Abstract
Gelatinous Chinese medicines made from mammalian skin or horn or reptile shell are a very important type of animal-derived Chinese medicine. They have been extensively used either as both hemopoietic and hemostatic agents to treat vertigo, palpitation, hematuria, and insomnia in traditional Chinese medicine clinics; consumed as a popular tonic for weaker persons such as the elderly or women after giving birth; or further manufactured to health supplements for certain populations. However, they cannot be discriminated from each other by only using the routine approach in the Chinese Pharmacopoeia, as it lacks enough specificity and, consequently, and the requirements can be met even by adding assayed ingredients. In this study, our efforts to differentiate three gelatinous Chinese medicines, Asini Corii Colla, Cervi Cornus Colla, and Testudinis Carapacis ET Plastri Colla, are presented, and a novel strategy based on enzymatic digestion followed by nano-flow liquid chromatography in tandem with orbitrap mass spectrum detector analysis is proposed herein. Fourteen diagnostic fragments identified from the digests of these medicines were exclusively selected for their discrimination. By taking advantage of the favorable features of this strategy, it is feasible and convenient to identify enzymatic-digested peptides originated from signature proteins in each medicine, which thus could be employed as potential biomarkers for their form of raw medicinal material, and the pulverized and the complex especially, that being the direct basis for authentication purpose.
Keywords: traditional Chinese medicine, animal-derived Chinese medicines, potential biomarkers, authentication, diagnostic fragments
Introduction
Traditional Chinese medicine (TCM), built upon a complete system of theory, is a broad range of medicinal practices generally incorporating common concepts which have been well developed and are based on experiences accumulated from different ethnic groups for 2000 years in the People’s Republic of China, involving various forms of application of natural products and other therapeutic methods. Typical of a number of herbs and mineral medicinals applied in TCM, the harness of animal-derived medicines also has a long history from the use of hirudo, ostreae concha, etc, documented in two ancient Chinese medical books of doctrine: Yellow Emperor’s Inner Canon1 and Treatise on Cold Damage.2 In addition, in the prevailing Chinese Pharmacopoeia (ChP),3 there are 51 raw animal-derived Chinese medicines (ACMs), 61 processed products from 36 of them, as well as 365 Chinese proprietary medicines composed of ACMs, accounting for 22.0% of all 2,165 Chinese medicines, which indicates their irreplaceable importance in the present day.4,5
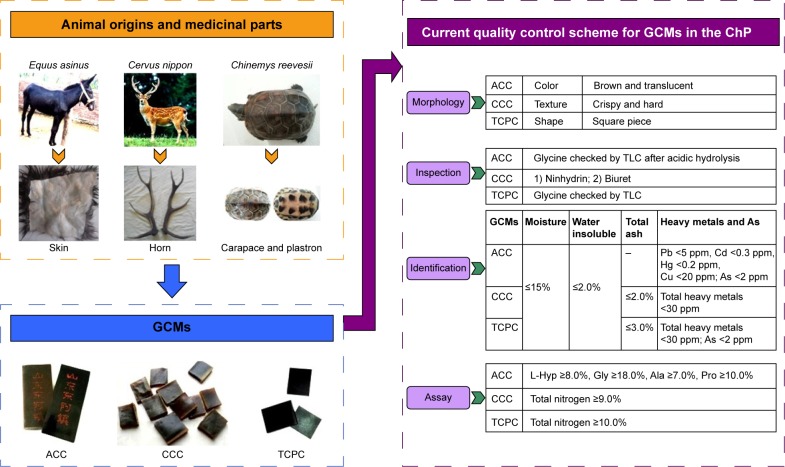
Gelatinous Chinese medicines (GCMs), made from mammalian skin or horn or reptile shell, are a very important type of ACM. They are extensively used as both hemopoietic and hemostatic agents to treat vertigo, palpitation, hematuria, and insomnia in TCM clinics; consumed as a popular tonic for weaker persons such as the elderly or women after giving birth; or further manufactured to health supplements for certain populations.6–13 In the ChP, there are three GCMs under direct regulation of the China Food and Drug Administration (CFDA) by the quality control scheme illustrated in Figure 1; the details of their animal origins, medicinal parts, etc, are summarized in Figure 1 and Table 1.
Figure 1.
Quality control scheme of GCMs in the ChP.
Abbreviations: ACC, Asini Corii Colla; CCC, Cervi Cornus Colla; ChP, Chinese Pharmacopoeia; GCM, gelatinous Chinese medicine; TCPC, Testudinis Carapacis ET Plastri Colla; TLC, thin layer chromatography.
Table 1.
Gelatinous Chinese medicines in the Chinese Pharmacopoeia3
| Latin name | Common names | Animal origins | Medicinal parts | Dosage (g/day) |
|---|---|---|---|---|
| Asini Corii Colla | Donkey-hide glue, E Jiao | Equus asinus L. | Skin | 3–9 |
| Cervi Cornus Colla | Deer-horn glue, Lu Jiao Jiao | Cervus elaphus Linnaeus, Cervus nippon Temminck | Ossified antlers or horn base (naturally detach after cutting pilose non-ossified antlers from deer) | 3–6 |
| Testudinis Carapacis ET Plastri Colla | Tortoise-shell glue, Gui Jia Jiao | Chinemys reevesii (Gray) | Carapace and plastron | 3–9 |
However, ACMs that are rich in protein and other molecules of biological information differ greatly from the herbs containing abundant secondary metabolites of small molecules or favorable cystine-rich peptides (cyclotides) discovered more and more extensively in recent decades.14–17 Which has greatly increased the difficulty of discriminating between them, and therefore the exploration of their discrimination was much fewer than other Chinese medicines so far, especially it is lack of specifically designed strategies and means for this purpose, and some cutting-edge MS technologies such as matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) and orbitrap extensively used for proteomics research have been rarely employed in the study of ACMs until the present. In this paper, our efforts to differentiate three GCMs from each other are presented, and, as shown in Figure 2, a novel strategy based on nano-flow liquid chromatography in tandem with orbitrap mass spectrum detection (NanoLC-orbitrap MSD) is proposed for their authentication, with some evidential differences explored on the GCM samples digested by specific enzyme. NanoLC-orbitrap MSD is distinguished by high resolution; great mass accuracy, specificity, sensitivity for analysis, and rapid conduction of MSn profiling for the peak of interest, making this technology one of the most powerful tools in current proteomics research.18 By taking advantage of these favorable features, it is feasible and convenient to identify the enzymatic-digested peptides originated from signature proteins present in each GCM, which thus could be employed as the potential biomarkers for ACMs, including their form of raw medicinal material, the pulverized material, and Chinese Proprietary medicines for authentication purposes.
Figure 2.
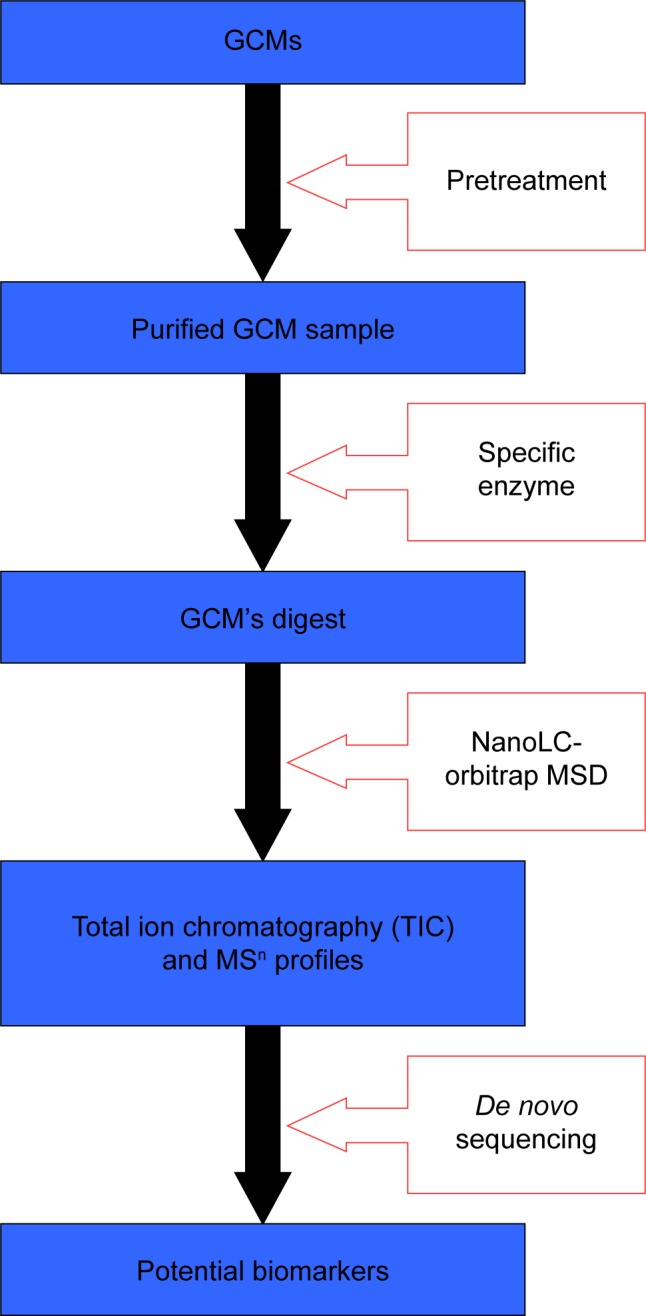
Novel strategy proposed for the discrimination of GCMs.
Abbreviations: GCM, gelatinous Chinese medicine; MSn, multi-stage mass spectrometry; NanoLC-orbitrap MSD, nano-flow liquid chromatography in tandem with orbitrap mass spectrum detection.
Materials and methods
Materials and chemicals
Asini Corii Colla (ACC) (B/N: 110916, 091204, 100235; Shandong Dong-E E-Jiao Co., LTD. Shandong, People’s Republic of China); Cervi Cornus Colla (CCC) (B/N: 20130922, 20120810, 20121201; Jilin Zhong-Ding Pharmaceutics Co. Jilin, People’s Republic of China); and Testudinis Carapacis ET Plastri Colla (TCPC) (B/N: 20131018, 20100529, 20120721; Shandong Dong-E E-Jiao Co., LTD). The voucher specimens were deposited and authenticated by Associate Professor Hongxia Chen, Pharmacognosy Research Center, Jiangsu University (Zhenjiang, People’s Republic of China). These raw materials were individually smashed into powder form by mortar in liquid nitrogen and passed through a 40-mesh sieve. The fine powder of each sample was then collected and stored in a desiccator at room temperature after lyophilization for 24 hours to remove the remaining moisture.
Tris base of molecular biology grade was obtained from Promega Corporation (Fitchburg, WI, USA). HCl solution (37%), acetonitrile, formic acid (FA), glacial acetic acid, NaCl, CaCl2, anhydrous Na2CO3, NH4HCO3, NaHCO3, and NaOH were purchased from EMD Millipore (Billerica, MA, USA). Ethylene glycol was provided by Alfa Aesar (Ward Hill, MA, USA). Ten percentage sodium dodecyl sulfate (SDS) solution, ammonium persulfate, tricine, 2× Laemmli sample buffer, glycine, 40% acrylamide/bis (19:1), 40% acrylamide/bis (29:1), 30% acrylamide/bis (29:1), and N,N,N′,N′-tetramethylethylenediamine TEMED are electrophoresis purity reagents purchased from Bio-Rad Laboratories Inc. (Hercules, CA, USA). Bovine serum albumin (BSA) (fraction V) was a product of GE Healthcare Bio-Sciences Corp. (Piscataway, NJ, USA). Exact-Pro broad range (10–245 kDa) prestained protein ladder (Bio-5150) was from First BASE Laboratories Sdn Bhd (Seri Kembangan, Selangor, Malaysia). AgNO3, Na2S2O3, N-[Tris(hydroxymethyl)methyl]-2-aminoethanesulfonic acid (TES), CuSO4⋅5H2O, Coomassie brilliant blue G-250 and R-250, H3PO4 solution (49%–51%), bicinchoninic acid (BCA) solution, formaldehyde solution (37%, in 10%–15% MeOH), 2-mercaptoethanol, gelatin from cold water fish skin, and collagenase from Clostridium histolyticum (type I) were all purchased from Sigma-Aldrich Co. (St Louis, MO, USA).
Total protein determination
Preparation of sample solutions
For the fine powder of each raw material, 0.100 g was precisely weighed into a beaker and 5.00 mL of distilled H2O was added to dissolve the sample in an 85°C water bath for 20 minutes, and the solution was completely transferred into a 10 mL volumetric flask. The solution was then centrifuged at 4,000 rpm for 30 minutes and the supernatant was collected, 1.00 mL of which was diluted to 10.0 mL with H2O for subsequent analysis by two conventional methods including both Bradford assay and BCA assay for comparison purposes.
BCA assay and Bradford assay
BCA assay was conducted on a UV transparent 96-well microplate and the absorbency was scanned at 560 nm using a microplate reader. Bradford assay was carried out in test tubes and the absorbency measurement was taken at 595 nm.19
Calculation of total protein content
The percentage of total protein content was calculated according to the following equation:
| (1) |
SDS polyacrylamide gel electrophoresis analysis
A Bio-Rad electrophoresis system was equipped with a PowerPac universal power supply and a Mini-PROTEAN Tetra cell for SDS polyacrylamide gel electrophoresis (SDS–PAGE) analysis.20 Glycine SDS–PAGE (5% stacking gel, 10% resolving gel, 1.0 mm) was performed at 80 V for 15 minutes followed by 110 V for another 75 minutes, and tricine SDS–PAGE (5% stacking gel, 18% resolving gel, 0.75 mm) was performed at 80 V for 10 minutes followed by 200 V until the loading dye front reached the bottom of the resolving gel. Then, the gel sandwich was disassembled and subject to either Coomassie brilliant blue staining or silver staining.
Preparation of sample solutions
Pretreatment of samples
One hundred milligrams of each GCM sample was dissolved in 10 mL Milli-Q water by vortex and sonication and then centrifuged at 12,000 rpm at 4°C for 30 minutes. The supernatant was collected for subsequent membrane ultra-filtration by centrifugal filter unit (5 kDa molecular weight cut-off; EMD Millipore) at 3,000 rpm at 4°C, according to the manufacturer’s instructions, to remove most of the small molecules existing in the solution. The remaining concentrated brown sample solution in the upper vessel of the unit was completely transferred to a glass container and subject to further lyophilization, resulting in a pale and light block mass which was then stored in a desiccator under room temperature for the following experiments.
Digestion by enzyme
Ten milligrams of individual pretreated GCM and gelatin were dissolved in 2.0 mL of 50 mM TES buffer (0.36 mM CaCl2, pH 7.4) for collagenase digestion to give the final concentration at 5.0 mg/mL. After sonication and vortexing to promote the dissolution, the samples were visually inspected for the presence of residual solid material and subject to centrifugation at 12,000 rpm for 30 minutes. Then, 100 μL of supernatant was denatured at 95°C for 10 minutes and cooled down to room temperature, and 2.0 μL of enzyme solution (0.10 mg/mL collagenase in TES buffer) was then added before the full digestion performed at 37°C over 24 hours.21,22 Few efforts were made to carefully enhance the activity of the enzyme, since the only purpose of this step was to digest the present proteins thoroughly, which could be simply achieved by the addition of excess enzymes, and then the completeness of the above digestions was ensured by means of SDS–PAGE with silver staining.
NanoLC-orbitrap MSD analysis conditions
Enzymatic-digested peptides were analyzed by NanoLC-MS/MS on an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) coupled with a Dionex UltiMate 3000 UHPLC system from Thermo Fisher Scientific. Spraying was done using a Michrom’s Thermo Captive Spray nanoelectrospray ion source (Bruker-Michrom Inc., Auburn, CA, USA) at 250°C capillary temperature and 1.5 kV source voltage. Separation was performed in a reverse-phase Acclaim PepMap RSL column (75 μm internal diameter ×15 cm, 2 μm particles), also from Thermo Fisher Scientific, which was maintained at 35°C throughout the analysis. Separation was done with a flow rate of 300 nL/min using 0.1% FA aqueous solution as eluent A and 0.1% FA in 90% acetonitrile aqueous solution as eluent B. A 60-minute gradient elution was used starting at 3% of eluent B for 1 minute, followed by 3%-35% of eluent B within 47 minutes, increased to 50% of eluent B within 4 minutes, again increased to 80% within 0.1 min, and kept isocratic for 1.3 minutes. Then, the mobile phases were returned to 3% of eluent B within 0.1 minute and maintained for 6.5 minutes for equilibrium.
Data acquisition was done in positive ion mode using the LTQ Tune Plus software (Thermo Fisher Scientific) alternating between full MS (350–1,600 m/z, resolution 60,000, with 1 μscan per spectrum) and FT-MS/MS (150–2,000 m/z, resolution 15,000, with 1 μscan per spectrum) for the ten most intense ions (with a 500-count threshold). Fragmentation was performed in high-energy collisional dissociation mode at 32% normalized collision energy. Automatic gain control target for both full MS and FT-MS/MS was set to 1e+06, and precursor ion charge state screening was activated. Dynamic exclusion list was enabled with an exclusion duration of 30 seconds and an exclusion list size of 500.
De novo sequencing of each peptide was conducted by PEAKS software automatically. Local confidence is the confidence (presented as a percentage) that a particular amino acid is present in the de novo peptide at a particular position. And the sum of the local confidence scores (0 to 1) from each amino acid in the peptide sequence was divided by the number of amino acids, which was presented to be average local confidence (ALC) used to assess the accuracy of the interpretation.
Results and discussion
Protein content
As shown in Table 2, high correlation coefficients (r2) of calibration curve were achieved in both assays, and the protein content of different GCMs was increased in the following order: TCPC < CCC < ACC. Meanwhile, the contents determined by BCA assay were much higher than in the Bradford assay, suggesting that the proteins in GCMs do not contain a large number of arginine and/or aromatic residues in their chemical structure.
Table 2.
Calibration curve and protein content
| Assays | Regression equations | r2 | Linearity ranges | Protein content (w/w)
|
||
|---|---|---|---|---|---|---|
| ACC | CCC | TCPC | ||||
| BCA | Y=0.0010X +0.0651 | 0.9993 | 62.50–2,000 μg/mL | 44.2% | 40.7% | 36.4% |
| Bradford | Y=0.0056X +0.0729 | 0.9995 | 20.00–100.0 μg/mL | 19.3% | 15.9% | 14.4% |
Abbreviations: ACC, Asini Corii Colla; BCA, bicinchoninic acid; CCC, Cervi Cornus Colla; TCPC, Testudinis Carapacis ET Plastri Colla; r2, correlation coefficient; w/w: weight/weight.
Our first attempt was to differentiate individual GCMs in terms of their protein content, which varied within approximately 10%–20% among different GCMs, implying the ambiguity of using these assays to help discriminate them.
SDS–PAGE analysis and enzymatic digestion
As another attempt to determine the difference in protein profile among ACC, CCC, and TCPC, their solutions (5 mg/mL) were loaded for electrophoresis examination on both 10% and 18% SDS–PAGE gels, which are preferred to profile proteins >10 kDa and small proteins from 1 to 20 kDa, respectively. However, by both commonly used staining methods, no visual bands were observed in the gels as desired, with the lanes being wholly stained in a dispersive manner all the way down from the top to the gel front, as shown in Figure 3. Contrastively, various and intensive bands ranging from 20 kDa to as much as >245 kDa were clearly observed in gelatin sample used as a reference. Considering the plentiful presence of proteins in the original animal medicinal parts used for GCMs production as well as the addition of some maltose, a favorable disaccharide having a reducing form as one of its two units may have an open-chain form with an aldehyde group; this result could be caused by the Maillard reaction, an important chemical reaction between the nucleophilic amino group of amino acids in protein and reactive carbonyl group of reducing sugars, which leads to the formation of a complex mixture of poorly characterized molecules responsible for a range of odors and flavors.
Figure 3.
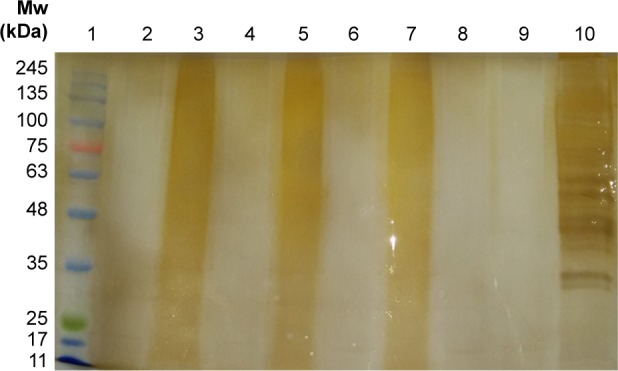
SDS–PAGE analysis of gelatinous Chinese medicines and their digests (1.0 mm, 5% stacking gel, 10% resolving gel, silver staining).
Notes: Lane 1: ladder. Lane 2: collagenase after incubation. Lane 3: ACC. Lane 4: ACC after in-solution digestion by collagenase. Lane 5: CCC. Lane 6: CCC after in solution digestion by collagenase. Lane 7: TCPC. Lane 8: TCPC after in solution digestion by collagenase. Lane 9: gelatin after in solution digestion by collagenase. Lane 10: gelatin.
Abbreviations: ACC, Asini Corii Colla; CCC, Cervi Cornus Colla; Mw, molecular weight; SDS–PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; TCPC, Testudinis Carapacis ET Plastri Colla.
During the production of GCMs in industry, long-term decoction has been employed for the extraction from an animal’s medicinal part or in the course of subsequent refinement and concentration with the addition of rice wine, soya bean oil, and rock sugar as ingredients, which caused an incomplete enzymatic digestion of the protein in the samples in the present study, so a necessary and convenient procedure for the purification of proteins was performed to remove them prior to the digestion step. It was also considered that all the investigated GCMs are derived from the skin of mammals or the bone of reptile, and collagens are primarily located in the extracellular matrix of animal skin, bone, and many other tissues. Accordingly, the use of bacterial-sourced collagenases for digestion can be regarded as a straightforward approach because these enzymes are capable of fragmenting different types of collagen from the cleavage site between ~Pro-X and Gly-Pro-Y, where X is most often a neutral amino acid and Y can be any nonspecific amino acid residue. As the collagenases from C. histolyticum are readily available in large amounts, are cheap, and also have satisfying cleavage efficiency, they are the enzymes of choice for digestion of the collagens into smaller peptides that are easily detectable by mass spectrum detection.
NanoLC-orbitrap MSD analysis
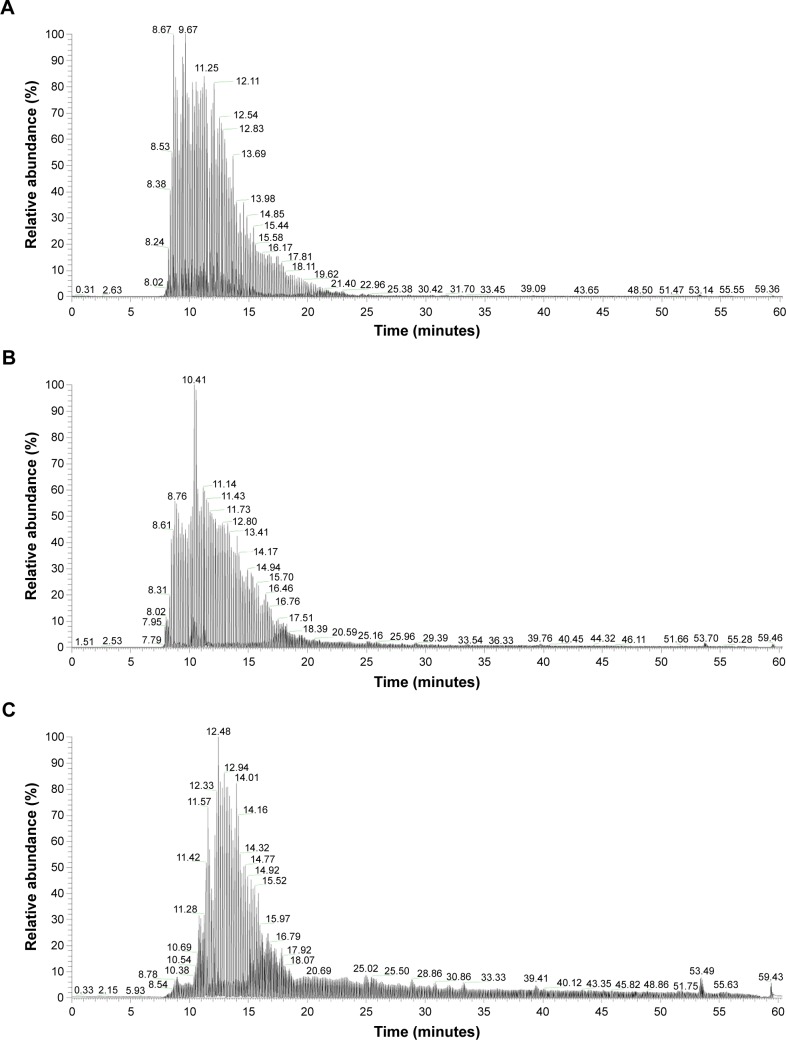
It was observed from the NanoLC-orbitrap MSD total ion chromatogram of the three GCMs’ digests, as shown in Figure 4, that most of the fragments generated were intensively eluted out of the capillary column from approximately 8 to 20 minutes. Also, the profiles of the peak clusters differed from each other slightly, although not much effort was made to optimize the liquid chromatography conditions for a better performance, as our major aim of these analyses was to identify the diagnostic biomarkers from the peptides for the differentiation of individual GCMs, which does not necessarily require a fine separation of the sample.
Figure 4.
NanoLC-orbitrap MSD TIC of gelatinous Chinese medicines’ digests.
Notes: NanoLC-orbitrap MSD TIC of (A) ACC (B/N: 110916), (B) CCC (B/N: 20130922), and (C) TCPC (B/N: 20131018).
Abbreviations: ACC, Asini Corii Colla; CCC, Cervi Cornus Colla; NanoLC-orbitrap MSD, nano-flow liquid chromatography in tandem with orbitrap mass spectrum detection; TCPC, Testudinis Carapacis ET Plastri Colla; TIC, total ion chromatogram; B/N, batch number; NL, normalized Level; RT, retention time.
The exact mass of the peptide ions was calculated according to the precursor mass and isotope pattern displayed in the MS spectrum at each time point, and the MS and MS/MS fragments obtained were all subject to de novo sequencing for further analysis by PEAKS Studio 7 (Bioinformatics Solutions Inc., Waterloo, ON, Canada). The cut-off ALC was set to be 90% for good peptide matching – much higher than the 55% ALC commonly recognized in usual practice, which must not ensure that the whole sequence is completely correct. In this way, 14 diagnostic fragments identified from the digests of these medicines were exclusively selected for their discrimination and are summarized in Table 3. All the fragments selected were within the range of 500–800 m/z of multiple charges at 2+ or 3+, which were composed of 10–19 amino acid residues. Among these, glycine contributed to more than one-third of the sequence in almost all the fragments, and proline was always at the C-terminal site of the sequence. In addition, some of the amino acids abundant in collagens were also incorporated in the sequence of the fragments, eg, glutamic acid, arginine, alanine, and threonine.
Table 3.
List of potential biomarkers for individual GCMs
| Codes | Potential biomarkers | GCMs | NanoLC
|
Orbitrap MSD
|
PEAKS
|
|||
|---|---|---|---|---|---|---|---|---|
| tR (min) | Charges | Precursor MH2+ | Precursor mass | Mass error | ALC | |||
| A1 | GFPGAVGAKGEGGGAP | ACC | 18.07 | 2+ | 664.8349 | 1,327.652 | 2.4 ppm | 95% |
| A2 | FGSGLDKADGGAGAP | ACC | 26.13 | 2+ | 660.3151 | 1,318.615 | 0.3 ppm | 94% |
| A3 | LAGAPGFRAGP | ACC | 16.87 | 2+ | 523.2746 | 1,044.535 | −0.5 ppm | 94% |
| A4 | FSGLDGSVAGGAGAP | ACC | 19.72 | 2+ | 631.8045 | 1,261.594 | 0.6 ppm | 93% |
| A5 | QTGFPGAAGGVVGPP | ACC | 17.53 | 2+ | 680.3351 | 1,358.647 | 6.8 ppm | 93% |
| C1 | GGPGSVFGGPGADGVAGKP | CCC | 14.26 | 3+ | 539.2655 | 1,614.764 | 6.8 ppm | 94% |
| C2 | QPGTPGPEGLATQGP | CCC | 17.17 | 2+ | 719.8500 | 1,437.674 | 8.2 ppm | 92% |
| C3 | VAGEPAAADGLGPGGP | CCC | 15.41 | 2+ | 684.3293 | 1,366.637 | 5.5 ppm | 91% |
| C4 | FSGLDGASVGGAGAP | CCC | 22.90 | 2+ | 631.8044 | 1,261.594 | 0.5 ppm | 90% |
| T1 | QTGFPGAAGRVGPP | TCPC | 17.93 | 2+ | 680.3376 | 1,358.658 | 2.2 ppm | 95% |
| T2 | QPGFPGARGPAGGAP | TCPC | 15.99 | 2+ | 692.8358 | 1,383.653 | 2.8 ppm | 95% |
| T3 | QVGPAGLNAVQGSGAGAP | TCPC | 16.00 | 2+ | 783.9045 | 1,565.78 | 9.3 ppm | 93% |
| T4 | LAGAPGFRQP | TCPC | 17.28 | 2+ | 523.2755 | 1,044.535 | 1.3 ppm | 92% |
| T5 | FSGLDGKDAGAGGAP | TCPC | 24.64 | 2+ | 660.3176 | 1,318.615 | 4.1 ppm | 92% |
Abbreviations: ACC, Asini Corii Colla; ALC, average local confidence; CCC, Cervi Cornus Colla; GCM, Gelatinous Chinese medicine; MSD, mass spectrum detection; NanoLC, nano-flow liquid chromatography; TCPC, Testudinis Carapacis ET Plastri Colla; tR, retention time.
In the present study, the term “potential biomarkers” was tentatively employed to acknowledge their potential value and at the same time to indicate their uncertainty, and more GCMs including the products made from other animal sources are yet to be analyzed furthermore.
Potential biomarkers’ sequence alignment and analysis
All of the sequences of the potential biomarkers were compared with those of other known proteins based on an online Basic Local Alignment Search Tool (BLAST) analysis, available on the National Center for Biotechnology Information (NCBI) website (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and were used as an input sequence for a Position-Specific Iterated BLAST (PSI-BLAST) search program against the non-redundant protein sequences databases (nrdb).23 As a consequence, a high degree of similarity was found to the sequences of predicted or previously published proteins, and most of them were collagens from mammals.
As summarized in Table 4, the highest sequence identity (82%) was found in the partial sequences of collagens from animals of the Equus genus, including Equus asinus L, which is the only medicinal animal origin for ACC, and collagen alpha-1 type I chain aligned with the sequence of the potential biomarker was one of 599 proteins discovered in this animal. The result accordingly suggested the certain reliability of the novel approach newly proposed in the current study. Although other potential biomarkers have been well aligned with the collagens from various animals, they could not be matched with the sequences of the proteins from their animal origins, namely Cervus elaphus, Cervus nippon, and Chinemys reevesii, which have only 914 proteins, 866 proteins, and 305 proteins incorporated under each entry in The National Centre for Biotechnology Information (NCBI) database, respectively. This could be largely caused by the very limited proteins discovered for each of the animal origins used for the production of the GCMs, which has greatly increased the difficulty level for further study.
Table 4.
List of sequences producing significant alignments by PSI-BLAST against nrdb
| GCM | Potentialbiomarker | Description | Max score | Total score | Query cover | Sequenceidentity | Accessions |
|---|---|---|---|---|---|---|---|
| ACC | GFPGAVGA KGEGGGAP |
Predicted: collagen alpha-1(I) chain (Equus caballus) | 37.1 | 79.5 | 100% | 82% | XP_005597538.1 |
| Predicted: collagen alpha-1(I) chain (Equus przewalskii) | 37.1 | 97.6 | 100% | 82% | XP_008516208.1 | ||
| Collagen alpha-1 type I chain (Equus asinus L) | 37.1 | 120 | 100% | 82% | ACM24774.1 |
Notes: Max score is the score of single best aligned sequence. Total score is sum of all aligned sequence scores. Query cover is what percent of query sequence is aligned.
Abbreviations: ACC, Asini Corii Colla; GCM, Gelatinous Chinese medicine; nrdb, non-redundant protein sequences databases; PSI-BLAST, Position-Specific Iterated Basic Local Alignment Search Tool.
Conclusion
It can be concluded that these three GCMs could not be well discriminated by using either the approach enforced in ChP for the quality control of those GCMs required by CFDA, as the methods lack sufficient specificity, or by using protein content determination and SDS–PAGE analysis as carried out as the first attempts in this study, since no crucial visual differences were observed.
The novel strategy proposed and developed in this study represents a simple and convenient method for the purpose of the discrimination of some GCMs by using a powerful proteomics tool, NanoLC-orbitrap MSD, after the enzymatic digestion by bacterial-sourced collagenases. This method retains the advantages of the highly specific method of qualitative analysis while avoiding the associated problems.
Acknowledgments
The research was supported by the Singapore National Research Foundation grant NRF-CRP8-2011-05; the Jiangsu Nature Science Foundation (BK2012290 and SBK2015042622); the Program for Graduate’s Innovative Research of Jiangsu (CXLX13_691); the Student Research Program of Jiangsu University (13A178); and the National Natural Science Foundation of China (81303174).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Zhicong Zhang. Yellow Emperor’s Inner Canon. Ha Er Bin: The North Literature and Art Publishing House; 2007. [Google Scholar]
- 2.Zhongjing Zhang. Treatise on Cold Damage. Beijing: China Press of Traditional Chinese Medicine; 2014. [Google Scholar]
- 3.Chinese Pharmacopoeia Committee . Chinese Pharmacopoeia. I.2010. Beijing: Chinese Medical Science and Technology Press; 2010. [Google Scholar]
- 4.Lee EJ, Jang KH, Im SY, et al. Physico-chemical properties and cytotoxic potential of Cordyceps sinensis metabolites. Nat Prod Res. 2015;29(5):455–459. doi: 10.1080/14786419.2014.948438. [DOI] [PubMed] [Google Scholar]
- 5.Zhu L, Liang Y, Lao D, Zhang T, Ito Y. Preparative separation of high-purity cordycepin from Cordyceps militaris(L.) link by high-speed countercurrent chromatography. J Liq Chromatogr Relat Technol. 2011;37(7):491–499. doi: 10.1080/10826076.2011.556965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zong Y, Wang Y, Li H, et al. Simultaneous quantification and splenocyte-proliferating activities of nucleosides and bases in Cervi cornu Pantotrichum. Pharmacogn Mag. 2014;10(40):391–397. doi: 10.4103/0973-1296.141757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D, Ru W, Xu Y, et al. Chemical constituents and bioactivities of Colla corii asini. Drug Discov Ther. 2014;8(5):201–207. doi: 10.5582/ddt.2014.01038. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y, He J, Huang Z, et al. Analysis of hexavalent chromium in Colla corii asini with on-line sample pretreatment valve-switching ion chromatography. J Chromatogr A. 2013;1305:171–175. doi: 10.1016/j.chroma.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 9.Choi HR, Nam KM, Kim DS, Huh CH, Na JI, Park KC. Cervi cornus Colla (deer antler glue) induce epidermal differentiation in the reconstruction of skin equivalents. Int J Cosmet Sci. 2013;35(3):281–285. doi: 10.1111/ics.12045. [DOI] [PubMed] [Google Scholar]
- 10.Kim J, Jeong HS, Li H, et al. Effects of Cervi cornus Colla (deer antler glue) in the reconstruction of a skin equivalent model. Arch Dermatol Res. 2013;305(1):85–89. doi: 10.1007/s00403-012-1283-8. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen PQ, Wang S, Kumar A, et al. Discovery and characterization of pseudocyclic cystine-knot α-amylase inhibitors with high resistance to heat and proteolytic degradation. FEBS J. 2014;281(19):4351–4366. doi: 10.1111/febs.12939. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen GK, Lian Y, Pang EW, Nguyen PQ, Tran TD, Tam JP. Discovery of linear cyclotides in monocot plant Panicum laxum of Poaceae family provides new insights into evolution and distribution of cyclotides in plants. J Biol Chem. 2013;288(5):3370–3380. doi: 10.1074/jbc.M112.415356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemu X, Qiu Y, Tam JP. Peptide macrocyclization through amide-to-amide transpeptidation. Tetrahedron. 2014;70:7707–7713. [Google Scholar]
- 14.Hemu X, Taichi M, Qiu Y, Liu DX, Tam JP. Biomimetic synthesis of cyclic peptides using novel thioester surrogates. Biopolymers. 2013;100:492–501. doi: 10.1002/bip.22308. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Zhang GJ, Sun SQ, Noda I. Study on similar traditional Chinese medicines cornu Cervi pantotrichum, cornu Cervi and cornu Cervi degelatinatum by FT-IR and 2D-IR correlation spectroscopy. J Pharm Biomed Anal. 2010;52:631–635. doi: 10.1016/j.jpba.2010.01.045. [DOI] [PubMed] [Google Scholar]
- 16.Wang D, Liu M, Cao J, et al. Effect of Colla corii asini (E’jiao) on D-galactose induced aging mice. Biol Pharm Bull. 2012;35:2128–2132. doi: 10.1248/bpb.b12-00238. [DOI] [PubMed] [Google Scholar]
- 17.Hijikata Y, Kano T, Xi L. Treatment for intractable anemia with the traditional Chinese medicines Hominis Placenta and Cervi Cornus Colla (deer antler glue) Int J Gen Med. 2009;2:83–90. doi: 10.2147/ijgm.s5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia W, Chu X, Ling Y, Huang J, Chang J. Multi-mycotoxin analysis in dairy products by liquid chromatography coupled to quadrupole orbitrap mass spectrometry. J Chromatogr A. 2014;1345:107–114. doi: 10.1016/j.chroma.2014.04.021. [DOI] [PubMed] [Google Scholar]
- 19.Allouni ZE, Gjerdet NR, Cimpan MR, Høl PJ. The effect of blood protein adsorption on cellular uptake of anatase TiO2 nanoparticles. Int J Nanomedicine. 2015;10:687–695. doi: 10.2147/IJN.S72726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franke K, Kettering M, Lange K, Kaiser WA, Hilger I. The exposure of cancer cells to hyperthermia, iron oxide nanoparticles, and mitomycin C influences membrane multidrug resistance protein expression levels. Int J Nanomedicine. 2013;8:351–363. doi: 10.2147/IJN.S37465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nimptsch A, Schibur S, Ihling C, et al. Quantitative analysis of denatured collagen by collagenase digestion and subsequent MALDI-TOF mass spectrometry. Cell Tissue Res. 2011;343:605–617. doi: 10.1007/s00441-010-1113-2. [DOI] [PubMed] [Google Scholar]
- 22.Eckhard U, Huesgen PF, Brandstetter H, Overall CM. Proteomic protease specificity profiling of clostridial collagenases reveals their intrinsic nature as dedicated degraders of collagen. J Proteomics. 2014;100:102–114. doi: 10.1016/j.jprot.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]