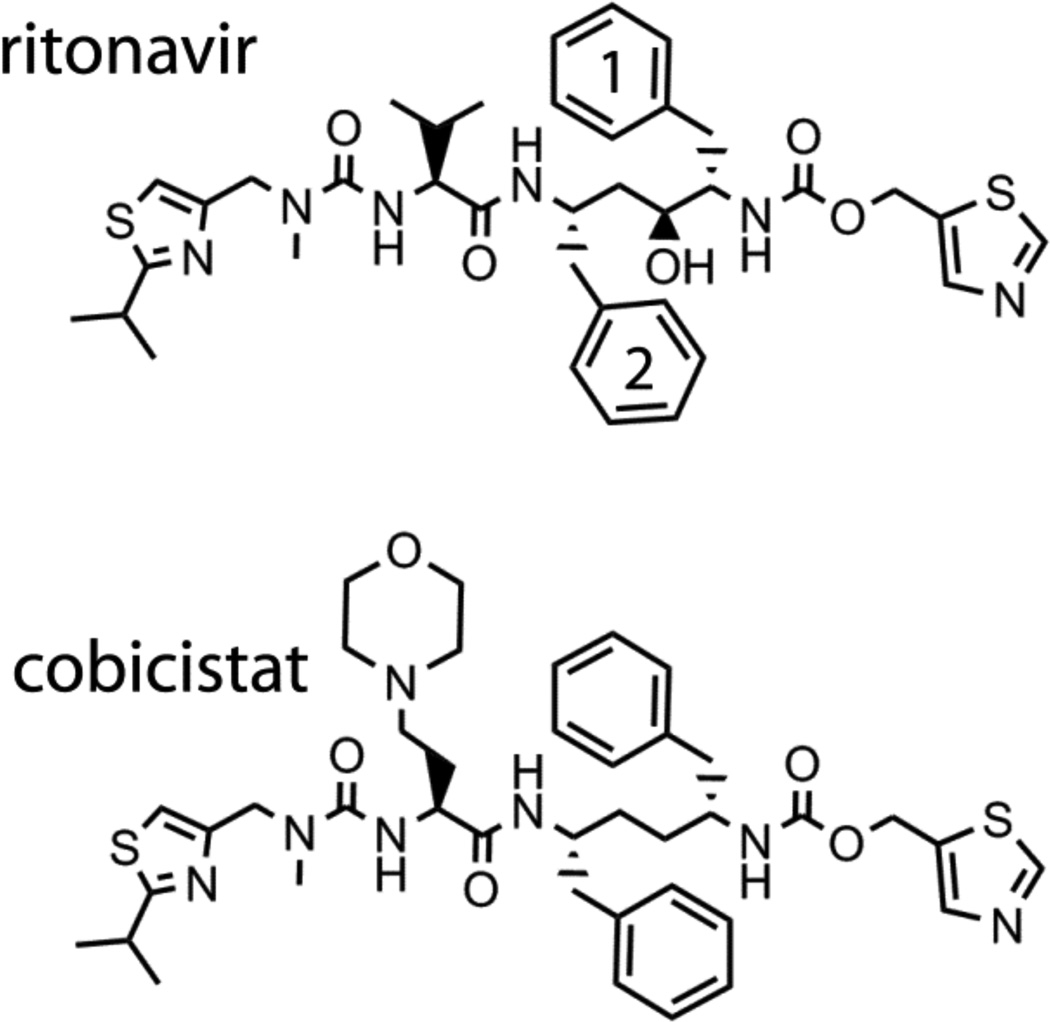
Figure 1.
Chemical structures of ritonavir and cobicistat.
2. Interaction of CYP3A4 with ritonavir
Ritonavir (ABT-538) is a peptidomimetic drug designed by Abbott Laboratories to inhibit HIV-1 protease [5]. It contains a thiazole head group, a mid-portion with two phenyl (Phe-1 and Phe- 2), one valine side groups, and the isopropyl-thiazole (IPT) end moiety (Fig. 1). In addition to acting as a PI, ritonavir potently inactivates the CYP3A family of enzymes [3]. Owing to this property, ritonavir is currently prescribed to HIV patients as a booster to enhance pharmacokinetics and clinical efficiency of other anti-HIV drugs metabolized by CYP3A4.
Earlier studies suggested that ritonavir is a mechanism-based inhibitor that upon oxidation converts into a reactive intermediate and selectively inactivates CYP3A4 by irreversibly attaching to the heme and/or active site amino acid residues. This was evidenced by a non- linear course of the ritonavir metabolism reaction, an increased potency (lower IC50) after preincubation with microsomes [6, 7] and formation of a covalent adduct absorbing at 450–460 nm [8]. The identity of reactive metabolites of ritonavir, however, remains unknown. Furthermore, the results of other studies argued against the mechanism-based CYP3A4 inhibition9 or indicated that ritonavir acts as a competitive [10] or mixed competitive-noncompetitive inactivator of CYP3A4 [11–13].
To resolve this controversy and gain functional and structural insights into the mechanism of CYP3A4 inhibition, we investigated the interaction of ritonavir with recombinant CYP3A4 [14]. Spectroscopic data showed that ritonavir is an irreversible, high affinity type II ligand that upon binding causes a red shift in the Soret band (416 to 421 nm) and lowers the heme redox potential by 20 mV. Ritonavir associates equally well to ferrous CYP3A4 (spectral dissociation constant (Ks) of 50 nM) and can displace substrates and other type II ligands bound to the active site. Another characteristic spectral feature is the 442 nm absorption of the ferrous ritonavir-bound CYP3A4, indicative of a strong σ-donor nitrogen ligation (Fig. 2A).
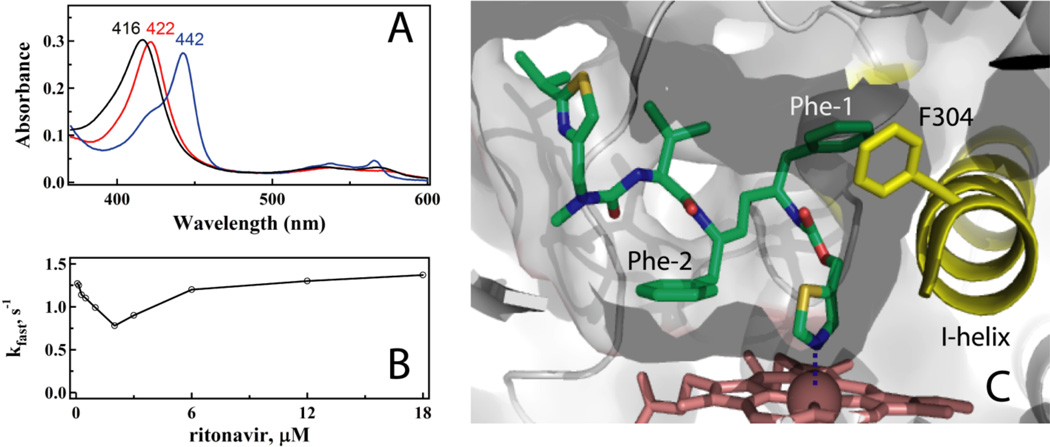
Figure 2.
A, Absorbance spectra of the ferric ligand-free and ritonavir-bound CYP3A4 (black and red, respectively) and its ferrous ritonavir-bound form (blue). Absorbance maxima for each species are indicated. B, Atypical (V-shaped) dependence of the rate constant for the ritonavir binding reaction on ligand concentration. C, Active site of ritonavir-bound CYP3A4.
Kinetics of CYP3A4-ritonavir binding was first investigated under conditions with the inhibitor in excess [14]. The reaction was biphasic, with the limiting rate constant for the fast phase (kfast) of 1.4 s−1 and the kinetic dissociation constant (Kd) of 0.9 µM. To better understand why there was a 18-fold difference between the Kd and Ks values, we reexamined the ritonavir binding reaction using a wider range of inhibitor concentrations that included both sub- and supra-equimolar ligand:protein ratios [15]. In this case, the kobs vs [ritonavir] plot was not hyperbolic but V-shaped, with the minimum at a ligand:protein ratio near unity (Fig. 2B). Such complex binding kinetics can be rationalized by the elongated shape of the drug, allowing entry into the active site of either thiazole or the isopropyl-thiazole first, or/and high the hydrophobicity and association with a peripheral site prior to translocating inside the active site cavity.
Importantly, ritonavir has no effect on NADPH consumption by cytochrome P450 reductase (CPR) but markedly decreases CPR-to-CYP3A4 electron flow [14]. Furthermore, the ratio between IC50s for the 7-benzyloxy-4-(trifluoromethyl) coumarin (BFC) hydroxylase activity of CYP3A4 measured under conditions of co- and pre-incubation with NADPH is close to unity [16]. These experimental data argue against the mechanism-based type of inhibition and rather suggest that ritonavir is a type II ligand that binds tightly and irreversibly both to ferric and ferrous CYP3A4 and, by decreasing the heme redox potential, prevents reduction by CPR.
The x-ray structure of the CYP3A4-ritonavir complex provided valuable insights into the ritonavir binding mode and protein-ligand interactions [14]. As spectrally predicted, ritonavir ligates to the heme via the thiazole nitrogen, with the Fe-N distance of 2.2–2.3 Å (Fig. 2C). Most of hydrophobic interactions are formed by the Phe-1 and Phe-2 groups. Phe-1 is embedded into a hydrophobic pocket near the I-helix (Phe-1 site) comprised by Phe108, Leu210, Leu211, Phe241, Ile 301, and Phe304. Clashing between Phe-1 and Phe304 leads to distortion in the I-helix. The second phenyl group orients parallel to the heme. However, since only the edges of the porphyrin and Phe-2 rings overlap, efficient π-π aromatic interactions cannot be established. Because of steric hindrance between Phe-2 and Ala371, the 369–371 peptide and the heme plane shift downwards by 2.0 and 0.6 Å, respectively. The valine side group and IPT are surrounded by the Tyr53, Phe57, Phe213 and Phe215 rings and almost fully sequestered from solvent. At 2.0 Å resolution, only six water molecules were detected in the active site, one of which connects the ITP nitrogen to a ‘polar umbrella’, a cluster of polar and charges residues (Asp61, Asp76, Arg106, Arg372 and Glu374). The aforementioned interactions allow ritonavir to associate with CYP3A4 very tightly although not in a perfect binding mode.
3. Critical importance of the heme ligating moiety
Since the nitrogen atom ligation does not necessarily lead to a potent CYP3A4 inhibition, we next attempted to elucidate what structural elements that contribute most to inhibition and the ligand binding process by using different sets of ritonavir analogues.
3.1 Effect of the thiazole nitrogen or entire thiazole group elimination Deaza-ritonavir (DAR)
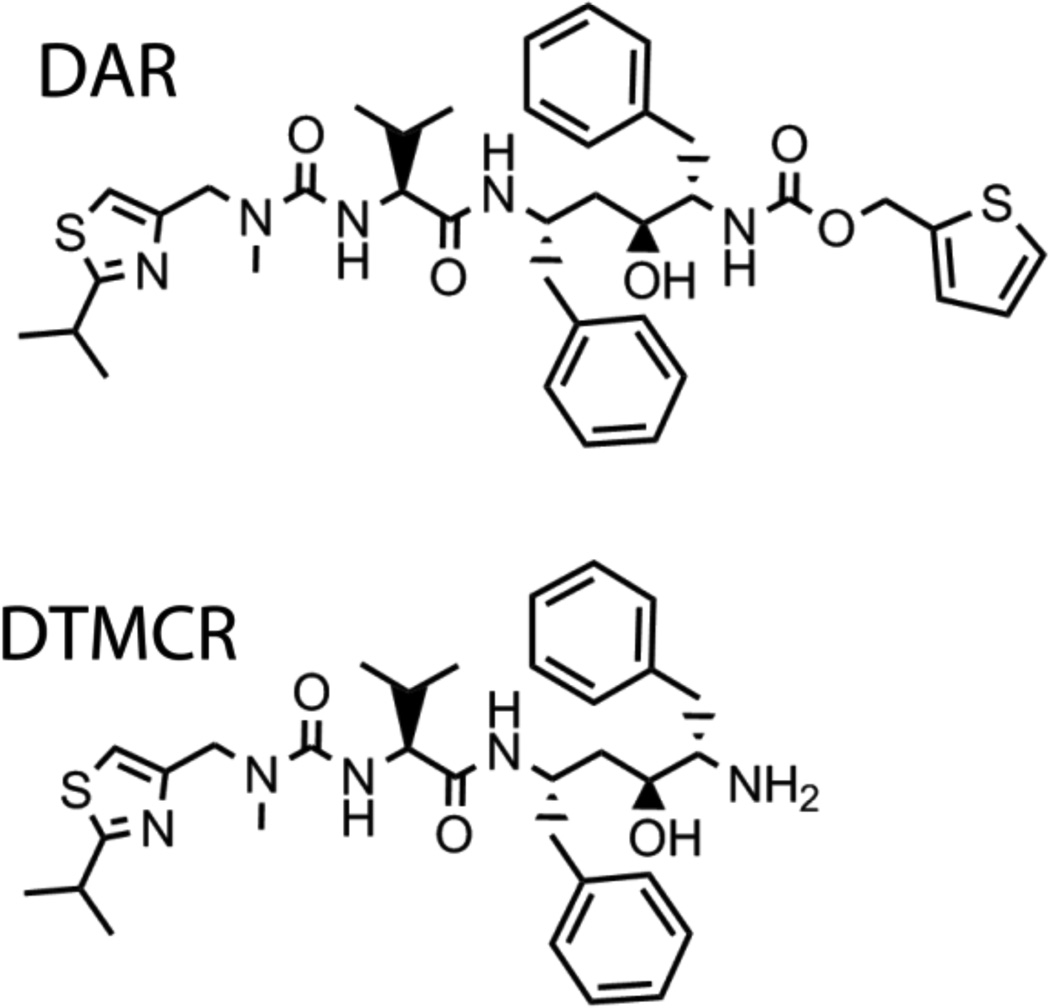
DAR lacks the heme ligating nitrogen atom (Fig. 3) and induces a partial 416 to 395 nm shift in CYP3A4 [15]. This type I spectral change indicates that DAR causes a low-to-high spin shift in the heme iron by displacing the distal water ligand. The DAR affinity for CYP3A4, however, is 10-fold lower than that of ritonavir (Ks of 0.5 µM). Another notable feature is the declining dependence of kfast on DAR concentration, suggesting that there is a rate-limiting process preceding heme ligation which becomes more dominant when the ligand concentration is increased. This phenomenon could not be solely explained by the DAR association to a peripheral docking site and may, in part, result from an increasing interference between the DAR molecules within the active site cavity. Unfortunately, we could not determine the CYP3A4-DAR complex structure because, similar to other type I ligands, DAR dissociates from CYP3A4 during crystallization.
Figure 3.
Chemical structures of DAR and DTMCR.
Desthiazolylmethyloxycarbonyl ritonavir (DTMCR)
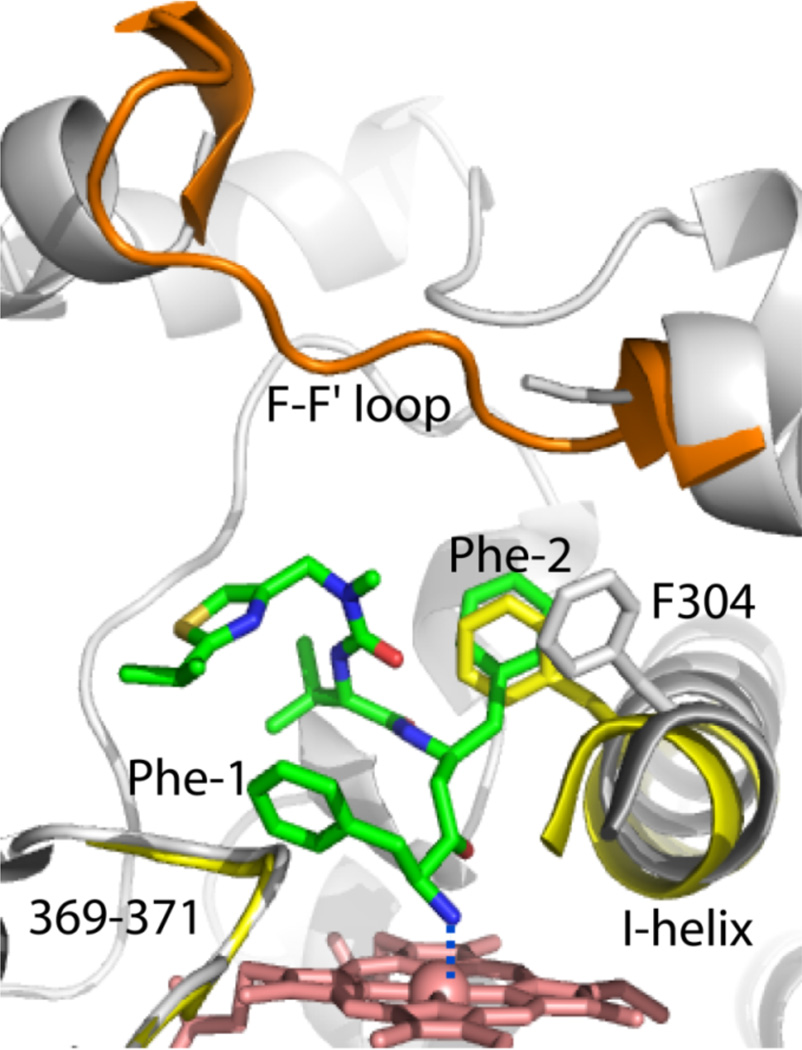
DTMCR is missing the entire thiazole head group (Fig. 3) but ligates to CYP3A4 via the primary amino group nearly as fast as ritonavir, albeit reversibly and 70-fold weaker (Ks of 0.4 µM) [15]. DTMCR inhibits the BFC hydroxylase activity of CYP3A4 with IC50 of 6.5 µM and remains bound to the active site during crystallization. One striking dissimilarity with the ritonavir binding mode is a 180° rotation of DTMCR around the heme-ligating nitrogen [15]. Such rearrangement is necessary for optimization of hydrophobic interactions established by the phenyl side groups (Fig. 4): Phe-2 occupies the Phe-1 site and displaces Phe304 and the I-helix to the same extent as Phe-1 in the ritonavir complex, whereas Phe-1 orients perpendicular to the heme plane and further from the 369–371 peptide. Since there is no steric clashing with Ala370 and the heme shift, the Fe-N bond becomes 0.1–0.2 Å shorter than in the ritonavir-bound structure. Overall, however, DTMCR establishes fewer hydrophobic contacts than ritonavir and no specific polar interactions. Since this analogue is shorter, its IPT cannot approach the ‘polar umbrella’ or the F-F’ loop, which leaves the 212–218 peptide disordered and the active site open to the solvent.
Figure 4.
Binding mode of DTMCR. DTMCR (green) ligates to CYP3A4 via the terminal amino group and rotates by 180° relative to ritonavir in order to place the Phe-2 group into the Phe-1 pocket. The F-F’ loop is well ordered in the ligand-free protein (shown in orange) but disordered in the CYP3A4-DTMCR complex. DTMCR does not clash with the 369–371 peptide but displaces the I-helix as much as ritonavir does. The I-helix and the 369–371 peptide of the ligand-free form are displayed in yellow.
Comparison of the ritonavir, DAR and DTMCR data allowed us to conclude that heme ligation is the major force that guides association of ritonavir-like inhibitors, whereas non-bonded contacts provided by the side groups define and optimize the orientation.
3.2 Effect of the backbone hydroxyl group elimination and thiazole substitution
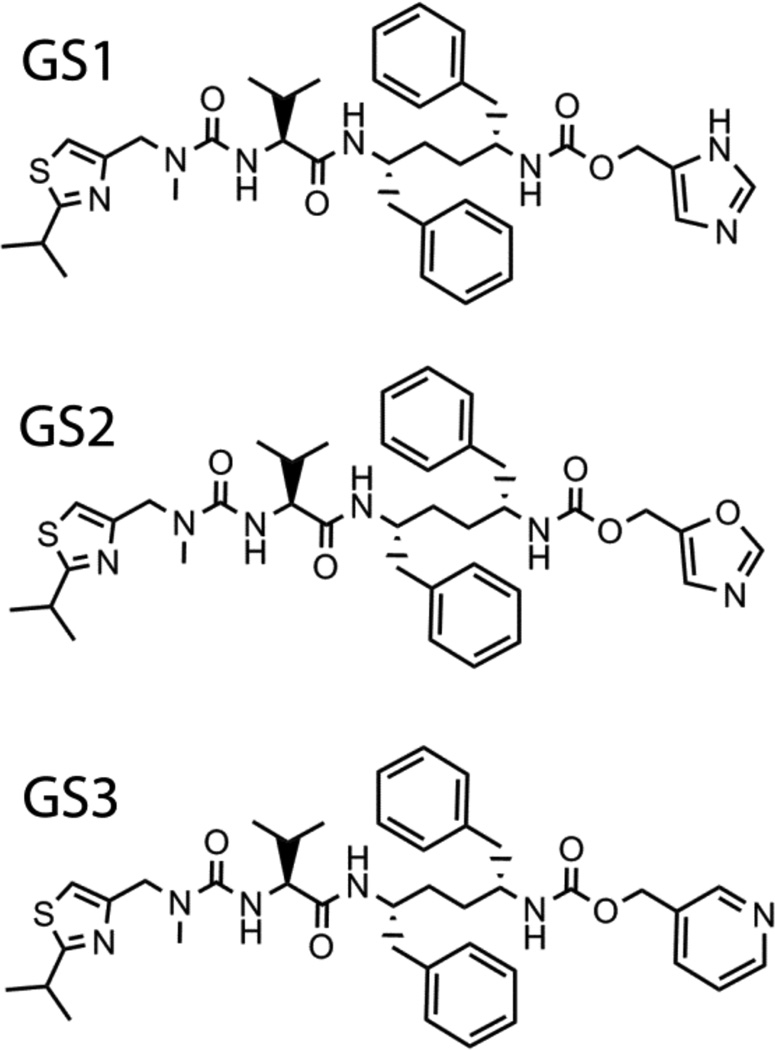
The ritonavir backbone hydroxyl group anchors the inhibitor to the catalytically important Asp25 of HIV-1 protease (Protein Data Base structure 2B60). Since ritonavir is prescribed in sub-therapeutic doses, its prolonged usage may lead to the development of drug-resistant HIV strains. To eliminate the undesired anti-HIV activity and other complications associated with ritonavir, Gilead Sciences developed another pharmacoenhacer, cobicistat (GS-9350) [4, 17], that has the desoxyritonavir backbone and a morpholine ring instead of the valine side group (Fig. 1). Cobicistat acts on the CYP3A enzymes via a similar mechanism but with a slightly lower effectiveness than ritonavir (kinact/Ki of 0.57 vs. 0.90 min−1 µM−1respectively) [4]. Spectral characteristics and the x-ray structure of the CYP3A4-cobicistat complex have not been reported. In our studies, we tested two sets of desoxyritonavir analogues, one of which contained compounds with the imidazole, oxazole or pyridine groups instead of the heme-ligating thiazole (GS1, GS2 and GS3, respectively; Fig. 5) [16].
Figure 5.
Chemical structures of GS1, GS2 and GS3.
GS1
Imidazole-containing GS1 induces the largest red shift in the Soret band (416 to 424 nm) and binds to CYP3A4 reversibly, with high affinity (Ks of 22 nM), and slightly faster than ritonavir (kfast of 1.9 s−1) [16]. Upon binding, GS1 causes a large decrease in the heme redox potential (>70 mV), due to which the GS1-bound species cannot be fully reduced by artificial or natural electron donors. Owing to this property, GS1 potently inhibits the BFC hydroxylase activity of CYP3A4 (IC50 of 280 nM). Similar to DAR, GS1 dissociates from CYP3A4 during crystallization.
GS2
Oxazole-containing GS2 is also a reversible type II ligand that has a 10-fold lower affinity and 6-fold lower inhibitory potency for CYP3A4 than ritonavir (Ks and IC50 are 0.6 and 3.4 µM, respectively) [16]. Although GS2 remains bound to the active site during crystallization, electron density is observed only for the heme-ligating oxazole and phenyl side groups, with the rest of the molecule being disordered.
GS3
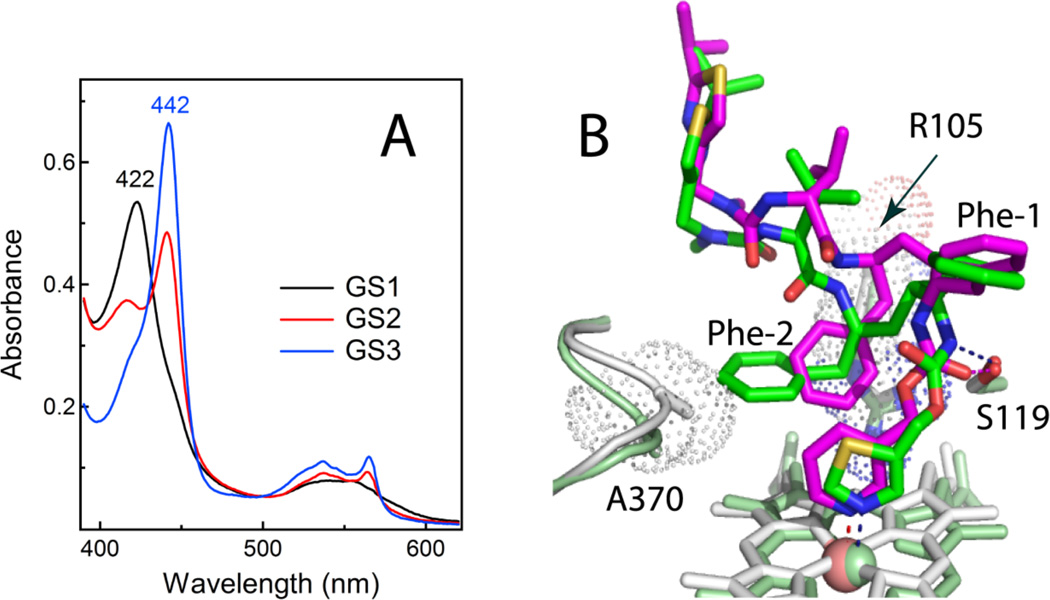
Pyridine-containing GS3 binds to CYP3A4 2-fold tighter and 4-fold faster than ritonavir (Ks and kfast are 25 nM and 7.0 s−1, respectively), and inhibits BFC hydroxylation with a 4-fold lower IC50 (130 nM) [16]. Similar to the ritonavir-bound form, the reduced CYP3A4-GS3 complex fully converts to the 442 nm absorbing species, as opposed to partial or negligible 442 nm absorption observed for the ferrous GS2- and GS1-bound protein (Fig. 6A). GS3 displaces GS1, GS2 and other type II and type I ligands from the active site, and co-crystallizes with CYP3A4.
Figure 6.
A, Absorbance spectra of the ferrous GS1-, GS2- and GS3-bound CYP3A4 (black, red and blue, respectively). B, Superposition of the ritonavir- and GS3-bound CYP3A4 structures (green/pale green and magenta/gray, respectively). GS3 has a more flexible backbone and places Phe-2 between the heme-ligating pyridine and the Arg105 guanidinium groups, thereby promoting π-π and cation-π interactions and preventing steric hindrance with the 369–371 peptide. Another notable difference with ritonavir is a peptide bond flip, due to which the GS3 carbonyl oxygen rather than amide nitrogen forms a hydrogen bond with Ser119 (shown as dotted lines).
In the crystal structure, GS3 adopts a more optimal conformation than ritonavir (Fig. 6B). Owing to a more flexible desoxyritonavir backbone, GS3 curves differently and positions Phe-2 further from the 369–371 peptide, thereby preventing steric clashing and elongation of the Fe-N bond. Most importantly, Phe-2 is now sandwiched between the near parallel heme-ligating pyridine and Arg105 guanidinium group. This promotes π-π and cation-π interactions that cannot be established by ritonavir. The GS3 conformation is further stabilized by a peptide bond flip that brings the amide nitrogen instead of the carbonyl oxygen closer to the Ser119 hydroxyl group, thereby leading to stronger hydrogen bonding (Fig. 6B). Mutagenesis data indicate that this Ser119-mediated interaction significantly affects the affinity and heme ligation kinetics [16]. There are no notable differences at the Phe-1 site, where Phe304 and the I-helix are displaced to the same extent as in the ritonavir-bound structure. Since the tail part of GS3 is not seen in the crystal structure, it is unclear how/whether IPT contributes to the ligand binding process.
It is evident from the experimental and structural data, however, that GS3 binds stronger and inhibits CYP3A4 more potently than ritonavir because of more favorable stereoelectronic properties of the pyridine nitrogen and a more flexible desoxyritonavir backbone, enabling GS3 to establish a stronger Fe-N bond and adopt a conformation that minimizes steric clashing and optimizes protein-ligand interactions.
4. Effect of substitution/elimination of the side groups and IPT removal
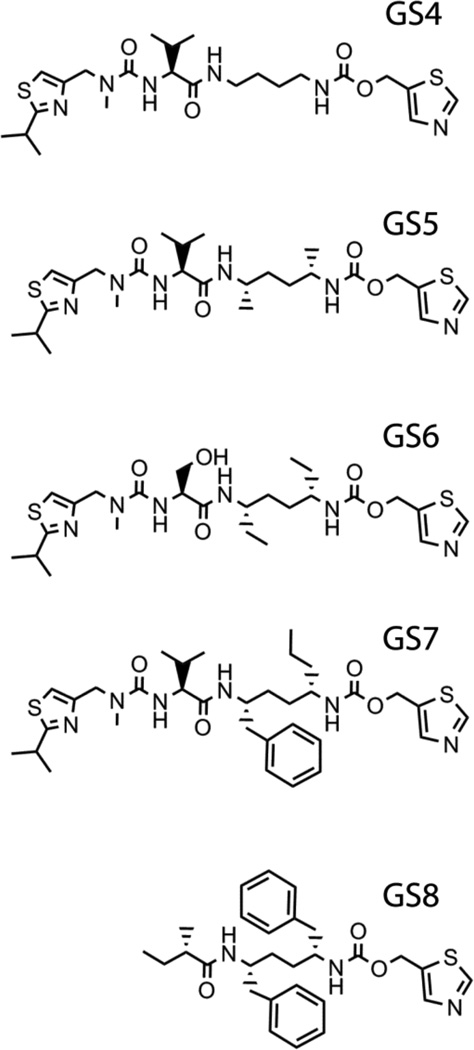
The second set of desoxyritonavir analogues to be examined includes GS4-GS8 compounds (Fig. 7) where either the phenyl side groups were substituted/eliminated or IPT removed [18]. All analogues induced type II spectral changes in CYP3A4 but significantly varied in the affinity and the binding mode. The heme binding reaction was biphasic for all ligands, with kfast close to that for ritonavir. The kobs on [ligand] dependence, however, was hyperbolic only for the side-group- lacking GS4 and atypical for other compounds.
Figure 7.
Chemical structures of the GS4-GS8 compounds.
GS4 and GS5
Elimination and methyl-group substitution of the side chains had the most drastic effect on the interaction with CYP3A4. Unlike other analogues, GS4 and GS5 have two binding sites, significantly differing in affinities (Ks of 1.5 µM/13 µM and 0.4 µM/7 µM, respectively), and distinct association modes.
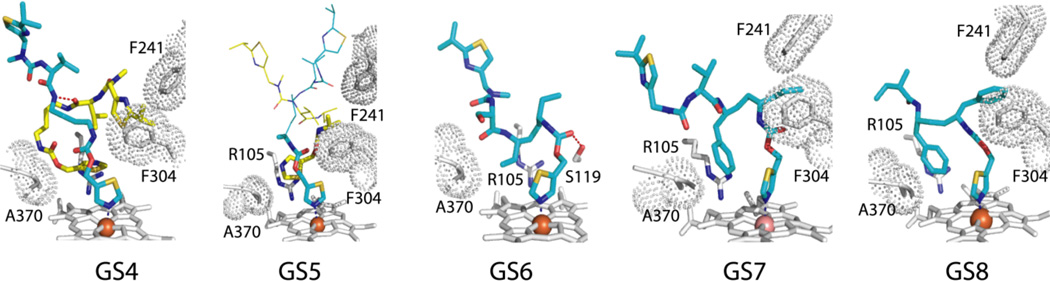
The CYP3A4-GS4 structure is most interesting, as both GS4 molecules, GS4-1 and GS4-2, are very well defined (Fig. 8). To accommodate GS4-2, GS4-1 binds in an extended conformation and rotates by 180° around the heme-ligating nitrogen. GS4-2, in turn, curls around the mid-portion of GS4-1 and fills the Phe-2 and Phe-1 pockets with the head and end groups, respectively. This orientation is fixed through three hydrogen bonds formed by GS4-2 with GS4- 1, Phe108 and Arg372. Another distinguishing feature is perpendicularly orientated GS4-1 and GS4-2 thiazoles, promoting S-π rather than π-π stacking interactions formed by other ligands. Notably, GS4-2 imposes no steric hindrance on the 369–371 peptide but displaces the I-helix as much as ritonavir and GS3 do.
Figure 8.
GS4-GS8 binding mode. Hydrogen bonds between GS4-1 and GS4-2 molecules and GS6 and Ser119 are shown as red dotted lines. The GS5-1 and GS5-2 segments shown as thin lines are not observed in the crystal structure and were modeled arbitrarily.
In the CYP3A4-GS5 complex, both GS5-1 and GS5-2 bind in an extended conformation, with the tail parts being completely disordered (Fig. 8). Similar to GS4-1, the heme-ligating GS5-1 rotates by 180° to allow GS5-2 to bind along and place the thiazole group and the mid-section into the Phe-2 and Phe-1 pockets, respectively. Again, the GS5-2 binding leads to a notable I-helix displacement but causes no hindrance with the 39–371 peptide.
GS6
In addition to two ethyl-side chains, GS6 has a valine-to-serine substitution (Fig. 7) and binds to CYP3A4 at a single site with Ks of 2.3 µM. Compared to other analogues, GS6 adopts a conformation with a deeper backbone curvature in order to fill the Phe-2 pocket with the ethyl and main chain atoms, leaving the Phe-1 site unoccupied (Fig. 8). Owing to higher polarity of the serine side group, the GS6 mid-portion shifts to form polar contacts with the main chain atoms of the nearby residues. GS6 is the only analogue in the set that forms an H-bond with Ser119.
GS7
There is only one phenyl-to-propyl side group substitution in GS7 (Fig. 7). The analogue has a high affinity for CYP3A4 (Ks of 40 nM) and adopts a GS3-like conformation, with the propyl group placed at the Phe-1 site (Fig. 8). This requires a 30° rotation of the thiazole ring and stretching in the connecting fragment which prevents H-bonding with Ser119. Despite the smaller size and higher flexibility of the propyl side group, GS7 distorts the I-helix similarly to GS3. Since IC50 for GS7 is several-fold higher than for GS3 and ritonavir, Phe-1 appears to be the element that imparts inhibitory potency.
GS8
GS8 lacks the IPT tail (Fig. 7) but has a relatively high affinity and inhibitory potency for CYPA4 (Ks and IC50 of 160 nM and 1.3 µM, respectively). The GS8 binding mode is highly similar to that of GS3, with nearly identical contacts established by the phenyl side groups (Fig. 8). Without the IPT-mediated interactions, however, the F-F’ loop becomes disordered and the active site opened to the solvent. This increases motional freedom of GS8 which, in turn, leads to a loss of the H-bond with Ser119 and lowers the binding affinity.
5. Strategies for a structure-guided CYP3A4 inhibitor design
Comparative analysis of spectral properties, binding affinity, heme ligation kinetics, and inhibitory potency of ritonavir analogues has revealed several important trends in the CYP3A4- ligand binding process that help define strategies for rational, structure-based inhibitor design. The first key finding was that strong and irreversible binding through the heme-ligating moiety is required for potent CYP3A4 inhibition and that the pyridine head group possesses more favorable stereoelectronic properties and establishes a stronger Fe-N bond than any other chemical groups tested.
Second, our studies showed that, in addition to elimination of the undesired anti-HIV protease activity4, the hydroxyl group removal from the ritonavir backbone has another beneficial effect which is an increased backbone flexibility that enables a better fit into the CYP3A4 active site. That neither of the investigated desoxyritonavir analogues imposes steric hindrance on the 369- 371 peptide suggests that this clash is undesirable and, therefore, should be avoided during drug design.
The next trend is a preferential occupancy of the Phe-2 site by all investigated ligands (Fig. 9A). Regardless of the side group nature and the number of molecules bound, this site is occupied first and, hence, is critically important. The aromatic moiety at the Phe-2 position is preferred because it not only fills the hydrophobic void but also establishes stabilizing π-π and cation-π interactions with the heme-ligating group and the guanidinium group of nearby Arg105. Since the Phe-2 site is spacious, the phenyl moiety can be substituted by a larger indole or bi-phenyl ring, which may provide more optimal aromatic stacking interactions with the heme-ligating group.
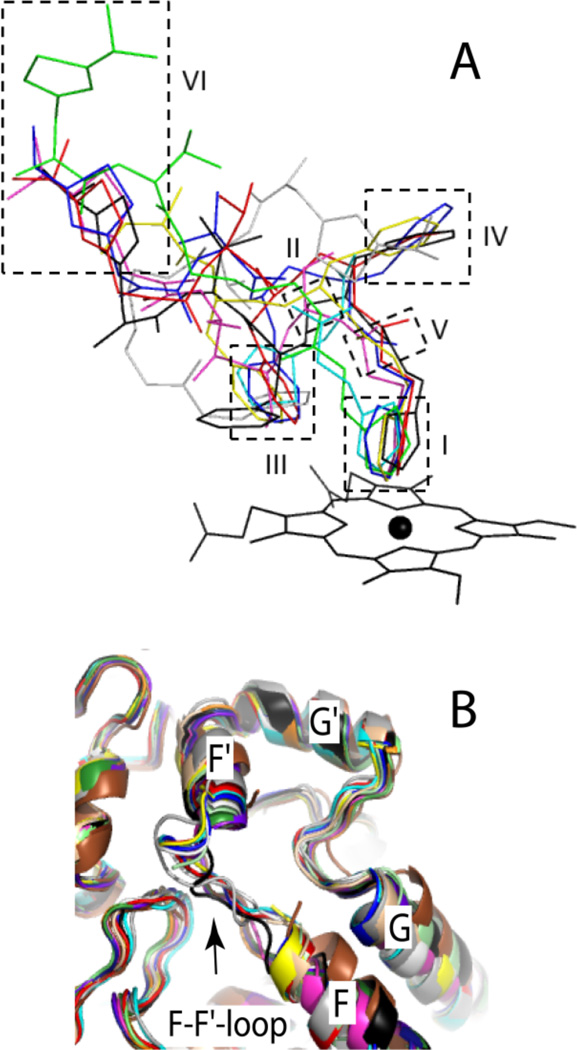
Figure 9.
A, Superposition of the ritonavir- and analogue-bound structures of CYP3A4. Ritonavir is shown in black; GS3 in blue; GS4-1 and GS4-2 are in green and gray, respectively; visible parts of GS5-1 and GS5-2 are in cyan; GS6 in magenta; GS7 in red; and GS8 in yellow. Molecular features defining the ligand binding affinity are boxed and numbered according to their contribution. I – the heme-ligating group is most important as it drives the ligand binding reaction and determines the strength and reversibility of the Fe-N bond. II – the desoxyritonavir backbone is preferable because it is more flexible and allows ligands to adopt a more optimal conformation in the active site. III – the aromatic moiety in Phe-2 position promotes π-π stacking interactions with the heme-ligating group and cation-π interactions with the Arg105 guanidinium group. IV – a bulky hydrophobic group at Phe-1 position is required for potent CYP3A4 inhibition. V – a hydrogen bond to Ser119 via the backbone amide nitrogen or carbonyl oxygen facilitates the CYP3A4-ligand association and stabilizes the ligand-bound form. VI – a poly-functional end-group can contribute to the binding affinity through polar, hydrophobic, π- π, S-π and possibly other interactions established with the residues of ‘polar umbrella’ and the F-F’-loop. B, Superposition of all available CYP3A4 structures showing that the F-F’-loop is very flexible and either adopts a unique conformation to optimize ligand-protein contacts or becomes disordered. Color coding: 1TQN – deep purple, 1WOE – green, 1WOF – orange, 1WOG – pale yellow, 2JOD – brown, 2VOM – gray, 3NXU – cyan, 3UA1 – purple, 3TJS – beige, 4I3Q – dark green, 4I4G – white, 4I4H – blue, 4K9T - black, 4K9U – pale green, 4K9V - magenta, 4K9W – red, and 4K9X – yellow. To simplify viewing, all ligands were excluded from the active site.
The Phe-1 pocket is also critical and filled by the ligands, when possible, even if it leads to I-helix distortion. The resulting hydrophobic interactions greatly improve inhibitory potency, with no apparent penalty for a conformational restraint. Since the phenyl ring penetrates the Phe-1 pocket as deeply as the smaller and more flexible propyl chain (Fig. 9) and is strictly required for potent CYP3A4 inhibition, whether its replacement with a more flexible hexane or smaller pentane ring would be more beneficial or not should be investigated.
Polar interactions with Ser119 is another factor that affects ligand affinity and association rate, regardless of whether the hydrogen bond with the ligand is established or not.16, 18 However, the H-bonding to Ser119 is desirable as it promotes binding and stabilizes the ligand-bound form. Unfortunately, the only way to check whether the H-bond is established or not is determination of the CYP3A4-ligand complex structure, which is not always possible. There are two quick and simple procedures though that could help identify strong CYP3A4 binders. During our studies, we found that ligand-dependent shifts in the CYP3A4 melting temperature and the amplitude of the 442 nm absorption of the ferrous ligand-bound forms are proportional to the binding affinity and the Fe-N bond strength.16 Therefore, thermal denaturation and spectral measurements on the ferrous ligand-bound CYP3A4 could be used as screening methods at initial stages of drug design.
At the moment, there is insufficient data for making conclusions on the relative role of IPT in the desoxyritonavir analogue binding process. The end group of ritonavir is known to be strictly required for potent CYP3A4 inhibition [5, 19] and could be responsible for production of reactive intermediates [6]. Our studies suggest that IPT does promote ligand association by mediating interactions with the ‘polar umbrella’ and the F-F’-loop but it is not as critical as the side groups. Depending on orientation, IPT can establish hydrophobic, polar, π-π and/or S-π interactions, through which the affinity and stability of the CYP3A4-inhibitor complex can be improved. The type and extent of protein-ligand interactions could be defined by the F-F’-loop orientation as well. As the x-ray data showed (Fig. 9B), this flexible peptide accommodates different conformations in response to structural changes in the ligand to maximally optimize protein-ligand contacts. Owing to the remarkable plasticity and adaptability of the F-F’ loop, it is not possible to predict the most optimal chemical group for the end-moiety without testing the CYP3A4 interaction with a wider range of desoxyritonavir analogues. Nonetheless, based on the available results, summarized in Figure 9A, we derived a pharmacophore (Fig. 10) that can guide a rational design of structure-based CYP3A4 inactivators.
Figure 10.
Pharmacophore derived from the structure/function studies on ritonavir and 10 analogues. Pharmacophoric features are: flexible backbone (the center carbon atom is shown in black), strong heme-ligating nitrogen donor (blue), hydrogen donor/acceptor (red), aromatic and hydrophobic moieties (cyan and green, respectively), and poly-functional end-group (yellow).
Acknowledgements
Financial support from the National Institute of General Medical Sciences (Grant GM33688), the California Center for Antiviral Drug Discovery, and Gilead Sciences is gratefully appreciated.
Abbreviations
- BFC
7-benzyloxy-4-(trifluoromethyl) coumarin
- CYP3A4
cytochrome P450 3A4
- CPR
cytochrome P450 reductase
- DAR
deaza-ritonavir
- DTMCR
desthiazolylmethyloxycarbonyl ritonavir
- IPT
isopropyl thiazole
- PI
protease inhibitor
References
- 1.Guengerich FP. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu. Rev. Pharmacol. Toxicol. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Sevrioukova IF, Poulos TL. Understanding the mechanism of cytochrome P450 3A4: recent advances and remaining problems. Dalton Trans. 2013;42(9):3116–3126. doi: 10.1039/c2dt31833d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kempf DJ, Marsh KC, Kumar G, Rodrigues AD, Denissen JF, McDonald E, Kukulka MJ, Hsu A, Granneman GR, Baroldi PA, Sun E, Pizzuti D, Plattner JJ, Norbeck DW, Leonard JM. Pharmacokinetic enhancement of inhibitors of the human immunodeficiency virus protease by coadministration with ritonavir. Antimicrob. Agents Chemother. 1997;41(3):654–660. doi: 10.1128/aac.41.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu L, Liu H, Murray B, Callebaut C, Lee MS, Hong A, Strickley RG, Tsai LK, Stray KM, Wang Y, Rhodes GR, Desai MC. Cobicistat (GS-9350): A potent and selective inhibitor of human CYP3A as a novel pharmacoenhancer. ACS Med. Chem. Lett. 2010;1:209–213. doi: 10.1021/ml1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kempf DJ, Marsh KC, Denissen JF, McDonald E, Vasavanonda S, Flentge CA, Green BE, Fino L, Park CH, Kong XP, et al. ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. Proc. Natl. Acad. Sci. U S A. 1995;92(7):2484–2488. doi: 10.1073/pnas.92.7.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koudriakova T, Iatsimirskaia E, Utkin I, Gangl E, Vouros P, Storozhuk E, Orza D, Marinina J, Gerber N. Metabolism of the human immunodeficiency virus protease inhibitors indinavir and ritonavir by human intestinal microsomes and expressed cytochrome P4503A4/3A5: mechanism-based inactivation of cytochrome P4503A by ritonavir. Drug. Metab. Dispos. 1998;26(6):552–561. [PubMed] [Google Scholar]
- 7.von Moltke LL, Durol AL, Duan SX, Greenblatt DJ. Potent mechanism-based inhibition of human CYP3A in vitro by amprenavir and ritonavir: comparison with ketoconazole. Eur. J. Clin. Pharmacol. 2000;56(3):259–261. doi: 10.1007/s002280000125. [DOI] [PubMed] [Google Scholar]
- 8.Ernest CS, Hall SD, Jones DR. Mechanism-based inactivation of CYP3A by HIV protease inhibitors. J. Pharmacol. Exp. Ther. 2005;312(2):583–591. doi: 10.1124/jpet.104.075416. 2nd. [DOI] [PubMed] [Google Scholar]
- 9.Sekiguchi N, Higashida A, Kato M, Nabuchi Y, Mitsui T, Takanashi K, Aso Y, Ishigai M. Prediction of drug-drug interactions based on time-dependent inhibition from high throughput screening of cytochrome P450 3A4 inhibition. Drug Metab. Pharmacokinet. 2009;24(6):500–510. doi: 10.2133/dmpk.24.500. [DOI] [PubMed] [Google Scholar]
- 10.Iribarne C, Berthou F, Carlhant D, Dreano Y, Picart D, Lohezic F, Riche C. Inhibition of methadone and buprenorphine N-dealkylations by three HIV-1 protease inhibitors. Drug. Metab. Dispos. 1998;26(3):257–260. [PubMed] [Google Scholar]
- 11.Eagling VA, Back DJ, Barry MG. Differential inhibition of cytochrome P450 isoforms by the protease inhibitors, ritonavir, saquinavir and indinavir. Br. J. Clin. Pharmacol. 1997;44(2):190–194. doi: 10.1046/j.1365-2125.1997.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar GN, Rodrigues AD, Buko AM, Denissen JF. Cytochrome P450-mediated metabolism of the HIV-1 protease inhibitor ritonavir (ABT-538) in human liver microsomes. J. Pharmacol. Exp. Ther. 1996;277(1):423–431. [PubMed] [Google Scholar]
- 13.Zalma A, von Moltke LL, Granda BW, Harmatz JS, Shader RI, Greenblatt DJ. In vitro metabolism of trazodone by CYP3A: inhibition by ketoconazole and human immunodeficiency viral protease inhibitors. Biol. Psychiatry. 2000;47(7):655–661. doi: 10.1016/s0006-3223(99)00176-6. [DOI] [PubMed] [Google Scholar]
- 14.Sevrioukova IF, Poulos TL. Structure and mechanism of the complex between cytochrome P4503A4 and ritonavir. Proc. Natl. Acad. Sci. U S A. 2010;107(43):18422–18427. doi: 10.1073/pnas.1010693107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sevrioukova IF, Poulos TL. Interaction of human cytochrome P4503A4 with ritonavir analogs. Arch. Biochem. Biophys. 2012;520(2):108–116. doi: 10.1016/j.abb.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sevrioukova IF, Poulos TL. Pyridine-substituted desoxyritonavir is a more potent cytochrome P450 3A4 inhibitor than ritonavir. J. Med. Chem. 2013;56(9):3733–3741. doi: 10.1021/jm400288z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathias AA, German P, Murray BP, Wei L, Jain A, West S, Warren D, Hui J, Kearney BP. Pharmacokinetics and pharmacodynamics of GS-9350: a novel pharmacokinetic enhancer without anti-HIV activity. Clin. Pharmacol. Ther. 2010;87(10):322–329. doi: 10.1038/clpt.2009.228. [DOI] [PubMed] [Google Scholar]
- 18.Sevrioukova IF, Poulos TL. Dissecting cytochrome P450 3A4-ligand interactions using ritonavir analogues. Biochemistry. 2013 doi: 10.1021/bi4005396. in press. [DOI] [PubMed] [Google Scholar]
- 19.Flentge CA, Randolph JT, Huang PP, Klein LL, Marsh KC, Harlan JE, Kempf DJ. Synthesis and evaluation of inhibitors of cytochrome P450 3A (CYP3A) for pharmacokinetic enhancement of drugs. Bioorg. Med. Chem. Lett. 2009;19(18):5444–5448. doi: 10.1016/j.bmcl.2009.07.118. [DOI] [PubMed] [Google Scholar]