Abstract
DNA methylation (the addition of a methyl group to a cytosine) is an important epigenetic event in mammalian cells because it plays a key role in regulating gene expression. Most previous methylation studies assume that DNA methylation occurs on both positive and negative strands. However, a few studies have reported that in some genes, methylation occurs only on one strand (ie, hemimethylation) and has clustering patterns. These studies report that hemimethylation occurs on individual genes. It is unclear whether hemimethylation occurs genome-wide and whether there are hemimethylation differences between cancerous and noncancerous cells. To address these questions, we have developed the first-ever pipeline, named hemimethylation pipeline (HMPL), to identify hemimethylation patterns. Utilizing the available software and the newly developed Perl and R scripts, HMPL can identify hemimethylation patterns for a single sample and can also compare two different samples.
Keywords: hemimethylation, NGS (next-generation sequencing), HMPL
Introduction
DNA methylation is an epigenetic modification in a cell. This modification adds a methyl group (CH3) to the 5′ position of cytosine in a DNA sequence and is inheritable through cell division.1,2 For mammalian cells, it occurs only at CpG sites (cytosines paired with guanines). DNA methylation plays an essential role for both normal and cancerous cell development3–6 and is closely related to significant processes, including X-chromosome inactivation, genomic imprinting, and tumor growth.7–10
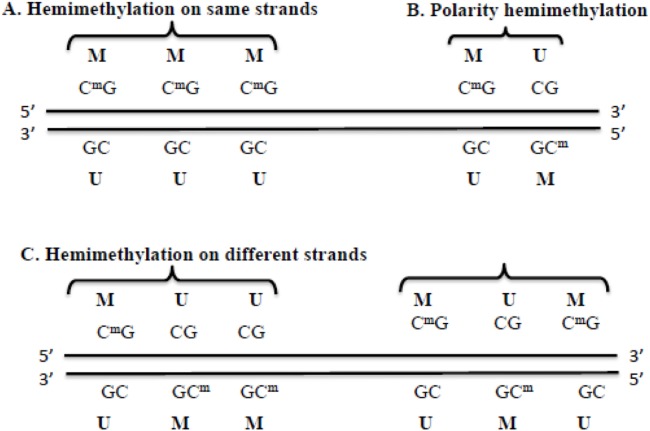
In a genome, there are different types of methylation patterns, including hypermethylation, hypomethylation, and hemimethylation. Hypermethylation occurs when samples in one group (eg, cancer patients) have more methylation than the samples in another group (eg, normal individuals). Hypomethylation occurs when samples in one group have less methylation than the samples in another group. Hemimethylation means that at a CpG site, only one strand of the DNA is methylated (denoted M in Fig. 1), and the other strand is unmethylated (denoted U in Fig. 1). Recent research studies11–13 show that, on a number of genes, hemimethylation may occur as a same-strand cluster (Fig. 1A), a polarity (or reverse) hemimethylation cluster (Fig. 1B) with only two CpG sites, or a different-strand cluster with more than two CpG sites (Fig. 1C). The cluster pattern means that two or more consecutive CpG sites are methylated only on one DNA strand, and not on the other strand, as shown in Figure 1A. The polarity (or reverse) hemimethylation clusters imply that, at the two consecutive CpG sites, the methylation patterns on the positive and negative strands are “MU and UM,” respectively, as shown in Figure 1B. “MU and UM” means that, at two adjacent CpG sites, the first site is hemimethylated as MU on the positive and negative strands, while the next adjacent site is hemimethylated as UM on two strands, which has a reversed pattern. The hemimethylation patterns shown in Figure 1 are like the footprints of DNA demethylation (ie, methyl groups are removed) in cancer.12 This “footprint” role exists because hemimethylation is a transitional state between being methylated and having no methylation at specific CpG sites. Therefore, the identification of hemimethylation is important for understanding both methylation events and the establishment of different methylation patterns.
Figure 1.
Examples of hemimethylation patterns. M and CmG represent a methylated site. U and CG represent an unmethylated site. A is an example of hemimethylation that occurs on the same strand. B is an example of polarity or reverse hemimethylation pattern with only two CpG sites. C is an example of hemimethylation on different strands with more than two CpG sites.
A major experimental limitation in hemimethylation studies has been the difficulty in obtaining methylation signals from the two complementary strands of DNA molecules for all CpG sites in an entire genome. Previous hemimethylation studies can only obtain the hemimethylation data for a few genes using the traditional Sanger sequencing and hairpin sequencing methods.11–13 Even though microarray technologies can obtain methylation levels genome-wide, they cannot produce methylation signals on two DNA strands separately. However, the next-generation sequencing (NGS) technology,14,15 combined with the bisulfite conversion technique (ie, C is converted to U and then becomes T), makes it possible to obtain methylation signals at the CpG site level on both DNA strands in an entire genome.16–19 During the last several years, a number of pioneering research groups have successfully used the bisulfite-converted methylation sequencing method on either Arabidopsis thaliana or human samples.16,18–25 Using bisulfite-converted methylation sequencing data, we can detect both the gain and loss of methylation by investigating hemimethylation patterns on two complementary strands. Nevertheless, the NGS technology produces a large amount of data.26 The quality of bisulfite sequencing data may be poor because of incomplete bisulfite conversion, genome variation, and sequencing errors.27 All of these features make processing and analyzing data challenging when identifying hemimethylation patterns. To address this challenge, we have developed the first-ever hemimethylation identification pipeline, HMPL. This pipeline can identify hemimethylated CpG sites and characterize different patterns in an entire genome. HMPL locates hemimethylated sites in each of the two different samples and then compares them. In the next section, we will explain the processes involved in HMPL in more detail.
Methods
The workflow of HMPL
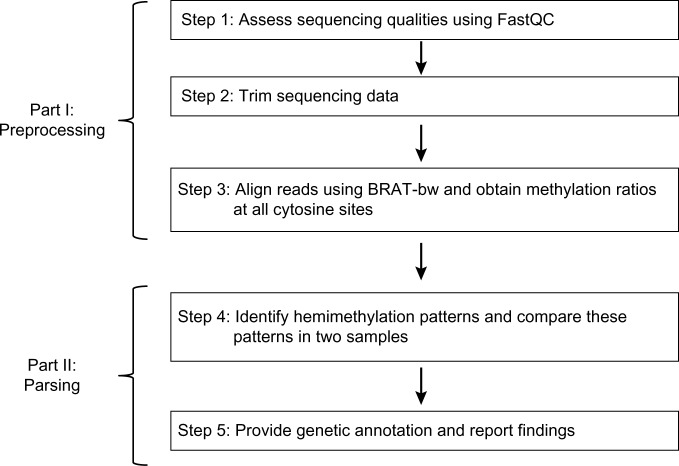
The workflow of our pipeline HMPL (see, Fig. 2) consists of two parts. Part I (preprocessing) utilizes available software packages in three steps: sequencing data quality assessment, trimming, and alignment (ie, Steps 1–3 of the workflow). Part II (parsing) is the new feature developed by our group. This part includes data parsing and summary reports (ie, Steps 4 and 5 of the work-flow). A more detailed description of these steps is introduced below. HMPL code and resource files can be downloaded from the following web link: http://hal.case.edu/~sun/HMPL/HMPL.zip.
Figure 2.
Workflow of the HMPL.
Step 1: assess sequencing qualities using FastQC.28
FastQC is a software package for assessing sequencing qualities by generating basic and informative diagnostic plots for sequencing data. This package provides a modular set of analyses for users to obtain a quick impression as to whether or not there are any obvious and serious problems before they start any downstream data analysis. FastQC produces basic statistics plots and summary reports for (1) per base sequence quality, (2) sequence quality scores, (3) summary of per base sequence content, (4) per sequence GC content, (5) per base N content, (6) sequence length distribution, (7) duplicate sequences, (8) overrepresented sequences, (9) adapter content, (10) Kmer (or K-base) content, and (11) per tile sequence quality.
Step 2: trim sequencing data.
Quite often, sequencing quality is very low at the 3′ end in sequencing reads, and raw reads may include adapter sequences. Therefore, HMPL has included the quality trimming and adapter-trimming step. In particular, dynamic trimming (the trim function provided in the software package BRAT29) and fixed-number-base trimming options are provided for quality trimming. The software package cutadapt30 is included for adapter trimming.
Step 3: align reads using BRAT-bw31 and obtain methylation ratios at all cytosine sites.
After trimming, alignment is done using BRAT-bw.31 After alignment, the methylation level (or ratio) at each cytosine (or C) site is obtained using the “acgt-count” function of the BRAT-bw package. The “acgt-count” function provides two options for generating output files: (1) the counts of “A,” “C,” “G,” and “T” at each cytosine site and (2) the methylation level for each cytosine site. In the HMPL package, we have chosen the second option of the “acgt-count” function. At each cytosine site, the methylation level is calculated as the ratio of the count of “C” (or the number of sequencing reads with methylated cytosine) to the count of “C” and “T” (or the total number of reads covering that site). The “acgt-count” function produces methylation levels for positive and negative strands in two separate output files. Each output file includes the following columns for each cytosine site (ie, each row): chromosome, start position, end position, total number of sequencing reads, methylation level, and DNA strand (“+” or “−”).
Step 4: identify hemimethylation patterns and compare these patterns in two samples.
In this step, all individual CpG sites that are hemimethylated are first identified. These sites are then classified into two groups: hemimethylated singleton and hemimethylated cluster. A hemimethylated CpG site is defined as a singleton if its adjacent CpG sites within d bases (eg, d = 100, a user-specified distance) are not hemimethylated. A hemimethylation cluster consists of at least two CpG sites that are all hemimethylated, and any two adjacent CpG sites in this cluster are within d bases. The following is a brief description of how to determine hemimethylated singletons and clusters. Starting from the first CpG site on a chromosome, for each hemimethylated CpG site, HMPL checks if the next CpG site is hemimethylated. If it is hemimethylated and is within a d-base region of the previous CpG site, these two are grouped together as a cluster and we continue to check the next (or third) CpG site; otherwise, this hemimethylated site is defined as a singleton. Hemimethylation clusters may have the following patterns: (1) consecutive CpG sites hemimethylated in the same DNA strand (eg, the three CpG sites in Fig. 1A), (2) the polarity or reverse hemimethylation cluster with only two CpG sites (as shown in Fig. 1B), and (3) consecutive CpG sites methylated on different strands and with more than two CpG sites, eg, the methylation of three CpG sites are MUU and UMM on the positive and negative strands, respectively (as shown in Fig. 1C). HMPL can also compare hemimethylated singleton sites and clusters of two samples.
Because high-throughput sequencing data may include sequencing and alignment errors, HMPL identifies a hemimethylated site using the following criteria. First, the user may determine a coverage cutoff value B (B > 0, eg, B = 5) depending on the sequencing quality and coverage level. On each strand, there must be at least B sequencing reads to cover a specific CpG site in order for HMPL to check whether or not this site is hemimethylated. Second, if the methylation level of a specific CpG site at one strand (eg, the positive strand) is larger than the cutoff value H0 (eg, H0 = 0.9), it is identified as M (methylated); if the methylation level is lower than the cutoff value L0 (eg, L0 = 0.1), it is identified as U (unmethylated). Using the above criteria, HMPL defines a CpG site as MU (ie, methylated on the positive strand and unmethylated on the negative strand), UM, MM, or UU. In the case of identifying hemimethylation clusters, HMPL uses the same criteria for two consecutive CpG sites. If a dataset has poor sequencing quality and low coverage, the results of using different cutoff values may be very different. Therefore, we recommend that users select very stringent cutoff values for H0 and L0 to reduce the false positive discovery rate because of poor sequencing quality and low coverage. For example, the user may use H0 = 0.9 or 0.95 instead of 0.8 and L0 = 0.1, or 0.05 instead of 0.2. If the user’s dataset has good sequencing quality and high coverage, changing the cutoff values may not affect the results significantly.
Step 5: provide genetic annotation and report findings
For each hemimethylated CpG site, HMPL provides the following genetic information.
Gene: If a hemimethylated CpG site is located on a gene, HMPL will report the name of that gene.
Pomoter: If a hemimethylated CpG site is within a D-base long region (eg, D = 1000) of the promoter of a gene (ie, D-base before the transcription starting site of a gene), HMPL will report the name of that gene.
Input and output
The HMPL uses raw sequencing reads (in FASTQ format) as input in Step 1 and Step 2. In Steps 3, 4, and 5, the input files are the output files from the previous step. More detailed information about the input and output files of HMPL can be found in the user manual, which can be downloaded from http://hal.case.edu/~sun/HMPL/HMPL.user.manual.pdf.
Usage, command options, and running time
HMPL is written in Perl32,33 and R34 scripts. It can be run as shown below in a LINUX or UNIX environment. The preprocessing step of HMPL (Part I) can be implemented with the following command (the command options of Pre.HMPL.pl are explained in Table 1).
Table 1.
The command options of HMPL Part I (Pre.HMPL.pl).
| OPTIONS | EXPLANATION |
|---|---|
| [−1<file>] | Required. FASTQ format single-end input file or pair-end input file 1, eg, −1 MCF7.fastq, which is the file name of a fastq dataset. |
| [−2<file>] | FASTQ format pair-end input file 2. By default, when there is no input 2, it only processes the input file 1 and processes it as a single-end file. |
| [-o <dir>] | The output directory. The default output directory is the user’s current directory. For example, if the current directory in which the user runs HMPL is ‘/home/user/check.folder/’, then when running HMPL command line without specifying ‘-o’, the user would have all the output files in ‘/home/user/check.folder’. |
| [-p <string>] | Required. The prefix written to the output file names. eg, –p MCF7, then the output file will have the prefix MCF7 (eg, MCF7.site, or MCF7.cluster). |
| [-r <file>] | The name of the file that lists the genome reference sequence (ie, *.fa) files that users will use to do alignment. Please note that this “-r” option must be provided whether or not the “-I” (ie, alignment index) option is provided. Otherwise, the “acgt-count” function in the BRAT-bw package will not generate proper output files. For example, we set it as “-r/home/reference/hg19/hg19.fa.filename.txt” This “hg19.fa.filename.txt” may include the following lines that show the location of the fasta files for chromosomes 10 and 11 (or other chromosomes) as shown below: /home/projects/data/reference/hg19/chr10.fa/home/projects/data/reference/hg19/chr11.fa |
| [-f <sanger or illumine>] | FASTQ format: HMPL accepts sanger or illumina format FASTQ files as input data; default is sanger. |
| [-a <yes or no>] | Adapter trimming: Users can select whether or not to utilize cutadapt for adapter trimming (default is no adapter trimming). |
| [-A <tirng>] | Adapter sequences: HMPL accepts two adapter sequence inputs, separated by a comma, and default is AGATCGGAAGAGCGGTTCAGCAG GAATGCCGAG, AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT. |
| [-T <fix or brat>] | Quality trim flag: Specifies whether to use BRAT dynamic trimming function (default is BRAT-trim) or the user can specify ‘fix’ to apply fixed quality trimming (ie, trim off a fixed number of bases). |
| [-N <int>] | Fixed quality trimming: Specifies the number of bases to be trimmed at the 5′ end (default is 5). |
| [-n <int>] | Fixed quality trimming: Specifies the number of bases to be trimmed at the 3′ end (default is 10). |
| [-Q <yes or no>] | Whether or not to do the quality assessment using FastQC (default is no). |
| [-I <dir>] | The index directory for BRAT-bw alignment. If the index folder is provided, it will be automatically used. Otherwise, it will build index, which is the default setting. |
| [-i <positive integer>] | To specify minimum insert size for paired-end mapping, the minimum distance allowed between the left-most ends of the mapped mates on the forward strand (default is 0). |
| [-m <positive integer>] | To specify maximum insert size for paired-end mapping, the maximum distance allowed between the left-most ends of the mapped mates on the forward strand (default is 1000). |
perl /<the_diretory_of_HMPL>/code/Pre.HMPL.pl −1 <FASTQ_input_file> -p <prefix> -r <reference_name> [OPTIONS]
The preprocessing pipeline ties the software and source code together with the appropriate dataflow to ensure that the correct output is achieved. Users need to have Perl, Python 2.6, R, FastQC, and BRAT-bw software installed on their system. They can run HMPL by entering commands in a Unix/Linux environment. If users have finished the hemimethylation pre-processing pipeline and have obtained the files of combined CpG sites, they may only run the parsing analysis using the Part II of HMPL (ie, “Parse.HMPL.pl”). The usage of Part II is given below (the command options of Parse.HMPL.pl are explained in Table 2).
Table 2.
The command options of HMPL Part II (Parse.HMPL.pl).
| OPTIONS | EXPLANATION |
|---|---|
| [−1 <file>] | Input file 1 is required. Note: For both Input 1 and Input 2 (see next row), the user can enter two kinds of inputs. One is the combined methylation level data (eg, “−1 MCF7.CG.combine”), and the other is the “acgt-count” output files, which includes uncombined methylation levels. If it is uncombined, that means the methylation levels on the forward and reverse strands are in two files and they should be separated by comma (,) when providing them as input files, eg, “−1 MCF7.CG.forward, MCF7CG.reverse”. |
| [−2 <file>] | Input file 2, optional. If specified, the pipeline will process both inputs and compare their final results. Default is only to process the input file 1, and not to do the comparing. Note: For both Input 1 and Input 2, the user can enter two kinds of inputs as explained in the above row. |
| [-o <dir>] | The output directory where all the output files are created and written. Default is “<current_dir>/final.results/.” |
| [-c <int>] | The value for selecting the methylation coverage is greater than B. (Default: B = 0). On each strand there must be at least B reads to cover a specific CpG site in order for HMPL to check if it is hemimethylated. Changing the “–c” value from a smaller value (eg, -c 5) to a larger value (eg, -c 10) will obtain a shorter list of hemimethylated sites and have a smaller false discovery rate. |
| [-l <real>] | The cutoff value for selecting low methylation level. (Default: 0.2, range: [0.05, 0.4]). This value corresponds to the “L0” mentioned in Step 4 of the pipeline. If the methylation level is less than this “-l” value, it will be claimed as unmethylated. Changing “-l” value from a smaller value (eg, -l 0.1) to a larger value (eg, -l 0.2) may give a longer list of hemimethylated sites, but there may be a larger false discovery rate. |
| [-h <real>] | The cutoff value for selecting high methylation level. (Default: 0.8, range: [0.6, 1]). This value corresponds to the “H0” mentioned in Step 4 of the pipeline. If the methylation level is greater than this “-h” value, it will be claimed as methylated. Changing “-h” value from a smaller value (eg, -h 0.7) to a larger value (eg, -h 0.9) may give a shorter list of hemimethylated sites, but there may be a smaller false discovery rate. |
| [-d <int>] | The maximum distance between two CpG sites to be selected as a cluster with default 50. If the maximum distance is changed from a smaller value (eg, -d 50) to larger value (eg, -d 100), the number of CpG sites in a cluster will be larger, but the total number of hemimethylation clusters will become smaller. |
| [-r<file>] | The reference gene file, not the genome reference sequence files. This file is used to provide genetic annotation (ie, gene names) to the hemimethylation sites. For example, we set it as “-r/home/reference/hg19/refGene.txt”. This “refGene.txt” file contains the gene names and gene information downloaded from the UCSC genome browser. |
| [-D <int>] | The distance of promoter region (Default: D = 1000). That is, if the transcript starting position is located at X = 5,000 bp on a chromosome, the promoter region of this gene is defined as from X-D = 4,000 to X = 5,000. |
perl /<the_diretory_of_HMPL>/code/Parse.HMPL.pl −1 <input 1> [OPTIONS].
In order for users to test the HMPL, we have prepared a small dataset with 5 million sequencing reads (a FASTQ *.fastq file), the human chromosome 22 reference sequence (a FASTA *.fa file), and the example scripts. Users may align these 5-million reads to chromosome 22 and then identify hemimethylated sites using both Part I and Part II of HMPL. Users simply need to download the data, install the HMPL package, and then change the data path/directory in the example script accordingly to run the HMPL. It takes about 11 minutes to run this small dataset using a Linux computer with 4 GB RAM. In addition, in order for users to explore the different options and arguments of HMPL, we have prepared a file named “README.” This file includes example scripts of running the HMPL using different command options and also explains to the user what to expect in the screen output. The small dataset, example scripts, and the “README” file can be downloaded from the following web link: http://hal.case.edu/~sun/HMPL/HMPL.zip.
In order to show the running time of HMPL in practice, we use two human example datasets. Each dataset has ~50 million raw sequencing reads, which will be introduced in detail in the Results section. Using the Linux server with dual quad-core 2.66 GHz Xeon E5430 processor that has 4 GB RAM for each core, it takes ~4 hours to run Part I (the pre-processing part) of the HMPL, Pre.HMPL.pl, if the reference index is provided. If the index file were not provided, it would first take about three to four additional hours to build a reference index for the whole human genome, which has about 3 billion bases (~3 GB data). Therefore, it is more efficient to first build a reference index for the alignment tool BRAT-bw before running the HMPL. It requires ~19 minutes to run Part II of HMPL (the parsing pipeline, Parse.HMPL.pl) when using uncombined input with the default coverage setting. The uncombined input means that positive and negative strand methylation levels of all CpG sites are provided as two separate files. If the positive and negative strand methylation data are combined, it will only take 15 minutes for HMPL to generate the results. If users have a faster Linux server or high-performance computing clusters that have more memory and computing power, it will take much less time (eg, a couple of hours) to run the HMPL preprocessing pipeline (Pre.HMPL.pl), and it will take just a few minutes to get the results of parsing and comparing two samples using the HMPL Part II (Parse. HMPL.pl).
Results
The HMPL pipeline can be used to compare any two samples of bisulfite sequencing data. In this paper, we demonstrate the use of HMPL using publicly available bisulfite-treated methylation sequencing datasets for cell lines MCF10A and MCF7.25 These two samples are breast cancer cell lines. Because a number of hypermethylated genes have been reported for breast cancer cells35 and there are hemimethylation patterns reported in individual genes,11–13 it is very likely that many CpG sites have been hemimethylated in these two cell lines. MCF10A is nontumorigenic and MCF7 is tumorigenic. We select these two cell lines because their hemimethylation patterns could be different. The bisulfite methylation sequencing data of MCF10A and MCF7 and more information about these two cell lines can be found from the corresponding references.25,36 The sequencing reads of MCF10A and MCF7 are generated using the reduced representative bisulfite sequencing (RRBS) protocol.16 There are 54,295,326 and 50,054,248 sequencing reads for MCF10A and MCF7, respectively, and the read length is 50-base for each dataset.
In order to identify hemimethylation singletons and clusters in both MCF10A and MCF7 and compare these two samples, we have run both Part I (Pre.HMPL.pl) and Part II (Parse.HMPL.pl) of the HMPL. In this section, we mainly focus on showing the hemimethylation result, which is the summary of the HMPL Part II (Parse.HMPL.pl) output. For a detailed description of the output files, see Table 3.
Table 3.
The description of HMPL Part II (Parse.HMPL.pl) output files.
| FILE NAME | CONTENTS |
|---|---|
|
*.grX eg, MCF10A.gr5 |
The CpG sites with coverage greater than X (X > 0). |
|
*.all.HM.sites eg, MCF10A.gr5.all.HM.sites |
The hemimethylated CpG sites identified by the high and low cutoff values. |
|
*.all.HM.sites.annotated eg, MCF10A.gr5.all.HM.sites.annotated |
The annotated hemimethylated sites (ie, gene names are provided). |
|
*.all.labelled.CG eg, MCF10A.gr5.all.labelled.CG |
The CpG sites with coverage greater than X and with the labels of methylation states (P: partially methylated, M: methylated, U: unmethylated). |
|
*. Summary eg, MCF10A.gr5.summary |
The summary file for all the methylation states of single hemimethylation sites and clusters. |
|
*.all.HMClusters eg, MCF10A.gr5.all.HM.Clusters |
All hemimethylated clusters. |
|
*.all.Rev.Clusters eg, MCF10A.gr5.all.Rev.Clusters |
All of the polarity (or reverse) clusters, including both consecutive and non-consecutive polarity clusters. |
|
*.non.Rev.Clusters eg, MCF10A.gr5.non.Rev.Clusters |
The hemimethylated clusters that are not polarity patterns. |
|
*.Singleton eg, MCF10A.gr5.Singleton |
Single hemimethylated CpG sites. |
|
*.consec.revs.Clusters eg, MCF10A.gr5.consec.revs.Clusters |
The consecutive polarity (or reverse) clusters (ie, with just two consecutive CpG sites). |
|
*.non.consec.revs.Clusters eg, MCF10A.gr5.non.consec.revs.Clusters |
The non-consecutive polarity clusters (ie, with two CpG sites that are not consecutive). |
|
*.compare eg, MM.gr5.all.HM.sites.annotated.compare MM.gr5.consec.revs.Clusters.compare MM.gr5.non.consec.revs.Clusters.compare MM.non.Rev.Clusters.compare MM.gr5.Singleton.compare |
The results of comparing two samples. (Note: To save space, we use “MM” to denote “MCF10A.MCF7” for the file names in the left column). |
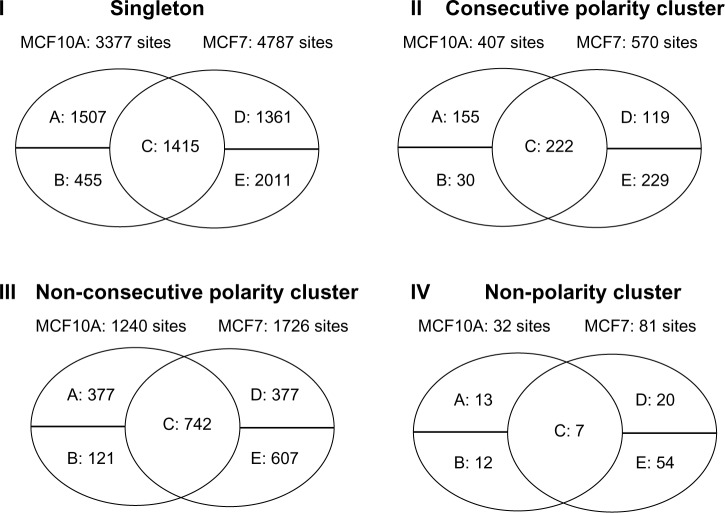
The hemimethylated singleton and cluster patterns are compared and summarized in Figure 3. In this figure, “I. Singleton” means comparing the singleton hemimethylated CpG sites in MCF10A and MCF7. “II. Consecutive Polarity Cluster” shows the results of comparing the polarity (or reverse) clusters that include two consecutive CpG sites; no other CpG sites are located between these two sites. “III. Non-Consecutive Polarity Cluster” means comparing the polarity (or reverse) clusters that include two CpG sites that are not consecutive. There is at least one CpG site located between these two sites, but there are either no sequencing reads (or data) or no hemimethylation sites between them. “IV. Non-Polarity Cluster” refers to the clusters that do not have the polarity (or reverse) pattern (eg, the patterns shown in Fig. 1A and C).
Figure 3.
MCF10A and MCF7 hemimethylation pattern comparison results. In each Venn diagram, the “A” entry means the number of CpG sites or clusters that are hemimethylated in the MCF10A sample, but not in the MCF7 sample. The “B” entry shows the number of CpG sites or clusters that are hemimethylated in the MCF10A sample, but there are no sequencing reads for these CpG sites in the MCF7 sample. The “C” entry represents the number of CpG sites or clusters that are hemimethylated in both MCF10A and MCF7. The “D” entry indicates the number of CpG sites or clusters that are hemimethylated in the MCF7 sample, but not in the MCF10A sample. The “E” entry means the number of CpG sites that are hemimethylated in the MCF7 sample, but there are no sequencing reads for these CpG sites in the MCF10A sample.
Figure 3 and Table 4 show the results of comparing MCF10A with MCF7, which are summarized based on the output files named “*compare” as shown in the last row of Table 3. The comparison results indicate that the hemimethylation patterns between the non-tumorigenic sample MCF10A and tumorigenic sample MCF7 are different at some CpG sites and/or genomic regions. Therefore, it is important that these CpG sites are investigated further. Our pipeline HMPL has provided gene annotation files for all CpG sites by giving names of genes in which hemimethylated CpG sites are located, as well as the names of genes in whose promoter regions the hemimethylated CpG sites are located.
Table 4.
The summary of MCF10A and MCF7 hemimethylation patterns.
| MCF10A | MCF7 | ||
|---|---|---|---|
| CLUSTER PATTERN | FREQUENCY | CLUSTER/PATTERN | FREQUENCY |
| MMMMMM-UUUUUU | 1 | MMMMM-UUUUU | 1 |
| MMMMM-UUUUU | 1 | MMMM-UUUU | 1 |
| MMM-UUU | 2 | MMM-UUU | 3 |
| MM-UU | 6 | MM-UU | 27 |
| MMU-UUM | 2 | MMU-UUM | 1 |
| MUM-UMU | 1 | MUM-UMU | 6 |
| MUMU-UMUM | 2 | MUMU-UMUM | 4 |
| MU-UM | 1643 | MU-UM | 2290 |
| UM-MU | 4 | MUU-UMM | 2 |
| UMU-MUM | 3 | UMM-MUU | 1 |
| UU-MM | 9 | UM-MU | 6 |
| UUU-MMM | 3 | UMU-MUM | 8 |
| UUUU-MMMM | 1 | UMUMU-MUMUM | 1 |
| UUUUU-MMMMM | 1 | UU-MM | 20 |
| UUU-MMM | 5 | ||
| UUUU-MMMM | 1 | ||
The results of comparing MCF10A and MCF7 show that there are more polarity (or reverse) clusters (eg, Fig. 1B) than the single-strand hemimethylation clusters (eg, Fig. 1A). This may be as a result of the fact that the methylation sequencing data we have used are generated by the RRBS protocol,16 which only sequences a small percentage of CpG sites in a human genome. In fact, there are ~5% of the CpG sites with at least 3× coverage in the RRBS data we have analyzed. If we use the whole genome bisulfite sequencing (WGBS) data, it is very likely that more single-strand hemimethylation clusters would be identified. Currently, we are not aware of any WGBS data for either MCF10A or MCF7. Therefore, we have used the available RRBS data to demonstrate the usage of HMPL. Even though RRBS data are not ideal for identifying all hemimethylation sites in an entire genome, we have found many hemimethylation singletons and clusters, which show the capability of our HMPL package. In fact, HMPL can be used to compare any two samples with data generated using either the RRBS or WGBS protocol.
Discussion
For the MCF7 sample, 532 genes have at least three hemimethylated CpG sites. In order to see if the genes with hemimethylated CpG sites are biologically important or meaningful, we have further investigated the 532 genes by comparing them with oncogenes, breast cancer methylated genes, and transcription factors. The comparison results show that seven of these genes are methylated, 17 are oncogenes, and 62 are transcription factors. We have also conducted the gene set enrichment analysis (GSEA) for these 532 genes using the GSEA software package and the molecular signature database provided by the Broad Institute.37 The analysis results show that 87 genes are significantly represented in (or overlapped with) 10 cancer modules (with P-value < 0.05), which are gene sets that are significantly changed in a variety of cancer conditions. These 87 genes and the 10 cancer modules they belong to are provided in Table 5. A detailed description of these 10 modules can be found online.38 The 532-gene list is included in the HMPL.zip file that can be downloaded from the following web link: http://hal.case.edu/~sun/HMPL/HMPL.zip.
Table 5.
Ten significant cancer modules identified using GSEA. The first column is the gene symbol, and the other columns indicate if a gene belongs to a specific cancer module. “X” shows that a gene belongs to that module, and a blank cell means that a gene does not belong to a specific module.
| GENE SYMBOL | CANCER MODULE NUMBER (OR ID) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 38 | 334 | 100 | 137 | 66 | 11 | 55 | 88 | 41 | 37 | |
| GRK5 | X | X | ||||||||
| TRPM2 | X | X | X | X | X | X | X | X | ||
| NRG2 | X | X | X | X | X | X | X | X | ||
| ALDH4A1 | X | X | X | X | X | X | X | |||
| PLXNA2 | X | X | X | X | X | X | ||||
| IQSEC1 | X | X | X | X | X | |||||
| COL6A2 | X | X | X | X | X | X | ||||
| SLMO1 | X | X | X | X | ||||||
| CNKSR1 | X | X | X | X | ||||||
| FBN2 | X | X | X | |||||||
| SLC38A10 | X | X | X | |||||||
| DNM2 | X | X | X | |||||||
| PLEC | X | X | X | |||||||
| ZBTB7A | X | X | X | |||||||
| TNFRSF10D | X | X | ||||||||
| PLXND1 | X | |||||||||
| LAMA5 | X | |||||||||
| TBX2 | X | |||||||||
| HOXB3 | X | |||||||||
| LTBP2 | X | |||||||||
| CDH15 | X | |||||||||
| KIAA0182 | X | |||||||||
| VAV2 | X | X | ||||||||
| DMD | X | |||||||||
| BMP6 | X | |||||||||
| FRMD4A | X | |||||||||
| LHX3 | X | |||||||||
| HS6ST1 | X | |||||||||
| OSBPL2 | X | |||||||||
| SNED1 | X | |||||||||
| MAP3K10 | X | |||||||||
| GRIN2C | X | |||||||||
| BAIAP2 | X | |||||||||
| CACNA1C | X | |||||||||
| GNG7 | X | X | X | X | X | X | X | |||
| CDH4 | X | X | X | X | X | X | X | |||
| PLCH2 | X | X | X | X | X | X | ||||
| SOX9 | X | X | X | X | X | X | ||||
| PTPRN2 | X | X | X | X | X | X | ||||
| CACNA2D2 | X | X | X | X | X | |||||
| GNG4 | X | X | X | X | X | |||||
| CDH22 | X | X | X | X | X | |||||
| GRIN1 | X | X | X | X | ||||||
| COL9A3 | X | X | X | X | ||||||
| CRMP1 | X | X | X | X | ||||||
| CTSF | X | X | X | X | ||||||
| ATP2B2 | X | X | X | X | ||||||
| PRPF6 | X | X | X | X | ||||||
| GABBR2 | X | X | X | X | ||||||
| DGCR2 | X | X | X | |||||||
| CA4 | X | X | X | |||||||
| ESR1 | X | X | X | |||||||
| SBNO2 | X | X | ||||||||
| KCNQ1 | X | X | ||||||||
| ATP11A | X | X | ||||||||
| ALDH1L1 | X | X | ||||||||
| TBCD | X | X | ||||||||
| COL18A1 | X | X | ||||||||
| CBS | X | X | ||||||||
| PEPD | X | X | ||||||||
| SLC22A1 | X | X | ||||||||
| GALNS | X | X | ||||||||
| MYBPC2 | X | |||||||||
| SLIT3 | X | |||||||||
| DIDO1 | X | |||||||||
| PCBP3 | X | |||||||||
| CAMK2B | X | |||||||||
| MLLT1 | X | |||||||||
| INTS9 | X | |||||||||
| PARD3 | X | |||||||||
| PLXNA3 | X | |||||||||
| GREB1 | X | |||||||||
| SDC3 | X | |||||||||
| ATP8A2 | X | |||||||||
| LMTK3 | X | |||||||||
| CCDC85C | X | |||||||||
| C9orf167 | X | |||||||||
| RNF220 | X | |||||||||
| BEGAIN | X | |||||||||
| ESPN | X | |||||||||
| OBSCN | X | |||||||||
| BCR | X | |||||||||
| PITPNC1 | X | |||||||||
| KIAA1522 | X | |||||||||
| RXRA | X | |||||||||
| LHPP | X | |||||||||
| MED24 | X | |||||||||
There are a number of alignment tools for bisulfite methylation sequencing data, such as BRAT,29 BRAT-bw,31 BSMAP,39 BS Seeker,40 Bismark,41 MethylCoder,42 RMAPBS,43 Pash,44 and BatMeth.45 A comprehensive list of these tools can be found at omictools.com.46 Among all these available tools, we have tested BRAT-bw, BRAT, and BSMAP. We have found that they provide similar results, but BRAT-bw is faster. BRAT-bw is user-friendly and has the following useful features: (1) it can align both single-end and paired-end reads, (2) it can produce the ACGT count for all cytosines in a genome, (3) it can account for overlapping paired-end reads, and (4) it can check strands. If users prefer another alignment tool, they can obtain the alignment results and methylation levels using their preferred alignment tool, and then run Part II of the HMPL to obtain hemimethylated singletons and clusters. If necessary, this can be done with some minor format changes of their alignment output files. Reformatting is easy because the parsing pipeline of HMPL (ie, Part II) only requires data with the following columns that most alignment tools provide for two DNA strands: chromosome, start position, end position, total number of sequencing reads, methylation level, and DNA strand.
The cutoff values used in HMPL, especially the ones provided in Table 2 for the parsing pipeline (ie, part II), should be determined depending on the sequencing quality and coverage. If the user has a sequencing dataset with very good quality and high coverage, changing the cutoff value may not significantly affect the results. However, if the sequencing dataset has low coverage and poor quality, changing the cutoff values may lead to very different results. For this case, we recommend that users set up very stringent cutoff values to reduce false discovery rates. We set up the default values based on findings in previous publications, some basic and common knowledge of methylation sequencing data, and our experience with bisulfite sequencing data; however, every dataset is different. We suggest that the users first determine the quality and coverage of their data, and then try different values to see if their results are dramatically different. If results are dramatically different, users may choose results that are obtained based on stringent cutoff values, especially when they plan to do experimental validation. Using results based on stringent cutoff values can ensure a high validation rate and then the user may expand their validation list by adding more hemimethylated singletons or clusters from the list obtained with less stringent cutoff values.
Ideally, it is best to have a list of known hemimethylated and nonhemimethylated sites to study the true and false discovery rates (or sensitivity and specificity) of HMPL. For the nonhemimethylated sites, we can choose the CpG sites that are located in the housekeeping genes, which are relatively stable and not likely to be methylated on either strand. In fact, housekeeping genes have been used for the purpose of “negative control” in previous methylation studies.47–49 For the example dataset MCF7, using the coverage cutoff of 5× (that is, at least five reads to cover each strand) and other default settings, there are 532 genes with at least three hemimethylated sites. We compare these 532 genes with the 205 known housekeeping genes used in a previous study,49 and there is no overlap; therefore, none of these 532 genes are housekeeping genes. As for the known hemimethylated sites, to the best of our knowledge, no available list of genes or sites can be used as a “positive control.” This is because genome-wide hemimethylation study is rarely done, and a few hemimethylated sites have been experimentally validated. Another possible way of validating the HMPL is to use simulated data. However, we have decided not to use this approach because there is little knowledge about genome-wide hemimethylation patterns. With little known information, a simulation would be very arbitrary and the simulated data would not reflect true unknown patterns. The purpose of our HMPL development is to provide an exploratory tool for this research topic. For genes identified with a number of hemimethylated CpG sites, users may do further investigation by studying their biological functions and relationships with other genes using pathway analyses. Users may also do experimental validation to discover novel methylated or hemimethylated genes.
Our pipeline has two limitations. First, the HMPL is designed for identifying hemimethylation only at CpG sites, but not at any non-CpG sites (eg, CHG and CHH sites, where H represents A, C, or T in a DNA sequence). If users are interested, our algorithms may be modified to study the hemimethylation of non-CpG sites. Second, our pipeline is developed for identifying hemimethylated sites (or clusters) by comparing two samples, but it is not designed for comparing multiple samples in two or more groups. Because the hemimethylation study is an area with little research, HMPL is good for preliminary studies. As for the topic of identifying hemimethylation patterns in multiple samples and in multiple groups, more sophisticated statistical methods may be used, and our group is working on projects related to this approach.
For the first step of the HMPL workflow, we have used the software package FastQC. Even though FastQC is not designed for bisulfite-treated methylation sequencing data, it can provide informative diagnostic plots for methylation sequencing data. If users find some serious sequencing quality issues in their data, we recommend that they check data more thoroughly using other available software packages, such as SAAP-RRBS,50 BSeQC,51 and MethyQA,27 before they interpret their HMPL results.
Conclusion
Hemimethylation patterns are useful for studying DNA methylation events. Therefore, it is important to develop a software package to identify such patterns. To address this need, we have developed a new software package, HMPL, which includes both preprocessing and data parsing. For two samples, each with 50 million reads, it takes a few hours for HMPL to align the sequencing reads, and it only takes a few minutes to process the methylation level data. If users have obtained their coverage and methylation ratio data, Part II of HMPL can identify hemimethylation patterns in minutes.
Acknowledgments
We appreciate the support provided by our IT colleagues at Case Western Reserve University and Texas State University.
Footnotes
ACADEMIC EDITOR: J.T. Efird, Editor in Chief
PEER REVIEW: Five peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1,552 words, excluding any confidential comments to the academic editor.
FUNDING: This work was supported by Dr. Shuying Sun’s start-up funds and the Research Enhancement Program provided by Texas State University. The authors confirm that the receipt of funding from Texas State University had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: SS. Analyzed the data: SS, PL. Wrote the first draft of the manuscript: SS. Contributed to the writing of the manuscript: SS, PL. Agreed with manuscript results and conclusions: SS, PL. Jointly developed the structure and arguments for the paper: SS, PL. Made critical revisions and approved the final version: SS, PL. Both authors reviewed and approved the final manuscript.
REFERENCES
- 1.Quon G, Lippert C, Heckerman D, Listgarten J. Patterns of methylation heritability in a genome-wide analysis of four brain regions. Nucleic Acids Res. 2013;41(4):2095–104. doi: 10.1093/nar/gks1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011;2(6):607–17. doi: 10.1177/1947601910393957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bestor TH, Edwards JR, Boulard M. Notes on the role of dynamic DNA methylation in mammalian development. Proc Natl Acad Sci U S A. 2015;112(22):6796–9. doi: 10.1073/pnas.1415301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senner CE. The role of DNA methylation in mammalian development. Reprod Biomed Online. 2011;22(6):529–35. doi: 10.1016/j.rbmo.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 5.Paulsen M, Ferguson-Smith AC. DNA methylation in genomic imprinting, development, and disease. J Pathol. 2001;195(1):97–110. doi: 10.1002/path.890. [DOI] [PubMed] [Google Scholar]
- 6.Jones PA, Buckley JD. The role of DNA methylation in cancer. Adv Cancer Res. 1990;54:1–23. doi: 10.1016/s0065-230x(08)60806-4. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA. DNA methylation and cancer. Cancer Res. 1986;46(2):461–6. [PubMed] [Google Scholar]
- 8.Sharp AJ, Stathaki E, Migliavacca E, et al. DNA methylation profiles of human active and inactive X chromosomes. Genome Res. 2011;21(10):1592–600. doi: 10.1101/gr.112680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feinberg AP, Cui H, Ohlsson R. DNA methylation and genomic imprinting: insights from cancer into epigenetic mechanisms. Semin Cancer Biol. 2002;12(5):389–98. doi: 10.1016/s1044-579x(02)00059-7. [DOI] [PubMed] [Google Scholar]
- 10.Plass C, Soloway PD. DNA methylation, imprinting and cancer. Eur J Hum Genet. 2002;10(1):6–16. doi: 10.1038/sj.ejhg.5200768. [DOI] [PubMed] [Google Scholar]
- 11.Flor I, Neumann A, Freter C, et al. Abundant expression and hemimethylation of C19MC in cell cultures from placenta-derived stromal cells. Biochem Biophys Res Commun. 2012;422(3):411–6. doi: 10.1016/j.bbrc.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Shao C, Lacey M, Dubeau L, Ehrlich M. Hemimethylation footprints of DNA demethylation in cancer. Epigenetics. 2009;4(3):165–75. doi: 10.4161/epi.4.3.8277. [DOI] [PubMed] [Google Scholar]
- 13.Xie FW, Peng YH, Chen X, et al. Regulation and expression of aberrant methylation on irinotecan metabolic genes CES2, UGT1A1 and GUSB in the in-vitro cultured colorectal cancer cells. Biomed Pharmacother. 2014;68(1):31–7. doi: 10.1016/j.biopha.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 14.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 15.Metzker ML. Sequencing technologies – the next generation. Nat Rev Genet. 2010;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 16.Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6(4):468–81. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 17.Lister R, Ecker JR. Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res. 2009;19(6):959–66. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lister R, Pelizzola M, Kida YS, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cokus SJ, Feng S, Zhang X, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452(7184):215–9. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu H, Bock C, Mikkelsen TS, et al. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods. 2010;7(2):133–6. doi: 10.1038/nmeth.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen KD, Timp W, Bravo HC, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43(8):768–75. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lister R, O’Malley RC, Tonti-Filippini J, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133(3):523–36. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Z, Asmann YW, Kalari KR, et al. Integrated analysis of gene expression, CpG island methylation, and gene copy number in breast cancer cells by deep sequencing. PLoS One. 2011;6(2):e17490. doi: 10.1371/journal.pone.0017490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mardis ER. The $1,000 genome, the $100,000 analysis? Genome Med. 2010;2(11):84. doi: 10.1186/gm205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun S, Noviski A, Yu X. MethyQA: a pipeline for bisulfite-treated methylation sequencing quality assessment. BMC Bioinformatics. 2013;14:259. doi: 10.1186/1471-2105-14-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrews S. FastQC. 2010. Available at: http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/
- 29.Harris EY, Ponts N, Levchuk A, Roch KL, Lonardi S. BRAT: bisulfite-treated reads analysis tool. Bioinformatics. 2010;26(4):572–3. doi: 10.1093/bioinformatics/btp706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17(1):10–2. [Google Scholar]
- 31.Harris EY, Ponts N, Le Roch KG, Lonardi S. BRAT-BW: efficient and accurate mapping of bisulfite-treated reads. Bioinformatics. 2012;28(13):1795–6. doi: 10.1093/bioinformatics/bts264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillespie T. Effective Perl programming. Libr J. 1998;123(4):121. [Google Scholar]
- 33.Gordon RS. Advanced Perl programming. Libr J. 2005;130(18):108. [Google Scholar]
- 34.R Core Team . R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2014. [Google Scholar]
- 35.Yang X, Yan L, Davidson NE. DNA methylation in breast cancer. Endocr Relat Cancer. 2001;8(2):115–27. doi: 10.1677/erc.0.0080115. [DOI] [PubMed] [Google Scholar]
- 36.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Available at: http://www.broadinstitute.org/gsea/msigdb/index.jsp.
- 38. Available at: http://robotics.stanford.edu/~erans/cancer/modules/
- 39.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen PY, Cokus SJ, Pellegrini M. BS Seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–2. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pedersen B, Hsieh TF, Ibarra C, Fischer RL. MethylCoder: software pipeline for bisulfite-treated sequences. Bioinformatics. 2011;27(17):2435–6. doi: 10.1093/bioinformatics/btr394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith AD, Chung WY, Hodges E, et al. Updates to the RMAP short-read mapping software. Bioinformatics. 2009;25(21):2841–2. doi: 10.1093/bioinformatics/btp533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coarfa C, Yu F, Miller CA, Chen Z, Harris RA, Milosavljevic A. Pash 3.0: a versatile software package for read mapping and integrative analysis of genomic and epigenomic variation using massively parallel DNA sequencing. BMC Bioinformatics. 2010;11:572. doi: 10.1186/1471-2105-11-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim JQ, Tennakoon C, Li G, et al. BatMeth: improved mapper for bisulfite sequencing reads on DNA methylation. Genome Biol. 2012;13(10):R82. doi: 10.1186/gb-2012-13-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Available at: http://omictools.com/bisulfite-mappers-c147-p1.html.
- 47.Sun S, Chen Z, Yan PS, Huang YW, Huang TH, Lin S. Identifying hypermethylated CpG islands using a quantile regression model. BMC Bioinformatics. 2011;12:54. doi: 10.1186/1471-2105-12-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun S, Huang YW, Yan PS, Huang TH, Lin S. Preprocessing differential methylation hybridization microarray data. BioData Min. 2011;4:13. doi: 10.1186/1756-0381-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun S, Yan PS, Huang TH, Lin S. Identifying differentially methylated genes using mixed effect and generalized least square models. BMC Bioinformatics. 2009;10:404. doi: 10.1186/1471-2105-10-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Z, Baheti S, Middha S, et al. SAAP-RRBS: streamlined analysis and annotation pipeline for reduced representation bisulfite sequencing. Bioinformatics. 2012;28(16):2180–1. doi: 10.1093/bioinformatics/bts337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin X, Sun D, Rodriguez B, et al. BSeQC: quality control of bisulfite sequencing experiments. Bioinformatics. 2013;29(24):3227–9. doi: 10.1093/bioinformatics/btt548. [DOI] [PMC free article] [PubMed] [Google Scholar]