Abstract
Digital PCR has developed rapidly since it was first reported in the 1990s. It was recently reported that an improved method facilitated the detection of genetically modified organisms (GMOs). However, to use this improved method, the samples must be pretreated, which could introduce inaccuracy into the results. In our study, we explored a pretreatment-free digital PCR detection method for the screening for GMOs. We chose the CaMV35s promoter and the NOS terminator as the templates in our assay. To determine the specificity of our method, 9 events of GMOs were collected, including MON810, MON863, TC1507, MIR604, MIR162, GA21, T25, NK603 and Bt176. Moreover, the sensitivity, intra-laboratory and inter-laboratory reproducibility of our detection method were assessed. The results showed that the limit of detection of our method was 0.1%, which was lower than the labeling threshold level of the EU. The specificity and stability among the 9 events were consistent, respectively. The intra-laboratory and inter-laboratory reproducibility were both good. Finally, the perfect fitness for the detection of eight double-blind samples indicated the good practicability of our method. In conclusion, the method in our study would allow more sensitive, specific and stable screening detection of the GMO content of international trading products.
Within the last several decades, an increasing number of GM crops have been received the approval of cultivation and commercialization from different countries. In 2014, the number of hectares planted with GM crops reached 181 million, having risen 3.4% since 2013, more than 100-fold since 1996 and covering 10% of the total agricultural areas1. The public awareness of GMO crops has increased with the increased development of such crops. To protect the right to know of consumers for the products containing GMO content, many groups and countries have instituted labeling laws stating that the products must be labeled when they are above a certain threshold. Different countries have established different thresholds, such as 0.9% in the European Union (EU) and 5% in Japan, whereas the USA has voluntary labeling and China has “yes-or-no” labeling. Most of the labeling laws depend on the quantitative detection of GM crops, and thus the stricter laws must rely on more sensitive detection methods. Currently, the most commonly recommended method in the detection-standard documents of different groups and countries is the Taqman-based quantitative polymerase chain reaction (qPCR) method2,3,4,5. Many detection standards relied on qPCR results were established because of its relatively high levels of precision and accuracy6. However, this method has obvious drawbacks. The results of qPCR are analyzed according to the amplification curve7, which are easily affected by many factors, including the PCR inhibitor8, the experience of the technicians and the matrix effect9 of the PCR tubes. These drawbacks make qPCR unsuitable for screening the products with low-abundance GMO content.
Digital PCR10 is a recently developed quantitative detection method based on limiting dilution and statistical analysis based on Poisson Distribution. For this method, the original PCR mixture is partitioned into a series of reaction samples. Additionally, the number of templates in the diluted PCR samples follows Poisson distribution, such that most of the partitioned samples contain zero copies of the template and others contain one or more copies. After amplification, the partitioned samples containing one or more copies of the templates would exhibit fluorescence signals. By calculating the number of signal-exhibiting partitions and combining with Poisson distribution, the absolute copy number of the target templates in the PCR reaction volume is determined. By performing limiting dilution, the volume of each PCR reaction can be decreased to as little as nL or pL level, such that the “matrix effect” of the PCR reaction can be mostly avoided. dPCR has been widely used for the analysis of clinical samples, such as the detection of allelic discrimination11,12, the determination of single cell expression profiles13,14, the detection of single nucleotide polymorphisms15 (SNPs), and the detection of low-copy targets16,17,18,19,20. Additionally, studies have shown that dPCR is more resistant to PCR inhibitors compared with the real-time PCR21. Thus, dPCR may be less reliant on the purity of the DNA template. Therefore, the technique is better suited for international-trade applications.
However, the most important limitation to the widespread use of dPCR is the need to pretreat the genome samples. Previous studies showed that dPCR samples always required pretreatment, including restriction enzyme digestion11,12,16,17,18,22 and hot-process denaturation23. A recent study23 showed that the pretreated group allowed better separation of the target, the reference and the quencher cluster than did the non-pretreated group. Recycling procedure should always be followed by the pretreatments, while the column recycling kit is regarding as the most popular method as its easy operation and lower cost. However, this may cause the unbalance recycling for the gene fragments of different length as the different binding activity causing by the column of the kits. Thus, the pretreatments can lead to inaccurate detection results. Additionally, in the case of large-scale detection assays, pretreatments greatly increase the necessary time and labor.
Screening method based on the detection of screening elements of most of GMO events could be used to determine the GMO content within a short period. To promote the use of dPCR in routine testing, we evaluated a validated detection method based on screening pretreatment-free dPCR. Moreover, the intra-laboratory sensitivity, repeatability and specificity of our method were systematically evaluated. Finally, we sent our samples and our protocol to six different laboratories to confirm the inter-laboratory reproducibility of our method.
Results
Optimization of the experimental conditions
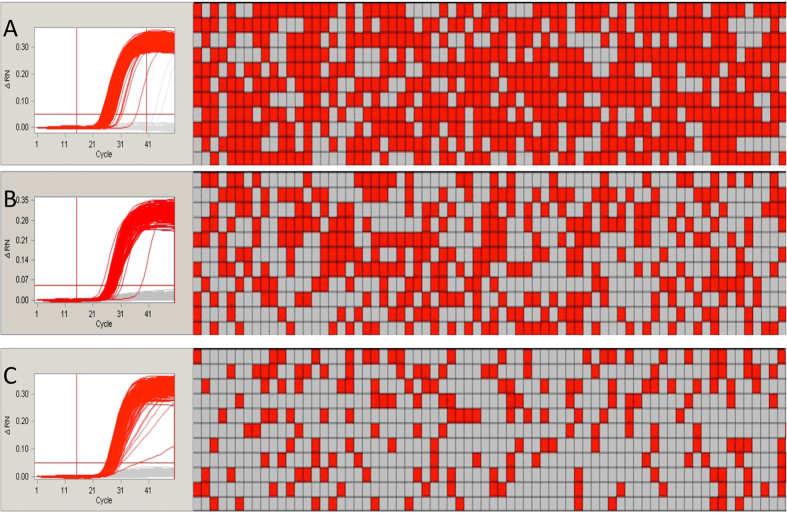
The thermal-cycling conditions, the concentration of probes and the mixing method were optimized in our assay. The final amplification plots and associated hot maps are shown in Fig. 1. Regarding the thermal-cycling conditions, studies24 have shown that the pre-denaturation duration may greatly affect the final results. Therefore, we chose a different pre-denaturation period for our PCR experiments: 120 s in our study and 300 s of officially recommendation. The amplification curves and hot maps showed that the shorter pre-denaturation time produced better final results. For the concentration of the probes, most of the validated real-time PCR detection methods used in China recommended a ratio of forward/reverse primers to probes of 2:2:1. To reduce the potential cost of our assay, we tested a forward/reverse primer to probes ratio of 9:9:2. The difference between the results of employing the two conditions was barely detectable. To improve the cost-efficiency of the method, we chose the ratio of 9:9:2 for further assays. For the aspect of mixing methods, two different methods were commonly utilized: pipetting or vortexing. We evaluated both of these methods and found that mixing by vortexing was more suitable for our experiments because pipetting can introduce inaccuracy due to DNA being left on the tips.
Figure 1. The amplification plots (left) and the panel hot map (right) of endogene and screening elements.
The read wells of the each hot map mean the positive amplification, the grey one mean no amplification. (A,B,C) represented the group of ZssIIb gene, CaMV35s promoter and NOS terminator, respectively.
Specificity and stability of the primers and probes
In our study, specificity was theoretically defined as the unique amplification of the events that contained the correct screening elements. Stability was defined as the stable amplification of all of the events that contained the matched elements. The MON810, MON863, TC1507, BT176, MIR604, MIR162, GA21, T25 and NK603 events were used to test the specificity and stability of the primers and probes in our assay. The amplification results are shown in Fig. 2. The experimental amplification results all perfectly matched the theoretical results.
Figure 2. The specificity and stability test of our method.
Determination of sensitivity
In our study, the limit of detection (LOD) was defined as the lowest percentage of GMO content that could be reliably detected. To determine the sensitivity of our method, samples contained different percentages of Bt176 and MIR162 events were prepared. The sensitivity levels observed are shown in Table 1. The results showed that the positive wells coule be stably seen from the 0.1% group. Over the course of several repeated experiments, the 0.1% group reliably showed positivity. Thus, the LOD of this detection method was determined to be 0.1%.
Table 1. The sensitivity and repeatability test.
| GMO content2 | A1 | B | The RSD of different operators for 35s | The RSD between different operators for NOS | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Three parallels of 35s | RSD | Three parallels of NOS | RSD | Three parallels of 35s | RSD | Three parallels of NOS | RSD | |||||||||||
| 5% | 142 | 136 | 138 | 2.20% | 103 | 110 | 118 | 6.80% | 145 | 142 | 149 | 2.42% | 115 | 114 | 99 | 8.20% | 3.02% | 6.16% |
| 1% | 19 | 19 | 21 | 5.87% | 15 | 14 | 17 | 9.96% | 18 | 16 | 22 | 16.37% | 14 | 12 | 12 | 9.12% | 10.18% | 12.37% |
| 0.5% | 6 | 4 | 6 | 21.65% | 6 | 8 | 7 | 14.29% | 5 | 5 | 7 | 20.38% | 6 | 6 | 8 | 17.32% | 17.41% | 13.13% |
| 0.4% | 5 | 5 | 4 | 12.37% | 5 | 4 | 4 | 13.32% | 5 | 4 | 5 | 12.37% | 4 | 6 | 4 | 24.74% | 10.10% | 16.97% |
| 0.3% | 4 | 2 | 3 | 33.33% | 4 | 3 | 2 | 33.33% | 5 | 2 | 5 | 43.30% | 4 | 2 | 5 | 41.66% | 35.95% | 33.17% |
| 0.2% | 3 | 3 | 2 | 21.65% | 4 | 2 | 2 | 43.30% | 2 | 2 | 3 | 24.74% | 3 | 5 | 2 | 45.83% | 20.00% | 38.49% |
| 0.1% | 1 | 1 | 2 | 43.30% | 1 | 1 | 2 | 43.30% | 1 | 3 | 2 | 50.00% | 2 | 1 | 1 | 43.30% | 44.72% | 35.36% |
1the A and B represents the different operators in our lab.
2the GMO content means the mass percentage of the GMO content.
Validation of the repeatability
Intra-laboratory repeatability validation were performed by two different operators using two different chambers to determine the GMO content of the same sample on two different days. The validation results are shown in Table 1. The data in the sheets showed that all the different concentration of GMO samples appeared posititve results. This indicated that this dPCR detection assay provided reliably repeatable results for the sample at both high and low concentrations.
Detection for the double-blind samples
To ascertain that this detection method was suitable for practical determinations of the GMO content, we conducted assays using eight double-blind samples that were mixed by coworkers in our laboratory. The GMO contents of the samples varied from 0.1% to 0.5%. The operators could not observe the theoretical data until the final practical results were available. The practical results and theoretical contents of the eight double-blind samples are shown in Table 2. The data showed that the practical results could be entirely fitted to the theoretical contents. Therefore, this dPCR detection method might be entirely suitable for the daily detection of GMO contents.
Table 2. The theoretical contents and practical results for the double-blind samples.
| Group number | Theoretical GMO contents | Experimental P-35s results | Experimental T-NOS results | Fitness |
|---|---|---|---|---|
| A | MON810 of 0.5% | Positive | Negative | 100% |
| B | MON810 of 1% | Positive | Negative | 100% |
| C | MON810 of 0.4% & MIR162 of 0.4% | Positive | Positive | 100% |
| D | MIR604 of 0.2% | Positive | Positive | 100% |
| E | MON810 of 0.3% & MIR604 of 0.2% | Positive | Positive | 100% |
| F | NK603 of 0.4% | Positive | Positive | 100% |
| G | Bt176 of 0.5% | Positive | Negative | 100% |
| H | Negative maize sample | Negative | Negative | 100% |
Validation of reproducibility by inter-laboratory repeats
Reproducibility is one of the most important elements of concern for validated detection methods. To verify the reproducibility of the results obtained using the dPCR detection method, we sent our samples, primers and probes, as well as the experimental protocol, to six different laboratories to determine the level of inter-laboratory reproducibility. The results were returned with an official seal on the documents, meaning that the results had legal validity. The results verified by the different laboratories are shown in Table 3. We found that the results obtained in different laboratories were totally similar to the theoretical results. The similar results obtained in the different laboratories conclusively demonstrated the high level of reproducibility of our detection method.
Table 3. The verified result by the different labs in China.
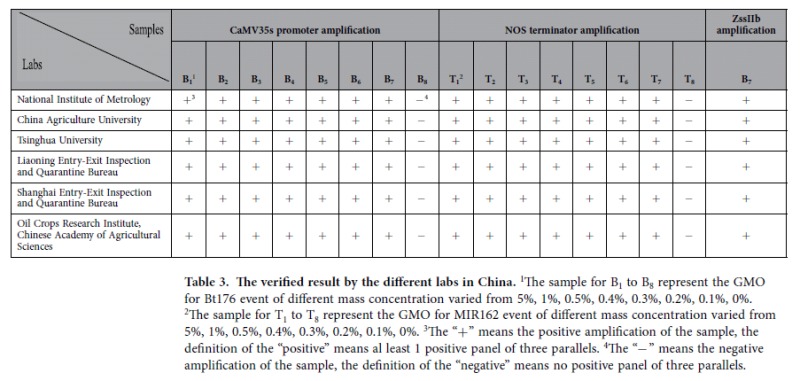
1The sample for B1 to B8 represent the GMO for Bt176 event of different mass concentration varied from 5%, 1%, 0.5%, 0.4%, 0.3%, 0.2%, 0.1%, 0%.
2The sample for T1 to T8 represent the GMO for MIR162 event of different mass concentration varied from 5%, 1%, 0.5%, 0.4%, 0.3%, 0.2%, 0.1%, 0%.
3The “+” means the positive amplification of the sample, the definition of the “positive” means al least 1 positive panel of three parallels.
4The “−” means the negative amplification of the sample, the definition of the “negative” means no positive panel of three parallels.
Validation of the repeatability by crossing-platform
The commercial platforms for conducting dPCR mainly involve two systems: the chamber-based digital PCR (cdPCR) and the droplet-based digital PCR (ddPCR) system. The cdPCR system is based on microfluidic chambers into which the reaction volume is partitioned, whereas the ddPCR system is based on droplets of water-in-oil emulsion. The different principles at play in these two systems may result in different detection results. We designed experiments to ascertain that our detection method was suited to these other dPCR platforms. The verification results are shown in Fig. 3. The results obtained using the ddPCR system indicated that the detection method we developed in this study had good inter-platform repeatability.
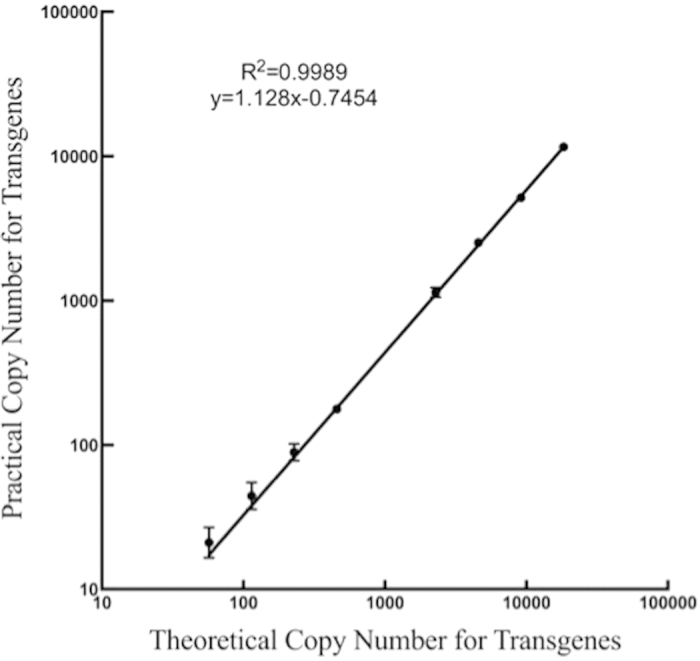
Figure 3. The linear curve between the theoretical copy number of transgenes and copy number of the screening elements.
Discussion
We developed an efficient GMO detection method based on dPCR that can be used to reliably detect a GMO content of 0.1% in genomic DNA samples that were not pre-treated. Having demonstrated the specificity, stability and repeatability of this method, as well as the reproducibility of its results at both the inter-laboratory and inter-platform levels, we have shown that our method is suitable for the detection of GMO content in international trading products.
Previous studies showed that pretreating genomic DNA samples was essential to achieving reliable dPCR results. A recent study8 showed that pretreatments could be omitted from the ddPCR protocol; however, some researchers17 were concerned that without pretreatments, the stability of the method would be uncertain. For practical detection applications, including pretreatment steps makes the assays inconvenient and may introduce uncertainty into the final results. To comply with China’s labeling laws, routine qualitative detection methods must have good stability and specificity. The dPCR is the best choice for the detection of the low-abundance targets due to the partitioning of the initial PCR sample. Therefore, in this study, a stable pretreatment-free dPCR method for GMO screening was explored. Due to the production of random breaks in the genomic DNA during sample extraction, quantitative detection cannot be achieved by the screening elements. Thus, the target samples were serially diluted to evaluate the stability and reliability of our method. The linear curve of the theoretical copy number of transgenes compared with the practical number of copies of the screening elements per microliter is shown in Fig. 4. Based on the correlation coefficient of this curve, the linearity value of our method was 0.9989, showed that the good isolation of undigested genomic DNA by chambers even the GMO content is extremly low. This result suggested that the pretreatment-free dPCR detection method developed in this study had good sensitivity and linearity even when the GMO content was very low.
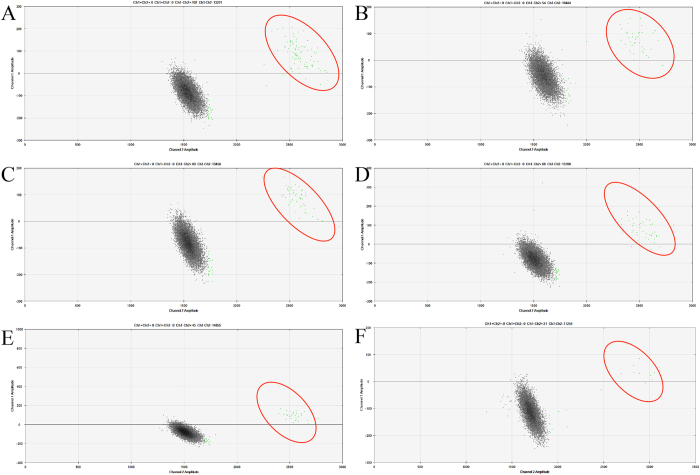
Figure 4. The amplification hot map of the verification of ddPCR with different GMO content.
(A–F) represented the different GMO content of 1%, 0.5%, 0.4%, 0.3%, 0.2%, and 0.1%. The positive droplets were highlighted by the red cycles.
Two dPCR systems, the chamber-based dPCR (cdPCR) system and the droplet-based dPCR (ddPCR) system, were developed since the commercialization of the dPCR. Different results11,16,17,24 obtained using these two methods have been published, attributed to various factors, including the different principles of partitioning, the differential detection limits and dynamic ranges. The dPCR detection method that we developed should be suitable for both the cdPCR and ddPCR systems. Thus, the repeatability of our method was validated as being suitable for both of two dPCR systems. We used the 48.770 chamber manufactured by Fluidigm in our assay to generate 770 individual wells for each panel, compared with the 20,000 partitions generated in the Bio-Rad ddPCR system. Thus, the dynamic range of the ddPCR system may be greater than that of the chamber-based dPCR system. However, qualitative detection was achieved through the serial dilution of samples with various GMO contents by using our detection system. Accordingly, the dynamic range might not be the barrier for the comparison of two platforms. Moreover, the final results obtained proved our hypothesis that the cdPCR and ddPCR systems both could achieve a sensitivity level of 0.1%. The results indicated that the dPCR detection method that we developed achieved stable amplification using both two dPCR platforms.
We chose the CaMV35s promoter and the NOS terminator as the screening elements for our experiments. Previous studies26 showed that these two elements covered 65.7% and 53.49%, respectively, of all GMO events, and together, they covered 81.4% of these events, indicating that most GMO events could be detected using these two elements. Thus, we chose the CaMV35s promoter and the NOS terminator to complete our investigation. Using this approach, in the future, we will attempt to improve upon the method described here by adding more elements to cover a larger percentage of GMO events.
The dPCR screening method that we developed in this study allowed the detection of the samples with 9 GMO maize events, using one non-GMO maize sample as a negative control, simutanously. Moreover, the LOD of our method, which was 0.1% of GMO mass content, was lower than that of the GMO labeling limit of EU for 0.9% and satisfied all of the quantification limits for GMO labeling in other countries. Thus, the experimental results of this study demonstrated that our method was suited for the routine detection of GMO content in internationally traded products.
Methods
GMO samples
The non-GMO samples utilized in our study were stored in our the Supervision, Inspection and Testing Center of Genetically Modified Organisms of the Ministry of Agriculture. The various GMO samples were kindly provided by established companies, such as the MIR162, Bt176, MIR604, and 3272 event-containing samples that were provided by Syngenta Seeds Inc. (California, US); the MON810, MON863, GA21 and NK603 event-containing samples that were provided by the Monsanto Company (St. Louis, Missouri, US); the TC1507 event-containing sample that was provided by DOW AgroSciences LLC (Indianapolis, Indiana, US); and the T25 event-containing sample that was provided by Bayer CropScience (Leverkusen, North Rhine-Westphalia, Germany). The samples containing Bt176 and MIR162 events were used as the positive control samples for detection based on the CaMV35s promoter and the NOS terminator, respectively. The screening elements containing in each GMO event are shown in Table 4.
Table 4. The screening elements contained in the events used in our study.
| GMO events | CaMV35s promoter | NOS terminator |
|---|---|---|
| MON810 | Positive | Negative |
| MON863 | Positive | Positive |
| TC1507 | Positive | Negative |
| MIR604 | Negative | Positive |
| MIR162 | Negative | Positive |
| GA21 | Negative | Positive |
| T25 | Positive | Negative |
| NK603 | Positive | Positive |
| Bt176 | Positive | Negative |
| Non-GMO | Negative | Negative |
Samples with different GMO contents
Before extracting the genomic DNA, the grains of the various GMO samples were ground using a Retsch® MM430 mixer. The grinding procedure consisted of 10 cycles of agitation for 20 s and resting for 1 min cooling to room temperature. The cooling steps protected the genomic DNA from heat-damage. After the grain samples were ground, samples with different GMO contents were prepared. In this study, the relative mass content of each GMO event was set to 50%, 20%, 5%, 1%, 0.5%, 0.4%, 0.3%, 0.2% and 0.1%. The mixed samples were then placed in Dynamic CM-200 mixer and shaken overnight to achieve equal distribution.
Extraction of genomic DNA
The genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Germany) from 50 μg of powdered samples containing various GMO events. The extraction procedure was conducted according to the official guidelines. The isolated genomic DNA was diluted using 60 μL of ddH2O. The concentration and purity of each sample were determined using NanoDrop N2000 (Thermo Fisher Scientific Inc, Wilmington, US) according to the OD260 and OD280 values. The samples were diluted to a final concentration of 50 ng/μL for our study.
Primers and probes
The primers and probes used in our assay are listed in Table 5. The TAMRA quencher that is generally used for real-time PCR was replaced by the Black Hole Quencher 1 (BHQ-1) to reduce the fluorescence baseline values. All of the primers and probes were synthesized by Invitrogen (Life Technologies, California, US).
Table 5. The primers and probes used in this study.
| Target genes | Primers/probes name | Primers/probes sequence | Reference |
|---|---|---|---|
| ZssIIb gene | ZssIIb-F | 5′-CTCCCAATCCTTTGACATCTGC-3′ | 29 |
| ZssIIb-R | 5′-TCGATTTCTCTCTTGGTGACAGG-3′ | ||
| ZssIIb-P | 5′-VIC-AGCAAAGTCAGAGCGCTGCAATGCA-BHQ1-3′ | ||
| CaMV35s promoter | P-35s-F | 5′- ATTGATGTGATATCTCCACTGACGT-3′ | 30 |
| P-35s-R | 5′- CCTCTCCAAATGAAATGAACTTCCT-3′ | ||
| P-35s-P | 5′-VIC- CCCACTATCCTTCGCAAGACCCTTCCT-BHQ1-3′ | ||
| NOS terminator | T-NOS-F | 5′-ATCGTTCAAACATTTGGCA-3′ | 25 |
| T-NOS-R | 5′-ATTGCGGGACTCTAATCATA-3′ | ||
| T-NOS-P | 5′-VIC-CATCGCAAGACCGGCAACAGG-BHQ1-3′ |
Digital PCR
Chamber-based digital PCR
Chamber-based PCR (cdPCR) was conducted using a BioMark system (Fluidigm, South San Francisco) equipped with a 48.770 digital array (Fluidigm). The 48.770 array had 48 individual panels, each of which contained 770 individual partitions. A 4-μL aliquot of the initial PCR reaction volume was added to each panel to achieve a final volume of approximately 654.5 nL (0.85 nL × 770) for cdPCR reaction.
The protocol of cdPCR consisted of four steps, including priming, transferring, loading and amplifying. All of the experimental procedures followed the official guidelines. The final 4 μL reaction volume contained 1× Taqman Gene Expression Mix containing the positive fluorescence signal ROX (Life Technologies, California, US), 2× sample loading reagent (Fluidigm, South San Francisco), forward and reverse screening or endogenous primers at a final concentration of 225 nM and 1 μL of 50 ng/μL undigested DNA template, with ddH2O for the rest of 4 μL, the concentration of the probes was optimized in the later experiments. The predicted number of copies of the endogenous gene in each panel was 3000 based on the average theoretical mass of Zea mays genome being 2.73 pg27. Three parallel assays of each sample were conducted. The final results were analyzed based on the results in all three parallel assays.
Droplet-based digital PCR
Droplet-based digital PCR was performed using Bio-Rad QX100 droplet system (Bio-Rad, Pleasanton, CA, USA). Many studies11,17 have shown that this droplet system provided good repeatability.
The ddPCR reaction volume included 10 μL of ddPCR Master Mix (Bio-Rad, Pleasanton, CA), 0.45 μL of the forward and reverse primers, 0.1 μL of each probe at the same concentration used for optimized cdPCR, and 5 μL of 50 ng/μL DNA template, with ddH2O added to bring the final volume to 20 μL.
The optimized cycling program used for both the cdPCR and ddPCR assays was as follows: 50 °C for 5 min, 95 °C for 5 min, and then 50 cycles of 95 °C for 1 min and 60 °C for 1 min, after which the fluorescence signals were collected.
Data analysis
After the PCR assays were performed, the curves were automatically analyzed using Digital PCR Analysis Software (Fluidigm, South San Francisco). The final fluorescence threshold manually set to 0.03 and the wells with Ct values of 25–45 regarded as positive.
We estimated the absolute copy number per panel according to the number of positive wells and the total number of partitions. According to the Poisson distribution, the copy number of each panel was calculated using the following equations27:
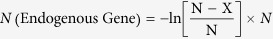 |
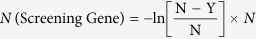 |
where the N (Endogenous Gene) value and N (Screening Gene) values were the estimated absolute copy number of the endogenous gene and the screening gene, respectively, and X and Y were the number of wells positive for the endogenous gene and the screening gene, respectively. N was the total number of separated partitions.
Conclusion
We developed a validated qualitative screening detection based on pretreatment-free dPCR method in this study that enabled the detection of most of the commercially significant GMO events. This detection method achieved a lower LOD as well as a higher specificity, repeatability and reproducibility than that of the commonly utilized qPCR method, and it largely avoided false-positive detection results. Further studies using more screening elements would be conducted to cover a larger percentage of the commercially significant GMO events in more crops such as soybeans, maize, potatoes, rice and wheat.
Additional Information
How to cite this article: Fu, W. et al. A highly sensitive and specific method for the screening detection of genetically modified organisms based on digital PCR without pretreatment. Sci. Rep. 5, 12715; doi: 10.1038/srep12715 (2015).
Acknowledgments
This work was supported by the National Grand Project of Science and Technology (2014ZX08012-001) and the basic scientific research foundation in the Chinese Academy of Inspection and Quarantine (2015JK005).
Footnotes
Author Contributions K.H., W.X., Z.D., Q.W., H.W. and S.Z. conceived and designed the experiments. P.Z., W.F., C.W. and W.T. performed the experiments. P.Z., W.F. and C.W. analyzed the data. W.F. and W.X. contributed reagents/materials/analysis tools. P.Z., W.F., W.X. and C.W. wrote the paper.
References
- James C. Global Status of Commercialized Biotech/ GM Crops: 2014. ISAAA. Brief 49 (2015) [Google Scholar]
- Xu W. et al. Event-specific detection of stacked genetically modified maize Bt11× GA21 by UP-M-PCR and Real-Time PCR. J Agric Food Chem. 57, 395–402 (2008). [DOI] [PubMed] [Google Scholar]
- Querci M. et al. Real-time PCR-based ready-to-use multi-target analytical system for GMO detection. Food Anal Meth 2, 325–336 (2009). [Google Scholar]
- Gašparič M. B. et al. Comparison of nine different real-time PCR chemistries for qualitative and quantitative applications in GMO detection. Anal Bioanal Chem. 396, 2023–2029 (2010). [DOI] [PubMed] [Google Scholar]
- Higuchi R., Fockler C., Dollinger G. & Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Bio/Technolgy. 11, 1026–1030 (1993). [DOI] [PubMed] [Google Scholar]
- Bustin S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 55, 611–622 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang N. et al. Event-specific qualitative and quantitative PCR detection of LY038 maize in mixed samples. Food Control. 22, 1287–1295 (2011). [Google Scholar]
- Kiselinova M. et al. Comparison of droplet digital PCR and seminested real-time PCR for quantification of cell-associated HIV-1 RNA. PloS one. 9, e85999 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger N. J. et al. “Limits of Control”–Crucial Parameters for a Reliable Quantification of Viable Campylobacter by Real-Time PCR. PloS one. 9, e88108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B. & Kinzler K. W. Digital PCR. Proc. Natl. Acad. Sci. USA. 96, 9236–9241 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson Benjamin J. et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 83, 8604–8610 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro Leonardo B. et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem 84, 1003–1011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G. et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell. 18, 675–685 (2010). [DOI] [PubMed] [Google Scholar]
- Yung T. K. et al. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non–small cell lung cancer patients. Clin Cancer Res. 15, 2076–2084 (2009). [DOI] [PubMed] [Google Scholar]
- Ludlow A. T. et al. Quantitative telomerase enzyme activity determination using droplet digital PCR with single cell resolution. Nucleic Acids Res. 42, e104–e104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R. et al. Evaluation of digital PCR for absolute DNA quantification. Anal Chem. 83, 6474–6484 (2011). [DOI] [PubMed] [Google Scholar]
- Morisset D. et al. Quantitative analysis of food and feed samples with droplet digital PCR. PLoS One. 8, e62583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M. et al. Single-molecule reverse transcription polymerase chain reaction using water-in-oil emulsion. J. Biosci. Bioeng. 99, 293–295 (2005). [DOI] [PubMed] [Google Scholar]
- Miotto E. et al. Quantification of Circulating miRNAs by Droplet Digital PCR: Comparison of EvaGreen-and TaqMan-Based Chemistries. Cancer Epidemiol. Biomarkers Prev. 23, 2638–2642 (2014). [DOI] [PubMed] [Google Scholar]
- Laurent-Puig P. et al. Clinical relevance of KRAS-mutated sub-clones detected with picodroplet digital PCR in advanced colorectal cancer treated with anti-EGFR therapy. Clin Cancer Res. 21, 1087–1097 (2014). [DOI] [PubMed] [Google Scholar]
- Dingle T. C., Sedlak R. H., Cook L. & Jerome K. R. Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin Chem. 59, 1670–1672 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns M. J., Burrell A. M. & Foy C. A. The applicability of digital PCR for the assessment of detection limits in GMO analysis. Eur Food Res Technol. 231, 353–362 (2010). [Google Scholar]
- Demeke T. et al. Assessment of droplet digital PCR for absolute quantification of genetically engineered OXY235 canola and DP305423 soybean samples. Food Control. 46, 470–474 (2014). [Google Scholar]
- Bhat S., McLaughlin J. L. & Emslie K. R. Effect of sustained elevated temperature prior to amplification on template copy number estimation using digital polymerase chain reaction. Analyst. 136, 724–732 (2011). [DOI] [PubMed] [Google Scholar]
- Xie J. J. et al. Detection of genetically modified plants and derived products— Qualitative PCR method for CaMV 35S promoter, FMV 35S promoter, NOS promoter, NOS terminator and CaMV 35S terminator. National standards of the People’s Republic of China. http://down.foodmate.net/standard/sort/3/38254.html. (2012) Date of access: 01 Sep. 2012.
- Wu Y. et al. Development of a general method for detection and quantification of the P35S promoter based on assessment of existing methods. Sci Rep. 4. 10.1038/srep07358 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganathan K. & Earle E. D. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 9, 208–218 (1991). [Google Scholar]
- Lun F. M. et al. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clin Chem. 54, 1664–1672 (2008). [DOI] [PubMed] [Google Scholar]
- Kuribara H. et al. Novel reference molecules for quantitation of genetically modified maize and soybean. J. AOAC Int. 85, 1077–1089 (2002). [PubMed] [Google Scholar]
- Lee S. H., Min D. M. & Kim J. K. Qualitative and quantitative polymerase chain reaction analysis for genetically modified maize MON863. J Agric Food Chem. 54, 1124–1129 (2006). [DOI] [PubMed] [Google Scholar]