Abstract
When transcription regulatory networks are compared among distantly related eukaryotes, a number of striking similarities are observed: a larger-than-expected number of genes, extensive overlapping connections, and an apparently high degree of functional redundancy. It is often assumed that the complexity of these networks represents optimized solutions, precisely sculpted by natural selection; their common features are often asserted to be adaptive. Here, we discuss support for an alternative hypothesis: the common structural features of transcription networks arise from evolutionary trajectories of “least resistance,” that is, the relative ease by which certain types of network structures are formed during their evolution.
Introduction
The complexity of cells continues to fascinate scientists. Two broad views are often advanced to account for such complexity. In one, it is assumed that any complexity must necessarily benefit the cell. Some cell and molecular biologists go even further and discuss how a particular mechanism was “designed” by evolution to be perfectly matched to its task. As with a machine, it is assumed that every molecular nut and bolt must have a purpose. Because this view seems intuitive and relatively simple (after all, examples abound of animals, plants, and microbes adapted to their environments), it is often invoked to explain any aspect of cell and molecular biology. A different view, the one we elaborate here, is embodied in Dobzhansky's famous line, now a cliché, “Nothing in biology makes sense except in the light of evolution.” According to this view, any rationalization of a modern cellular mechanism depends critically on understanding its evolutionary history. We argue that this emphasis on evolutionary history is appropriate for analyzing transcription circuits and for rationalizing their structures. This view has explanatory power in that it can readily account for some of the more bewildering and counterintuitive features of modern transcription circuits; it also gives us insight into the best ways to describe and study such circuits.
In this article, we first review common features of transcription network structures—observed across diverse species—and argue that these similarities cannot be the result of descent from a single ancestral circuit possessing these characteristics. Next, we consider key biochemical and biophysical properties of transcription regulators and cis-regulatory sequences that make certain evolutionary pathways much more probable than others, in part because they circumvent fitness barriers. Finally, we argue that many aspects of transcription circuits, particularly those that seem overly complex and counterintuitive, can be understood as relatively crude products of high-probability evolutionary trajectories rather than as highly optimized, specific solutions.
The arguments discussed in this perspective rely heavily on prior ideas advanced by evolutionary biologists, particularly those ideas concerning the role of non-adaptive mutations in generating complexity (Covello and Gray, 1993; Doolittle, 2013; Force et al., 1999; Gray et al., 2010; Lukes et al., 2011; Lynch, 2007a; 2007b; Lynch et al., 2014; Stoltzfus, 1999; Zuckerkandl, 1997). Although sometimes dismissed as unimportant (or uninteresting), non-adaptive mutations have a profound role in generating evolutionary novelty. Of particular importance is the idea, sometimes called “constructive neutral evolution,” that changes that arise neutrally can open up new evolutionary pathways; in some cases, changes that arose non-adaptively can become essential for function if they are incorporated into subsequent layers of evolutionary change. Through this sequence of events, molecular and organismal complexity can be increased through non-adaptive mutations. As we discuss, the biochemical and biophysical properties of transcription network components support the idea that their evolutionary trajectories—which depend on mutation, selection, and genetic drift—lead to specific types of structures, particularly those that alter circuits without breaking them. Because their components are highly conserved across eukaryotes, we argue that it is inevitable that networks across a wide variety of species tend to converge on similar structures. We propose that these common structures are not likely to represent optimized solutions, but are, in a sense, “default” evolutionary products.
Depictions of Transcription Networks
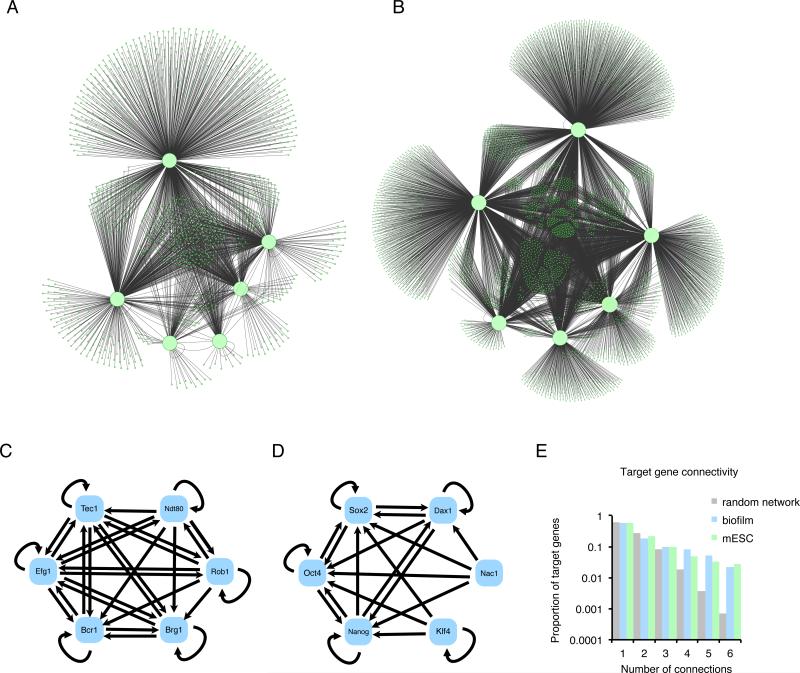
For the most part, genome-wide studies of transcriptional network structures have been largely descriptive, often culminating in large “hairball” diagrams such as those depicted in Fig. 1. Their complexity has made it difficult to formulate simple conclusions regarding the logic or outputs of these networks, particularly since quantitative parameters and dynamic measurements are typically lacking.
Fig. 1.
Typical depictions of transcription regulatory networks. (A) The C. albicans biofilm network (Nobile et al., 2012) and (B) the M. musculus embryonic stem cell network (Kim et al., 2008) are depicted as graphs where balls represent genes and lines represent the binding of transcription regulators to intergenic regions. Master transcription regulators (defined in the text) are shown as large balls and “target genes” are shown as small balls. For the stem cell network, only the six most heavily connected transcription regulators are shown. (C and D) Close-up of the core of each network, showing only the binding connections between the master transcription regulators. Directionality of the connection is indicated by arrows. Note that the arrows refer only to binding connections and do not imply that the connection activates the recipient gene. (C) C. albicans biofilm, (D) mouse stem cell networks. (E) The degree of connectivity for nodes in the two networks. The two biological networks show a larger proportion of nodes with high connectivity than would be found in a random network (Lee et al., 2002).
Although there are many components of gene expression networks, we will focus here on only two key elements, transcription regulators and cis-regulatory sequences. We define transcription regulators as sequence-specific DNA binding proteins that control the transcription of specific genes by binding to cis-regulatory sequences, short (typically 6-15 nucleotides) DNA sequences. It is the distribution of these cis-regulatory sequences across the genome that largely specifies the time, place, and rate of each gene's transcription; this information is “read” by transcription regulators, whose binding to DNA specifies, often through a complex series of downstream steps, the rate of transcription of the gene. Although in many eukaryotic species, cis-regulatory sequences are typically located within several thousand nucleotide pairs of the genes they control, in plants and animals, they can be spread out over hundreds of thousands of nucleotide pairs. Nearly all eukaryotic genes are directly controlled by more than one transcription regulator, and most genes respond to dozens of regulators, specified by the identity and arrangement of their cis-regulatory sequences. We also know, from decades of “promoter bashing” experiments, that cis-regulatory sequences can be moved from one gene to another (and from one species to another) and still retain much of their specificity to direct transcription. Finally, transcription regulators typically bind cooperatively to DNA, a fundamental property that, as we shall discuss, has important implications for network evolution.
Many additional proteins besides transcription regulators are needed to transcribe a gene (for example, RNA polymerase and chromatin remodeling complexes), but it is a useful simplification to consider a transcription network as being composed of direct binding connections between transcription regulators and genes (or more precisely, the cis-regulatory sequences of that gene). This information is summarized in diagrams such as those in Fig. 1.
If a given transcription regulator occupies the cis-regulatory sequences associated with a gene in vivo (as determined, for example, by a chromatin immunoprecipitation experiment), we will refer to that gene as a target gene of the transcription regulator. We realize that this convention does not require that the binding of the regulator to DNA be proven to be functional in the organism. There are three reasons for nonetheless including these connections in diagrams such as those in Figure 1. 1) The “function” of a given connection has been demonstrated in only a small number of cases; for the great majority of reliable binding data, no direct test has been performed. 2) Although many approaches (e.g. conservation across species or experimental mutation of the cis-regulatory sequence) can provide strong evidence for function, it is impossible to rigorously establish that a binding connection is non-functional under all possible conditions. 3) The DNA binding properties of transcription regulators predict that in vivo, there will be some degree of non-functional binding (Lin and Riggs, 1975). Such “non-functional” binding events are nonetheless real properties of evolving transcription networks.
Depictions of transcription networks based on these conventions often show “master transcription regulators” and target genes as nodes (balls) and regulatory interactions as edges (lines) between these nodes (Fig. 1 A and B). Although the term master transcription regulator is used in many different ways in the literature (Chan and Kyba, 2013), we define it, for the purpose of this article, as a transcription regulator (1) whose presence is required to carry out the specific biological process controlled by the network and (2) whose ectopic expression alone or in combination with other regulators, can trigger the biological process even in the absence of the appropriate developmental or environmental signals (Halder et al., 1995; Takahashi and Yamanaka, 2006; Tapscott et al., 1988; Tursun et al., 2011; Vierbuchen et al., 2010; Zordan et al., 2007).
Common Features of Transcription Networks
We first compare two transcription networks from two different species and that coordinate two different biological processes, but were deduced by similar methodologies. The network specifying the embryonic stem cell state (pluripotency) was chosen because it has been studied extensively by numerous labs and is supported by multiple studies (Boyer et al., 2005; Kim et al., 2008). For comparison, we chose the circuit controlling biofilm development in the pathogenic yeast C. albicans, a network this lab has studied extensively (Nobile et al., 2012). The two networks are depicted in Fig. 1 A and B using a similar graphical format.
These two circuits were chosen, in part, because they might be expected from first principles to have little in common. Mammals and yeast diverged from a common ancestor approximately 1.5 billion years ago (Wang et al., 1999), and there is little conceptual similarity between biofilm formation and pluripotency. Moreover, the two networks appear to have evolved independently, well after the two lineages split (see below). Yet, the overall structures of the two networks, as depicted in the figure, appear remarkably similar. Both C. albicans biofilm development and mouse embryonic stem cell pluripotency are controlled by a set of master transcription regulators that form binding connections among themselves (Fig. 1 C and D) and to the regulatory regions of over a thousand target genes, with multiple master regulators typically binding to the same targets (Fig. 1 A, B, and E, Table 1). In both cases a substantial proportion of the target genes are other transcription regulators, indicating substantial indirect regulation of additional genes. The C. albicans genome is significantly smaller than the mouse genome, yet each network comprises about one fifth of the genes in their respective genomes.
Table 1.
Metrics comparing C. albicans biofilm and mouse embryonic stem cell networks.
| Biofilm | mESC | |
|---|---|---|
| Master transcription regulators | 6 | 6 |
| Connections | 2018 | 7234 |
| Target genes | 1037 | 3968 |
| Fraction of genome in network | 0.17 | 0.21 |
| Binding feed forward loops | 3145 | 6886 |
| Nonfunctional binding events | 1207 | unknown |
Connections and genes were determined by whole-genome chromatin immunoprecipitation . “Non-functional binding” was defined as genes whose expression does not change when a direct regulator is deleted from the genome.
Although the two networks control very different processes, their master regulators have similar properties. In both networks, these regulators contain sequence-specific DNA binding domains such as homeodomains, MADS domains, and zinc fingers (Weirauch and Hughes, 2011). In some cases the cis-regulatory sequence recognized by a given transcription regulator has not changed significantly since the divergence of yeast and mammals (Hayes et al., 1988). Moreover, transcription regulators from one species (e.g. Gal4 from brewer's yeast) can control transcription in many different species (e.g. Fischer et al., 1988; Kakidani and Ptashne, 1988). Key aspects of the C. albicans biofilm circuit were formed well after C. albicans diverged from closely related, non-pathogenic yeasts (Nobile et al., 2012), providing additional support for the conclusion that the structure of the yeast and mouse networks evolved independently—even though the master transcription regulators were present in the common ancestor of both species. These ideas are consistent with the generalization that, although the transcription regulators and their recognition sequences are often deeply conserved, transcriptional networks themselves are rewired at a rapid pace during evolution (reviewed in Li and Johnson, 2010; Tuch et al., 2008b; Weirauch and Hughes, 2010; Wray et al., 2003). (Like most generalizations in biology, this one has important exceptions. See for example (Baker et al., 2011; Sayou et al., 2014) for cases where the DNA-binding specificity of a regulator has changed dramatically over relatively short periods of evolutionary history.) In any case, it is highly unlikely that any of the connections between regulators and target genes in the mouse pluripotency and yeast biofilm networks are conserved from a common ancestor, despite the deep conservation of the DNA-binding properties of the master transcription regulators.
If transcription networks evolve rapidly, why do the embryonic stem cell and biofilm networks appear structurally similar? One hypothesis is that elaborate and interconnected networks such as these represent optimized solutions for organizing biological processes. According to this view, the similarities between these networks result primarily from selection and reflect the same underlying requirements for transcriptional logic, for example modularity or robustness. Some features of the circuits (for example, the large number of direct and indirect feedback loops) may well reflect these requirements in a general way, but the similarities seem too great to be readily explained this way. We propose instead that circuit architecture is dominated by severe constraints on the evolutionary trajectories available for network evolution. Allowable trajectories, we argue, must (A) be probable from a biochemical and biophysical standpoint and (B) avoid fitness barriers; that is, the allowable trajectories will typically not pass through stages in which the circuit becomes broken and non-functional (Carroll, 2008; Stern and Orgogozo, 2009; Wagner, 2003).
The components of circuits (DNA-binding proteins and cis-regulatory sequences) and their properties (for example, cooperative binding) are common to fungi and mammals, and we suggest that the available trajectories for evolutionary change rely heavily on these properties coupled with the avoidance of fitness barriers. According to this view, the similarities among independently derived transcription networks arise primarily from the “low-energy” pathways of evolution rather than the selective pressures specific to one circuit or another. In the following sections, we examine specific properties of networks in more detail and consider the extent to which this idea can account for them.
Size Accrues
One surprising feature of many transcriptional networks is their large size (Borneman et al., 2006; Hernday et al., 2013; Iyer et al., 2001; Junion et al., 2012; Kim et al., 2008; Liang and Biggin, 1998; MacArthur et al., 2009; Mastick et al., 1995; Nobile et al., 2012; Novershtern et al., 2011). As mentioned above, the yeast biofilm network and the mouse embryonic stem cell network, as depicted in Fig. 1, incorporate approximately one fifth of the protein-coding genes in their respective genomes. Although a few examples have been described where eukaryotic transcription networks appear small (e.g. the mating type specification circuit (Galgoczy et al., 2004) and the galactose regulatory circuit (Ren et al., 2000), both from S. cerevisiae), the majority of networks that have been carefully studied using full-genome methods appear larger and more complex than might have been expected.
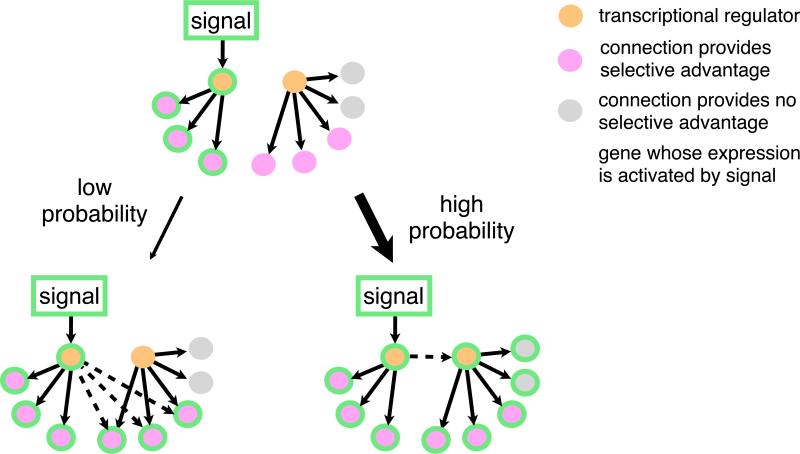
Why is the typical network so large? In contrast to a model where every connection in a network serves a specific function in that network, we propose that many target genes in networks are incorporated non-adaptively during the formation of the network. Fig. 2 shows a hypothetical example in which a new response to a signal evolves under selection. If there is an advantage of gaining regulation of multiple target genes in response to the signal, it is much more probable to gain a binding site upstream of a single transcription regulator of those genes than to gain binding sites for each individual target gene (Gerhart and Kirschner, 1997; Raff and Kaufman, 1983). Moreover, because most proteins work in groups, any selective advantage of incorporating a new gene into an existing circuit may not be realized until several genes are brought into the circuit, making the “gene-by-gene” model even less probable. The “regulator-first” model would result in a new regulator being incorporated into the old circuit along with all the pre-existing target genes of this regulator. Some of these target genes may be extraneous with respect to the new circuit, but, if the original function of the regulator is retained, these connections would nonetheless be maintained by purifying selection. According to this simple idea, newly formed networks would be expected to contain connections nonessential to that network and would therefore be predicted to be larger than strictly necessary.
Fig. 2.
Pathways for evolving a new transcriptional response to a signal. In this hypothetical scenario, incorporation of three additional genes into the signaling pathway confers a selective advantage. Two alternative paths are possible: (1) The genes could be incorporated one by one through independent changes in their cis-regulatory sequences. (2) The new genes could be incorporated through a single incorporation of the transcription regulator that already controls them. If the incorporation of multiple target genes is needed to confer an increase in fitness, gain of regulation of the transcription regulator will be more probable than the gain of each individual target. As the number of target genes increases, the difference in probability will be greater. Note that the second scenario will likely incorporate additional genes non-adaptively.
Experimental evidence suggests that the “regulators first” scenario is common; that is, networks often form by incorporating new regulators rather than by incorporating individual target genes (Frankel et al., 2012; Monteiro, 2012; Pires et al., 2013). For example, the red wing color in Heliconius butterflies takes place through repeated rewiring of the expression pattern of the transcription regulator optix rather than one-by-one incorporation of individual target genes (Reed et al., 2011). A second example is found in networks regulating morphological transitions in different yeast species; the regulator Tec1 and its target genes have been incorporated into environmental response networks multiple times (Mösch and Fink, 1997; Nobile et al., 2012; Schweizer et al., 2000). Thus, the regulators-first model of transcription network formation is predicted to lead to the expansion of circuit size beyond that strictly required for the new response. Although this model might be expected to create detrimental pleiotropic effects of expressing many extraneous genes at once, modeling and experimental evidence suggests that this pleiotropy can be alleviated gradually over time (Pavlicev and Wagner, 2012; Qian et al., 2012) or even avoided altogether (Stern and Orgogozo, 2009). It is important to note that these observations probably do not apply to the regulatory networks of smaller genomes in species with very large population sizes where selection is the dominant evolutionary force. For example, the regulatory network of lambda phage is small and each component and connection contributes to the function of the circuit (Little, 2010).
Gains in Interconnectedness
Another common feature of transcription networks across diverse species is the degree of connectivity between different transcription regulators and between these regulators and their targets (Borneman et al., 2006; Boyle et al., 2014; Junion et al., 2012; Kim et al., 2008; MacArthur et al., 2009; Nobile et al., 2012; Novershtern et al., 2011; Reece-Hoyes et al., 2013). We define this degree as the number of connections made between the master transcription regulators and a given target gene. For example, if a given target gene in the C. albicans biofilm network is bound by three different master transcription regulators, the degree of connection of that target gene is three. The degree distributions for the yeast and mouse cases show a similar profile (Fig. 1 E), one that shows a higher degree of connection than would be predicted for a randomly distributed network (Featherstone and Broadie, 2002; Guelzim et al., 2002).
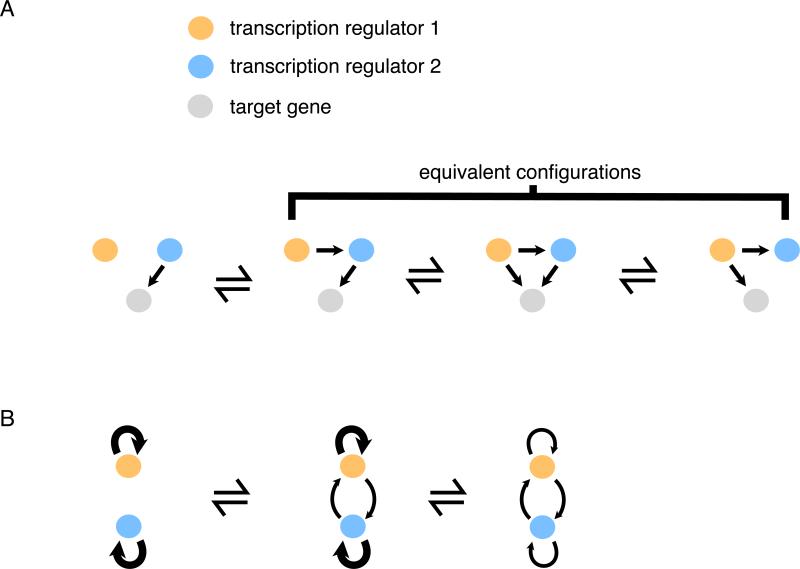
Rather than speculating what this high degree of interconnectedness might “do for the cell,” we subscribe to the simpler hypothesis that it results from the neutral (i.e. non-adaptive) gains of regulatory connections that inevitably occur over time, particularly in small populations (Lynch, 2007a; Stone and Wray, 2001). This idea can be explained by considering a simple situation, one that would be predicted to arise often by the “regulator-first” model (Fig. 3A). Here, one transcription regulator (blue) regulates the target gene (grey). In the regulators first model, a second transcription regulator (orange) gains control of the blue regulator, and indirectly, the grey target gene. Next, the interconnectedness in this simple scheme would increase if the orange regulator gained direct control of the grey target gene.
Fig. 3.
The tendency for co-expressed regulators to become interconnected. (A) Once the orange regulator gains control of the blue regulator, causing them to be expressed at the same time, target genes can, through neutral evolution, rapidly gain and lose binding sites for the two regulators. (B) Two regulators expressed at the same time each have positive feedback. Subsequently, neutral gains of reciprocal regulation between the regulators can occur while preserving the overall positive feedback control. Over evolutionary time, positive feedback distributed over both regulators (rather than purely autonomous loops) is predicted to occur.
Why would this happen? It has been argued, using population genetic models, that such connections are predicted to form non-adaptively, that is, without selection for improvement of the circuit (Lynch, 2007a). According to this view, the additional connections do not disrupt the existing regulation, and they arise through random mutations that produce a new DNA binding site for a transcription regulator. Thus, this increase in total number of connections is predicted to occur, in essence, because nothing stops it. Such a change, even though it arose non-adaptively, can become fixed if subsequent evolutionary changes in the network render its loss detrimental.
Although it might seem counterintuitive that new circuit connections can form non-adaptively, the biochemical features of transcription regulators and cis-regulatory sequences predict this. As has been pointed out many times, because cis-regulatory sequences are usually short and somewhat degenerate, there is a significant probability that new point mutations will readily create matches with existing transcription regulators. Given that target genes often have long intergenic regions in which cis-regulatory sequences can function, many target genes would be predicted to develop multiple connections (Lynch, 2007a; Paixão and Azevedo, 2010; Stone and Wray, 2001). Although these additional binding sites may not be under purifying selection (unless the original connection is lost or some other change in the network renders their loss detrimental), they would be predicted to form at a high enough frequency to ensure an appreciable steady state level of such connections, despite the losses due to mutation.
These same forces are also predicted to lead to the high interconnectedness observed between the master regulators themselves (Fig. 1B). Many transcription regulators control their own transcription (Bateman, 1998; Kiełbasa and Vingron, 2008; Lee et al., 2002). When two such regulators function at the same time and place (although not necessarily in the same biological function), over time they may acquire regulation of each other through the gain of cis-regulatory sequences (Fig. 3B). This reciprocal regulation would be redundant (at least in a general sense) with the auto-regulation of each of the transcription regulators themselves and could partially replace it over time, resulting in interlocking, auto-regulatory master regulators of the type we see in Fig. 1.
Various types of simulations both support these ideas and highlight additional features that promote high degrees of circuit connectivity. For example long regulatory regions and high recombination rates promote the evolution of multiple cis-regulatory sites by non-adaptive mechanisms (Lynch, 2007a; Ruths and Nakhleh, 2012). Similarly, the greater the permissible degeneracy of cis-regulatory sequences, the greater is the probability of multiple connections (Paixão and Azevedo, 2010).
Support for these ideas also comes from direct observation of transcription circuits in different species. First, as previously pointed out, independently evolved circuits show similar, high degrees of connectivity. Recent studies of transcription networks by the Encyclopedia Of DNA Elements (ENCODE) project have greatly increased the number of examples where network structure is observed to be highly similar across organisms, in this case humans, mice, Caenorhabditis elegans, and Arabadopsis thaliana (Boyle et al., 2014; Stergachis et al., 2014; Sullivan et al., 2014). Although some similarities (e.g. the same master regulator controlling the same biological process in two different species) are clearly conserved from a common ancestor, we argue that similarities in overall network structure are largely due to the pathways we have outlined, in which non-adaptive evolution is a major force.
Second, there are several documented examples where evolutionary rewiring of an entire network has occurred without apparent changes in the output (Baker et al., 2012; Lavoie et al., 2010; Ludwig et al., 2000; Moses et al., 2006; Schmidt et al., 2010; Tanay et al., 2005; Tsong et al., 2003; 2006). These studies indicate that, even as the output of a circuit is maintained by stabilizing selection, the individual connections may be free to drift to new configurations.
Third, many connections in networks appear unimportant as assessed by conventional experiments (Fisher et al., 2012; Whitfield et al., 2012). Although it is virtually impossible to prove that a connection is non-functional under all conceivable conditions, several types of experiments suggest that parts of circuits may be functionally unimportant. For example, many direct target genes show no change in transcript levels when a regulator that binds to the gene is deleted or reduced in expression. This behavior describes the majority of the C. albicans biofilm network: 60% of binding events do not elicit expression changes when the regulator is deleted, with the provision that biofilms were monitored under a narrow range of conditions. Moreover, many target genes, when deleted, do not appear to compromise the output of the circuit. Although these results can be explained away by circuit compensation, redundancy, inability to monitor a wide variety of conditions, and the like, we suggest it is highly plausible, based on the arguments made above, that many circuit connections simply do not contribute to the output. In any case, many observations made on modern circuits are consistent with a model whereby much of the interconnectedness of transcription circuits have arisen non-adaptively, simply as a consequence of the ease of forming new connections.
Cooperative binding produces connectivity
Cooperative binding is a near-universal feature of eukaryotic transcription regulators, and next we discuss how this property increases the ease of forming new circuit connections and thereby shapes circuit structures. We use the term cooperative binding to mean that the binding of one transcription regulator to a cis-regulatory sequence increases the probability that another will occupy a nearby sequence. Mechanistically, this can occur through three distinct means: 1) competitive displacement of nucleosomes, through which binding of one transcription regulator to DNA can increase the accessibility of DNA to a second regulator, thereby increasing its occupancy (Polach and Widom, 1996); 2) a direct, weak, favorable, physical interaction between the two regulators (Johnson et al., 1979); and 3) physical interactions with additional non-DNA-binding proteins that stabilize binding of both of the transcription regulators on DNA (Ptashne and Gann, 2002).
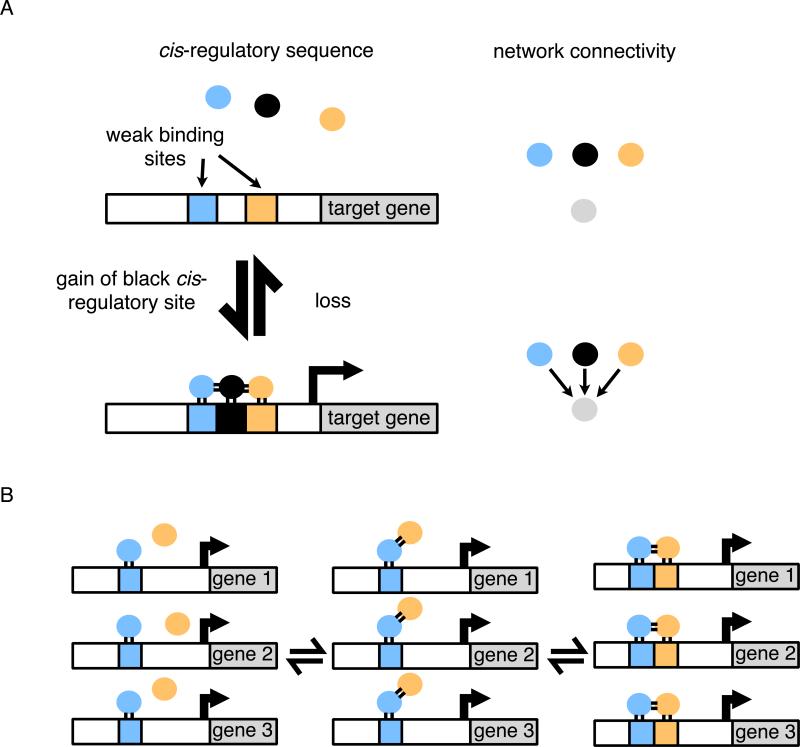
All three forms of cooperative binding would favor the drift of circuits into states of high connectivity by relaxing the cis-regulatory sequence requirements needed for a second transcription regulator to be added to a target gene. This idea has an additional implication: cooperativity means that a single change in a cis-regulatory sequence or a regulatory protein can establish or eliminate numerous connections. For example, gain of a cis-regulatory site for one regulator may allow other regulators to occupy nearby, previously existing weak sites (Fig. 4A). Acquisition of a new, favorable protein-protein interaction between transcriptional regulators can have an even more profound effect. Here, cooperative binding can, at least in principle, catalyze the rewiring of an entire set of genes (Tuch et al., 2008a). In this scenario, the gain of a protein-protein interaction leads to cooperative binding of two regulators when a binding site for only one of the regulators is present (Fig. 4B). Following this gain, there can be gene-by-gene gains of cis-regulatory sequences for the second regulator. The dually regulated set of genes can then diversify, loosening their connections with the original regulator and strengthening the new ones. In this way, gene sets can be “handed off” from one regulator to another in the course of evolution, a type of change that seems common (Baker et al., 2012; Martchenko et al., 2007; Tanay et al., 2005; Tsong et al., 2006).
Fig. 4.
Gain of multiple regulatory connections through cooperative binding. (A) Cooperativity between regulators allows binding energy to be shared between protein-DNA and protein-protein interactions. When a strong binding site is gained for one regulator, this may increase the occupancy of regulators on nearby weak binding sites that would otherwise be unoccupied. The effect is a concerted increase in connectivity of that target gene. (B) The gain of a protein-protein interaction between the blue and orange regulators results in a concerted rewiring of the entire set of genes. As shown in the third panel, direct binding sites for the orange regulator can be gained step-wise at each gene individually without disrupting the circuit. Finally (not shown), the circuit can diversify by moving between equivalent configurations.
The important point is, to influence transcription, transcription regulators must occupy cis-regulatory sequence in the cell, but the energy needed for this occupancy can be shared out between protein-DNA and protein-protein interactions. As individual interactions are strengthened and weakend over evolutionary time, the circuit configuration can drift between different “energy-sharing” solutions. Cooperative binding, combined with the ease of strengthening and weakening cis-regulatory sequences by random mutations, predicts that networks will drift to high degrees of connectivity—a prediction that is supported experimentally (Baker et al., 2012; Lynch and Wagner, 2008; Stefflova et al., 2013; Tsong et al., 2006). Thus, any two regulators that overlap in their expression would be predicted to share a fraction of their targets under the conditions they are both expressed (Lynch, 2007a), unless these additional connections are specifically selected against.
Formation of Common Network Motifs
One strategy to simplify and understand the function of complex transcription networks has been to search for network motifs—configurations of regulators and target genes that occur repeatedly within networks (Alon, 2007; Davidson, 2010). One of the most common motifs is a simple feed-back loop, whereby a transcription regulator controls (directly or indirectly) its own rate of synthesis (Bateman, 1998; Lee et al., 2002). Feedback is a hallmark of many different processes in biology, and it seems likely that, in its most general form (but not necessarily in its detailed mechanism), it is often under purifying selection.
But what about motifs other than positive feedback loops? A more complex network motif known as a feed forward loop (in which one regulator controls a second regulator and both control the same target gene) is overrepresented in biological networks (Milo et al., 2002). Depending on the parameters of binding, a given feed-forward loop can, in principle, perform logic operations such as pulse detection or expression delay (Alon, 2007; Davidson, 2010). However, it is currently unclear whether the majority of naturally occurring feed forward loops meet the types of parameter requirements needed for these behaviors.
There are thousands of feed-forward loops embedded in the two networks of Fig. 1. We note that feed-forward loops are common byproducts of the evolutionary paths diagramed in Figs. 2 and 3, and thus, the preponderance of feed-forward loops in biological networks may be a result of the same non-adaptive processes that result in large network size and interconnectedness. Indeed, it has been explicitly proposed that many network motifs have arisen as a result of neutral evolutionary processes rather than selection for a particular function of the motif itself (Cordero and Hogeweg, 2006; Ingram et al., 2006; Ruths and Nakhleh, 2013; Ward and Thornton, 2007). These ideas contrast with models where each feed-forward loop in the network possesses optimized parameters that specify a particular transcriptional input-output relationship.
We note that feed-forward loops may also represent non-adaptive intermediates between alternative forms of transcriptional regulation (Fig. 5). Rewiring of transcription networks, at least in some cases, proceeds through intermediates that are regulated by both the ancestral and derived mechanisms (Li and Johnson, 2010), allowing the regulatory output to be preserved during the rewiring. Although they might arise non-adaptively, feed-forward loops can serve as redundant intermediates between the ancestral and derived states, and thus many observed examples of transcription network rewiring may be a simple consequence of the high frequency with which feed forward loops are formed by neutral evolution (Lynch, 2007a).
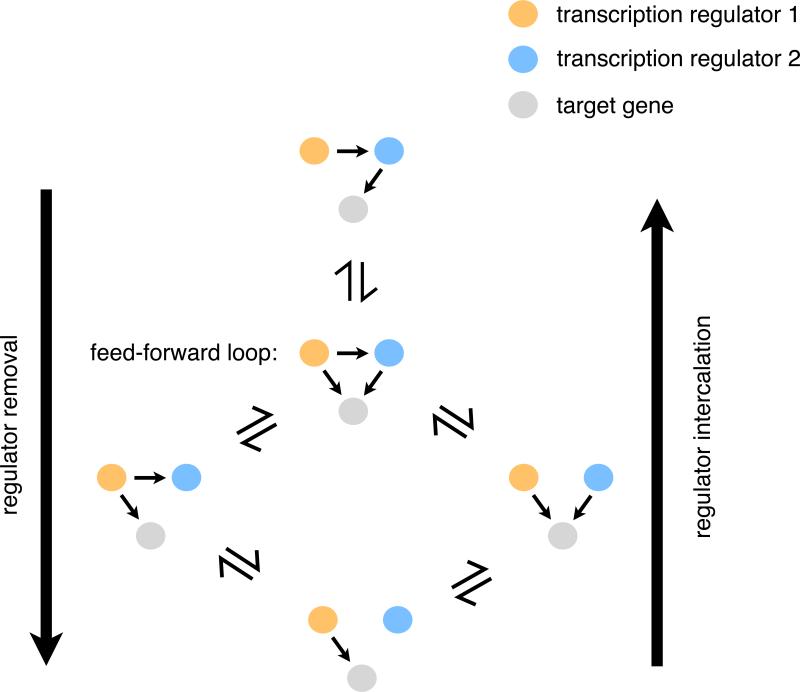
Fig. 5.
Pathways for incorporation or removal of a transcription regulator without breaking the network. Removal of the blue regulator from the linear regulatory pathway shown in the top network diagram can proceed by first forming a feed-forward loop. Subsequent loss of the connections between the red and blue regulator and between the blue regulator and the target gene will completely remove the blue regulator from the network as shown in the bottom diagram. The opposite process starting from the bottom diagram and proceeding to the top results in intercalation of the blue regulator into the pathway. If the functions of the blue and red regulators are redundant in the context of the network, the network can drift between these configurations over evolutionary time without compromising the output of the circuit.
Conclusion
Genomes evolve under selective pressure, but we no longer expect their structures to be orderly, logical affairs dictated by underlying design principles. Here we have argued that there is no reason to expect transcription circuit networks to be any different. We argue that the drift of transcription networks to steady-state levels of high complexity and interconnection is consistent with the biochemical and biophysical properties of regulatory proteins and cis-regulatory sequences, particularly the cooperative binding of transcription regulators to DNA. Combined with universal processes of evolution such as mutation, genetic drift, and selection, network complexity is predicted, from first principles, to be a natural consequence. Complex structures, even if they arise non-adaptively, can nonetheless serve as substrates for future evolutionary innovations, or be locked in place by secondary changes; thus, they can be retained by purifying selection even though they arose non-adaptively and are not optimized solutions. If transcription circuits are considered as relatively crude products whose structures are dominated by high-probability evolutionary pathways, many of their more baffling features—size, similarity across diverse species, complexity, redundancy, interconnectedness, for example—begin to make conceptual sense.
Acknowledgements
We would like to thank C. Nobile, E. Mancera, S. Coyle, and V. Hanson-Smith for helpful comments on the manuscript. We would also like to thank K. Carniol and two anonymous reviewers for providing critical feedback on the content and language. The work was supported by grant R01 GM037049 from the National Institutes of Health. T.R.S. was supported by a Graduate Research Fellowship from the National Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Trevor R Sorrells, Department of Biochemistry & Biophysics, Tetrad Graduate Program University of California, San Francisco San Francisco, CA 94158, USA.
Alexander D Johnson, Department of Microbiology & Immunology, Tetrad Graduate Program University of California, San Francisco San Francisco, CA 94158, USA.
References
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Baker CR, Booth LN, Sorrells TR, Johnson AD. Protein modularity, cooperative binding, and hybrid regulatory States underlie transcriptional network diversification. Cell. 2012;151:80–95. doi: 10.1016/j.cell.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CR, Tuch BB, Johnson AD. Extensive DNA-binding specificity divergence of a conserved transcription regulator. Proc Natl Acad Sci USA. 2011;108:7493–7498. doi: 10.1073/pnas.1019177108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman E. Autoregulation of eukaryotic transcription factors. Prog Nucleic Acid Res Mol Biol. 1998;60:133–168. doi: 10.1016/s0079-6603(08)60892-2. [DOI] [PubMed] [Google Scholar]
- Borneman AR, Leigh-Bell JA, Yu H, Bertone P, Gerstein M, Snyder M. Target hub proteins serve as master regulators of development in yeast. Genes Dev. 2006;20:435–448. doi: 10.1101/gad.1389306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Araya CL, Brdlik C, Cayting P, Cheng C, Cheng Y, Gardner K, Hillier LW, Janette J, Jiang L, et al. Comparative analysis of regulatory information and circuits across distant species. Nature. 2014;512:453–456. doi: 10.1038/nature13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Chan SS-K, Kyba M. What is a Master Regulator? J Stem Cell Res Ther. 2013;3 doi: 10.4172/2157-7633.1000e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero OX, Hogeweg P. Feed-forward loop circuits as a side effect of genome evolution. Mol Biol Evol. 2006;23:1931–1936. doi: 10.1093/molbev/msl060. [DOI] [PubMed] [Google Scholar]
- Covello PS, Gray MW. On the evolution of RNA editing. Trends Genet. 1993;9:265–268. doi: 10.1016/0168-9525(93)90011-6. [DOI] [PubMed] [Google Scholar]
- Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF. Is junk DNA bunk? A critique of ENCODE. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1221376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone DE, Broadie K. Wrestling with pleiotropy: genomic and topological analysis of the yeast gene expression network. Bioessays. 2002;24:267–274. doi: 10.1002/bies.10054. [DOI] [PubMed] [Google Scholar]
- Fischer JA, Giniger E, Maniatis T, Ptashne M. GAL4 activates transcription in Drosophila. Nature. 1988;332:853–856. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- Fisher WW, Li JJ, Hammonds AS, Brown JB, Pfeiffer BD, Weiszmann R, MacArthur S, Thomas S, Stamatoyannopoulos JA, Eisen MB, et al. DNA regions bound at low occupancy by transcription factors do not drive patterned reporter gene expression in Drosophila. Proc Natl Acad Sci USA. 2012;109:21330–21335. doi: 10.1073/pnas.1209589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Wang S, Stern DL. Conserved regulatory architecture underlies parallel genetic changes and convergent phenotypic evolution. Proc Natl Acad Sci USA. 2012;109:20975–20979. doi: 10.1073/pnas.1207715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgoczy DJ, Cassidy-Stone A, Llinás M, O'Rourke SM, Herskowitz I, DeRisi JL, Johnson AD. Genomic dissection of the cell-type-specification circuit in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2004;101:18069–18074. doi: 10.1073/pnas.0407611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart J, Kirschner M. Cells, Embryos and Evolution. Wiley-Blackwell; 1997. [Google Scholar]
- Gray MW, Lukes J, Archibald JM, Keeling PJ, Doolittle WF. Cell biology. Irremediable complexity? Science. 2010;330:920–921. doi: 10.1126/science.1198594. [DOI] [PubMed] [Google Scholar]
- Guelzim N, Bottani S, Bourgine P, Képès F. Topological and causal structure of the yeast transcriptional regulatory network. Nat Genet. 2002;31:60–63. doi: 10.1038/ng873. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Hayes TE, Sengupta P, Cochran BH. The human c-fos serum response factor and the yeast factors GRM/PRTF have related DNA-binding specificities. Genes Dev. 1988;2:1713–1722. doi: 10.1101/gad.2.12b.1713. [DOI] [PubMed] [Google Scholar]
- Hernday AD, Lohse MB, Fordyce PM, Nobile CJ, DeRisi JL, Johnson AD. Structure of the transcriptional network controlling white-opaque switching in Candida albicans. Mol Microbiol. 2013;90:22–35. doi: 10.1111/mmi.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram PJ, Stumpf MPH, Stark J. Network motifs: structure does not determine function. BMC Genomics. 2006;7:108. doi: 10.1186/1471-2164-7-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Meyer BJ, Ptashne M. Interactions between DNA-bound repressors govern regulation by the lambda phage repressor. Proc Natl Acad Sci USA. 1979;76:5061–5065. doi: 10.1073/pnas.76.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junion G, Spivakov M, Girardot C, Braun M, Gustafson EH, Birney E, Furlong EEM. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell. 2012;148:473–486. doi: 10.1016/j.cell.2012.01.030. [DOI] [PubMed] [Google Scholar]
- Kakidani H, Ptashne M. GAL4 activates gene expression in mammalian cells. Cell. 1988;52:161–167. doi: 10.1016/0092-8674(88)90504-1. [DOI] [PubMed] [Google Scholar]
- Kiełbasa SM, Vingron M. Transcriptional autoregulatory loops are highly conserved in vertebrate evolution. PLoS ONE. 2008;3:e3210. doi: 10.1371/journal.pone.0003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie H, Hogues H, Mallick J, Sellam A, Nantel A, Whiteway M. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 2010;8:e1000329. doi: 10.1371/journal.pbio.1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Li H, Johnson AD. Evolution of transcription networks--lessons from yeasts. Curr Biol. 2010;20:R746–R753. doi: 10.1016/j.cub.2010.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Biggin MD. Eve and ftz regulate a wide array of genes in blastoderm embryos: the selector homeoproteins directly or indirectly regulate most genes in Drosophila. Development. 1998;125:4471–4482. doi: 10.1242/dev.125.22.4471. [DOI] [PubMed] [Google Scholar]
- Lin S, Riggs AD. The general affinity of lac repressor for E. coli DNA: implications for gene regulation in procaryotes and eucaryotes. Cell. 1975;4:107–111. doi: 10.1016/0092-8674(75)90116-6. [DOI] [PubMed] [Google Scholar]
- Little JW. Evolution of complex gene regulatory circuits by addition of refinements. Curr Biol. 2010;20:R724–R734. doi: 10.1016/j.cub.2010.06.028. [DOI] [PubMed] [Google Scholar]
- Ludwig MZ, Bergman C, Patel NH, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- Lukes J, Archibald JM, Keeling PJ, Doolittle WF, Gray MW. How a neutral evolutionary ratchet can build cellular complexity. IUBMB Life. 2011;63:528–537. doi: 10.1002/iub.489. [DOI] [PubMed] [Google Scholar]
- Lynch M. The evolution of genetic networks by non-adaptive processes. Nat Rev Genet. 2007a;8:803–813. doi: 10.1038/nrg2192. [DOI] [PubMed] [Google Scholar]
- Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci USA. 2007b;104:8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Field MC, Goodson HV, Malik HS, Pereira-Leal JB, Roos DS, Turkewitz AP, Sazer S. Evolutionary cell biology: two origins, one objective. Proceedings of the National Academy of Sciences. 2014;111:16990–16994. doi: 10.1073/pnas.1415861111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch VJ, Wagner GP. Resurrecting the role of transcription factor change in developmental evolution. Evolution. 2008;62:2131–2154. doi: 10.1111/j.1558-5646.2008.00440.x. [DOI] [PubMed] [Google Scholar]
- MacArthur S, Li X-Y, Li J, Brown JB, Chu HC, Zeng L, Grondona BP, Hechmer A, Simirenko L, Keränen SVE, et al. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 2009;10:R80. doi: 10.1186/gb-2009-10-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martchenko M, Levitin A, Hogues H, Nantel A, Whiteway M. Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr Biol. 2007;17:1007–1013. doi: 10.1016/j.cub.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastick GS, McKay R, Oligino T, Donovan K, López AJ. Identification of target genes regulated by homeotic proteins in Drosophila melanogaster through genetic selection of Ultrabithorax protein-binding sites in yeast. Genetics. 1995;139:349–363. doi: 10.1093/genetics/139.1.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Monteiro A. Gene regulatory networks reused to build novel traits: co-option of an eye-related gene regulatory network in eye-like organs and red wing patches on insect wings is suggested by optix expression. Bioessays. 2012;34:181–186. doi: 10.1002/bies.201100160. [DOI] [PubMed] [Google Scholar]
- Moses AM, Pollard DA, Nix DA, Iyer VN, Li X-Y, Biggin MD, Eisen MB. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput Biol. 2006;2:e130. doi: 10.1371/journal.pcbi.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösch HU, Fink GR. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. A Recently Evolved Transcriptional Network Controls Biofilm Development in Candida albicans. Cell. 2012;148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paixão T, Azevedo RBR. Redundancy and the evolution of cis- regulatory element multiplicity. PLoS Comput Biol. 2010;6:e1000848. doi: 10.1371/journal.pcbi.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlicev M, Wagner GP. A model of developmental evolution: selection, pleiotropy and compensation. Trends Ecol Evol (Amst) 2012 doi: 10.1016/j.tree.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Pires ND, Yi K, Breuninger H, Catarino B, Menand B, Dolan L. Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proceedings of the National Academy of Sciences. 2013;110:9571–9576. doi: 10.1073/pnas.1305457110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polach KJ, Widom J. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J Mol Biol. 1996;258:800–812. doi: 10.1006/jmbi.1996.0288. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Genes and Signals. CSHL Press; 2002. [Google Scholar]
- Qian W, Ma D, Xiao C, Wang Z, Zhang J. The Genomic Landscape and Evolutionary Resolution of Antagonistic Pleiotropy in Yeast. Cell Rep. 2012 doi: 10.1016/j.celrep.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff RA, Kaufman TC. Embryos, Genes, and Evolution. 1983 [Google Scholar]
- Reece-Hoyes JS, Pons C, Diallo A, Mori A, Shrestha S, Kadreppa S, Nelson J, Diprima S, Dricot A, Lajoie BR, et al. Extensive rewiring and complex evolutionary dynamics in a C. elegans multiparameter transcription factor network. Mol Cell. 2013;51:116–127. doi: 10.1016/j.molcel.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RD, Papa R, Martin A, Hines HM, Counterman BA, Pardo-Diaz C, Jiggins CD, Chamberlain NL, Kronforst MR, Chen R, et al. optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science. 2011;333:1137–1141. doi: 10.1126/science.1208227. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Ruths T, Nakhleh L. ncDNA and drift drive binding site accumulation. BMC Evol Biol. 2012;12:159. doi: 10.1186/1471-2148-12-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruths T, Nakhleh L. Neutral forces acting on intragenomic variability shape the Escherichia coli regulatory network topology. Proc Natl Acad Sci USA. 2013;110:7754–7759. doi: 10.1073/pnas.1217630110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayou C, Monniaux M, Nanao MH, Moyroud E, Brockington SF, Thévenon E, Chahtane H, Warthmann N, Melkonian M, Zhang Y, et al. A promiscuous intermediate underlies the evolution of LEAFY DNA binding specificity. Science. 2014;343:645–648. doi: 10.1126/science.1248229. [DOI] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer A, Rupp S, Taylor BN, Röllinghoff M, Schröppel K. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol Microbiol. 2000;38:435–445. doi: 10.1046/j.1365-2958.2000.02132.x. [DOI] [PubMed] [Google Scholar]
- Stefflova K, Thybert D, Wilson MD, Streeter I, Aleksic J, Karagianni P, Brazma A, Adams DJ, Talianidis I, Marioni JC, et al. Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell. 2013;154:530–540. doi: 10.1016/j.cell.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Sandstrom R, Haugen E, Reynolds AP, Zhang M, Byron R, Canfield T, Stelhing-Sun S, Lee K, et al. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature. 2014;515:365–370. doi: 10.1038/nature13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323:746–751. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus A. On the possibility of constructive neutral evolution. J Mol Evol. 1999;49:169–181. doi: 10.1007/pl00006540. [DOI] [PubMed] [Google Scholar]
- Stone JR, Wray GA. Rapid evolution of cis-regulatory sequences via local point mutations. Mol Biol Evol. 2001;18:1764–1770. doi: 10.1093/oxfordjournals.molbev.a003964. [DOI] [PubMed] [Google Scholar]
- Sullivan AM, Arsovski AA, Lempe J, Bubb KL, Weirauch MT, Sabo PJ, Sandstrom R, Thurman RE, Neph S, Reynolds AP, et al. Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Rep. 2014;8:2015–2030. doi: 10.1016/j.celrep.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tanay A, Regev A, Shamir R. Conservation and evolvability in regulatory networks: the evolution of ribosomal regulation in yeast. Proc Natl Acad Sci USA. 2005;102:7203–7208. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242:405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- Tsong AE, Miller MG, Raisner RM, Johnson AD. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- Tsong AE, Tuch BB, Li H, Johnson AD. Evolution of alternative transcriptional circuits with identical logic. Nature. 2006;443:415–420. doi: 10.1038/nature05099. [DOI] [PubMed] [Google Scholar]
- Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 2008a;6:e38. doi: 10.1371/journal.pbio.0060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuch BB, Li H, Johnson AD. Evolution of eukaryotic transcription circuits. Science. 2008b;319:1797–1799. doi: 10.1126/science.1152398. [DOI] [PubMed] [Google Scholar]
- Tursun B, Patel T, Kratsios P, Hobert O. Direct conversion of C. elegans germ cells into specific neuron types. Science. 2011;331:304–308. doi: 10.1126/science.1199082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Does selection mold molecular networks? Sci STKE. 20032003:PE41. doi: 10.1126/stke.2003.202.pe41. [DOI] [PubMed] [Google Scholar]
- Wang DYC, Kumar S, Hedges SB. Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. Proc Biol Sci. 1999;266:163–171. doi: 10.1098/rspb.1999.0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JJ, Thornton JM. Evolutionary models for formation of network motifs and modularity in the Saccharomyces transcription factor network. PLoS Comput Biol. 2007;3:1993–2002. doi: 10.1371/journal.pcbi.0030198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Hughes TR. A catalogue of eukaryotic transcription factor types, their evolutionary origin, and species distribution. Subcell. Biochem. 2011;52:25–73. doi: 10.1007/978-90-481-9069-0_3. [DOI] [PubMed] [Google Scholar]
- Weirauch MT, Hughes TR. Conserved expression without conserved regulatory sequence: the more things change, the more they stay the same. Trends Genet. 2010;26:66–74. doi: 10.1016/j.tig.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Whitfield TW, Wang J, Collins PJ, Partridge EC, Aldred SF, Trinklein ND, Myers RM, Weng Z. Functional analysis of transcription factor binding sites in human promoters. Genome Biol. 2012;13:R50. doi: 10.1186/gb-2012-13-9-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E. Neutral and nonneutral mutations: the creative mix--evolution of complexity in gene interaction systems. J Mol Evol 44 Suppl. 1997;1:S2–S8. doi: 10.1007/pl00000048. [DOI] [PubMed] [Google Scholar]