Abstract
Objectives
To test the hypothesis that multiple constituents of the apical plasma membrane residing alongside the causal CF Transmembrane Conductance Regulator (CFTR) protein, including known cystic fibrosis (CF) modifiers SLC26A9, SLC6A14, and SLC9A3, would be associated with prenatal exocrine pancreatic damage as measured by newborn screened (NBS) IRT levels.
Study design
NBS IRT measures and genome-wide genotype data were available on 111 subjects from Colorado, 37 subjects from Wisconsin, and 80 subjects from France. Multiple linear regression was used to determine whether any of eight SNPs in SLC26A9, SLC6A14 and SLC9A3 were associated with IRT and whether other constituents of the apical plasma membrane contributed to IRT.
Results
In the Colorado sample, three SLC26A9 SNPs were associated with NBS IRT (min P = 1.16 × 10−3; rs7512462), but no SLC6A14 or SLC9A3 SNPs were associated (P > 0.05). The rs7512462 association replicated in the Wisconsin sample (P = 0.03) but not in the French sample (P = 0.76). Furthermore, rs7512462 was the top ranked apical membrane constituent in the combined Colorado and Wisconsin sample.
Conclusions
NBS IRT is a biomarker of prenatal exocrine pancreatic disease in patients with CF, and a SNP in SLC26A9 accounts for significant IRT variability. This suggests SLC26A9 as a potential therapeutic target to ameliorate exocrine pancreatic disease.
Keywords: immunoreactive trypsinogen (trypsinogen), exocrine pancreas, Modifier genes, newborn screening (neonatal screening or Newborn infant screening)
Cystic fibrosis (CF), a life-shortening, autosomal recessive disorder caused by mutations in the CF Transmembrane Conductance Regulator (CFTR) gene1 affects multiple organs, including the pancreas. The CF pancreas is characterized by reduced luminal fluid secretion and concentrated acinar proteins that accumulate in the small ducts2, 3. Pancreatic ducts and acini show signs of obstruction from proteinaceous secretions in utero4, and damage continues in early life5. Postnatally, exocrine pancreatic tissue is replaced with fibrotic tissue and fat6, 7.
CFTR genotype is correlated with pancreatic disease severity, with pancreatic insufficiency primarily occurring in individuals with two severe CFTR genotypes8. However individuals with the same CFTR genotype have variable pancreatic disease at birth9, suggesting a role for additional genetic contributors. Serum levels of the pancreatic enzyme precursor, trypsinogen, measured by an immunoreactive trypsinogen (IRT) assay, are often elevated near birth in infants10 with two CF-causing CFTR mutations11. Elevated IRT levels at birth are the primary biomarker of CF in newborn screening programs.
In individuals with CFTR genotypes associated with pancreatic insufficiency, the persistent IRT elevation in infancy is often followed by a progressive decline, with 95% of these individuals with CF having lower IRT levels than a non-CF population by age seven12. This is likely due to progression of pancreatic damage, reduced acinar mass, and reduced capacity to produce digestive enzymes such as trypsin. Thus, extrapolating backwards in an IRT-elevated CF population, lower IRT near birth is thought to reflect more severe pancreatic damage13. This is supported by the findings that lower IRT levels at birth is associated with the CF intestinal obstruction14 meconium ileus15, and increased risk of CF-related diabetes13. However, explicitly evaluating this hypothesis in vivo would require human studies of CF pancreatic tissue.
Decline in IRT levels can be used to monitor acinar destruction postnatally and to predict the development of pancreatic insufficiency12, 16. Thus, IRT levels may represent a biomarker of exocrine pancreatic damage in CF. Measures of IRT in early infancy are heritable15, suggesting that NBS IRT may provide a tool to identify the genetic architecture of prenatal exocrine pancreatic damage in CF.
With loss of CFTR function, variation in multiple constituents of the apical plasma membrane contribute to the risk of meconium ileus, including 8 SNPs in three solute carriers, SLC26A9, SLC6A14, and SLC9A317. In this study, we asked whether the meconium ileus risk-alleles in SLC26A9, SLC6A14, and SLC9A3 were associated with early exocrine pancreatic damage as measured by NBS IRT and whether multiple apical plasma membrane constituents contribute to prenatal exocrine pancreatic damage.
METHODS
All subjects were part of the Colorado, Wisconsin, or French sites of the International Cystic Fibrosis Gene Modifier Consortium, which collected DNA and clinical data from CF patients. All protocols were approved by institutional review boards. All participants provided written informed consent.
Individuals included for analysis were of European descent, had two severe CFTR mutations known to confer pancreatic insufficiency, and had an NBS IRT measurement. NBS IRT values were measured by Children’s Hospital Denver (1982–1987) or the Colorado Department of Public Health and Environment (1987-current)11, the Wisconsin State Laboratory of Hygiene18, or regional screening laboratories in France19. All subjects at each site who had an NBS IRT were eligible to participate in the study.
Quality Control (QC) and DNA Collection
Standard QC measures were performed as previously described17, 20. We did not identify any cryptic relatedness, sex incongruity, or non-European ancestry as determined by principal component analysis using Eigenstrat21. There were six sibling pairs genotyped in the Colorado sample, five in the Wisconsin sample, and no siblings in the French sample. All pairwise relationships were confirmed using identity by descent estimation in PLINK22, and one sibling from each pair was removed at random. The final analysis included 111 individuals from Colorado, and two replication samples of 37 and 80 individuals from Wisconsin and France, respectively. Commercially available IRT kits use different antibodies23, and Lafont et al note that these antibodies can have different specificity for trypsinogen or bind to different forms of trypsinogen present in serum24. Because of this, IRT measurements across sites may not be comparable. Hence, each of the three samples was analyzed separately. Colorado was the largest sample, and was treated as the discovery sample, with Wisconsin and France each used as a replication sample.
DNA was extracted from whole blood or transformed lymphocytes and quantified with fluorimetry. DNA of the French sample was genotyped using two genotyping platforms with different subsets of genetic markers, the Illumina CNV 370-Duo BeadChip or the Illumina 6620W-Quad BeadChip (660W). DNA of the Colorado and Wisconsin samples was genotyped simultaneously using the 660W. Individuals from Colorado or Wisconsin with missing genotype rate >2% (n = 22) were re-genotyped on the Illumina Omni5-Quad BeadChip (Omni5). There were 431,504 SNPs on the 660W and Omni5 that overlapped and passed QC.
Genotype imputation uses a reference set of population haplotypes to infer un-typed genotypes in a sample of interest. We used MaCH and Minimac25 to impute the 22 individuals genotyped on the Omni5 at 79,159 SNPs unique to the 660W, and all French samples to the SNPs on the 660W (Figure 1; available at www.jpeds.com). SLC9A3 SNP rs17563161 was not present on either genotyping platform, and genotype calls for this SNP were imputed in the full sample. Imputed SNPs were required to have R2 > 0.3 and minor allele frequency ≥ 5.0%.
Figure 1.
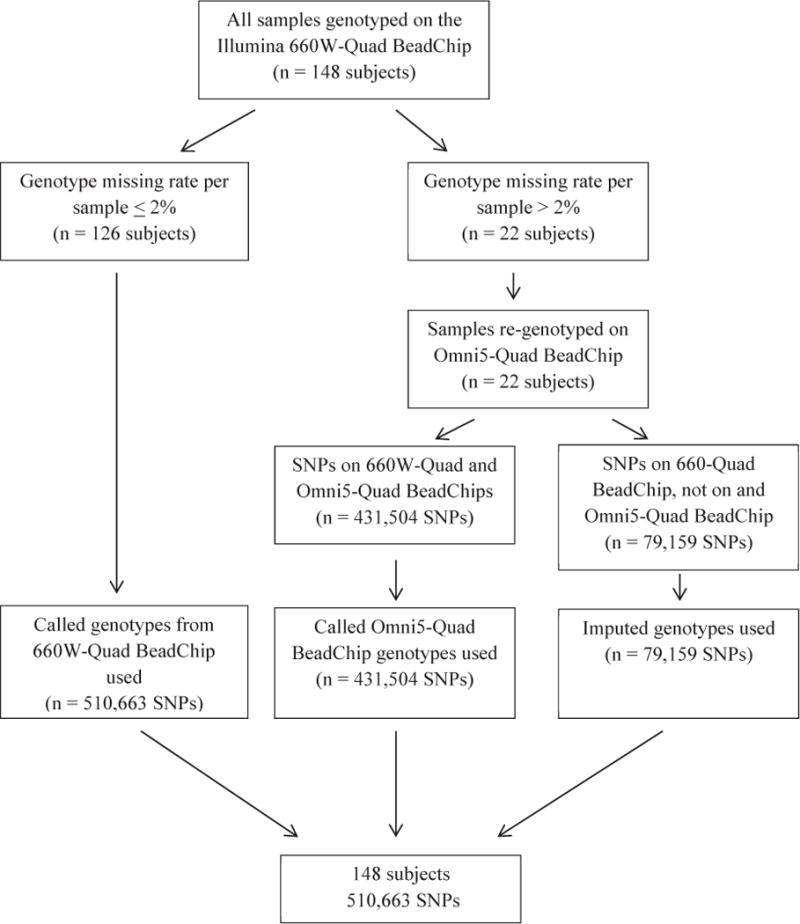
Methodology for combining genotypes for individuals from Colorado and Wisconsin genotyped on the Illumina 660W-Quad BeadChip and re-genotyped on the Illumina Omni5-Quad BeadChip. Standard quality control was performed on all genotypes.
NBS IRT Measurement and Assay
NBS for CF has been in place in Colorado since 198211. In Wisconsin NBS for CF was available as a clinical trial from 1985 to 199426, and available to all infants after 199418. Nationwide NBS has been in place in France since 200219, but previously existed in Normandy (1980)27 and Brittany (1989)28. In Colorado and Wisconsin, IRT is measured by a single state laboratory; in France, it is measured at one of 22 regional laboratories19. In all three locations, newborn infants’ blood is collected via a heel stick and placed on filter cards. IRT is extracted and measured from the dried blood spots.
The assays used to measure IRT have changed during the course of NBS at all three sites (Figure 2; available at www.jpeds.com). Additional details of the assays and changes are published elsewhere11, 18, 19, 27, 29. The assay used in France until 2002 required all newborn infants with IRT ≥ 900 ng/ml undergo follow-up testing for CF. The IRT assay changed in 2002, and the new assay had a positive CF-screening value of 60 ng/ml. In France, IRT values from the older assay are divided by 15 in order to make them comparable with values from the new assay.
Figure 2.
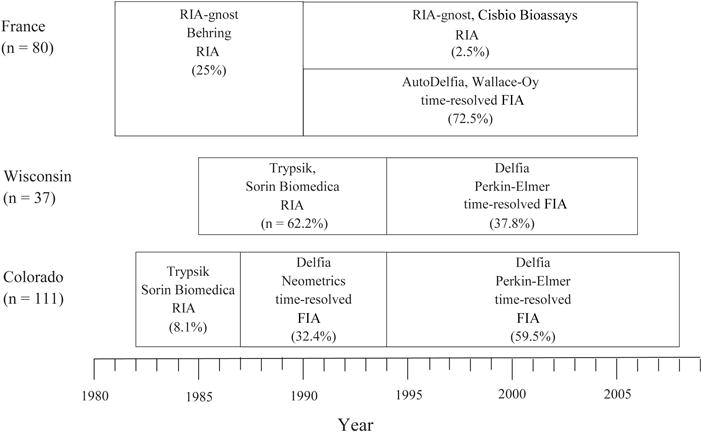
Changes to the IRT assay over time in each site. The x-axis represents year of measurement, and each box indicated the type of assay used, including the (1) assay name, (2) manufacturer, (3) type of assay, and (4) percent of subjects in each site that used that assay.
Statistical Analyses
All analyses were conducted using R30. Kruskal-Wallis tests were used to determine if IRT varied by assay in any of the sites, varied between sites, or varied with other covariates of interest. We used Spearman’s correlation to test for a correlation between IRT and age of IRT measurement. Chi-square tests were used to determine if descriptive characteristics (i.e. sex, meconium ileus) varied significantly (P < 0.05) by site.
In the Colorado sample, 8 SNPs were tested for associations with IRT individually using multiple linear regression. SNPs were coded additively as 0, 1, or 2, corresponding to the number of risk alleles defined by meconium ileus findings17. For chromosome X SNPs, males were coded as 0 or 2. Regression models for each SNP were adjusted for each of the following covariates separately: meconium ileus status, sex, CFTR genotype (delF508 homozygotes versus all others), IRT assay, and population structure using principal components. Inclusion of these covariates did not impact the results, and unadjusted results are presented, with adjusted results for the most significantly associated SNP presented in Tables V and VI (available at www.jpeds.com). The effective number of independent tests across the 8 SNPs was estimated as 6 using the Genetics Type I Error Calculator software, which takes into account correlation between markers31. To adjust for multiple hypothesis testing, we required a significance level of α = 0.0083 (0.05/6) to conclude statistical significance.
Table 5.
Covariate adjustment for association of rs7512462 with NBS IRT in the French sample (n = 80).
| Covariate in Model | pSNP | βSNP (95% CI) | pcovariate | R2 model |
|---|---|---|---|---|
| None (rs7512462 only) | 0.76 | 3 (−18, 24) | NA | 0.00 |
| Sex | 0.78 | 3 (−18, 24) | 0.35 | 0.01 |
| CFTRa | 0.72 | 4 (−18, 25) | 0.58 | 0.01 |
| IRT assay | 0.67 | 5 (−17, 26) | 0.38 | 0.02 |
| Meconium ileusb | 0.83 | 3 (−20, 25) | 0.33 | 0.01 |
| Principal componentsc | 0.83 | 2 (−19, 24) | 0.64 | 0.06 |
: Subjects are classified as delF508 homozygotes versus all other mutations.
: Based on a total sample of 140; 8 subjects are missing Meconium ileus status.
: Adjusted for population structure, based on the first four principal components.
Table 6.
Top ranked apical plasma membrane constituents in the combined Colorado and Wisconsin samples (n = 148).
| SNP | Chromosome | Positiona | P value | Gene |
|---|---|---|---|---|
| rs7512462 | 1 | 205899595 | 6.77 × 10−5 | SLC26A9 |
| rs242050 | 1 | 4789179 | 3.72 × 10−4 | AJAP1 |
| rs12047830 | 1 | 205916699 | 7.61 × 10−4 | SLC26A9 |
| rs7415921 | 1 | 205910883 | 8.80 × 10−4 | SLC26A9 |
| rs7232775 | 18 | 43202404 | 1.42 × 10−3 | SLC14A2 |
| rs9675236 | 17 | 58226800 | 1.70 × 10−3 | CA4 |
| rs4077468 | 1 | 205914757 | 2.24 × 10−3 | SLC26A9 |
| rs4077469 | 1 | 205914885 | 2.24 × 10−3 | SLC26A9 |
| rs2627907 | 17 | 58218881 | 2.63 × 10−3 | CA4 |
| rs1321507 | 6 | 51826281 | 3.41× 10−3 | PKHD1 |
: Based on Genome Reference Consortium Human Reference 37/Human Genome 19.
We assessed the evidence for replication of a SNP-IRT association identified in the Colorado sample, using the Wisconsin and French samples. We used the same statistical models and required the same risk allele to be associated, although all reported p-values are two-sided.
To determine if IRT has a complex genetic inheritance with multiple constituents of the apical plasma membrane contributing to pancreatic damage, we conducted a gene-set analysis of the apical constituents and a single-SNP analysis to assess significance at the individual markers annotated to the gene-set. As in the report by Sun et al17, a list of 157 genes that localize to the apical plasma membrane was identified. Because the majority of the study participants were genotyped on the 660W, we limited analysis to SNPs on this platform that passed QC with a minor allele frequency ≥ 2.0%. The distribution of IRT was similar between Colorado and Wisconsin (P = 0.57); thus the two samples were combined (n = 148) for the gene-set analysis with 3,614 SNPs. We repeated the gene-set analysis in the French sample (3,567 SNPs). We used permutation analysis to assess statistical significance of the Wald Chi-squared sum statistic across the 3,614 SNPs (Colorado and Wisconsin) or 3,567 SNPs (France) in the gene-set.
RESULTS
The three samples are described in Table I. In France, all NBS blood samples were collected at three days of age, whereas in Colorado and Wisconsin the timing of the blood collection varied. The three sites are comparable in distribution of sex, percent with meconium ileus, and occurrence of delF508 mutations. IRT distribution does not differ between Colorado and Wisconsin, but is lower in France (P < 0.0001) than either North American sample (Figure 3; available at www.jpeds.com).
Table 1.
Characteristics of 228 CF individuals with a NBS IRT value by site.
| Site
|
|||
|---|---|---|---|
| Colorado (n = 111) |
Wisconsin (n = 37) |
France (n = 80) |
|
| IRT, ng/ml, mean (SD) | 255 (116) | 236 (101) | 151 (68) |
| Age at IRT measurement, days, median (IQR) | 2 (1–3) | 3 (2–4) | 3 (NA)a |
| Year of birth, range | 1983–2008 | 1986–2006 | 1981–2006 |
| Male, n (%) | 46 (41) | 21 (57) | 41 (51) |
| Meconium ileus positive, n (%) | 23 (22)b | 9 (24) | 14 (18)c |
| Number p.Phe508del mutations, n (%)d | |||
| 0 | 4 (4) | 1 (3) | 3 (4) |
| 1 | 45 (41) | 9 (24) | 24 (30) |
| 2 | 62 (56) | 27 (73) | 53 (66) |
SD, standard deviation; IQR, inter-quartile range
: All blood samples are drawn at 3 days of age in this sample
: Based on a sample size of 103; 8 subjects missing meconium ileus status.
: Based on a sample size of 76; 4 subjects missing meconium ileus status.
: Cells may not sum to 100% due to rounding.
Figure 3.
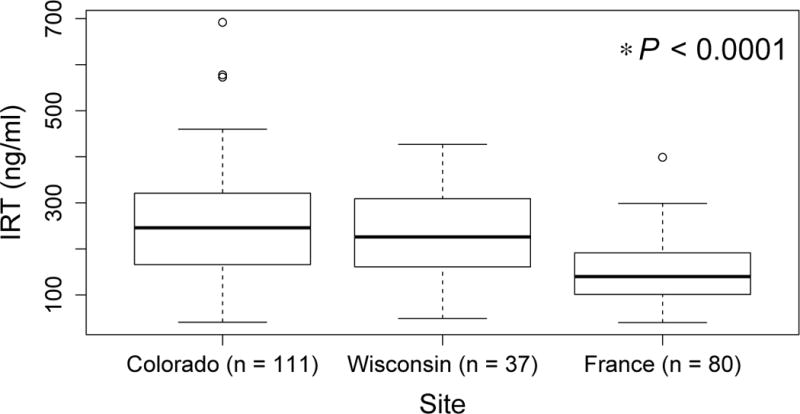
Distribution of NBS IRT in Colorado, Wisconsin and French samples. Box represents 25th, 50th (median), and 75th percentile. Whiskers represent 5th and 95th percentiles, and open circles represent values above the 95th percentile.
IRT does not vary by sex or CFTR genotype () in any of the three sites. IRT values are not correlated with age at IRT measurement (Colorado and Wisconsin; Figure 4 available at www.jpeds.com). In Wisconsin, IRT levels were greater when measured with the radio-immunoassay than the time-resolved fluorometric assay (P = 0.008; Figure 4). IRT is similar by assay in Colorado and France. IRT is lower in individuals with meconium ileus (Colorado: P = 0.002; Wisconsin: P = 0.03), though this does not reach statistical significance in the French sample (P = 0.09; Figure 4).
Figure 4.
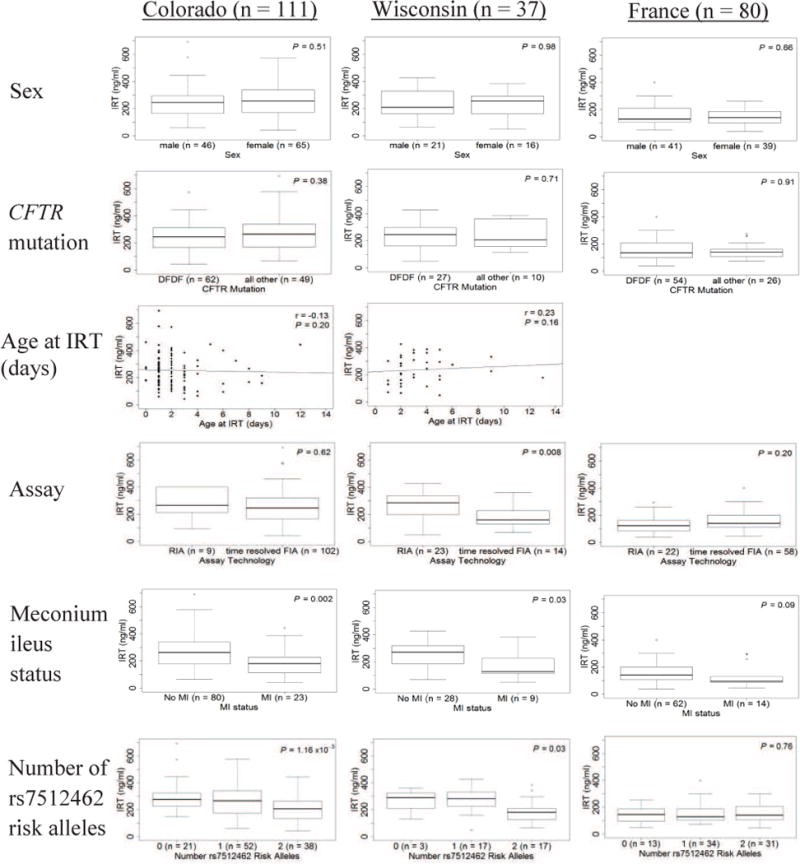
Distribution of NBS IRT is compared, within each site, by the following variables of interest: Sex (row 1): males versus female. CFTR mutation (row 2): Subjects are classified as delF508 homozygotes (DFDF) versus all others. Age at IRT (row 3): Correlation of age at IRT with IRT value (Colorado and Wisconsin only; In France, IRT is measured at 3 days of age). Assay (row 4): Categorized by the technology used to measure IRT, either radio-immunoassay (RIA), or a time-resolved fluorometric assay (FIA). Meconium ileus status (row 5): subjects with meconium ileus (MI) versus those without MI. Total n may be reduced as some subjects are missing MI status. Number of rs7512462 risk alleles (row 6): 0, 1, or 2 risk alleles. P-values for sex, assay, MI status and CFTR determined using Kruskal-Wallis tests. P-value for rs7512462 determined using linear regression. Age at IRT correlation determined using Spearman’s correlation. Box represents 25th, 50th (median), and 75th percentile. Whiskers represent 5th and 95th percentiles, and open circles represent values above the 95th percentile.
In the Colorado sample, three SNPs in SLC26A9 were associated with NBS IRT (min P = 1.16 × 10−3 at rs7512462; Table II). No SNPs in SLC9A3 (P = 0.50) or SLC6A14 (min P = 0.21) showed evidence of association (Table II). Each additional copy of the rs7512462 meconium ileus risk allele17 was associated with lower NBS IRT (i.e. worse pancreatic disease) of 49 ng/ml (95% Confidence Interval (CI): (−78, −20)); this SNP explained 9% of the variability in NBS IRT. The distribution of IRT by number of rs7512462 risk alleles is presented in Figure 4. Limiting the analysis to individuals homozygous for the CFTR delF508 mutation (n = 62) resulted in similar findings (P = 6.65 × 10−3; β = −50 ng/ml). The three associated SNPs in SLC26A9 are in reasonably high linkage disequilibrium (Figure 5).32 After adjusting for the effect of rs7512462, the other SNPs were no longer associated (P = 0.45 and 0.19 for rs4077468 and rs12047830, respectively) suggesting the three SNPs tag the same underlying genetic cause.
Table 2.
Association of eight candidate SNPs with NBS IRT values in 111 subjects from Colorado.
| SNP | Chromosome | Positiona | Gene | Risk Alleleb | Risk Allele Frequency | P value | β (95% CI) | R2 |
|---|---|---|---|---|---|---|---|---|
| rs7512462 | 1 | 204166218 | SLC26A9 | T | 0.57 | 1.16 × 10−3 | −49 (−78, −20) | 0.09 |
| rs4077468 | 1 | 204181380 | SLC26A9 | T | 0.55 | 7.20 × 10−3 | −38 (−66, −11) | 0.06 |
| rs12047830 | 1 | 204183322 | SLC26A9 | C | 0.50 | 1.92 × 10−3 | −47 (−77, −18) | 0.08 |
| rs7419153 | 1 | 204183932 | SLC26A9 | T | 0.37 | 0.02 | −34 (−63, −5) | 0.05 |
| rs17563161 | 5 | 550624 | SLC9A3 | T | 0.28 | 0.50 | 13 (−24, 49) | 0.00 |
| rs12839137 | X | 115479578 | SLC6A14 | C | 0.84 | 0.87 | −3 (−39, 32) | 0.00 |
| rs5905283 | X | 115479909 | SLC6A14 | C | 0.63 | 0.21 | −16 (−42, 10) | 0.01 |
| rs3788766 | X | 115480867 | SLC6A14 | T | 0.73 | 0.41 | −12 (−40, 17) | 0.01 |
CI, Confidence Interval
: Based on Genome Reference Consortium Human Reference 37/Human Genome 19.
: Meconium ileus risk allele from Sun et al16.
Figure 5.
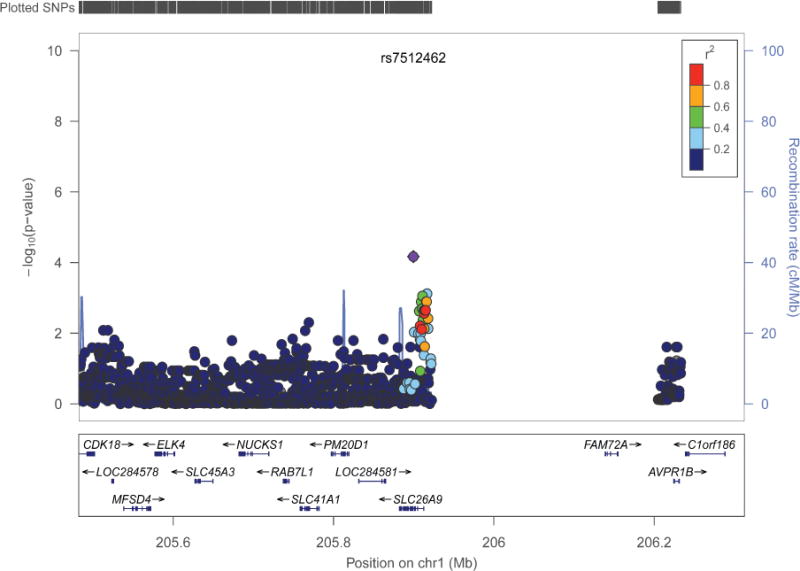
Regional plot for SLC26A9 in the combined Colorado and Wisconsin sample. LocusZoom viewer32 was used to show association evidence around SLC26A9 based on human genome build 19. Symbol coloring represents LD r2 values of the 1000 genomes European sample with the most significant SNP.
In the Wisconsin sample, rs7512462 was significantly associated with NBS IRT (P = 0.03, β = −57 ng/ml, 95% CI: −108, −6; Table III) and explained ~13% of the variability in IRT. Evidence for association was similar in the delF508 homozygous group (n = 27; p = 0.023; β = −66 ng/ml). There was no evidence that rs7512462 contributed to variability in the French measure of NBS IRT (P = 0.76, β = 3 ng/ml, 95% CI: (−18, 24)), and this was similar when limiting to the delF508 homozygous group (n = 52, p = 0.76, β = 4 ng/ml). When the two North American samples were combined, evidence for association between NBS IRT and rs7512462 was similar to the site specific samples (P = 6.77 × 10−5; β = −51 ng/ml, 95% CI: (−76, −26)); rs7512462 explained ~10% of the variance in IRT. Evidence for association was similar in the delF508 homozygous group (n = 89, p = 6.65 × 10−3, β = −55 ng/ml). Covariate adjustment did not impact these results (Tables IV and V; Table IV available at www.jpeds.com). Imputation of the SLC26A9 region did not identify any additional SNPs that were more strongly associated with NBS IRT than rs7512462 (Figure 5).
Table 3.
Association of SLC26A9 rs7512462 with NBS IRT values by site.
| Site | N | rs7512462 Risk Allele Frequency | P value | β (95% CI) | R2 |
|---|---|---|---|---|---|
| Colorado | 111 | 0.57 | 1.16 × 10−3 | −49 (−78, −20) | 0.09 |
| Wisconsin | 37 | 0.69 | 0.03 | −57 (−108, −6) | 0.13 |
| France | 80 | 0.62 | 0.76 | 3 (−18, 24) | 0.00 |
| Colorado + Wisconsin | 148 | 0.60 | 6.77 × 10−5 | −51 (−76, −26) | 0.10 |
CI, Confidence Interval
Table 4.
Covariate adjustment for association of SLC26A9 SNP rs7512462 with NBS IRT in the Colorado and Wisconsin combined samples (n = 148).
| Covariate in Model | pSNP | βSNP (95% CI) | pcovariate | R2 model |
|---|---|---|---|---|
| None (rs7512462 only) | 6.77 × 10−5 | −51 (−76, −26) | N/A | 0.10 |
| Sex | 7.30 × 10−5 | −51 (−76, −26) | 0.72 | 0.10 |
| CFTRa | 2.02 × 10−5 | −56 (−80, −31) | 0.06 | 0.13 |
| Age at IRT measurement (days) | 1.56 × 10−4 | −51 (−77, −25) | 0.95 | 0.10 |
| IRT assay | 4.85 × 10−5 | −52 (−77, −28) | 0.16 | 0.12 |
| Meconium ileusb | 1.37 × 10−4 | −48 (−72, −24) | 1.90 × 10−4 | 0.19 |
| Principal componentsc | 2.32 × 10−5 | −56 (−81, −31) | 0.15 | 0.13 |
| Site | 9.87 × 10−5 | −51 (−77, −25) | 0.74 | 0.10 |
: Subjects are classified as delF508 homozygotes versus all other mutations.
: Based on a total sample of 140; 8 subjects are missing Meconium ileus status.
: Adjusted for population structure, based on the first four principal components.
We did not find significant evidence of a complex genetic architecture for exocrine pancreatic disease as measured by NBS IRT, with multiple constituents of the apical plasma membrane contributing to early exocrine pancreatic damage (p = 0.26; Figure 6 available at www.jpeds.com). Further, when we examine the association evidence from the individuals SNPs in this group, rs7512462 in SLC26A9 is the top ranked SNP (Table VI) with no other SNP reaching an experiment-wise (0.05/3614 = 1.38 × 10−5) significance level.
Figure 6.
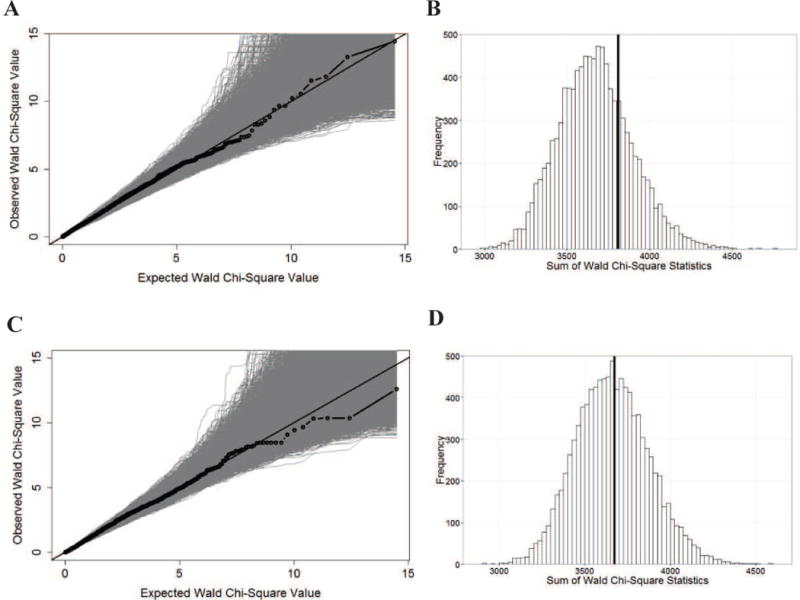
There is no evidence of association between NBS IRT and the apical plasma membrane constituents in the Colorado and Wisconsin samples (n = 148) or in the French samples (n = 80). As in Sun et al.17, a list of 157 genes that localize to the apical plasma membrane was identified. We included 3,614 SNPs (Colorado and Wisconsin) or 3,567 SNPs (French) from the Illumina 660W-Quad BeadChip with minor allele frequency >2% that were annotated to +/−10 kb of the boundaries of the 155 genes. Two genes were not tagged by any GWAS SNPs. (A) Quantile-quantile plot in the combined Colorado and Wisconsin samples. The observed association statistics (black dotted line) and the statistics calculated from 10,000 IRT permuted replicates (gray) are shown. (B) Statistical significance of the apical plasma membrane hypothesis in the combined Colorado and Wisconsin samples. P-value (0.26) was established via a statistic that summed over the association evidence (Wald chi-squared statistic) of all the 3,614 SNPs with the observed sum statistic shown as a vertical line (black) and 10,000 permutation-based sum statistics shown as a histogram. (C) Quantile-quantile plot in the French replication sample. (D) Statistical significance of the apical membrane hypothesis in the French replication sample (P = 0.46).
Similarly, in the French sample, we did not find any evidence that multiple constituents contributed to early exocrine pancreatic damage (P = 0.46; Figure 6), and there are no individual SNPs annotated to the 155 genes that reach experiment-wise significance (Table VII; available at www.jpeds.com).
Table 7.
Top ranked apical plasma membrane constituents in the French sample (n = 80).
| SNP | Chromosome | Positiona | P value | Gene |
|---|---|---|---|---|
| rs35806 | 10 | 79165830 | 6.58 × 10−4 | KCNMA1 |
| rs776385 | 8 | 31736625 | 1.88 × 10−3 | NRG1 |
| rs4458837 | 8 | 32304383 | 1.90 × 10−3 | NRG1 |
| rs1425802 | 11 | 35158401 | 1.96 × 10−3 | CD44 |
| rs673195 | 10 | 79164857 | 2.61 × 10−3 | KCNMA1 |
| rs11802794 | 1 | 197372250 | 2.95 × 10−3 | CRB1 |
| rs353625 | 11 | 35168546 | 3.45 × 10−3 | CD44 |
| rs6718187 | 2 | 44082362 | 4.69 × 10−3 | ABCG8 |
| rs4148179 | 2 | 44056856 | 4.72 × 10−3 | ABCG5 |
| rs4148178 | 2 | 44057030 | 4.72 × 10−3 | ABCG5 |
: Based on Genome Reference Consortium Human Reference 37/Human Genome 19.
DISCUSSION
We found a strong association between SLC26A9 rs7512462 and prenatal exocrine pancreatic disease in a sample of individuals with CF from Colorado. This association replicated in a sample of individuals with CF from Wisconsin, but not in a sample from France. In both the Colorado and Wisconsin samples, the rs7512462 meconium ileus risk allele was associated with more severe pancreatic disease, as indicated by a lower newborn screened IRT value; provided similar effect size estimates; and accounted for a similar and substantial amount of the variability in NBS IRT. We did not find any SNPs in SLC6A14 or SLC9A3 that were associated with early exocrine pancreatic disease severity. This supports previous findings that SLC6A14 and SLC9A3 are associated with CF pulmonary phenotypes, but not pancreatic phenotypes33, and is consistent with known expression data. Based upon a study of human adult tissues34, SLC9A3 is abundantly expressed in kidney and small intestine, and SLC6A14 is abundantly expressed in human lung and salivary gland. Lohi et al report expression of SLC26A9 in the human pancreas35. Pancreatic expression was not reported in a study of adult human tissues34, although the importance of SLC26A9 function in the gastrointestinal tract of very young mice has been reported36. SLC26A9 encodes an anion transporter that can mediate chloride-bicarbonate exchange and sodium-coupled transport37. It can physically interact with CFTR and be influenced by CFTR activity38, 39. In CF, with the loss of CFTR function at the apical plasma membrane, SLC26A9 may augment ion transport and fluid flow, explaining its modifying effect. From a clinical perspective, individuals diagnosed with CF through newborn screening could be screened for rs7512462. Individuals who carry the risk allele may have worse pancreatic damage and could be monitored more closely for failure to thrive and the need to start enzyme treatments sooner.
We were unable to find evidence of a complex genetic model for CF exocrine pancreatic disease with multiple apical constituents contributing to NBS IRT, unlike for other CF comorbidities such as meconium ileus and lung disease17, 33. However, for other CF comorbidities, individual associated SNPs accounted for much less than 10% of trait variability, which is consistent with a complex inheritance model.
Multiple factors could explain the lack of association with rs7512462 in the French sample. It is possible that a different constituent of the apical plasma membrane contributes to exocrine pancreatic damage in the French sample, and we were underpowered to detect this. Another explanation is that the French sample differed fundamentally from the American samples. IRT was assayed at one of 22 regional laboratories in France, whereas the Wisconsin and Colorado sites each have centralized measurements of IRT, introducing more heterogeneity in the French sample. Moreover, different commercial IRT kits use different antibodies23, which have different specificity for trypsinogen or bind to different forms of trypsinogen present in serum24. Further, there is no certified IRT reference material, so the IRT provided as a reference can vary in level in different kits40. Finally, the stability of IRT measured from blood spots may be affected by transport time from the hospital to the testing facility and the climate of the area41. All of these factors could affect the IRT levels measured in the French sample, leading to the observed difference in IRT distribution across the samples and the lack of rs7512462 association. We were unable to link the specific laboratory corresponding to individual IRT measurements in the French data to determine if the IRT values differed by site.
A limitation of this study is the relatively small sample size available, which precluded interrogation of the whole genome for markers associated with prenatal exocrine pancreatic damage. However, the observed difference in IRT distribution between the French and American samples provide a note of caution for future studies that may wish to use the NBS IRT values from multiple sites/laboratories in order to increase their sample sizes. Not all CF IRT-based newborn screening protocols result in equally interpretable biomarkers. A second limitation is that the study was restricted to individuals of European ancestry, and therefore our results only generalize to that population.
Despite changes to the assay, the ratio of signal to noise was strong enough that we were able to use the NBS IRT value from the Colorado and Wisconsin samples as a biomarker to reflect prenatal exocrine pancreatic disease severity in CF and identify a gene modifier, suggesting that NBS IRT has utility beyond a threshold for determining CF. Further investigation is required to disentangle which assays may be more or less useful for this purpose outside of those considered here.
Prenatal exocrine pancreatic damage in CF is influenced by the solute carrier SLC26A9 with one SNP in this gene accounting for a substantial percentage of the variability in NBS IRT. The robust finding that SLC26A9 is a CF gene modifier and a marker of CF pancreatic disease severity suggests a potential therapeutic target to slow the progression of exocrine pancreatic damage in CF.
Acknowledgments
The authors would like to thank the patients and families who participated in the study. We would also like to thank Jaclyn Stonebraker for assistance in sample processing, data handling, and useful discussion; Jacques Brouard, Michel Roussey, Valérie David, Christophe Marguet, and Pauline Touche for patient recruitment and the data collection of the French sample; Melissa Reske and Rachel Bersie for patient recruitment and data collection of the Wisconsin sample; and Meg Anthony for patient recruitment and study design of the Colorado sample.
Funding support information is available at www.jpeds.com (Appendix).
Funded by the Canadian Institutes of Health Research (MOP-258916 [to L.S.]), Cystic Fibrosis Canada (2626 [to L.S.]), the National Institutes of Health/National Heart Lung and Blood Institute (R01 HL068890-10 [to M.K.] and DP20D007031 [to H.L.]), National Institute of Diabetes and Digestive and Kidney Diseases (1RO1 DK61886-01 [to F.A.]), the Children’s Hospital of Wisconsin Research Institute (to H.L.), and the United States Cystic Fibrosis Foundation (Sontag07AO and Miller13A0). Funds for genome-wide genotyping of North American participants were generously provided by the United States Cystic Fibrosis Foundation. H.C. and P.-Y. B. are supported by the Institut National de la Santé et de la Recherche Médicale, Assistance Publique-Hôpitaux de Paris, Université Pierre et Marie Curie Paris, Agence Nationale de la Recherche (R09186DS), DGS, Association Vaincre La Mucoviscidose, Chancellerie des Universités (Legs Poix), Association Agir Informer Contre la Mucoviscidose, and GIS-Institut des Maladies Rares. M.M., W.L., and D.S. are fellows of the CIHR Strategic Training for Advanced Genetic Epidemiology and Statistical Genetics.
ABBREVIATIONS
- CF
Cystic Fibrosis
- CFTR
Cystic Fibrosis Transmembrane Conductance Regulator
- CI
Confidence Interval
- IRT
Immunoreactive Trypsinogen
- NBS
newborn screened/newborn screening
- Omni5
Illumina Omni5-Quad BeadChip
- QC
quality control
- 660W
Illumina 660W-Quad BeadChip
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- 1.Kerem B, Rommens J, Buchanan J, Markiewicz D, Cox T, Chakravarti A, et al. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–80. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 2.Kopelman H, Corey M, Gaskin K, Durie P, Weizman Z, Forstner G. Impaired chloride secretion, as well as bicarbonate secretion, underlies the fluid secretory defect in the cystic fibrosis pancreas. Gastroenterology. 1988;95:349–55. doi: 10.1016/0016-5085(88)90490-8. [DOI] [PubMed] [Google Scholar]
- 3.Kopelman H, Durie P, Gaskin K, Weizman Z, Forstner G. Pancreatic fluid secretion and protein hyperconcentration in cystic fibrosis. N Engl J Med. 1985;312:329–34. doi: 10.1056/NEJM198502073120601. [DOI] [PubMed] [Google Scholar]
- 4.Boue A, Muller F, Nezelof C, Oury JF, Duchatel F, Dumez Y, et al. Prenatal diagnosis in 200 pregnancies with a 1-in-4 risk of cystic fibrosis. Hum Genet. 1986;74:288–97. doi: 10.1007/BF00282551. [DOI] [PubMed] [Google Scholar]
- 5.Imrie JR, Fagan DG, Sturgess JM. Quantitative evaluation of the development of the exocrine pancreas in cystic fibrosis and control infants. Am J Pathol. 1979;95:697–708. [PMC free article] [PubMed] [Google Scholar]
- 6.Oppenheimer EH, Esterly JR. Pathology of cystic fibrosis review of the literature and comparison with 146 autopsied cases. Perspect Pediatr Pathol. 1975;2:241–78. [PubMed] [Google Scholar]
- 7.Iannucci A, Mukai K, Johnson D, Burke B. Endocrine pancreas in cystic fibrosis: an immunohistochemical study. Hum Pathol. 1984;15:278–84. doi: 10.1016/s0046-8177(84)80191-4. [DOI] [PubMed] [Google Scholar]
- 8.Ahmed N, Corey M, Forstner G, Zielenski J, Tsui L-C, Ellis L, et al. Molecular consequences of cystic fibrosis transmembrane regulator (CFTR) gene mutations in the exocrine pancreas. Gut. 2003;52:1159–64. doi: 10.1136/gut.52.8.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bronstein MN, Sokol RJ, Abman SH, Chatfield BA, Hammond KB, Hambidge KM, et al. Pancreatic insufficiency, growth, and nutrition in infants identified by newborn screening as having cystic fibrosis. J Pediatr. 1992;120:533–40. doi: 10.1016/s0022-3476(05)82478-3. [DOI] [PubMed] [Google Scholar]
- 10.Crossley JR, Elliot RB, Smith PA. Dried-blood spot screening for cystic fibrosis in the newborn. The Lancet. 1979;313:472–4. doi: 10.1016/s0140-6736(79)90825-0. [DOI] [PubMed] [Google Scholar]
- 11.Sontag MK, Hammond KB, Zielenski J, Wagener JS, Accurso FJ. Two-tiered immunoreactive trypsinogen-based newborn screening for cystic fibrosis in Colorado: screening efficacy and diagnostic outcomes. J Pediatr. 2005;147:S83–S8. doi: 10.1016/j.jpeds.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Durie PR, Forstner GG, Gaskin KJ, Moore DJ, Cleghorn GJ, Wong SS, et al. Age-related alterations of immunoreactive pancreatic cationic trypsinogen in sera from cystic fibrosis patients with and without pancreatic insufficiency. Pediatr Res. 1986;20:209–13. doi: 10.1203/00006450-198603000-00002. [DOI] [PubMed] [Google Scholar]
- 13.Soave D, Miller M, Keenan K, Li W, Gong J, Ip W, et al. Evidence for a causal relationship between early exocrine pancreatic disease and cystic fibrosis-related diabetes: A Mendelian randomization study. Diabetes. 2014;63:2114–9. doi: 10.2337/db13-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welsh MJ, Ramsey BW, Accurso FJ, Cutting GR. Cystic fibrosis. In: Scrivner CR, Sly WS, Childs B, et al., editors. The metabolic and molecular bases of inherited disease. III. New York: McGraw-Hill, Inc; 2001. pp. 5121–5188. [Google Scholar]
- 15.Sontag MK, Corey M, Hokanson JE, Marshall JA, Sommer SS, Zerbe GO, et al. Genetic and physiologic correlates of longitudinal immunoreactive trypsinogen decline in infants with cystic fibrosis identified through newborn screening. J Pediatr. 2006;149:650–7.e2. doi: 10.1016/j.jpeds.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 16.Couper RTL, Corey M, Moore DJ, Fisher LJ, Forstner GG, Durie PR. Decline of exocrine pancreatic function in cystic fibrosis patients with pancreatic sufficiency. Pediatr Res. 1992;32:179–82. doi: 10.1203/00006450-199208000-00011. [DOI] [PubMed] [Google Scholar]
- 17.Sun L, Rommens JM, Corvol H, Li W, Li X, Chiang TA, et al. Multiple apical plasma membrane constituents are associated with susceptibility to meconium ileus in individuals with cystic fibrosis. Nat Genet. 2012;44:562–9. doi: 10.1038/ng.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rock MJ, Hoffman G, Laessig RH, Kopish GJ, Litsheim TJ, Farrell PM. Newborn screening for cystic fibrosis in Wisconsin: nine-year experience with routine trypsinogen/DNA testing. J Pediatr. 2005;147:S73–S7. doi: 10.1016/j.jpeds.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Munck A, Dhondt J-L, Sahler C, Roussey M. Implementation of the French nationwide cystic fibrosis newborn screening program. J Pediatr. 2008;153:228–33.e1. doi: 10.1016/j.jpeds.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 20.Wright FA, Strug LJ, Doshi VK, Commander CW, Blackman SM, Sun L, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet. 2011;43:539–46. doi: 10.1038/ng.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindau-Shepard BA, Pass KA. Newborn screening for cystic fibrosis by use of a multiplex immunoassay. Clin Chem. 2010;56:445–50. doi: 10.1373/clinchem.2009.132480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lafont P, Guy-Crotte O, Paulin C, Galvain D, Mertani S, Figarella C, et al. A specific immunoradiometric assay of cationic trypsin(ogen) that does not recognize trypsin-alpha-1-proteinase inhibitor complex. Clin Chim Acta. 1995;235:197–206. doi: 10.1016/0009-8981(95)06030-x. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fost N, Farrell PM. A prospective randomized trial of early diagnosis and treatment of cystic fibrosis: a unique ethical dilemma. Clin Res. 1989;37:495–500. [PubMed] [Google Scholar]
- 27.Brouard J, Lecoq I, Viel JF, Guillot M, Laurans M, Laroche D, et al. Evaluation of diagnosis and follow-up in screened children with cystic fibrosis in Normandy. Arch Pediatr. 2001;8(Suppl 3):603–9. doi: 10.1016/s0929-693x(01)80015-4. [DOI] [PubMed] [Google Scholar]
- 28.Scotet V, de Braekeleer M, Roussey M, Rault G, Parent P, Dagorne M, et al. Neonatal screening for cystic fibrosis in Brittany, France: assessment of 10 years’ experience and impact on prenatal diagnosis. The Lancet. 2000;356:789–94. doi: 10.1016/S0140-6736(00)02652-0. [DOI] [PubMed] [Google Scholar]
- 29.Farrell PM, Kosorok MR, Laxova A, Shen G, Koscik RE, Bruns WT, et al. Nutritional benefits of neonatal Screening for cystic fibrosis. N Engl J Med. 1997;337:963–9. doi: 10.1056/NEJM199710023371403. [DOI] [PubMed] [Google Scholar]
- 30.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 31.Li M-X, Yeung JY, Cherny S, Sham P. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet. 2012;131:747–56. doi: 10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W, Soave D, Miller M, Keenan K, Lin F, Gong J, et al. Unraveling the complex genetic model for cystic fibrosis: pleiotropic effects of modifier genes on early cystic fibrosis-related morbidities. Hum Genet. 2013:1–11. doi: 10.1007/s00439-013-1363-7. [DOI] [PubMed] [Google Scholar]
- 34.Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lohi H, Kujala M, Makela S, Lehtonen E, Kestila M, Saarialho-Kere U, et al. Functional characterization of three novel tissue-specific anion exchangers SLC26A7, -A8, and -A9. J Biol Chem. 2002;277:14246–54. doi: 10.1074/jbc.M111802200. [DOI] [PubMed] [Google Scholar]
- 36.Liu X, Li T, Riederer B, Lenzen H, Ludolph L, Yeruva S, et al. Loss of Slc26a9 anion transporter alters intestinal electrolyte and HCO3 – transport and reduces survival in CFTR-deficient mice. Pflugers Arch. 2014:1–15. doi: 10.1007/s00424-014-1543-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang MH, Plata C, Zandi-Nejad K, Sindic A, Sussman CR, Mercado A, et al. Slc26a9–anion exchanger, channel and Na+ transporter. J Membr Biol. 2009;228:125–40. doi: 10.1007/s00232-009-9165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertrand CA, Zhang R, Pilewski JM, Frizzell RA. SLC26A9 is a constitutively active, CFTR-regulated anion conductance in human bronchial epithelia. J Gen Physiol. 2009;133:421–38. doi: 10.1085/jgp.200810097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avella M, Loriol C, Boulukos K, Borgese F, Ehrenfeld J. SLC26A9 stimulates CFTR expression and function in human bronchial cell lines. J Cell Physiol. 2011;226:212–23. doi: 10.1002/jcp.22328. [DOI] [PubMed] [Google Scholar]
- 40.Li L, Zhou Y, Bell CJ, Earley MC, Hannon WH, Mei JV. Development and characterization of dried blood spot materials for the measurement of immunoreactive trypsinogen. J Med Screen. 2006;13:79–84. doi: 10.1258/096914106777589623. [DOI] [PubMed] [Google Scholar]
- 41.Therrell BL, Jr, Hannon WH, Hoffman G, Ojodu J, Farrell PM. Immunoreactive trypsinogen (IRT) as a biomarker for cystic fibrosis: Challenges in newborn dried blood spot screening. Mol Genet Metab. 2012;106:1–6. doi: 10.1016/j.ymgme.2012.02.013. [DOI] [PubMed] [Google Scholar]


