Abstract
Objective
Huntington’s disease (HD) is a rare neurodegenerative disease caused by the expansion of an N-terminal repeat in the huntingtin protein. The protein is expressed in all cells in the body; hence, peripheral tissues, such as blood, may recapitulate processes in the brain. The plasma metabolome may provide a window into active processes that influence brain health and a unique opportunity to noninvasively identify processes that may contribute to neurodegeneration. Alterations in metabolic pathways in brain have been shown to profoundly impact HD. Therefore, identification and quantification of critical metabolomic perturbations could provide novel biomarkers for disease onset and disease progression.
Methods
We analyzed the plasma metabolomic profiles from 52 premanifest (PHD), 102 early symptomatic HD, and 140 healthy controls (NC) using liquid chromatography coupled with a highly sensitive electrochemical detection platform.
Results
Alterations in tryptophan, tyrosine, purine, and antioxidant pathways were identified, including many related to energetic and oxidative stress and derived from the gut microbiome. Multivariate statistical modeling demonstrated mutually distinct metabolomic profiles, suggesting that the processes that determine onset were likely distinct from those that determine progression. Gut microbiome-derived metabolites particularly differentiated the PHD metabolome, while the symptomatic HD metabolome was increasingly influenced by metabolites that may reflect mutant huntingtin toxicity and neurodegeneration.
Interpretation
Understanding the complex changes in the delicate balance of the metabolome and the gut microbiome in HD, and how they relate to disease onset, progression, and phenotypic variability in HD are critical questions for future research.
Introduction
Huntington’s disease (HD) is an autosomal dominant inherited neurodegenerative disorder characterized by progressive motor, psychiatric, cognitive, and metabolic dysfunction. HD is caused by the abnormal expansion of a polymorphic triplet (CAG) repeat in the N-terminus of the Huntington gene leading to an excessive and toxic polyglutamine sequence in the huntingtin protein. The mutant huntingtin protein is expressed ubiquitously throughout the body but causes its greatest harm to neurons, especially in the striatum and cerebral cortex, where dysfunction and neurodegeneration cause the most consequential clinical symptoms of the disease. Aberrant interactions between mutant huntingtin, or its proteolytic fragments, and many other proteins, as well as downstream effects have been identified, which collectively play roles in neurodegeneration and which have become therapeutic targets for disease modification. Because HD is highly variable and slowly progressive clinically, there is an urgent need for useful biomarkers to help detect disease activity, monitor progression, and assess the pharmacodynamic effects and potential efficacy of experimental therapies. Since blood is easily and repeatedly accessible clinically and since its collection and processing is readily standardized, we have sought to discover markers of HD in blood that could be useful clinically.
Metabolomics is a global approach to understanding metabolic pathways and metabolic networks, including the precursors and products of all cellular biochemical pathways. The metabolome reflects dynamic interactions between the genome, transcriptome, proteome, and environment and provides information about the chemical “state” at a particular time. Metabolomic profiling has tremendous potential to provide critical information about “when” a system is perturbed, information about “which” specific molecular pathways might be implicated, and about “how” profiles change with disease. These are all difficult questions that remain largely unanswered in HD; identifying affected pathways could provide markers of disease onset or progression and may represent pathogenic pathways that could be targets for treatment and provide pharmacodynamic markers of potential treatments. As the huntingtin protein is present ubiquitously, analyzing the plasma metabolome is a less invasive way of investigating biochemical changes taking place in the presence of the mutant protein that may reflect centrally acting processes.
We therefore applied a targeted approach to metabolomic profiling to identify global biochemical changes in HD in plasma samples derived from a cohort of premanifest subjects (PHD), early symptomatic HD patients (HD), and age- and gender-matched healthy controls (NC). We used high-performance liquid chromatography coupled with highly sensitive electrochemical detection to profile plasma metabolites and focused on tryptophan, tyrosine, and purine pathway constituents. These biochemical pathways have been previously implicated as relevant to neurodegeneration in HD,1–3 and may reflect cellular events involving mutant huntingtin, oxidative stress, inflammation, mitochondrial dysfunction, synaptic dysfunction, and cell death.
Materials and Methods
Patients and sample processing
Blood samples were collected prospectively from 140 healthy controls (NC, F:M 68:72; age 50.8 ± 8.8), 102 patients with early symptomatic HD (HD, F:M 58:44; age 47 ± 8.8; CAG repeat 44.6 ± 2.9), and 52 subjects known to carry the trinucleotide expansion but who were without clinical symptoms (premanifest) of HD (PHD, F:M 33:19; age 43 ± 9.3; CAG 42.2 ± 2.0) at the MGH HD Center of Excellence as part of the REVEAL-HD translational biomarker program. A detailed history was obtained for each subject, including age, medications, and total functional capacity assessment. Procedures were explained and consent obtained according to the Declaration of Helsinki (BMJ 1991; 302:1194). Study protocols were approved by the Partners Human Research Committee.
Blood was collected by venipuncture into tubes containing ethylenediaminetetraacetic acid as an anticoagulant and kept on ice until centrifugation, which occurred within 3 h of collection, first at 1000g for 10 min to remove red blood cells, and then at 15,800g for 20 min. The plasma was aliquoted into 500 μL aliquots and subsequently frozen at −80°C until analyzed.
Sample preparation and metabolomics analysis
Plasma samples were prepared for analysis using extraction/precipitation of 125 μL of plasma with 500 μL of acetonitrile/0.4% acetic acid, centrifugation, centrifugal evaporation of the supernatant, and reconstitution in the Liquid Chromatography Electrochemical Array running buffer for injection. Samples were analyzed in a blinded fashion using an established well-validated Liquid Chromatography Electrochemical Array long gradient method.4–10 This method has been used previously for defining markers of caloric restriction,9–11 signatures of neurodegenerative disease12–15 and response to therapy in depression.15 Samples were randomized and run along with a standard containing 60 metabolites of interest. Duplicate samples were interleaved for quality control. All chromatograms were time normalized (stretched) to the chromatogram of a sample pool run in the middle of the run (sequence sets of 14) to 0.5 sec for major peaks and 1.5–2 sec for minor peaks. Data were then response normalized to the average pool value for each sequence.
Statistical analyses
Although ∼1300 metabolites are resolved on the Liquid Chromatography Electrochemical Array platform, we restricted this initial analysis to the 29 known metabolites related to the pathways of interest (Table S1).
Bivariate analyses of individual compounds were carried out using t-tests and analysis of variance models to test differences in mean metabolite levels between gene-expanded subjects (including both PHD and early HD) as compared to age- and gender-matched healthy controls as well as between the PHD and early HD groups. Differences amongst the groups in product/substrate ratios within the pathways as potential indicators of altered enzymatic processes were assessed by calculating their respective Pearson correlations. Correlations between metabolites were obtained by calculating their respective Pearson’s coefficients and are reported along with unadjusted P-values based on two-sided Student’s t-tests. Heat maps and network graphs are used to graphically represent the correlations within each group.
To further evaluate differences between the metabolomic profile of subjects comprising each of the groups we employed a number of multivariate techniques. The discrimination performance (measured by area under receiver operating curve, ROC) of the metabolite combinations was evaluated using well-validated receiver operator curve methodology. Models were validated internally using bootstrap techniques.16,17 The calibration of a prediction model measures how well the predicted probabilities agree with the actual observed outcome. The Hosmer–Lemeshow test was used to evaluate the calibration of the models.
Composite metabolomic profiles were used to construct partial least square discriminant analysis models, k-nearest neighbor hierarchical cluster analysis (including k-NN1 and k-NN3) to evaluate the categorical separation of the three study groups and to identify metabolites that contributed most to discriminating amongst groups. Correlation frequency distribution analysis was used to assess the degree to which relationships among compounds discriminated among the groups. Scoring using correct frequency discriminant analysis of the ratios underlying these correlations among and within pathways asks the question “to what extent do the metabolic relationships and sources among and within these pathways separate one state from another?” This approach differs fundamentally from the information obtained from the degree of separation given by individual compound concentrations using hierarchical cluster analysis, partial least square analysis, or linear discriminant analysis (LDA), for example. It relates both to the underlying enzymatic processes driving metabolic transitions and to the relative efficacy of sources of those compounds.
Results
Participant demographics
The demographic characteristics of the groups were compared; there were no significant differences with respect to age or gender amongst the three groups. The mean CAG repeat length for the mutant Huntington allele in the PHD subjects and symptomatic HD patients did not differ. Nine NC subjects, ten PHD subjects and 31 symptomatic HD patients reported taking selective serotonin reuptake inhibitors (SSRI’s) of these, nine NC, seven PHD subjects, and 17 symptomatic HD patients had evidence of plasma serotonin re-uptake inhibitor metabolites at the time of sample acquisition. No other medications (e.g., antihistamines, analgesics, antihypertensives) were used with a frequency or regularity that permitted analysis or modeling and were considered essentially as part of the inherent variability in any human study.
Individual and pathway metabolite analyses to evaluate differences between NC, PHD and early symptomatic HD subjects
We used several multivariate analyses to determine the extent to which the entire data determined the category of an individual and the specific metabolite and relationship amongst them most responsible for the categorization. Table1 provides a summary of the mean, and standard deviations, metabolite concentrations for each group and comparisons between NC versus PHD subjects, NC versus HD patients, and PHD versus HD patients including both the uncorrected and FDR corrected significance levels. Several metabolites from the tyrosine, tryptophan, purine, and tocopheral pathways demonstrated significant differences between groups, the changes were not necessarily linear between PHD and HD groups; in some cases, metabolites were higher in the PHD and lower in the HD group (or vice versa) as compared to NC, suggesting dynamic changes through the disease progression process. While several of the individual compounds studied did not reach significance when corrected for multiple comparisons, taken together, they compose unique descriptive metabotypes for PHD and HD subjects.
Table 1.
Metabolite concentrations (ng/ml) in controls (NC), premanifest (PHD) and early symptomatic Huntington’s disease (HD)
| Pathway | Metabolite | Mean (SD) | P-values | ||||
|---|---|---|---|---|---|---|---|
| NC | PHD | HD | NC versus PHD | NC versus HD | PHD versus HD | ||
| Tryptophan | Tryptophan (TRP) | 4225.2 (872.3) | 4225.6 (781.1) | 4238.2 (953.1) | 0.998 | 0.913 | 0.993 |
| Serotonin (5HT) | 17.5 (30.8) | 8.7 (12.9) | 6.9 (14.5) | 0.006** | <0.001** | 0.427 | |
| N-acetylserotonin (NA5HT) | 12.1 (7.31) | 15.9 (5.93) | 17.2 (8.93) | <0.001** | <0.001** | 0.308 | |
| 5-hydroxytryptophan (5HTP) | 4 (3.2) | 4.7 (2.76) | 3.3 (2.22) | 0.128 | 0.044 | 0.002** | |
| 5-hydroxyindoleacetate (5HIAA) | 18 (10.2) | 14.5 (8.22) | 18.4 (15.2) | 0.014** | 0.855 | 0.042** | |
| Indole-3-acetic acid (I3AA) | 227 (144.1) | 253.7 (137.6) | 240.8 (160.5) | 0.243 | 0.491 | 0.606 | |
| Kynurinine (KYN) | 149.1 (38.6) | 145.6 (33.1) | 138.5 (36.9) | 0.537 | 0.031** | 0.225 | |
| 3-hydroxyanthranilic acid (3OHAN) | 3.3 (2.7) | 3.9 (3.6) | 2.6 (2.5) | 0.284 | 0.028** | 0.019** | |
| 3-hydroxykynurenine (3OHKY) | 6.2 (3.8) | 6.6 (2.5) | 6.1 (3.0) | 0.381 | 0.765 | 0.231 | |
| Indole-3-lactic acid (ILA) | 31.5 (10.8) | 28.6 (9.99) | 30 (9.8) | 0.081* | 0.253 | 0.41 | |
| N-methyltryptamine (NMTRYP) | 13 (6.50) | 11 (3.8) | 12 (4.7) | 0.013** | 0.186 | 0.173 | |
| Indole-3-propionic acid (I3PA) | 191.1 (199.2) | 138.5 (130.1) | 107.7 (87.8) | 0.035** | <0.001** | 0.128 | |
| Tyrosine | Tyrosine (TYR) | 3570.8 (1128.5) | 3954.5 (980.4) | 3809.3 (1253.8) | 0.023** | 0.128 | 0.431 |
| Homovanillic acid (HVA) | 21.6 (9.8) | 20.4 (6.70) | 24.7 (10.52) | 0.339 | 0.021** | 0.003** | |
| Homogentisic acid (HGA) | 2.8 (2.45) | 3.6 (2.36) | 3.2 (2.59) | 0.045** | 0.268 | 0.31 | |
| Vanillylmandelic acid (VMA) | 23.6 (11.04) | 23.6 (10.70) | 21.4 (14.5) | 0.961 | 0.204 | 0.275 | |
| 2-hydroxyphenylacetate (2HPAC) | 104.6 (102.3) | 120.7 (136.2) | 78.7 (89.4) | 0.441 | 0.037** | 0.048** | |
| 3-hydroxyphenylacetate (3HPAC) | 124.2 (150.8) | 120.7 (110.7) | 175.4 (208.1) | 0.862 | 0.035** | 0.034** | |
| 4-hydroxyphenylacetate (4HPAC) | 79.2 (110.1) | 88.5 (52.06) | 91.4 (75.7) | 0.429 | 0.304 | 0.77 | |
| 4-hyroxyphenyllactic acid (4HPLA) | 155 (60.8) | 145 (38.9) | 127.2 (42.8) | 0.18 | <0.001** | <0.01** | |
| Purine | Hypoxanthine (HX) | 285.3 (399.9) | 406.8 (498.8) | 309.2 (373.9) | 0.119 | 0.634 | 0.217 |
| Guanosine (GR) | 56.9 (51.67) | 73.4 (69.5) | 55.3 (60.12) | 0.13 | 0.823 | 0.12 | |
| Xanthine (XAN) | 122.6 (61.6) | 98.3 (35.9) | 103.7 (45.7) | 0.001** | 0.007* | 0.419 | |
| Xanthosine (XANTH) | 57.7 (19.3) | 57.5 (17.3) | 53.3 (16.47) | 0.938 | 0.055* | 0.15 | |
| Urate (URATE) | 40,099.7 (1001) | 41,701 (6770.2) | 38,936 (8475.0) | 0.209 | 0.331 | 0.03* | |
| Paraxanthine (PARAXAN) | 68.9 (60.5) | 70.1 (63.0) | 77.1 (70.6) | 0.909 | 0.341 | 0.528 | |
| Antioxidant | Gama tocopherol (GTOCO) | 898.8 (585.2) | 887.7 (537.2) | 1077 (721.2) | 0.901 | 0.041** | 0.068* |
| Alpha tocopherol (ATOCO) | 2699 (1562.4) | 3307.5 (1471.7) | 3166 (1836.1) | 0.014** | 0.038** | 0.605 | |
| Methionine | Methionine; (MET) | 1409.1 (666.5) | 1709.3 (594.0) | 1484.6 (611.0) | 0.003** | 0.361 | 0.03** |
P < 0.1
P < 0.05, FDR corrected.
Tryptophan pathway
Serotonin was reduced in both PHD and HD groups as compared to NC. In contrast, N-acetylserotonin was increased in both groups. 5-hydroxytryptophan, 3-hydroxyanthranilic acid, and kynurenine were significantly reduced in the HD group but were not significantly different in PHD as compared to NC groups; however, differences between PHD and HD groups were significant except for kynurenine. In contrast, 5-hydroxyindoleactetate and N-methyltryptamine were significantly reduced in the PHD group but not significantly different between the HD and NC groups; differences between PHD and HD groups were significant for 5-hydroxyindoleacetate. The tryptophan metabolite, indole-3-propionic acid was significantly reduced in both the PHD and HD groups. There were no significant differences amongst groups in the concentrations of indole-3-acetic acid, 3-hydroxykynurenine or in tryptophan itself.
Tyrosine pathway
Tyrosine was increased in the PHD and symptomatic HD groups compared to NC group but the increase was significant only in the PHD group. Homogentisic acid was significantly increased in the PHD group, but was not significantly different in the HD group. Although 2-hydoxyphenylacetate was higher in the PHD group compared to the NC group, the difference was not significant; in contrast, 2-hydroxyphenylacetate was significantly reduced in symptomatic HD patients. 4-hydroxyphenyllactic acid was significantly reduced in HD in comparison to both NC and PHD subjects. 3-hydoxyphenylacetate was significantly higher in HD as compared to both NC and PHD subjects. Homovanillic acid was significantly higher in HD but reduced in PHD subjects. There were no differences in the concentrations of 4-hydoxyphenylacetate or vanillylmandelic acid concentrations amongst the groups.
Purine pathway
Xanthine was significantly reduced in both HD and PHD groups as compared to the NC group; there was no significant difference between the HD and PHD groups. Xanthosine was significantly lower in the HD group as compared to the NC group. Guanosine, hypoxanthine, and paraxanthine were not significantly different between groups. Urate concentrations were significantly lower in HD as compared to the PHD group.
Tocopherols
The tyrosine pathway-related antioxidant gamma tocopherol was significantly increased in HD; alpha tocopheral was increased in both HD and PHD subjects.
Methionine
Methionine was significantly increased in PHD subjects only; this difference was significant as compared to both the NC and HD groups.
Multivariate analysis of the NC, PHD, and HD groups
Receiver operator curves
To determine whether the metabolite profiles discriminate amongst groups, we used Receiver Operator Curves. The area under the curve (AUC) provides the discriminating power of a given variable, or group of variables to differentiate between NC and PHD subjects, between NC and HD patients and between PHD subjects and HD patients (Fig.1).
Figure 1.
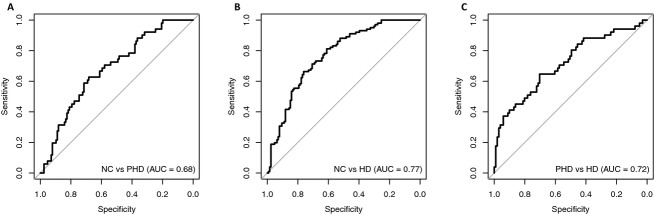
Receiver operator curves. (A) NC versus PHD. (B) NC versus HD. (C) PHD versus HD. In each case, the AUC demonstrated excellent sensitivity and specificity, demonstrating clear separation between groups and suggesting specific effects on the HD plasma metabolome. AUC, area under the curve.
The AUC values for sensitivity and specificity (index corrected) for the entire set of metabolites were between 74% and 80%. Finally, to validate the models and obtain an unbiased estimate of future model performance we used the Bootstrap method proposed by Efron.18 The bootstrap adjusted AUC values are provided in Table2, further supporting group differences.
Table 2.
Metabolite area under the curve values
| Pathway | Comparison | Index corrected |
|---|---|---|
| All metabolites | NC versus PHD | 0.74 |
| NC versus HD | 0.80 | |
| PHD versus HD | 0.77 | |
| Indole-3-propionic acid | NC versus PHD | 0.58 |
| NC versus HD | 0.67 | |
| PHD versus HD | 0.55 | |
| Purine | NC versus PHD | 0.69 |
| NC versus HD | 0.60 | |
| PHD versus HD | 0.60 | |
| Tyrosine | NC versus PHD | 0.64 |
| NC versus HD | 0.73 | |
| PHD versus HD | 0.73 | |
| Tryptophan | NC versus PHD | 0.71 |
| NC versus HD | 0.74 | |
| PHD versus HD | 0.70 |
Partial least squares-discriminant analysis
We used projection on latent structures discriminate analysis models with one out testing to define the level of categorical separation. Strong group separation was achieved between the groups; PHD versus NC; symptomatic HD versus NC, and PHD versus HD (Fig.2).
Figure 2.
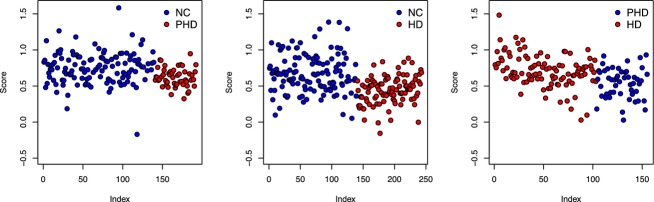
One out testing scores using PLSDA models for NC versus PHD, NC versus HD and PHD versus HD, using the 29 chosen compounds from the tyrosine, tryptophan, purine pathways and markers of oxidative progression (from Table1). The CCR are summarized in Table3. PLSDA, partial least squares discriminant analysis; HD, Huntington’s disease; CCR, correct classification rates.
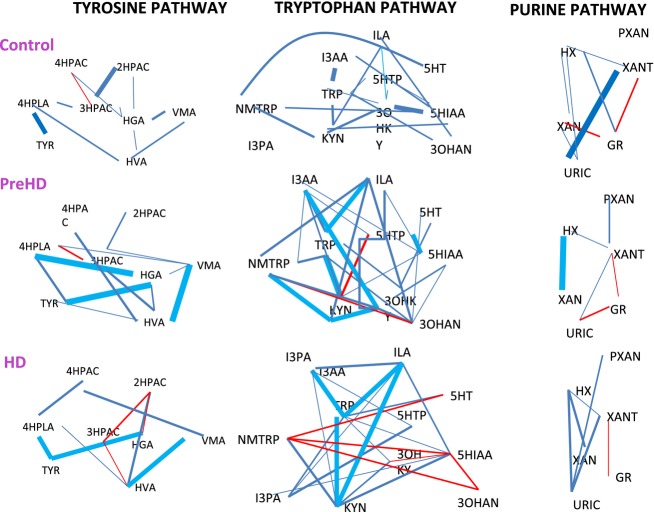
Comparative discriminant models were developed using LDA, hierarchical cluster analysis (kNN1 and kNN3) and LDA; the relationships amongst compounds as discriminators among groups were evaluated using correlation frequency distribution analysis (shown in Fig.3 as a heat map and in Fig.4 as a network graph.)
Figure 3.
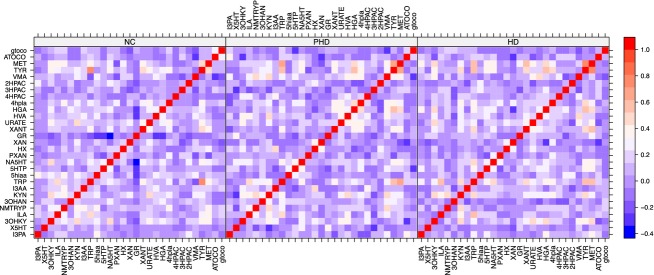
Heat map demonstrating altered correlations in the cross-metabolite correlations. Red demonstrates a positive correlation, blue a negative correlation, between pairs of metabolites.
Figure 4.
Network graph representing Pearson correlations between metabolites for NC, PHD, and HD groups (r = ±0.7 to ±0.2) for each pathway. Red lines represent negative correlations while blue lines represent positive correlations. These results demonstrate altered relationships amongst metabolites across disease groups, suggesting unique alterations in the feedback control of key enzymatic processes that change with HD progression.
The correlation frequency distribution analysis of these correlations is shown in Figure5.
Figure 5.
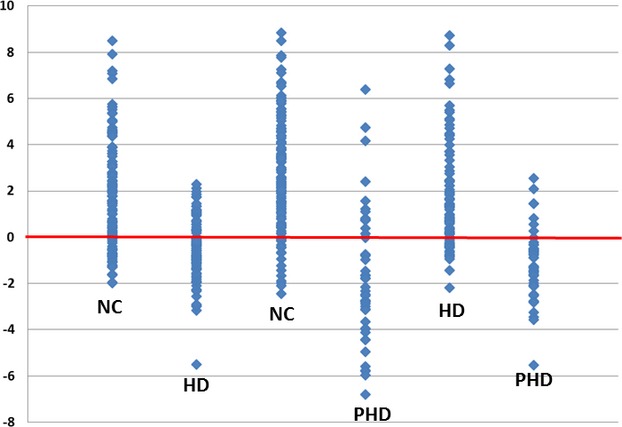
Summation of one-out scoring. The graph is presented with 0.5 subtracted from all values to center around 0 and scaled by a factor of 100 for visualization. From the left, the first two columns score NC versus HD with NC as category A. The third and fourth columns score NC versus PHD with NC as category A. The fifth and sixth columns score HD versus PHD with HD as category A.
The correct classification rates among the three categories NC, PHD subjects, and HD patients are summarized in Table3, for all discriminant models tested. We validated the performance of each model using one out scoring. The training and validation sets were iterated until each subject was included in a validation set.
Table 3.
Correct classification rates using distinct models
| NC versus HD | NC versus PHD | PHD versus HD | |
|---|---|---|---|
| PLSDA | 0.744 | 0.656 | 0.721 |
| LDA | 0.731 | 0.629 | 0.662 |
| KNN 1 | 0.626 | 0.618 | 0.591 |
| KNN | 0.668 | 0.688 | 0.636 |
| CFDA | 0.712 | 0.701 | 0.706 |
PLSDA, partial least squares discriminant analysis; LDA, linear discriminant analysis; kNN2, kNN3, k-nearest neighbor hierarchical cluster analysis; CFDA, correlation frequency distribution analysis.
Table4 provides the metabolite ratios that have the strongest effect on discrimination between group pairs. In particular, N-acetylserotonin/xanthine, N-acetylserotonin/indole lactic-3-acid, 4-hydroxyphenyllactic acid/5-hydroxyindoleactetate, N-acetylserotonin/kynurenine, 5-hydroxyindoleacetate/N-acetylserotonin, 5-hydroxytryptophan/homovanillic acid, indole-3-propionic acid/homovanillic acid, N-acetylserotonin/xanthosine, 4-hydroxyphenyllactic acid/homovanillic acid; indole-3-propionic acid/N-acetyl serotonin, urate/homovanillicacid, indole-3-propionic acid/N-acetylserotonin, homovanillic acid/methionine.
Table 4.
Associations between compounds that best separate groups (CFDA)
| NA5HT/KYN | 5HIAA/NA5HT | 4HPLA/5HIAA |
| I3PA/HVA | NA5HT/XANTH | 5HTP/HVA |
| NA5HT/XAN | I3PA/NA5HT | 4HPLA/HVA |
| I3PA/NA5HT | HVA/MET | URATE/HVA |
CFDA, correlation frequency distribution analysis; NA5HT, N-Acetyl-serotonin; KYN, Kynurinine; I3PA, Indole-3-propionic acid; HVA, Homovanillic acid; ILA, Indole-3-lactic acid; 5HIAA, 5-hydroxyindoleacetate; XANTH, Xanthosine; MET, Methionine; 4HPLA, 4-hydroxyphenyllactic acid; URATE, Urate, XAN, Xanthine.
Controlling for drug effects
In order to determine if concomitant SSRI’s might have an impact or explain reduced serotonin, as has been previously reported, or might exert an effect on other metabolites in the tyrosine pathway, we independently evaluated the metabolomics profiles of those participants who were on SSRI’s (nine NC, ten PHD subjects, and 31 HD patients). No subject was on Haldol. We were able to identify peaks corresponding to known metabolites for the parent drug in all NC, 65% of PHD subjects who reported taking these medications and in 60% of symptomatic HD patients who were prescribed SSRI’s, suggesting that some of these individuals were not actually taking these medications. We found a reduction in the mean concentration of serotonin (8.68 vs. 6.98 ng/mL) in SSRI users, which was not significant (P = 0.09). Removing those individuals from the original analyses did not significantly change the results (Fig.6).
Figure 6.
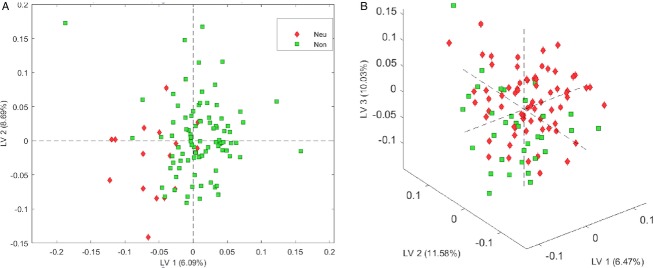
PLSDA modeling of effects of medications on metabolomics profiling. (A) There was no difference in the metabolomic profiles of PHD and HD individuals taking SSRI’s (Red) and those who were not on SSRI’s (Green). (B) There was no difference in the metabolomic profiles of PHD and HD individuals taking Neuroleptics (Green) and those not on neuroleptics (Red). This suggests that the HD metabotype is sufficiently robust to be insignificantly affected by these medications.
Discussion
Although the cardinal symptoms of HD are due to neurodegeneration in the central nervous system, mutant huntingtin protein is expressed throughout all tissues. The plasma metabolome integrates the effects of neurodegeneration, as leaked into the plasma; the systemic effects of the widespread expression of the mutant protein; and the global effects of disease on systemic metabolism. We identified distinct alterations in tyrosine, tryptophan, and purine pathways in prodromal and early HD, many related to oxidative stress and cellular metabolism. Selective dysregulation of some pathways and paradoxically increased regulation of other pathways suggests complex alterations in the feedback control of the underlying enzymes, proteins, or genes in HD. Remarkably, the normal control, PHD, and early HD plasma metabolomes were mutually distinct rather than differing along a continuum suggesting differing influences during the prodromal and symptomatic stages of the disease. Many of the metabolites differentiating the control from the PHD and HD metabolomes are highly linked to the gut microflora,19 suggesting that the HD genetic mutation favors a distinct microbiome or “enterotype”.20,21 Enteric mutant huntingtin22 or systemic effects of HD on the gut could influence energy homeostasis, the supply of vitamins and metabolites, and neuroimmune function23–27 and impact the clinical expression of HD. Our data also suggests that the PHD metabolome is more influenced by the gut microbiome than the HD metabolome, perhaps due to increasing effects of mutant huntingtin (mthtt) toxicity and neurodegeneration.
Tryptophan pathway
We found indole-3-propionic acid, a product of tryptophan metabolism through indole-3-pyruvic acid and indole-3-lactic acid, to be significantly reduced in premanifest subjects and in HD. Indole-3-propionic acid is a potent hydroxyl radical scavenger normally found in the plasma and cerebrospinal fluid (CSF) that is produced exclusively by the commensal gut bacteria Clostridium sporogenes.19,28 Its primary metabolite, kynuric acid, was undetectable at levels of pg/mL even after concentrating plasma up to a factor of 20 arguing against increased metabolism. Consumption by covalent binding to oxidized protein residues is also inconsistent with the modest increase of bound indole-3-propionic acid found in HD (Matson, pers. comm.). Thus, reduced production or transport through the gut best explains the indole-3-propionic acid reductions we have observed and suggests an HD microbiome that discriminates against Clostridium sporogenes. Interestingly, indole-3-propionic acid, is protective against A-Beta-mediated toxicity in vitro29 and iron-mediated oxidative damage30 and is under development as a possible treatment for Alzheimer’s disease.31 As levels of indole-3-propionic acid, and other circulating antioxidants diminish in HD, the level of the oxidative damage to proteins likely increases.
Quinolinic acid-induced excitotoxic striatal lesions led to the hypothesis that the tryptophan pathway could play an important role in HD pathophysiology.32 Tryptophan is converted to, kynurenine, which is reduced in HD patients, and thence to kynurenic acid, a glutamate receptor antagonist and endogenous neuroprotectant. Its metabolite, 3-hydroxyanthranilic acid, is reduced in HD, as is the 3-hydroxyanthranilic/anthranilate ratio, as previously reported.2 This reduction in plasma 3-hydroxyanthranilic acid could be due to modulation of 3-hydroxyanthranilic acid oxidase in favor of reducing the downstream production of quinolinic acid. If brain levels of 3-hydroxyanthranilic acid are also low, kynurenine 3-monooxygenase inhibitors, a potential treatment for HD,33 may simply shift the production and metabolism of 3-hydroxyanthranilic acid to other pathways and may not have a major effect on quinolinic acid levels.34 Interestingly, 3-hydroxyanthranilic levels were not correlated with its precursor 3-hydroxykynurenine in any group, suggesting alternate routes in its production.
In controls, kynurenine correlated with its precursor tryptophan and with the subsequent pathway from kynurenine through 3-hydroxykynurenine and 3-hydroxyanthranilinic acid. With phenoconversion, kynurenine became more strongly correlated with tryptophan and with its product, 3-hydroxykynurenine (and not 3-hydroxyanthranilic acid) in HD. This suggests that the pathway through 3-hydroxyanthranilic acid to quinolinate, and eventually NAD/NADH, becomes more dominant with symptom onset. Although the kynurenine branch product kynurenic acid was not measured in this study, the shift of regulation is consistent with progressive loss of protection from kynurenic acid and with previous findings of lower levels in HD.35 Alternatively, 3-hydroxyanthranilic acid could arise from the tryptophan pathway leading through indole-3-acetic acid to anthranilate and to 3-hydroxyanthranilic acid, either through enzymatic processes or direct hydroxyl radical attack on anthranilate. The strong correlation between tryptophan and indole-3-acetic acid in HD patients but not in healthy controls supports this.
Other metabolites comprising the tryptophan pathway were also altered. Serotonin was markedly reduced in both PHD and HD and its primary metabolite 5-hydroxyindoleacetate was significantly reduced in PHD subjects. Since plasma serotonin levels reflect release from enterochromaffin cells in the gut wall, selective uptake by platelets, and metabolism to 5-hydroxyindoleacetate by the liver, the reductions observed are certainly peripheral in origin. Despite the reduction in serotonin, N-acetyl serotonin, a neuroprotective antioxidant metabolite,36 and precursor for melatonin, was significantly increased in both PHD and HD. Reduced metabolism, would be consistent with reports of reduced melatonin release in brain HD,37 even though serum levels of melatonin are not altered in HD.38,39 A more likely explanation is increased production of N-acetyltryptophan in the gut with direct conversion to N-acetylserotonin. These findings, which were not explained by SSRI’s, may be relevant clinically for HD given the importance of serotonin levels for mood, cognition, appetite, and gastrointestinal motility.
Tyrosine pathway
Alterations in the gut microflora could also underlie the tyrosine elevations that were observed via excess production from the essential dietary amino acid phenylalanine. 2-hydroxyphenylacetate, which was increased in HD; 3-hydroxyphenylacetate, which was increased in HD; and 4-hydroxyphenylacetate, which was reduced in HD, are polyphenol metabolites of phenylacetate, which is derived from dietary phenylalanine in the gut.40 These polyphenols are largely formed nonenzymatically from direct hydroxyl radical attack on the free amino acid or on residues in proteins that are subsequently degraded. Interestingly, the gut microflora forms these compounds through the microbial styrene degradation pathway by which 2-hydroxyphenylacetate and 3-hydroxyphenylacetate are converted to homogentisic acid, which we also find to be increased in premanifest plasma. 4-hydroxyphenyllactic acid, which was reduced progressively from PHD to HD, is a tyrosine metabolite formed by gut microbes, such as lactobacilli41 or consumed in dietary plant material.42 Interestingly, 4-hydroxyphenyllactic acid decreases reactive oxygen species production in both mitochondria and neutrophils and like indole-3-propionic acid, is an antioxidant supplied by the gut microbiome.43 Its diminution in HD could further indicate an HD/gut interaction or a change in mitochondrial status in HD.
Tyrosine metabolism in the gut may also relate to the increases in alpha tocopherol we observed in both premanifest and symptomatic groups and to increases in gamma tocopherol (vitamin E) we observed in symptomatic HD. The tocopherols are absorbed by the intestine into the lymphatic system and released into the plasma by the liver after consumption in the diet. While most vitamin E in humans originates from ingested plant material in the form of dietary vegetable oil,44 it is also synthesized by microbes in a pathway through tyrosine and homogentisic acid.45 We hypothesize that the Huntington enterotype that leads to an increase in tyrosine and homogentisic acid also increases production and absorption of the tocopherols. Further research will be necessary to determine whether an HD enterotype significantly modulates the onset and course and thus might also be a therapeutic target as has been suggested for other disorders.46
More widespread alterations in the tyrosine pathway were suggested. HVA, the primary metabolite of dopamine was increased in HD. A significant percentage of the HVA in plasma is believed to originate from the central nervous system.47,48 Higher HVA levels in CSF have been correlated with psychotic symptoms in schizophrenia while lower levels have been correlated with depression49,50; in HD CSF levels have been reported to be unchanged,51 so higher levels of HVA in the plasma may represent reduced metabolism in the periphery. Although reduced dopamine beta hydroxylase activity could also explain the increase, there are several reports that plasma dopamine beta hydroxylase activity is normal in HD.52 Elevated monoamine oxidase activity, which has been reported in platelets from HD patients,53 is an alternative explanation that would be consistent with the reductions in 5-hydroxyindoleacetic acid we observed.
Purine pathway
Alterations in the purine pathway were also significant. While its precursors hypoxanthine and guanosine were unchanged and xanthosine trended down in HD, xanthine was markedly reduced in both PHD and HD groups. In contrast, there were no significant differences between controls and PHD or HD in its metabolite, the systemic antioxidant urate (although there were significant differences between HD and PHD groups). Perhaps urate oxidase activity is increased to maintain levels of urate in HD, resulting in precursor depletion. The purine pathway is also closely linked to cellular energetics and mitochondrial function through adenosine metabolism and ADP/ATP dynamics, which are altered in HD transgenic mouse models. Mitochondrial dysfunction would spill ADP into the production of hypoxanthine, decoupling it from its immediate product xanthine, consistent with the elevated hypoxanthine/xanthine ratio we observed in the premanifest group. Both xanthine and hypoxanthine are normally about 10-fold higher in ventricular CSF than in plasma, so alterations in purine metabolism in brain could also effect peripheral levels. Lower plasma levels of urate have been associated with Parkinson’s disease while higher levels may be associated with slower progression of HD.54,55 Inosine treatment to raise urate levels is currently being tested in clinical trials for Parkinson disease.56,57 Levels of paraxanthine, the principle metabolite of caffeine were not altered amongst groups.
We observed an increase in plasma methionine in premanifest subjects. This is consistent with other suggestions of altered one-carbon metabolism, such as the increases in DNA and histone methylation.58 Since there is normally an equilibrium between methionine and cysteine, this finding could also be connected to alterations in L-cysteine levels that have been reported in HD59 and related to oxidative mechanisms of pathogenesis.
Limitations
This work is based on one of the largest plasma data sets available for HD and is based on the use of several models and while cross-validation and boot-strap methods were used in an effort to validate the models, a follow-up study, using an independent sample set is warranted.
Conclusion
HD is a nervous system disorder that demonstrates complex brain-body perturbations, including dysregulation of bioenergetic pathways and of the gut microbiome. Collectively, our findings identify specific and selective alterations of the plasma metabolome that could provide useful markers of prodromal and symptomatic HD patients; confirm that HD involves important metabolic pathways; demonstrate that mthtt affects systemic as well as central nervous system biochemistry; and identifies significant interactions between HD and the gut that could influence disease onset and progression and have important implications for treatment.
To determine the predictive potential of the compounds, these models were evaluated using leave-one-out cross-validation to assess the predictive performance of these models and to further refine the variables included in prediction analyses. By using cross-validation, we were able to estimate a prediction error from the withheld data used in the validation process. While such an analysis is not as powerful in assessing the true predictive performance of a model, it does provide a reasonable estimate of the predictive performance.
Because of the high dimensional and sparse nature of the data, to try to assess whether the resulting models were better than would be expected by chance, we performed permutation testing to ascribe statistical significance to the resulting models. The best correlation coefficient and AUC for each permuted data set were recorded, and an empirical distribution of model fit statistics was generated across the permuted data sets. The values from the real data analysis were the compared to the empirical distribution to generate an empirical P-value.
We also used a network approach to evaluate differences amongst groups. Categorizing disease based on the correlations in the metabolomics network is a different strategy representing a different biological phenomenon from the use of ROCs of partial least squared discriminant analysis (receiver operator curve or partial least squares discriminant analysis), which rely on the levels of compounds in the data set. Correlation networks reflect the aggregate influence of the underlying enzymes and sources of input among categories and the information for discriminating categories is inherent in those correlations.
Receiver operator curve, k-nearest neighbor hierarchical cluster analysis, and LDA analyses were performed using R (R Core Team [2013]). Partial least squares discriminant analyses were performed using Matlab. Correlation frequency distribution analysis was performed using an in house program and approach.60
Acknowledgments
This work was supported by the National Institutes of Health (NS042861, NS058793, AT000613, and FD003359 to H. D. R.; NS058793 to G. D.; NS058793, NS042861, AT000613, and FD003359 to S. M. H.)
Author Contributions
H. D. R. was responsible for the study design, conception and protocol development, clinical assessments, data analyses, and interpretation and authoring the manuscript. G. D. was the study statistician responsible for the data analysis and interpretation and contributed to authoring and reviewing the manuscript and generation of figures. S. B. was involved in the sample preparation, analysis, metabolomic analysis, figure preparation modeling, and reviewing of this manuscript. B. T. was involved in the sample preparation and reviewing of this manuscript. S. G. was involved in the study design, data management, and reviewing of the data. K. M. was involved in the protocol development, data analysis, and reviewing of this manuscript. W. M. was responsible for the metabolomics analysis and contributed to the editing of this manuscript. S. M. H. was the Principal investigator and was responsible for obtaining funding, study design and conception, data interpretation, and played a key role in authoring this manuscript. Other: We are very grateful to the individuals who participated in this study, who so generously contributed their time and energy to this work, and without whom it would not have been possible. We thank the individuals who helped recruit participants for this study, including: Susan Maya, Caleb Dresser, Jessica Meyer, Puja Turakhia, Rachel Goldstein, Lauren Woo, Alex Bender, Angela Hu, and Jennifer Lee and who helped with sample processing, including Samantha Matson.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1.List of known compounds quantified by the LCECA (Liquid Chromatography Electrochemical Array) platform.
References
- Beal MF, Matson WR, Swartz KJ, et al. Kynurenine pathway measurements in Huntington’s disease striatum: evidence for reduced formation of kynurenic acid. J Neurochem. 1990;55:1327–1339. doi: 10.1111/j.1471-4159.1990.tb03143.x. [DOI] [PubMed] [Google Scholar]
- Stoy N, Mackay GM, Forrest CM, et al. Tryptophan metabolism and oxidative stress in patients with Huntington’s disease. J Neurochem. 2005;93:611–623. doi: 10.1111/j.1471-4159.2005.03070.x. [DOI] [PubMed] [Google Scholar]
- Pearson SJ, Reynolds GP. Increased brain concentrations of a neurotoxin, 3-hydroxykynurenine, in Huntington’s disease. Neurosci Lett. 1992;144:199–201. doi: 10.1016/0304-3940(92)90749-w. [DOI] [PubMed] [Google Scholar]
- Kristal BS, Shurubor YI, Kaddurah-Daouk R, Matson WR. Metabolomics in the study of aging and caloric restriction. Methods Mol Biol. 2007;371:393–409. doi: 10.1007/978-1-59745-361-5_25. [DOI] [PubMed] [Google Scholar]
- Kristal BS, Shurubor YI, Kaddurah-Daouk R, Matson WR. High-performance liquid chromatography separations coupled with coulometric electrode array detectors: a unique approach to metabolomics. Methods Mol Biol. 2007;358:159–174. doi: 10.1007/978-1-59745-244-1_10. [DOI] [PubMed] [Google Scholar]
- Kristal BS, Vigneau-Callahan KE, Matson WR. Simultaneous analysis of the majority of low-molecular-weight, redox-active compounds from mitochondria. Anal Biochem. 1998;263:18–25. doi: 10.1006/abio.1998.2831. [DOI] [PubMed] [Google Scholar]
- Kristal BS, Vigneau-Callahan KE, Moskowitz AJ, Matson WR. Purine catabolism: links to mitochondrial respiration and antioxidant defenses? Arch Biochem Biophys. 1999;370:22–33. doi: 10.1006/abbi.1999.1387. [DOI] [PubMed] [Google Scholar]
- Schiavo S, Ebbel E, Sharma S, et al. Metabolite identification using a nanoelectrospray LC-EC-array-MS integrated system. Anal Chem. 2008;80:5912–5923. doi: 10.1021/ac800507y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Paolucci U, Vigneau-Callahan KE, et al. Development of biomarkers based on diet-dependent metabolic serotypes: practical issues in development of expert system-based classification models in metabolomic studies. OMICS. 2004;8:197–208. doi: 10.1089/omi.2004.8.197. [DOI] [PubMed] [Google Scholar]
- Shurubor YI, Matson WR, Willett WC, et al. Biological variability dominates and influences analytical variance in HPLC-ECD studies of the human plasma metabolome. BMC Clin Pathol. 2007;7:9. doi: 10.1186/1472-6890-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolucci U, Vigneau-Callahan KE, Shi H, et al. Development of biomarkers based on diet-dependent metabolic serotypes: concerns and approaches for cohort and gender issues in serum metabolome studies. OMICS. 2004;8:209–220. doi: 10.1089/omi.2004.8.209. [DOI] [PubMed] [Google Scholar]
- Rozen S, Cudkowicz ME, Bogdanov M, et al. Metabolomic analysis and signatures in motor neuron disease. Metabolomics. 2005;1:101–108. doi: 10.1007/s11306-005-4810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Matson WR, Wang L, et al. Metabolomic profiling to develop blood biomarkers for Parkinson’s disease. Brain. 2008;131:389–396. doi: 10.1093/brain/awm304. [DOI] [PubMed] [Google Scholar]
- Johansen KK, Wang L, Aasly JO, et al. Metabolomic profiling in LRRK2-related Parkinson’s disease. PLoS One. 2009;4:e7551. doi: 10.1371/journal.pone.0007551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddurah-Daouk R, Boyle SH, Matson W, et al. Pretreatment metabotype as a predictor of response to sertraline or placebo in depressed outpatients: a proof of concept. Transl Psychiat. 2011;1:e26. doi: 10.1038/tp.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell FE. Regression modeling strategies: with applications to linear models, logistic regression, and survival analysis. New York, NY: Springer; 2001. p. xxii. Springer series in statistics. 568 p. [Google Scholar]
- Steyerberg EW. SpringerLink (online service) In: Gail M, Samet JM, Tsiatis A, Wong W, editors. Statistics for biology and health. New York, NY: Springer; 2009. p. xxviii. 497 p. [Google Scholar]
- Efron B. How biased is the apparent error rate of a prediction rule? J Am Stat Assoc. 1986;81:461–470. [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaarala O, Atkinson MA, Neu J. The “perfect storm” for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes. 2008;57:2555–2562. doi: 10.2337/db08-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C, Sarvetnick N. The incidence of type-1 diabetes in NOD mice is modulated by restricted flora not germ-free conditions. PLoS One. 2011;6:e17049. doi: 10.1371/journal.pone.0017049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulet D, Cicchetti F. The role of immunity in Huntington’s disease. Mol Psychiatry. 2011;16:889–902. doi: 10.1038/mp.2011.28. [DOI] [PubMed] [Google Scholar]
- De Ponti F. Drug development for the irritable bowel syndrome: current challenges and future perspectives. Front Pharmacol. 2013;4:7. doi: 10.3389/fphar.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politis M, Su P, Piccini P. Imaging of microglia in patients with neurodegenerative disorders. Front Pharmacol. 2012;3:96. doi: 10.3389/fphar.2012.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkqvist M, Wild EJ, Thiele J, et al. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington’s disease. J Exp Med. 2008;205:1869–1877. doi: 10.1084/jem.20080178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. published online EpubJun 8. [DOI] [PubMed] [Google Scholar]
- Resta SC. Effects of probiotics and commensals on intestinal epithelial physiology: implications for nutrient handling. J Physiol. 2009;587:4169–4174. doi: 10.1113/jphysiol.2009.176370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley RJ, Fredricks WW, Smith OH. Effect of tryptophan analogs on derepression of the Escherichia coli tryptophan operon by indole-3-propionic acid. J Bacteriol. 1978;136:219–226. doi: 10.1128/jb.136.1.219-226.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyan YJ, Poeggeler B, Omar RA, et al. Potent neuroprotective properties against the Alzheimer beta-amyloid by an endogenous melatonin-related indole structure, indole-3-propionic acid. J Biol Chem. 1999;274:21937–21942. doi: 10.1074/jbc.274.31.21937. [DOI] [PubMed] [Google Scholar]
- Karbownik M, Reiter RJ, Garcia JJ, et al. Indole-3-propionic acid, a melatonin-related molecule, protects hepatic microsomal membranes from iron-induced oxidative damage: relevance to cancer reduction. J Cell Biochem. 2001;81:507–513. [PubMed] [Google Scholar]
- Bendheim PE, Poeggeler B, Neria E, et al. Development of indole-3-propionic acid (OXIGON) for Alzheimer’s disease. J Mol Neurosci. 2002;19:213–217. doi: 10.1007/s12031-002-0036-0. [DOI] [PubMed] [Google Scholar]
- Coyle JT, Schwarcz R. Lesion of striatal neurones with kainic acid provides a model for Huntington’s chorea. Nature. 1976;263:244–246. doi: 10.1038/263244a0. [DOI] [PubMed] [Google Scholar]
- Thevandavakkam MA, Schwarcz R, Muchowski PJ, Giorgini F. Targeting kynurenine 3-monooxygenase (KMO): implications for therapy in Huntington’s disease. CNS Neurol Disord Drug Targets. 2010;9:791–800. doi: 10.2174/187152710793237430. [DOI] [PubMed] [Google Scholar]
- Zwilling D, Huang SY, Sathyasaikumar KV, et al. Kynurenine 3-monooxygenase inhibition in blood ameliorates neurodegeneration. Cell. 2011;145:863–874. doi: 10.1016/j.cell.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF, Matson WR, Storey E, et al. Kynurenic acid concentrations are reduced in Huntington’s disease cerebral cortex. J Neurol Sci. 1992;108:80–87. doi: 10.1016/0022-510x(92)90191-m. [DOI] [PubMed] [Google Scholar]
- Zhou H, Wang J, Jiang J, et al. N-acetyl-serotonin offers neuroprotection through inhibiting mitochondrial death pathways and autophagic activation in experimental models of ischemic injury. J Neurosci. 2014;34:2967–2978. doi: 10.1523/JNEUROSCI.1948-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinali DP, Pagano ES, Scacchi Bernasconi PA, et al. Melatonin and mitochondrial dysfunction in the central nervous system. Horm Behav. 2013;63:322–330. doi: 10.1016/j.yhbeh.2012.02.020. [DOI] [PubMed] [Google Scholar]
- Aziz NA, Pijl H, Frolich M, et al. Delayed onset of the diurnal melatonin rise in patients with Huntington’s disease. J Neurol. 2009;256:1961–1965. doi: 10.1007/s00415-009-5196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofides J, Bridel M, Egerton M, et al. Blood 5-hydroxytryptamine, 5-hydroxyindoleacetic acid and melatonin levels in patients with either Huntington’s disease or chronic brain injury. J Neurochem. 2006;97:1078–1088. doi: 10.1111/j.1471-4159.2006.03807.x. [DOI] [PubMed] [Google Scholar]
- Mohamed Mel S, Ismail W, Heider J, Fuchs G. Aerobic metabolism of phenylacetic acids in Azoarcus evansii. Arch Microbiol. 2002;178:180–192. doi: 10.1007/s00203-002-0438-y. [DOI] [PubMed] [Google Scholar]
- Mu W, Yang Y, Jia J, et al. Production of 4-hydroxyphenyllactic acid by Lactobacillus sp. SK007 fermentation. J Biosci Bioeng. 2010;109:369–371. doi: 10.1016/j.jbiosc.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Petersen M, Abdullah Y, Benner J, et al. Evolution of rosmarinic acid biosynthesis. Phytochemistry. 2009;70:1663–1679. doi: 10.1016/j.phytochem.2009.05.010. doi: 10.1016/j.phytochem.2009.05.010 ; published online Epub Oct-Nov. [DOI] [PubMed] [Google Scholar]
- Beloborodova N, Bairamov I, Olenin A, et al. Effect of phenolic acids of microbial origin on production of reactive oxygen species in mitochondria and neutrophils. J Biomed Sci. 2012;19:89. doi: 10.1186/1423-0127-19-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayden HJ, Traber MG. Absorption, lipoprotein transport, and regulation of plasma concentrations of vitamin E in humans. J Lipid Res. 1993;34:343–358. [PubMed] [Google Scholar]
- Hughes PE, Tove SB. Occurrence of alpha-tocopherolquinone and alpha-tocopherolquinol in microorganisms. J Bacteriol. 1982;151:1397–1402. doi: 10.1128/jb.151.3.1397-1402.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, Delzenne NM. The gut microbiome as therapeutic target. Pharmacol Ther. 2011;130:202–212. doi: 10.1016/j.pharmthera.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Sternberg DE, Heninger GR, Roth RH. Plasma homovanillic acid as an index of brain dopamine metabolism: enhancement with debrisoquin. Life Sci. 1983;32:2447–2452. doi: 10.1016/0024-3205(83)90370-3. [DOI] [PubMed] [Google Scholar]
- Amin F, Davidson M, Davis KL. Homovanillic acid measurement in clinical research: a review of methodology. Schizophr Bull. 1992;18:123–148. doi: 10.1093/schbul/18.1.123. [DOI] [PubMed] [Google Scholar]
- Russo S, Kema IP, Bosker F, et al. Tryptophan as an evolutionarily conserved signal to brain serotonin: molecular evidence and psychiatric implications. World J Biol Psychiatry. 2009;10:258–268. doi: 10.1080/15622970701513764. [DOI] [PubMed] [Google Scholar]
- Russo S, Kema IP, Haagsma EB, et al. Irritability rather than depression during interferon treatment is linked to increased tryptophan catabolism. Psychosom Med. 2005;67:773–777. doi: 10.1097/01.psy.0000171193.28044.d8. [DOI] [PubMed] [Google Scholar]
- Kurlan R, Goldblatt D, Zaczek R, et al. Cerebrospinal fluid homovanillic acid and parkinsonism in Huntington’s disease. Ann Neurol. 1988;24:282–284. doi: 10.1002/ana.410240221. [DOI] [PubMed] [Google Scholar]
- Ziegler MG, Kennedy B, Holland OB, et al. The effects of dopamine agonists on human cardiovascular and sympathetic nervous systems. Int J Clin Pharmacol Ther Toxicol. 1985;23:175–179. [PubMed] [Google Scholar]
- Norman TR, Chiu E, French MA. Platelet monoamine oxidase activity in patients with Huntington’s disease. Clin Exp Pharmacol Physiol. 1987;14:547–550. doi: 10.1111/j.1440-1681.1987.tb01511.x. [DOI] [PubMed] [Google Scholar]
- Ascherio A, LeWitt PA, Xu K, et al. Urate as a predictor of the rate of clinical decline in Parkinson disease. Arch Neurol. 2009;66:1460–1468. doi: 10.1001/archneurol.2009.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzschild MA, Schwid SR, Marek K, et al. Serum urate as a predictor of clinical and radiographic progression in Parkinson disease. Arch Neurol. 2008;65:716–723. doi: 10.1001/archneur.2008.65.6.nct70003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson Study Group S.-P. D. I. Schwarzschild MA, Ascherio A, et al. Inosine to increase serum and cerebrospinal fluid urate in Parkinson disease: a randomized clinical trial. JAMA Neurol. 2014;71:141–150. doi: 10.1001/jamaneurol.2013.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wu G, Schwarzschild MA. Urate in Parkinson’s disease: more than a biomarker? Curr Neurol Neurosci Rep. 2012;12:367–375. doi: 10.1007/s11910-012-0282-7. doi: 10.1007/s11910-012-0282-7 ; published online Epub Aug. [DOI] [PubMed] [Google Scholar]
- Thomas B, Matson S, Chopra V, et al. A novel method for detecting 7-methyl guanine reveals aberrant methylation levels in Huntington disease. Anal Biochem. 2013;436:112–120. doi: 10.1016/j.ab.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JH, Connor T, Stiles M, et al. Cysteine oxidation within N-terminal mutant huntingtin promotes oligomerization and delays clearance of soluble protein. J Biol Chem. 2011;286:18320–18330. doi: 10.1074/jbc.M110.199448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson W U. S. Patents. 2001. Method of diagnosing or categorizing disorders from biochemical profiles.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.List of known compounds quantified by the LCECA (Liquid Chromatography Electrochemical Array) platform.