Abstract
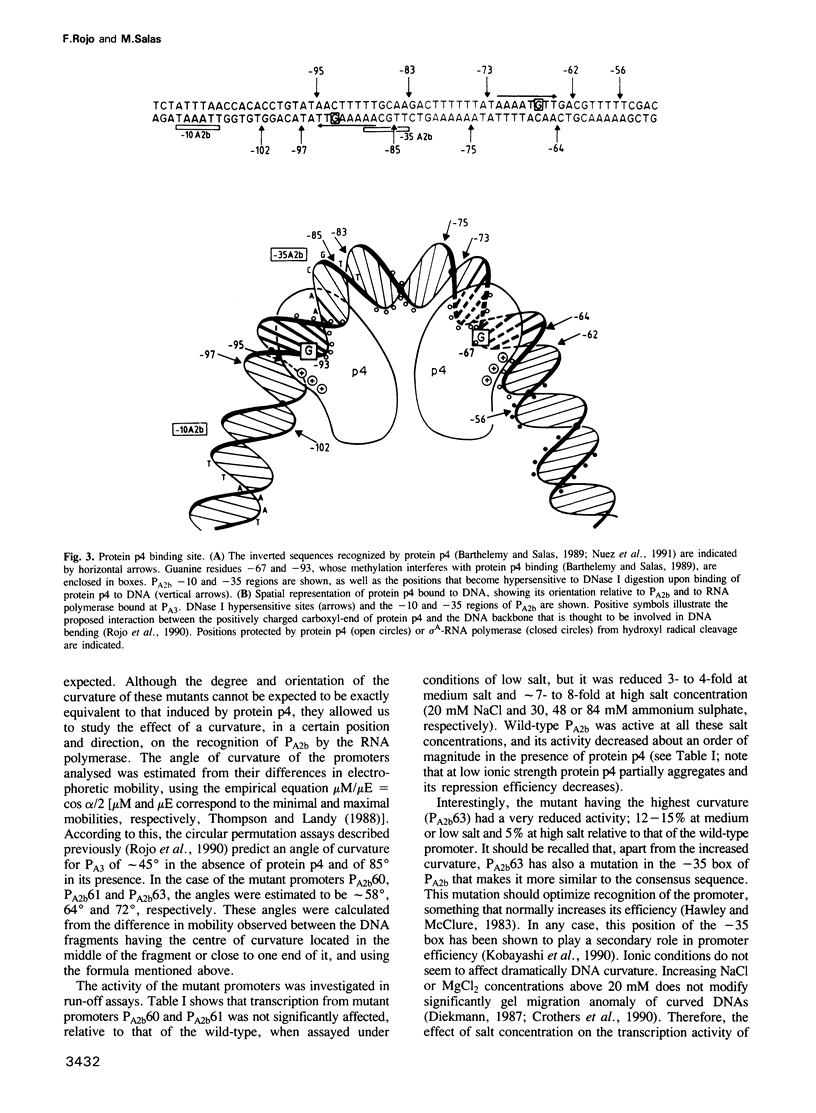
Binding of phage phi 29 regulatory protein p4 to its target sequences produces a strong bend in the DNA that is important for activation of the late A3 promoter (PA3). Protein p4 binding site in PA3 overlaps with the divergently transcribed main early promoter. PA2b, which suggested that p4 could also act as a repressor. We show that protein p4 both excludes Bacillus subtilis sigma A-RNA polymerase from PA2b and directs it to the divergently transcribed A3 promoter. Although steric hindrance is likely to be involved in the repression process, we have also analysed the effect on PA2b activity of a sequence-dependent curvature that simulates that induced by protein p4. A progressive increase in the DNA curvature of protein p4 binding region, performed by site-directed mutagenesis, has indicated that a static DNA curvature by itself can inhibit transcription from PA2b, both by impairing the binding of sigma A-RNA polymerase to the promoter and by reducing its ability to form transcriptionally active open complexes. These results indicate that bending promoter sequences in a direction unfavourable for RNA polymerase binding can repress transcription. Protein p4-induced DNA bending could therefore participate in PA2b repression by producing a DNA structure not recognized as a promoter by sigma A-RNA polymerase.
Full text
PDF



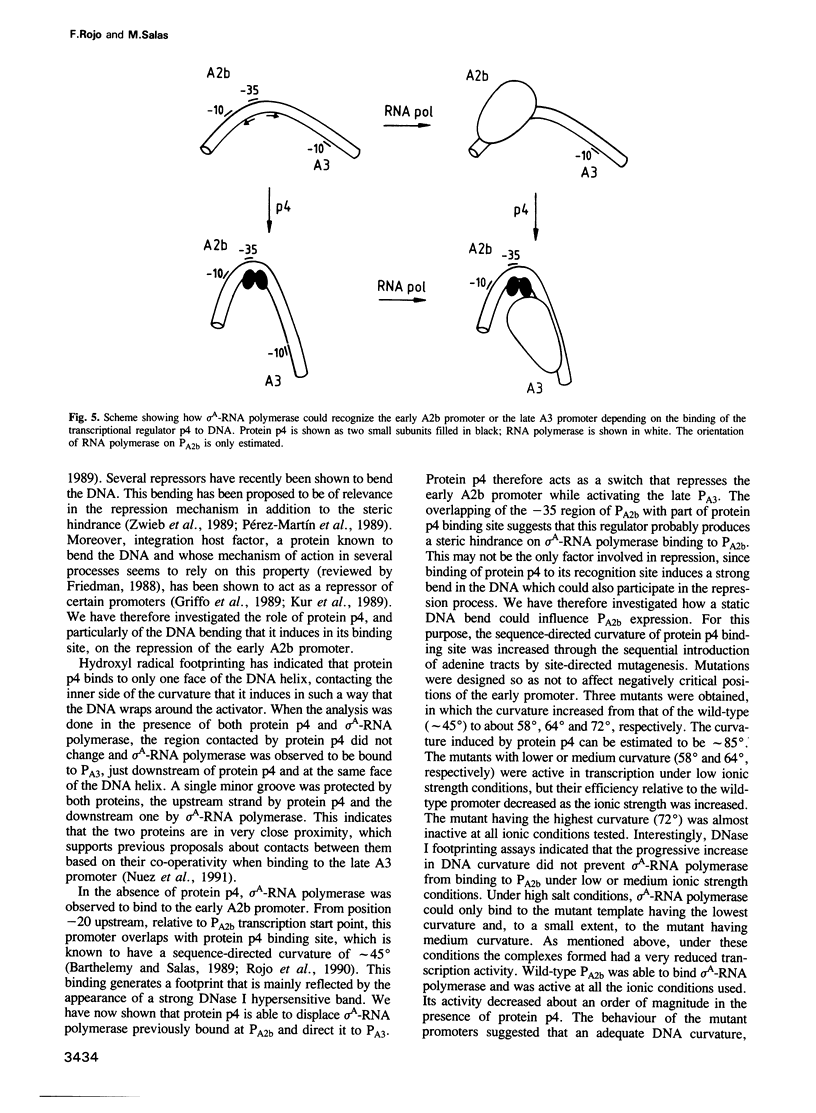
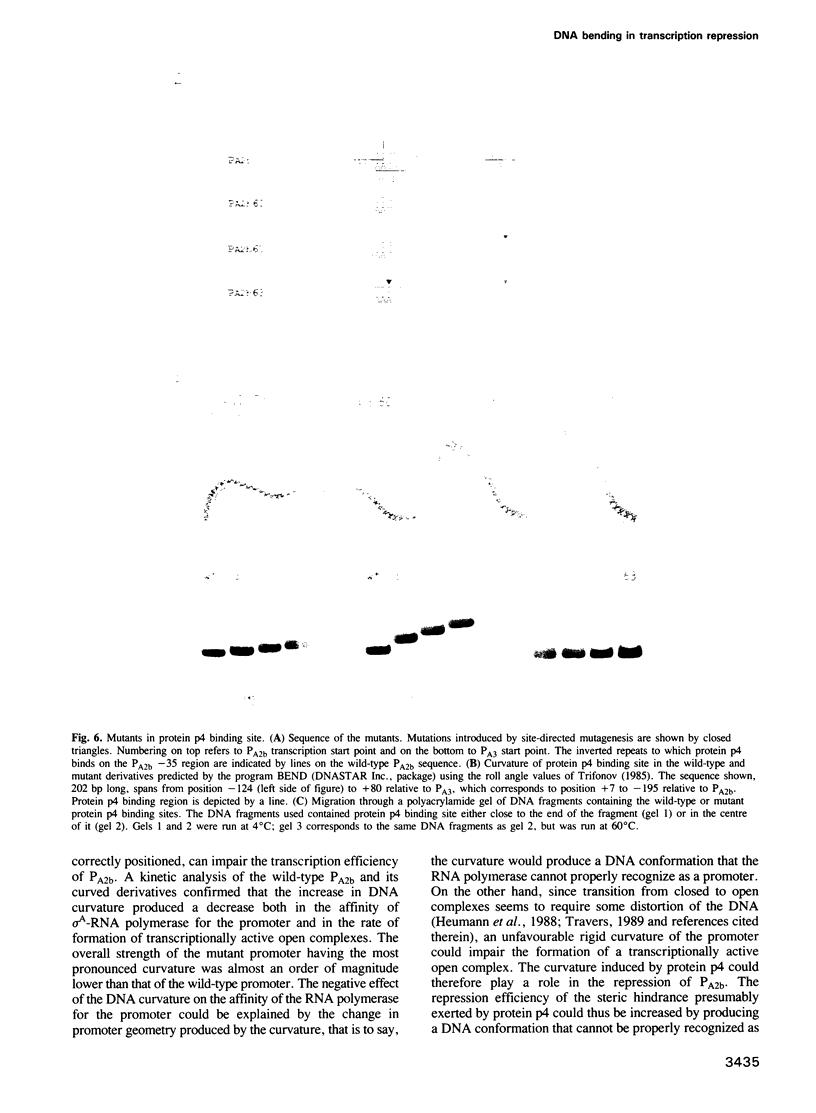



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S. Multipartite genetic control elements: communication by DNA loop. Annu Rev Genet. 1989;23:227–250. doi: 10.1146/annurev.ge.23.120189.001303. [DOI] [PubMed] [Google Scholar]
- Barthelemy I., Lázaro J. M., Méndez E., Mellado R. P., Salas M. Purification in an active form of the phage phi 29 protein p4 that controls the viral late transcription. Nucleic Acids Res. 1987 Oct 12;15(19):7781–7793. doi: 10.1093/nar/15.19.7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelemy I., Mellado R. P., Salas M. Symmetrical transcription in bacteriophage phi 29 DNA. Biochimie. 1988 May;70(5):605–609. doi: 10.1016/0300-9084(88)90243-x. [DOI] [PubMed] [Google Scholar]
- Barthelemy I., Salas M. Characterization of a new prokaryotic transcriptional activator and its DNA recognition site. J Mol Biol. 1989 Jul 20;208(2):225–232. doi: 10.1016/0022-2836(89)90384-7. [DOI] [PubMed] [Google Scholar]
- Barthelemy I., Salas M., Mellado R. P. In vivo transcription of bacteriophage phi 29 DNA: transcription initiation sites. J Virol. 1986 Dec;60(3):874–879. doi: 10.1128/jvi.60.3.874-879.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracco L., Kotlarz D., Kolb A., Diekmann S., Buc H. Synthetic curved DNA sequences can act as transcriptional activators in Escherichia coli. EMBO J. 1989 Dec 20;8(13):4289–4296. doi: 10.1002/j.1460-2075.1989.tb08615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crothers D. M., Haran T. E., Nadeau J. G. Intrinsically bent DNA. J Biol Chem. 1990 May 5;265(13):7093–7096. [PubMed] [Google Scholar]
- Diekmann S. Temperature and salt dependence of the gel migration anomaly of curved DNA fragments. Nucleic Acids Res. 1987 Jan 12;15(1):247–265. doi: 10.1093/nar/15.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giladi H., Gottesman M., Oppenheim A. B. Integration host factor stimulates the phage lambda pL promoter. J Mol Biol. 1990 May 5;213(1):109–121. doi: 10.1016/S0022-2836(05)80124-X. [DOI] [PubMed] [Google Scholar]
- Griffo G., Oppenheim A. B., Gottesman M. E. Repression of the lambda pcin promoter by integrative host factor. J Mol Biol. 1989 Sep 5;209(1):55–64. doi: 10.1016/0022-2836(89)90169-1. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. In vitro comparison of initiation properties of bacteriophage lambda wild-type PR and x3 mutant promoters. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6381–6385. doi: 10.1073/pnas.77.11.6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heumann H., Ricchetti M., Werel W. DNA-dependent RNA polymerase of Escherichia coli induces bending or an increased flexibility of DNA by specific complex formation. EMBO J. 1988 Dec 20;7(13):4379–4381. doi: 10.1002/j.1460-2075.1988.tb03336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inciarte M. R., Lázaro J. M., Salas M., Vińuela E. Physical map of bacteriophage phi29 DNA. Virology. 1976 Oct 15;74(2):314–323. [PubMed] [Google Scholar]
- Kim J., Zwieb C., Wu C., Adhya S. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene. 1989 Dec 21;85(1):15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Nagata K., Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase: effect of base substitutions in the promoter -35 region on promoter strength. Nucleic Acids Res. 1990 Dec 25;18(24):7367–7372. doi: 10.1093/nar/18.24.7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kur J., Hasan N., Szybalski W. Physical and biological consequences of interactions between integration host factor (IHF) and coliphage lambda late p'R promoter and its mutants. Gene. 1989 Sep 1;81(1):1–15. doi: 10.1016/0378-1119(89)90331-4. [DOI] [PubMed] [Google Scholar]
- Lanzer M., Bujard H. Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Johnson H. N., Gartenberg M. R., Crothers D. M. The DNA binding domain and bending angle of E. coli CAP protein. Cell. 1986 Dec 26;47(6):995–1005. doi: 10.1016/0092-8674(86)90814-7. [DOI] [PubMed] [Google Scholar]
- Lund P. A., Ford S. J., Brown N. L. Transcriptional regulation of the mercury-resistance genes of transposon Tn501. J Gen Microbiol. 1986 Feb;132(2):465–480. doi: 10.1099/00221287-132-2-465. [DOI] [PubMed] [Google Scholar]
- Malan T. P., Kolb A., Buc H., McClure W. R. Mechanism of CRP-cAMP activation of lac operon transcription initiation activation of the P1 promoter. J Mol Biol. 1984 Dec 25;180(4):881–909. doi: 10.1016/0022-2836(84)90262-6. [DOI] [PubMed] [Google Scholar]
- Malan T. P., McClure W. R. Dual promoter control of the Escherichia coli lactose operon. Cell. 1984 Nov;39(1):173–180. doi: 10.1016/0092-8674(84)90203-4. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mellado R. P., Barthelemy I., Salas M. In vitro transcription of bacteriophage phi 29 DNA. Correlation between in vitro and in vivo promoters. Nucleic Acids Res. 1986 Jun 25;14(12):4731–4741. doi: 10.1093/nar/14.12.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuez B., Rojo F., Barthelemy I., Salas M. Identification of the sequences recognized by phage phi 29 transcriptional activator: possible interaction between the activator and the RNA polymerase. Nucleic Acids Res. 1991 May 11;19(9):2337–2342. doi: 10.1093/nar/19.9.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Halloran T. V., Frantz B., Shin M. K., Ralston D. M., Wright J. G. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989 Jan 13;56(1):119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- Parkhill J., Brown N. L. Site-specific insertion and deletion mutants in the mer promoter-operator region of Tn501; the nineteen base-pair spacer is essential for normal induction of the promoter by MerR. Nucleic Acids Res. 1990 Sep 11;18(17):5157–5162. doi: 10.1093/nar/18.17.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martín J., Espinosa M. The RepA repressor can act as a transcriptional activator by inducing DNA bends. EMBO J. 1991 Jun;10(6):1375–1382. doi: 10.1002/j.1460-2075.1991.tb07657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martín J., del Solar G. H., Lurz R., de la Campa A. G., Dobrinski B., Espinosa M. Induced bending of plasmid pLS1 DNA by the plasmid-encoded protein RepA. J Biol Chem. 1989 Dec 15;264(35):21334–21339. [PubMed] [Google Scholar]
- Reitzer L. J., Magasanik B. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell. 1986 Jun 20;45(6):785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- Rojo F., Zaballos A., Salas M. Bend induced by the phage phi 29 transcriptional activator in the viral late promoter is required for activation. J Mol Biol. 1990 Feb 20;211(4):713–725. doi: 10.1016/0022-2836(90)90072-t. [DOI] [PubMed] [Google Scholar]
- Ross W., Thompson J. F., Newlands J. T., Gourse R. L. E.coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990 Nov;9(11):3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schickor P., Metzger W., Werel W., Lederer H., Heumann H. Topography of intermediates in transcription initiation of E.coli. EMBO J. 1990 Jul;9(7):2215–2220. doi: 10.1002/j.1460-2075.1990.tb07391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleif R. DNA binding by proteins. Science. 1988 Sep 2;241(4870):1182–1187. doi: 10.1126/science.2842864. [DOI] [PubMed] [Google Scholar]
- Serrano M., Barthelemy I., Salas M. Transcription activation at a distance by phage phi 29 protein p4. Effect of bent and non-bent intervening DNA sequences. J Mol Biol. 1991 Jun 5;219(3):403–414. doi: 10.1016/0022-2836(91)90182-6. [DOI] [PubMed] [Google Scholar]
- Sogo J. M., Inciarte M. R., Corral J., Viñuela E., Salas M. RNA polymerase binding sites and transcription map of the DNA of Bacillus subtilis phage phi29. J Mol Biol. 1979 Feb 5;127(4):411–436. doi: 10.1016/0022-2836(79)90230-4. [DOI] [PubMed] [Google Scholar]
- Spassky A., Busby S., Buc H. On the action of the cyclic AMP-cyclic AMP receptor protein complex at the Escherichia coli lactose and galactose promoter regions. EMBO J. 1984 Jan;3(1):43–50. doi: 10.1002/j.1460-2075.1984.tb01759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W., Porter S., Kustu S., Echols H. DNA-looping and enhancer activity: association between DNA-bound NtrC activator and RNA polymerase at the bacterial glnA promoter. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin J. F., Schleif R. F. Purification and properties of RhaR, the positive regulator of the L-rhamnose operons of Escherichia coli. J Mol Biol. 1990 Jan 5;211(1):75–89. doi: 10.1016/0022-2836(90)90012-B. [DOI] [PubMed] [Google Scholar]
- Travers A. A. DNA conformation and protein binding. Annu Rev Biochem. 1989;58:427–452. doi: 10.1146/annurev.bi.58.070189.002235. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. Curved DNA. CRC Crit Rev Biochem. 1985;19(2):89–106. doi: 10.3109/10409238509082540. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolberger C., Dong Y. C., Ptashne M., Harrison S. C. Structure of a phage 434 Cro/DNA complex. Nature. 1988 Oct 27;335(6193):789–795. doi: 10.1038/335789a0. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zwieb C., Kim J., Adhya S. DNA bending by negative regulatory proteins: Gal and Lac repressors. Genes Dev. 1989 May;3(5):606–611. doi: 10.1101/gad.3.5.606. [DOI] [PubMed] [Google Scholar]