Abstract
Centronuclear myopathy (CNM) is a rare congenital muscle disease characterized by fibers with prominent centralized nuclei in muscle biopsies. The disease is clinically heterogeneous, ranging from severe neonatal hypotonic phenotypes to adult-onset mild muscle weakness, and can have multiple modes of inheritance in association with various genes, including MTM1, DNM2, BIN1 and RYR1. Here we analyzed 18 sporadic patients with clinical and histological diagnosis of CNM and sequenced the DNM2 gene, which codes for the dynamin 2 protein. We found DNM2 missense mutations in two patients, both in exon 8, one known (p.E368K) and one novel (p.F372C), which is found in a position of presumed pathogenicity and appeared de novo. The patients had similar phenotypes characterized by neonatal signs followed by improvement and late childhood reemergence of slowly progressive generalized muscle weakness, elongated face with ptosis and ophthalmoparesis, and histology showing fibers with radiating sarcoplasmic strands (RSS). These patients were the only ones in the series to present this histological marker, which together with previous reports in the literature suggest that, when RSS are present, direct sequencing of DNM2 mutation hot spot regions should be the first step in the molecular diagnosis of CNM, even in sporadic cases.
Keywords: centronuclear myopathy, DNM2, dynamin 2, congenital myopathy
Centronuclear myopathy (CNM) is a rare, clinically heterogeneous, structural congenital myopathy characterized by the presence of centralized nuclei in the majority of muscle fibers in muscle biopsies that are not linked to excessive regeneration (Zanoteli et al., 1998). Three inheritance patterns are described: (i) X-linked recessive, which gives rise to a form also referred to as myotubular myopathy (MIM #310400), with a severe neonatal presentation marked by profound hypotonia and respiratory failure; this pattern is associated with the MTM1 gene (MIM *300415) (Laporte et al., 1996); (ii) autosomal dominant, causing a milder, neonatal to adult-onset muscle involvement (MIM #160150); this is related with mutations in the DNM2 gene (MIM *602378) (Bitoun et al., 2005); and (iii) autosomal recessive, with intermediate severity compared to the other forms (MIM #255200); the latter is caused by mutations in the genes BIN1 (MIM *601248) (Nicot et al., 2007) or RYR1 (MIM *180901) (Wilmshurst et al., 2010; Bevilacqua et al., 2011). In addition, heterozygous variants in the genes CCDC78 (MIM *614666) and MTMR14 (MIM *611089) were proposed to be associated with autosomal dominant early-onset centronuclear myopathy (Tosch et al., 2006; Majczenko et al., 2012), and more recently the genes TTN (MIM *188840) and SPEGwere put forward as a cause of recessive cases (Ceyhan-Birsoy et al., 2013; Agrawal et al., 2014). Disease severity and, more importantly, inheritance pattern dictate which genes should be studied to establish a molecular diagnosis. However, sporadic cases with both mild and severe phenotypes can presumably have any of the genes involved and thus present a diagnostic challenge.
Peculiar findings in muscle biopsies can help further to pinpoint the probably implicated gene (Romero, 2010), potentially reducing the time to molecular diagnosis. Large centralized nuclei within rounded muscle fibers similar to fetal myotubes and increased oxidative activity in the center of fibers are typical of cases associated with mutations in the MTM1 gene. The finding of fibers with radiating sarcoplasmic strands (aspect of “spoke of wheels”) suggests the DNM2 gene as the culprit. A majority of rounded fibers with clusters of centralized nuclei is suggestive of involvement of the BIN1 gene, while the presence of areas of myofibrillar disorganization devoid of mitochondria, such as moth-eaten or core-like structures, points to the RYR1 gene.
We screened a cohort of 18 Brazilian patients with sporadic CNM for mutations in the DNM2 gene. To select our CNM cohort, we analyzed sequential muscle biopsy reports from the last five years taken from two large neuromuscular center biopsy banks in Sao Paulo, Brazil (LIM15/FMUSP and Setor de Doenças Neuromusculares/UNIFESP), and screened for diagnoses of CNM, myotubular myopathy, or descriptions which included internal nuclei as a prominent feature, as long as the biopsy did not show a dystrophic pattern. A muscle pathology specialist then reviewed the biopsy slides to confirm the findings and to note specific features. Subsequently, patients were recruited for a clinical consultation and drawing of peripheral blood for genomic DNA extraction, after parents signed an informed consent approved by HC-FMUSP Ethics Committee (CAPPesq 0734/11). Following PCR amplification of its exons and intron-exon boundaries using specifically designed pairs of oligonucleotides (available upon request), the DNM2 gene was Sanger sequenced using an ABI 3130 sequencer (Applied Biosystems, Foster City, CA, USA). Sequences were compared to the wild-type DNM2 sequence (RefSeq NM_001005360.2), and variants identified were matched to a database of DNM2 variants in 100 control individuals in the Brazilian population, data from the Exome Variant Server (EVS) of NHLBI, and the variants database of the 1000 Genomes Project, to exclude polymorphisms. Potential mutations were compared to DNM2 mutation databases curated at the Leiden Open Variation Database (LOVD), at the Universal Mutation Database (UMD), and at the Human Gene Mutation Database (HGMD).
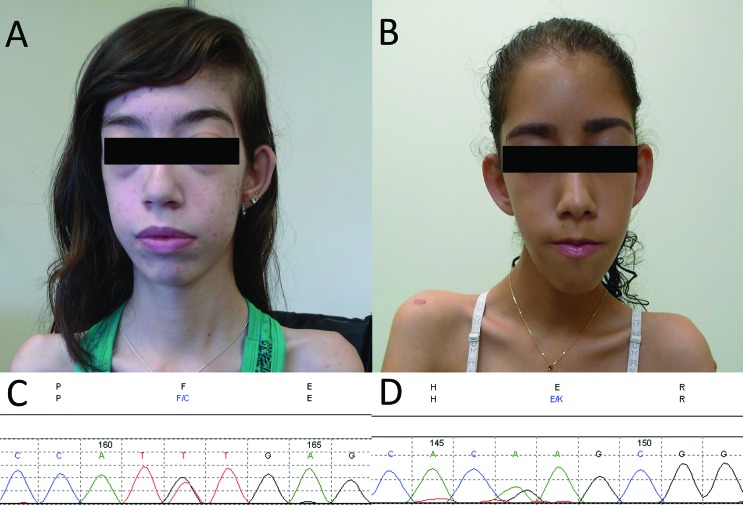
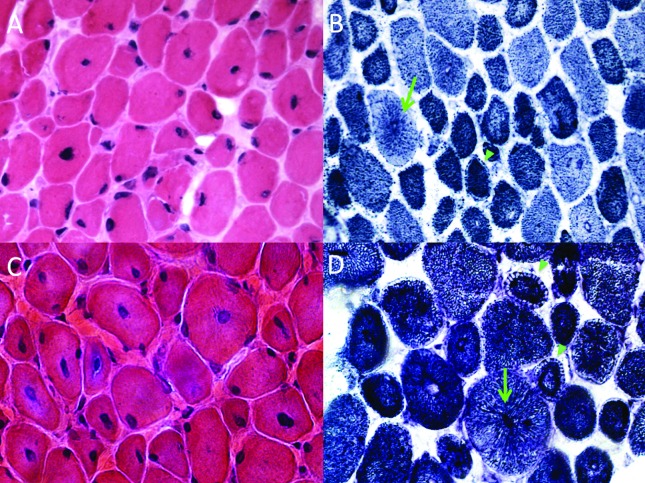
Two patients were found to carry DNM2 mutations. Patient 1 was a 20 year-old female born at term on cesarean section from a normal pregnancy. She was the fifth and last child and the only one affected of a non-consanguineous couple. From early on, the child was hypotonic, with weak suction and cry. However, she achieved normal infantile developmental milestones, having walked at 1 year and 2 months, and gaining ability to run and bike. At age 6, she started having feet deformity and scoliosis, which prompted medical investigation, and at the same time difficulty in climbing stairs and raising from low positions became apparent, with slow progression. At age 12, she became wheelchair bound, mainly due to the equinovarus feet deformities. Her motor ability remained stable thereafter. She recently started to show respiratory compromise, manifested with dyspnea upon exertion, morning headache and excessive daytime sleepiness, confirmed as a severe restrictive pattern in a pulmonary function test (FVC 18% of predicted for age and sex). Despite being wheelchair bound and not walking due to the bilateral feet deformity, she is able to keep standing if put in position with the aid of prosthetics. The physical exam showed an elongated face, asymmetric ptosis, bilateral ophthalmoparesis with eye movement restriction in all directions, bilateral facial weakness, high-arched palate and micrognathia (Figure 1A). There was generalized muscle atrophy, pectus excavatum, marked scoliosis, fixed bilateral knee and heel contractures. Multiple serum CK level measurements were normal, as well as echocardiography and EKG-Holter. Muscle biopsy performed at age 16 on the biceps brachii muscle showed increased fiber size variability with numerous centralized and internalized nuclei, moderate increase of endomysial connective tissue, in addition to a type 1 fiber predominance (Figure 2A). The NADH-TR oxidative reaction showed a typical radiating sarcoplasmic strands (“spoke of wheels”) fiber pattern (Figure 2B). Sanger sequencing of DNM2 uncovered the mutation c.1115T > G, p.F372C, in exon 8, in a heterozygous state (Figure 1C). Sanger sequencing of this exon in both parents was normal, indicating a de novo mutation.
Figure 1. Patients’ clinical features and DNM2 mutations. Both patients (A, Patient 1; B, Patient 2) presented an elongated face, facial weakness and bilateral ptosis. Patient 1 also had asymmetrical ophthalmoparesis. (C) heterozygous DNM2 mutation c.1115TG, p.F372C found in Patient 1. (D) heterozygous DNM2 mutation c.1102G > A, p.E368K found in Patient 2.
Figure 2. Muscle biopsy findings in the patients with DNM2 mutations. Patient 1: (A) histology showed moderate variability of muscle fiber size, fibers with prominence of centralized nuclei and mild increase of connective tissue (H&E stain, 40X); (B) fiber type architecture was abnormal, characterized, after oxidative stains, by fibers with radiating sarcoplasmic strands (aspect of “spoke of wheels”, green arrow), and ring-like fibers with a peripheral clear area (arrowheads) (NADH-TR, 40X). Patient 2: (C) as in Patient 1, the muscle biopsy showed a prominence of fibers with centralized nuclei and variable size, in addition to increased connective tissue (H&E stain, 40X); (D) oxidative stains also showed typical fibers with radiating sarcoplasmic strands (green arrow) and ring-like fibers (green arrowheads) (NADH-TR, 40X).
Patient 2, a 16-year old female, single child of non-consanguineous parents, was born at 8 months post amenorrhea after an uneventful pregnancy in which the mother noticed reduced fetal movements. She had low weight (1500 g) and mild neonatal hypoxia, requiring incubator therapy and O2 administration for one month. After hospital discharge, she had normal infantile development, acquiring motor and cognitive milestones at the expected ages. Until the age of 9 the patient had minimal motor involvement. Thereafter, she started to manifest muscle weakness in her lower limbs, with frequent falls and difficulty in climbing stairs, with continuous, albeit slow, worsening. She is now unable to rise from the floor or chair without help, but is ambulant if put in a standing position. She is slow to chew, but has no difficulty swallowing.
She has a typical congenital myopathy elongated face with ptosis, in addition to high-arched palate and slight facial weakness (Figure 1B). There is generalized muscular atrophy, with appendicular weakness predominantly in the lower limbs, proximal more than distal (MRC 4− vs.MRC 4+), marked fixed bilateral ankle contractures, and mild bilateral knee contractures. While she has normal head control, cervical flexors are involved (MRC 4). Serum CK level was always normal, EMG showed a myopathic pattern with short duration, small amplitude and polyphasic MUAPs. At the age of 13, she had a muscle biopsy performed in the deltoid muscle, which showed a typical centronuclear myopathy pattern with increased centralized and internalized nuclei in the majority of fibers (Figure 2C). There was also mild proliferation of endomysial connective tissue. The NADH-TR oxidative reaction showed fibers with radiated sarcoplasmic strands (Figure 2D). Sanger sequencing of DNM2 showed the heterozygous mutation c.1102G > A, p.E368K, already known to be pathogenic in dominant families (Figure 1D). Further sequencing of the parents confirmed the mutation as de novo.
The DNM2 gene, mapped to chromosome 19 at 19p13.2, codes for dynamin 2, a GTPase involved in intracellular vesicle trafficking and cytoskeleton remodeling with ubiquitous tissue expression (Bitoun et al., 2005). In addition to its association to autosomal dominant and sporadic forms of centronuclear myopathy, DNM2 is also mutated in axonal and dominant intermediate forms of Charcot-Marie-Tooth neuropathy (MIM #606482) (Bitoun et al., 2007, 2009; Züchner et al., 2005) and a recessive mutation is implicated in a lethal congenital contracture syndrome (MIM #615368) (Koutsopoulos et al., 2013). So far a total of 19 DNM2 mutations have been deemed responsible for the CNM phenotype, concentrated in hot spot regions encompassing exons 8, 11 and 16, followed by exons 14 and 15. These exons code for either the stalk or the pleckstrin homology (PH) protein domains (Böhm et al., 2012). Patients with mutations in exon 16 tend to present as a severe neonatal phenotype with profound hypotonia, whereas mutations in exon 8 are linked to mild or intermediate courses with neonatal to childhood onset.
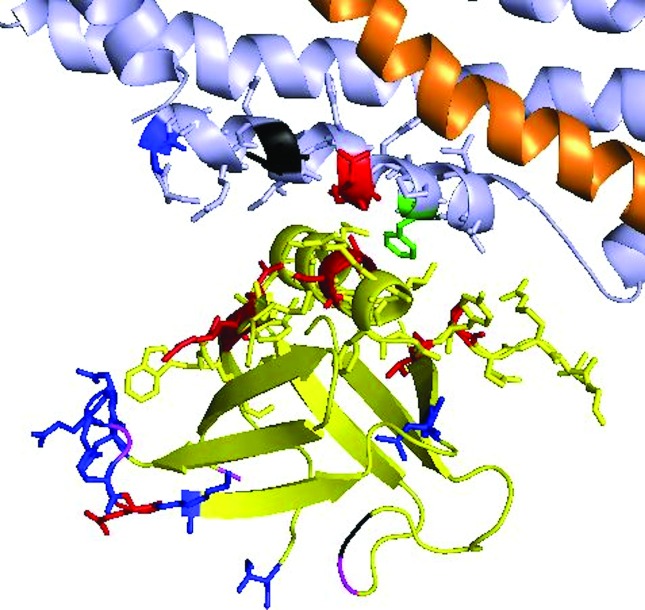
The mutation found in Patient 2 (p.E368K, in exon 8) is the second most frequent in association with autosomal dominant CNM, giving rise to an invariable intermediate severity phenotype with neonatal or infantile onset of diffuse weakness including facial and periorbital muscles in 19 families (Böhm et al., 2012). The mutation is located in the middle/stalk domain of the dynamin 2 protein, and is expected to disrupt its binding interface with the pH domain, according to the dynamin 1 crystal structure model (Faelber et al., 2011). The novel mutation found in Patient 1 (p.F372C, in exon 8) changes phenylalanine to cysteine three amino acids down the coding sequence from the common mutation of Patient 2, in a position that also sits in the short region of the middle domain that composes the interface with the pH domain (Figure 3). This interface is made up of the positions 368, 369 (in both of which CNM mutations have been described), 372 (first patient of this report), and 373, 374 and 376, for which no mutation has been reported so far. Moreover, this novel p.F372C appeared de novo, sustaining its pathogenicity.
Figure 3. Dynamin 2 tridimensional structure prediction. The middle/stalk domain is shown at the top, in grey, and the pH domain at the bottom, in yellow. Residues shown in red represent the mutations described in association with CNM. These cluster at the interface between the pH domain and the middle domains, whereas mutations implicated in Charcot-Marie-Tooth neuropathies occur outside this interface, in the residues shown in blue. The novel mutation described in Patient 1 affects the F372 residue, shown in green, and this is predicted to directly disturb the interface structure.
In this Brazilian cohort, the two patients with DNM2 mutations were the only ones to present typical fibers with radial arrangement of sarcoplasmic strands, a consistent finding described in various works (Jeannet et al., 2004; Bitoun et al., 2005; Romero, 2010). Direct sequencing of the mutation hot spots in the DNM2 gene thus seems a promising approach in both autosomal dominant CNM cases and sporadic CNM cases when radiated sarcoplasmic strands are a prominent feature.
Acknowledgments
OA was supported by a fellowship from CAPES Foundation, Ministry of Education of Brazil, process number 1286/51-2.
Footnotes
Associate Editor: Angela M. Vianna-Morgante
References
- Agrawal PB, Pierson CR, Joshi M, Liu X, Ravenscroft G, Moghadaszadeh B, Talabere T, Viola M, Swanson LC, Haliloglu G, et al. SPEG interacts with myotubularin, and its deficiency causes centronuclear myopathy with dilated cardiomyopathy. Am J Hum Genet. 2014;95:218–226. doi: 10.1016/j.ajhg.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua JA, Monnier N, Bitoun M, Eymard B, Ferreiro A, Monges S, Lubieniecki F, Taratuto AL, Laquerrière A, Claeys KG, et al. Recessive RYR1 mutations cause unusual congenital myopathy with prominent nuclear internalization and large areas of myofibrillar disorganization. Neuropathol Appl Neurobiol. 2011;37:271–284. doi: 10.1111/j.1365-2990.2010.01149.x. [DOI] [PubMed] [Google Scholar]
- Bitoun M, Maugenre S, Jeannet PY, Lacène E, Ferrer X, Laforêt P, Martin JJ, Laporte J, Lochmüller H, Beggs AH, et al. Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat Genet. 2005;37:1207–1209. doi: 10.1038/ng1657. [DOI] [PubMed] [Google Scholar]
- Bitoun M, Bevilacqua JA, Prudhon B, Maugenre S, Taratuto AL, Monges S, Lubieniecki F, Cances C, Uro-Coste E, Mayer M, et al. Dynamin 2 mutations cause sporadic centronuclear myopathy with neonatal onset. Ann Neurol. 2007;62:666–670. doi: 10.1002/ana.21235. [DOI] [PubMed] [Google Scholar]
- Bitoun M, Bevilacqua JA, Eymard B, Prudhon B, Fardeau M, Guicheney P, Romero NB. A new centronuclear myopathy phenotype due to a novel dynamin 2 mutation. Neurology. 2009;72:93–95. doi: 10.1212/01.wnl.0000338624.25852.12. [DOI] [PubMed] [Google Scholar]
- Böhm J, Biancalana V, Dechene ET, Bitoun M, Pierson CR, Schaefer E, Karasoy H, Dempsey MA, Klein F, Dondaine N, et al. Mutation spectrum in the large GTPase dynamin 2, and genotype-phenotype correlation in autosomal dominant centronuclear myopathy. Hum Mutat. 2012;33:949–959. doi: 10.1002/humu.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceyhan-Birsoy O, Agrawal PB, Hidalgo C, Schmitz-Abe K, DeChene ET, Swanson LC, Soemedi R, Vasli N, Iannaccone ST, Shieh PB, et al. Recessive truncating titin gene, TTN, mutations presenting as centronuclear myopathy. Neurology. 2013;81:1205–1214. doi: 10.1212/WNL.0b013e3182a6ca62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faelber K, Posor Y, Gao S, Held M, Roske Y, Schulze D, Haucke V, Noé F, Daumke O. Crystal structure of nucleotide-free dynamin. Nature. 2011;477:556–560. doi: 10.1038/nature10369. [DOI] [PubMed] [Google Scholar]
- Jeannet PY, Bassez G, Eymard B, Laforêt P, Urtizberea JA, Rouche A, Guicheney P, Fardeau M, Romero NB. Clinical and histologic findings in autosomal centronuclear myopathy. Neurology. 2004;62:1484–1490. doi: 10.1212/01.wnl.0000124388.67003.56. [DOI] [PubMed] [Google Scholar]
- Koutsopoulos OS, Kretz C, Weller CM, Roux A, Mojzisova H, Böhm J, Koch C, Toussaint A, Heckel E, Stemkens D, et al. Dynamin 2 homozygous mutation in humans with a lethal congenital syndrome. Eur J Hum Genet. 2013;21:637–642. doi: 10.1038/ejhg.2012.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte J, Hu LJ, Kretz C, Mandel JL, Kioschis P, Coy JF, Klauck SM, Poustka A, Dahl N. A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat Genet. 1996;13:175–182. doi: 10.1038/ng0696-175. [DOI] [PubMed] [Google Scholar]
- Majczenko K, Davidson AE, Camelo-Piragua S, Agrawal PB, Manfready RA, Li X, Joshi S, Xu J, Peng W, Beggs AH, et al. Dominant mutation of CCDC78 in a unique congenital myopathy with prominent internal nuclei and atypical cores. Am J Hum Genet. 2012;91:365–371. doi: 10.1016/j.ajhg.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot AS, Toussaint A, Tosch V, Kretz C, Wallgren-Pettersson C, Iwarsson E, Kingston H, Garnier JM, Biancalana V, Oldfors A, et al. Mutations in amphiphysin 2 (BIN1) disrupt interaction with dynamin 2 and cause autosomal recessive centronuclear myopathy. Nat Genet. 2007;39:1134–1139. doi: 10.1038/ng2086. [DOI] [PubMed] [Google Scholar]
- Romero NB. Centronuclear myopathies: A widening concept. Neuromuscul Disord. 2010;20:223–228. doi: 10.1016/j.nmd.2010.01.014. [DOI] [PubMed] [Google Scholar]
- Tosch V, Rohde HM, Tronchère H, Zanoteli E, Monroy N, Kretz C, Dondaine N, Payrastre B, Mandel JL, Laporte J. A novel PtdIns3P and PtdIns(3,5)P2 phosphatase with an inactivating variant in centronuclear myopathy. Hum Mol Genet. 2006;15:3098–3106. doi: 10.1093/hmg/ddl250. [DOI] [PubMed] [Google Scholar]
- Wilmshurst JM, Lillis S, Zhou H, Pillay K, Henderson H, Kress W, Muller CR, Ndondo A, Cloke V, Cullup T, et al. RYR1 mutations are a common cause of congenital myopathies with central nuclei. Ann Neurol. 2010;68:717–726. doi: 10.1002/ana.22119. [DOI] [PubMed] [Google Scholar]
- Zanoteli E, Oliveira AS, Schmidt B, Gabbai AA. Centronuclear myopathy: Clinical aspects of ten Brazilian patients with childhood onset. J Neurol Sci. 1998;158:76–82. doi: 10.1016/s0022-510x(98)00091-4. [DOI] [PubMed] [Google Scholar]
- Züchner S, Noureddine M, Kennerson M, Verhoeven K, Claeys K, De Jonghe P, Merory J, Oliveira SA, Speer MC, Stenger JE, et al. Mutations in the pleckstrin homology domain of dynamin 2 cause dominant intermediate Charcot-Marie-Tooth disease. Nat Genet. 2005;37:289–94. doi: 10.1038/ng1514. [DOI] [PubMed] [Google Scholar]
Internet Resources
- Online Mendelian Inheritance in Man (OMIM) [(Accessed July 2014)]. http://www.omim.org/
- NHLBI Exome Sequencing Project (ESP) Exome Variant Server (EVS) [(Accessed July 2014)]. http://evs.gs.washington.edu/EVS/
- 1000 Genomes, A Deep Catalog of Human Genetic Variation. [(Accessed July 2014)]. http://www.1000genomes.org/
- Leiden Muscular Dystrophy Pages Dynamin 2 (DNM2) Database. [(Accessed July 2014)]. http://www.dmd.nl/nmdb/home.php?select_db=DNM2.
- The UMD-DNM2-isoform1 mutations database. [(Accessed July 2014)]. http://www.umd.be/DNM2/
- The Human Gene Mutation Database at the Institute of Medical Genetics in Cardiff. [(Accessed July 2014)]. http://www.hgmd.org/