Abstract
The increase of methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE) poses a worldwide and serious health threat. Although new antibiotics, such as daptomycin and linezolid, have been developed for the treatment of infections of Gram-positive pathogens, the emergence of daptomycin-resistant and linezolid-resistant strains during therapy has now increased clinical treatment failures. In the past few years, studies using quantitative proteomic methods have provided a considerable progress in understanding antibiotic resistance mechanisms. In this review, to understand the resistance mechanisms to four clinically important antibiotics (methicillin, vancomycin, linezolid, and daptomycin) used in the treatment of Gram-positive pathogens, we summarize recent advances in studies on resistance mechanisms using quantitative proteomic methods, and also examine proteins playing an important role in the bacterial mechanisms of resistance to the four antibiotics. Proteomic researches can identify proteins whose expression levels are changed in the resistance mechanism to only one antibiotic, such as LiaH in daptomycin resistance and PrsA in vancomycin resistance, and many proteins simultaneously involved in resistance mechanisms to various antibiotics. Most of resistance-related proteins, which are simultaneously associated with resistance mechanisms to several antibiotics, play important roles in regulating bacterial envelope biogenesis, or compensating for the fitness cost of antibiotic resistance. Therefore, proteomic data confirm that antibiotic resistance requires the fitness cost and the bacterial envelope is an important factor in antibiotic resistance.
Keywords: quantitative proteomics, methicillin resistance, vancomycin resistance, linezolid resistance, daptomycin resistance
Introduction
Antibiotic resistance has posed a serious threat to the worldwide public health in the past two decades. The gradual increase in resistance rates of several important pathogens, including methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus (VRE), multidrug-resistant (MDR) Pseudomonas aeruginosa, imipenem-resistant Acinetobacter baumannii, and third-generation cephalosporin-resistant Escherichia coli and Klebsiella pneumonia, has become an increasingly severe problem in many hospitals worldwide (Lee et al., 2013). However, the decline in novel antibiotics that are introduced in the market weakens the hope of overcoming this threat by the development of new antibiotics. Most of the antibiotic classes used in hospitals today were discovered during the period 1930–1960. Only two new systemic classes of antibiotics that were developed during the past 30 years were linezolid and daptomycin, which are used only in the treatment of Gram-positive pathogens (Lee et al., 2013). Because many Gram-positive pathogens increasingly develop resistance against currently available antibiotics such as methicillin and vancomycin, these new antibiotics have become valuable for the treatment of various infections of methicillin- or vancomycin-resistant S. aureus and Streptococcus pneumonia (Ament et al., 2002; Mendes et al., 2014). However, the emergence of daptomycin-resistant or linezolid-resistant strains has recently been described in some Gram-positive pathogens (Fischer et al., 2011; Mendes et al., 2014). In this review, we summarize resistance mechanisms to four clinically important antibiotics (methicillin, vancomycin, linezolid, and daptomycin) used in the treatment of Gram-positive pathogens, and highlights recent important studies using comparative proteomic tools to understand resistance mechanisms of these antibiotics in more detail.
Action and resistance mechanisms of methicillin, vancomycin, linezolid, and daptomycin resistance
Methicillin
Methicillin is a narrow-spectrum β-lactam antibiotic of the penicillin class. Like other β-lactam antibiotics, methicillin prevents the synthesis of bacterial cell walls by inhibiting peptidic cross-linkage between the linear peptidoglycan polymer chains, which provides rigidity to the cell wall of Gram-positive bacteria (Chambers, 1997) (Table 1). Methicillin and other β-lactam antibiotics are structural analogs of D-Ala-D-Ala, which is the terminus of a short amino acid chain attached in N-acetylmuramic acids; so, they interact with and irreversibly inhibit the transpeptidase enzyme [also called penicillin-binding protein (PBP)] that crosslinks the linear peptidoglycan polymer chains (Lee et al., 2012). This process leads to loss of osmotic integrity and makes the bacterial cells susceptible to lysis. Although most β-lactam antibiotics are inhibited by bacterial enzymes that hydrolyze the β-lactam ring (named β-lactamases), due to a modification of the original penicillin structure methicillin is resistant to β-lactamases (Lee et al., 2012). Therefore, since the late 1950s when methicillin was first introduced in markets, this antibiotic has been used to treat infections caused by Staphylococcus pathogens such as Staphylococcus aureus, most of which produces β-lactamase (Newsom, 2004).
Table 1.
Modes of action of four clinically important antibiotics (methicillin, vancomycin, linezolid, and daptomycin) and resistance mechanisms to these antibiotics.
| Antibiotics | Target | Mechanism of action | Resistance mechanisms found by non-proteomic approaches |
|---|---|---|---|
| Methicillin | Transpeptidase enzyme [penicillin-binding protein (PBP)] | Inhibition of peptidoglycan biosynthesis | Expression of penicillin-binding protein 2a (MecA), efflux pump |
| Vancomycin | D-Ala-D-Ala dipeptide terminus of the nascent peptidoglycan | Inhibition of peptidoglycan biosynthesis | Alteration of the D-Ala-D-Ala dipeptide |
| Linezolid | 23S rRNA | Inhibition of translation | Alteration of 23S rRNA |
| Daptomycin | Cell membrane | The formation of holes that leak intracellular ions | Remained to be elucidated |
Today, methicillin is not as effective against these organisms due to resistance (Cordwell et al., 2002; Newsom, 2004). Although the resistance phenotype of methicillin is influenced by numerous factors, including mecA, glmM, fmtAB, murE, llm, β-lactamase (bla) regulatory elements, and fem factors (Chambers, 1997; Cordwell et al., 2002; Hao et al., 2012), one major reason for methicillin resistance is the expression of the mecA gene, encoding penicillin-binding protein 2a (PBP 2a) that is not inhibited by classical β-lactam antibiotics including methicillin (Katayama et al., 2004) (Table 1). PBP 2a works in a similar manner to other PBPs, but it is bound by β-lactams with very low affinity (Katayama et al., 2004). Expression of PBP 2a confers resistance to all β-lactams. A variety of factors such as MecI and MecR1 controlled the mecA expression (Chambers, 1997). Resistance to methicillin exhibited by strains lacking the mecA gene is associated with modifications in native PBPs, β-lactamase hyperproduction, or possibly a methicillinase (Chambers, 1997). In pathogenesis, it has been reported that some virulence factors (Panton-Valentine leukocidin, phenol-soluble modulin, arginine catabolic mobile element, and other toxin elements) and two-component regulation systems (agr, saeRS, and vraRS) involved in pathogenesis can enhance the fitness of methicillin-resistant pathogens (Hao et al., 2012).
Vancomycin
Vancomycin made by the soil bacterium Amycolatopsis orientalis is a member of the glycopeptide antibiotic class and has an important role in the treatment of serious infections caused by Gram-positive bacteria such as Staphylococcus and Streptococcus (Woodford, 1998). It is a complex compound consisting of a branched tricyclic glycosylated peptide and is a rare example of a halo-organic natural compound containing two covalently bonded chlorine atoms (Levine, 2006). Vancomycin inhibits the peptidoglycan synthesis by binding at the D-Ala-D-Ala dipeptide terminus of the nascent peptidoglycan in Gram-positive bacteria (Healy et al., 2000; Levine, 2006). This binding of vancomycin to the D-Ala-D-Ala prevents the peptidic cross-linking between the linear peptidoglycan polymer chains by inhibiting the proper interaction with the transpeptidase enzyme (Healy et al., 2000) (Table 1).
Most Gram-negative bacteria are intrinsically resistant to vancomycin because it cannot penetrate the outer membrane of Gram-negative bacteria. In Gram-positive bacteria, one mechanism of resistance to vancomycin is the alteration of the terminal amino acid residues (D-Ala-D-Ala), to which vancomycin binds (Table 1). The D-Ala-D-Ala dipeptide terminus of the nascent peptidoglycan is replaced by D-Ala-D-Lac or D-Ala-D-Ser. The D-Ala-D-Lac variation results in a 1000-fold decrease in the affinity between vancomycin and the peptide, and the D-Ala-D-Ser variation causes a 6-fold loss of affinity, most likely due to steric hindrance (Courvalin, 2005). These alterations of the D-Ala-D-Ala dipeptide terminus require the coordinate action of several enzymes encoded by the van genes. Alternative ligases catalyze the formation of the D-Ala-D-Lac peptide (VanA, B, and D type enzymes) or D-Ala-D-Ser peptide (VanC, E, and G type enzymes) in peptidoglycan synthesis. VanH protein (α-keto acid reductase) reduces pyruvate to D-Lac, and the D,D-dipeptidase VanX selectively removes the D-Ala-D-Ala produced by the native ligase to enhance the incorporation of the D-Ala-D-Lac or D-Ala-D-Ser into the peptidoglycan precursor. VanR and VanS constitute a two-component regulatory system that activates the transcription of the van gene cluster (Marcone et al., 2010).
Linezolid
Linezolid is a first synthetic oxazolidinone antibiotic used to treat infections caused by VRE and MRSA. Although the mechanism of action of linezolid is not fully understood, it seems to bind to the 50S subunit of the bacterial ribosome through interaction with the central loop of the 23S rRNA and block the formation of protein synthesis initiation complexes (Swaney et al., 1998; Ament et al., 2002) (Table 1). Because linezolid binds to the 23S portion of the 50S subunit different from the binding sites of other ribosome-binding antibiotics such as chloramphenicol, cross-resistance between linezolid and other protein synthesis inhibitors is highly rare (Herrmann et al., 2008). The crystal structures of linezolid bound to the 50S subunit in 2008 showed that linezolid binds to the A site of the 50S ribosomal subunit and induces a conformational change perturbing the correct positioning of tRNAs on the ribosome (Ippolito et al., 2008; Wilson et al., 2008).
Most Gram-negative bacteria have an intrinsic resistance to linezolid due to the high activity of efflux pumps, which actively pump linezolid out of the cell (Schumacher et al., 2007). In Gram-positive bacteria, the acquired resistance to linezolid was first reported in 1999 in multidrug-resistant Enterococcus faecium (Mendes et al., 2014). High-resolution structures of linezolid with the 50S ribosomal subunit showed that it binds to a deep cleft that is surrounded by the central loop of domain V of 23S rRNA (Long and Vester, 2012). Therefore, the most common resistance mechanism of Gram-positive bacteria to linezolid was a point mutation known as G2576T, in which the G2576 position of 23S ribosomal RNA is converted to thymine (Mendes et al., 2014). In addition to mutations in 23S rRNA, other mechanisms have been identified in Gram-positive bacteria, including a six base pair deletion in the ribosomal protein L4, mutations in the ribosomal protein L3, mutations in an RNA methyltransferase (encoded by the cfr gene) that methylates G2445 of the 23S rRNA, and mutations causing increased expression of ABC transporter genes (patA and patB).
Daptomycin
Daptomycin is a lipopeptide antibiotic consisting of a lipid molecule conjugated with anionic peptide and is a natural compound found in the soil bacterium Streptomyces roseosporus (Miao et al., 2005). Daptomycin absolutely requires Ca2+ for activity, making this agent a cationic antimicrobial peptide functionally (Baltz, 2009). The poorly calcium-decorated form of daptomycin is 10 times less active microbiologically than the heavily calcium-decorated form (Baltz, 2009). The calcium-bound daptomycin interacts with phosphatidylglycerol in the bacterial membrane and inserts into the cell membrane, leading to the formation of holes that leak intracellular ions (Pogliano et al., 2012). A loss of membrane potential causes inhibition of protein, DNA, and RNA synthesis, which results in bacterial cell death (Pogliano et al., 2012). Because of a distinct mechanism of action of daptomycin, it is used in the treatment of life-threatening infections caused by multiple drug-resistant Gram-positive bacteria (Baltz, 2009). Because vancomycin and daptomycin have molecular weight (MWs) of more than 1000 Da (vancomycin of 1449 Da and daptomycin of 1620 Da), they cannot penetrate the outer membrane of Gram-negative bacteria (Lee et al., 2013). Therefore, two antibiotics are used to control infections caused by Gram-positive bacteria.
Although daptomycin was clinically introduced in 2003, clinical treatment failures by the emergence of daptomycin-resistant strains during therapy have now been described (Hobbs et al., 2008; Fischer et al., 2011). Up to now, specific genetic determinant of the daptomycin-resistant strain remained to be elucidated, despite the finding of several phenotypic and genetic determinants (altered phospholipid synthesis, thickened cell walls, alteration of cell membrane fluidity, and the acquisition of mutations within the mprF or yycG gene) (Mishra et al., 2009; Fischer et al., 2011). The mprF gene encodes a dual functional enzyme that catalyzes the coupling of lysine to phosphatidylglycerol (PG) and transfers the lysyl-PG (LPG) to the outer leaflet of the membrane. The LPG is less acidic than PG, and membranes lacking LPG are more acidic than those containing PG and LPG (Baltz, 2009). Daptomycin-resistant strains with mprF mutations have membranes with increased levels of LPG (Jones et al., 2008). Therefore, the increased positive charge caused by increased LPG in the mprF mutant (gain-of-function) reduces the binding of Ca2+-bound daptomycin to bacterial membranes by a less favorable electrostatic interaction. YycG is a membrane spanning sensor histidine kinase of a two-component signal transduction system that partners with the YycF response regulator. YycFG functions as a master regulatory system for cell wall metabolism and biofilm formation and is the only two-component system required for viability in many Gram-positive bacteria (Winkler and Hoch, 2008; Baltz, 2009).
Comparative proteomic analyses of methicillin, vancomycin, linezolid, and daptomycin resistance
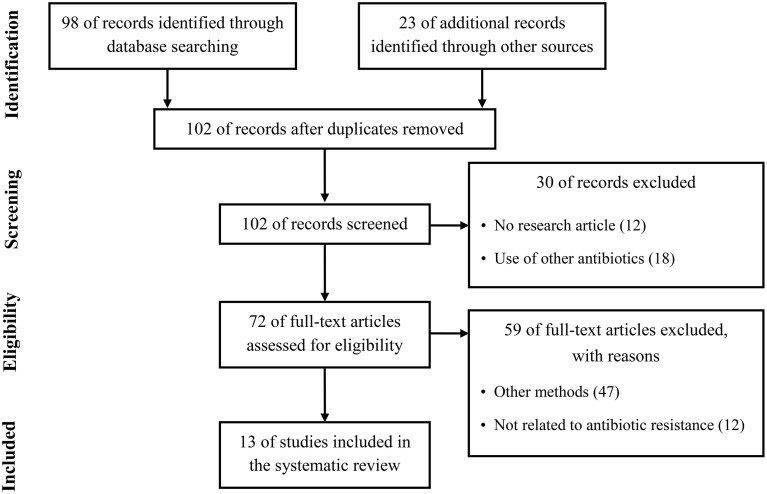
Quantitative proteomics have been considerably improved during the past decade and have been employed for investigation of the differences in whole protein expression dynamics of cells grown under a variety of growth conditions or stress conditions such as antibiotics (Radhouani et al., 2012). Therefore, by studies using quantitative proteomic approaches in the past few years, a considerable progress has recently been made in the study of antibiotic resistance mechanism. To summarize recent updates to understand the resistance mechanism to four clinically important antibiotics used in the treatment of Gram-positive pathogens, we used the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) in our review (Figure 1) (Moher et al., 2009). We conducted a systematic literature search in the following databases: Medline via PubMed and Embase. We used keywords as search terms. We combined terms for selected indications (methicillin, vancomycin, linezolid, daptomycin, and proteomics). The literature search included all studies published in English between 2000 and 2015. We identified 13 proteomics studies comparing proteomic profiles in antibiotic-resistant and antibiotic-sensitive strains or exploring proteomic profiles in cells treated with or without antibiotics.
Figure 1.
Literature selection process (PRISMA flow diagram).
Methicillin
Two studies exploring proteomic profiles of methicillin-susceptible S. aureus (MSSA) and methicillin-resistant S. aureus (MRSA) in the absence of methicillin were reported (Cordwell et al., 2002; Enany et al., 2014). Cordwell et al. compared the protein profiles between S. aureus strains COL (methicillin-resistant) and 8325 (methicillin-susceptible) in the absence of methicillin (Cordwell et al., 2002). Interestingly, among proteins previously known as resistance-related factors (e.g., mecA, glmM, fmtAB, murE, llm, bla, and fem factors), only FemA protein, which is known as a host-mediated factor essential for methicillin resistance in S. aureus (Berger-Bächi et al., 1989), was more highly expressed in methicillin-resistant cells (Cordwell et al., 2002). However, upon growth of both strains in the presence of Triton X-100 (TX-100), a detergent that has been shown to reduce methicillin resistance, no difference on the production of the essential methicillin-resistance factor FemA was detected (Cordwell et al., 2002). Instead, expression levels of stress-related proteins including cold-shock proteins (CspABC) and alkaline-shock protein 23 (Asp23) increased in the methicillin-resistant S. aureus strain COL (Cordwell et al., 2002). Notably, the amount of CspB, CspC, and Asp23 proteins was affected in cases of vancomycin and daptomycin antibiotics, despite being down-regulated in the vancomycin-resistant strain and up-regulated in the daptomycin-resistant strain (Table 7). This study also showed that three proteins linked to the alternative sigma factor σB, Asp23, anti-anti- σB factor RsbV, and conserved hypothetical protein SA0772, were also present at significantly higher levels in methicillin-resistant cells (Cordwell et al., 2002). In the presence of TX-100 weakening the methicillin resistance, the comparative proteomic analysis showed that proteins of the σB and SarA (a regulator of virulence genes) regulons are involved in methicillin resistance of S. aureus (Cordwell et al., 2002). The level of SarA protein also increased in vancomycin-resistant and daptomycin-resistant cells (Table 7). This study also showed that the stage V sporulation protein G (SpoVG), originally identified in Bacillus subtilis as being involved in the formation of the spore cortex (Matsuno and Sonenshein, 1999), was up-regulated in the methicillin-resistant S. aureus strain COL. In the non-sporulating S. aureus, SpoVG contributes to stimulate capsule synthesis, and was recently shown to regulate a small σB-subregulon comprising mainly excreted virulence factors including the highly up-regulated virulence factor EsxA (Schulthess et al., 2012). Recently, it has been reported that SpoVG was involved in resistance mechanisms to methicillin and glycopeptide (Schulthess et al., 2009). Together with this report, a comparative proteome analysis showed that the expression level of SpoVG increased in strains resistant to methicillin, vancomycin, and daptomycin (Table 5), indicating that SpoVG may be involved in resistance mechanisms to other antibiotics as well as methicillin and glycopeptide.
Another report explored proteome profiles of extracellular proteins in methicillin-sensitive and methicillin-resistant S. aureus (Enany et al., 2014). They identified some proteins increased in MRSA; Asp23 (10-fold more in MRSA than MSSA), alkyl hydroperoxide reductase subunit C (AhpC) (2-fold), D-lactate dehydrogenase (LdhD) (2-fold), general stress protein 20U (3-fold), L-lactate dehydrogenase (LdhA) (2-fold), pyruvate dehydrogenase E1 component beta subunit (PdhB) (2-fold), superoxide dismutase (SodA) (2-fold), triacylglycerol lipase precursor (LipA) (2-fold), triosephosphate isomerase (TpiA) (2-fold), and universal stress protein family protein (7-fold) (Enany et al., 2014). Notably, among them, most proteins (AhpC, SodA, LdhA, LipA, and TipA) also have altered expression levels in other antibiotic-resistant strains (Table 7). In addition, elongation factor G (encoded by the fusA gene) was also increased in MRSA. Our analysis showed that PusA is one of the three proteins affected in all four antibiotic-resistant strains (Table 5). Although elongation factor G is a major target of fusidic acid which has been used as a topical agent for skin infection and for some systemic infections caused by S. aureus (Howden and Grayson, 2006), and had a contribution to fusidic acid resistance mechanisms evolved in MRSA (Koripella et al., 2012), the relationship between elongation factor G and resistance mechanisms of other antibiotics has not yet been identified.
Vancomycin
There were two studies exploring proteomic profiles in vancomycin-susceptible S. aureus (VSSA) and vancomycin-intermediate S. aureus (VISA) with a minimal inhibitory concentration (MIC) of 4–8 μg/ml, one study exploring proteomic profiles in VSSA and heterogeneous vancomycin-intermediate S. aureus (hVISA) with a vancomycin MIC of ≤2 μg/ml, one study exploring proteomic profiles in VISA and vancomycin-resistant S. aureus (VRSA) with MIC of ≥8 μg/ml, one study analyzing global proteomes of vancomycin stress in S. aureus, and two studies examining vancomycin-induced proteomes of Enterococcus faecalis under vancomycin treatment (Pieper et al., 2006; Scherl et al., 2006; Drummelsmith et al., 2007; Wang et al., 2010; Chen et al., 2013; Hessling et al., 2013; Ramos et al., 2015). Many proteins previously known as resistance-related factors, including VanA, VanB, VanX, and VanR, were also identified in comparative proteomic analyses (Table 2). Scherl et al. (2006) showed that a total of 155 proteins are differentially expressed between two vancomycin-susceptible S. aureus strains (MRGR3 and 14-4Rev) and the vancomycin-intermediate S. aureus strain 14-4, and most proteins play a role in energy metabolism, cell envelope biosynthesis, protein turnover, amino acids transport, and metabolism, and inorganic ion transport. Genes or gene products known to be involved in resistance mechanisms to different antibiotics, such as PBP 2a (MecA), O-nucleotidyltransferase(9) [Ant(9)], UDP-N-acetylmuramyl tripeptide synthetase (MurE), and penicillin-binding methicillin resistant-related protein (FmtA), were up-regulated in the VISA strain (Scherl et al., 2006). All of them are involved in peptidoglycan biosynthesis. Levels of many other proteins involved in peptidoglycan metabolism also increased in the VISA strain, such as glycosyltransferase (SgtB) and CHAP (Cysteine, Histidine-dependent Amidohydrolases/Peptidases)-domain amidase (SsaA). SsaA belongs to the CHAP amidase family, members of which such as LysK and LytA have been shown to have D-alanyl-glycyl endopeptidase activity, cleaving between the crossbridge and the stem peptide (Delaune et al., 2011), and protein levels of SsaA were also changed in cases of methicillin and linezolid (Table 6), indicating the importance of this protein on peptidoglycan metabolism and antibiotic resistance.
Table 2.
Differentially expressed proteins identified by the quantitative proteomic approach: proteins involved in resistance mechanisms.
| Biological process | Protein name | Gene | Antibiotics | Regulation | Frequency of difference | References | Protein description |
|---|---|---|---|---|---|---|---|
| Antibiotic inactivation | Bleomycin resistance protein | ble | Van | Down | 1 | Pieper et al., 2006 | Inhibition of bleomycin by a direct interaction |
| Kanamycin nucleotidyltransferase | knt | Van | Down | 1 | Pieper et al., 2006 | Modification of kanamycin | |
| Vancomycin resistance protein | vanA | Van | Up | 2 | Wang et al., 2010; Ramos et al., 2015 | Alteration of the D-Ala-D-Ala dipeptide | |
| O-nucleotidyltransferase (9) | ant(9) | Van | Up | 1 | Scherl et al., 2006 | Modification of vancomycin |
They also identified several regulatory systems contributing to the VISA phenotype, such as the two-component system (VraSR) regulating expression of a set of genes involved in the cell wall biosynthesis or degradation (Boyle-Vavra et al., 2013), the signal transduction protein TRAP acting on quorum sensing (Gov et al., 2004), the DivIVA protein known to regulate cell division in B. subtilis (Perry and Edwards, 2004), and putative transcription factors SA2296 and SarH1. VraSR (vancomycin resistance associated regulator) was up-regulated under vancomycin treatment (Kuroda et al., 2003) and in the VISA strain when compared with an isogenic vancomycin-susceptible strain (Kuroda et al., 2000). In addition, inactivation of the vraSR gene increased vancomycin susceptibility (Kuroda et al., 2003). Interestingly, VraSR was also induced by other antibiotic classes that target the cell wall, including β-lactam (Gardete et al., 2006; Yin et al., 2006), mersacidin (Sass et al., 2008), certain cationic peptides (Pietiäinen et al., 2009), and daptomycin (Muthaiyan et al., 2008). Inactivation of the vraSR gene attenuates resistance to various antibiotics, such as vancomycin (Kuroda et al., 2003; Gardete et al., 2006), daptomycin (Mehta et al., 2012), and β-lactams (Kuroda et al., 2003; Boyle-Vavra et al., 2006; Gardete et al., 2006). The expression of many genes, such as ctpA, drp35, fmtA, opuD, pbp2, prsA, sgtB, and vraX, is regulated by VraSR (Utaida et al., 2003; McAleese et al., 2006; Dengler et al., 2011). Among them, FmtA is typically known as a factor involved in methicillin-resistant phenotype of S. aureus (Fan et al., 2007), and PrsA (foldase precursor) was recently reported to be involved in both glycopeptide and oxacillin resistance in S. aureus (Jousselin et al., 2012). Similarly, at three independent studies of comparative proteomic analysis, it has been proven that the expression level of PrsA is up-regulated in VISA when compared with VSSA (Table 5), indicating that proteomic studies can support the identification of targets involved in antibiotic resistance. They also identified another important protein VraX (a hypothetical protein which encodes a 55-amino acids protein) differentially expressed between vancomycin-susceptible S. aureus strains and the vancomycin-intermediate S. aureus strain 14-4 (Scherl et al., 2006). This gene was up-regulated by multiple cell wall and/or membrane active compounds (bacitracin, d-cycloserine, oxacillin, tunicamycin, flavomycin, fosfomycin, teicoplanin, vancomycin, daptomycin, lysostaphin, epicatechin gallate, ranalexin, and antimicrobial peptides) (Utaida et al., 2003; Pietiäinen et al., 2009; Dengler et al., 2011; Cuaron et al., 2013). The vraX gene belongs to the vra operon together with the vraA gene encoding for a long chain fatty acid-CoA ligase, which was up-regulated in the VISA. Additionally, this gene seems to be involved in resistance mechanism to vancomycin (Hanaki et al., 1998; Buntaran et al., 2013). Finally, stress-related proteins such as proteinases (CtpA), methionine sulfoxide reductase A (MsrA2), and the methionine sulfoxide reductase regulator MsrR, were over-expressed in the vancomycin-intermediate S. aureus strain 14-4 (Scherl et al., 2006). In other studies, MsrA2 was also up-regulated in hVISA (Chen et al., 2013).
Table 5.
Differentially expressed proteins identified by the quantitative proteomic approach: proteins involved in replication, cell division, transcription, translation, and protein turnover.
| Biological process | Protein name | Gene | Antibiotics | Regulation | Frequency of difference | References | Protein description |
|---|---|---|---|---|---|---|---|
| Replication, recombination and repair | Initiation-control protein | yabA | Van | Up | 1 | Ramos et al., 2015 | Replication |
| Topoisomerase IV subunit B | parE | Van | Down | 1 | Pieper et al., 2006 | Replication | |
| DNA gyrase subunit B | gyrB | Van | Down | 1 | Pieper et al., 2006 | Replication | |
| Single-stranded DNA binding protein | traM | Van | Down | 1 | Pieper et al., 2006 | Replication | |
| Formamidopyrimidine-DNA glycosylase | mutM | Lin | Up | 1 | Feng et al., 2011 | DNA repair | |
| Single-stranded DNA-binding protein | ssbB | Lin | Down | 1 | Feng et al., 2011 | Replication | |
| DNA-entry nuclease | endA | Lin | Up | 1 | Feng et al., 2011 | DNA repair | |
| Recombinase A | recA | Van | Up | 1 | Wang et al., 2010 | Recombination | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Endonuclease IV | nfo | Van | Up | 1 | Wang et al., 2010 | DNA repair | |
| Dap | Down | 1 | Fischer et al., 2011 | ||||
| Sporulation and cell division | Anti-anti-σB factor | rsbV | Met | Up | 1 | Cordwell et al., 2002 | Sporulation |
| Cell division protein | mraZ | Van | Up | 1 | Pieper et al., 2006 | Cell division | |
| Cell division protein | divIVA | Van | Up | 1 | Scherl et al., 2006 | Cell division | |
| Cell division protein | ftsZ | Van | Up | 1 | Wang et al., 2010 | Cell division | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Regulatory protein SpoVG | spoVG | Met | Up | 1 | Cordwell et al., 2002 | Sporulation | |
| Van | Up | 1 | Pieper et al., 2006 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Transcription | DNA-directed RNA polymerase subunit delta | rpoE | Lin | Up | 1 | Feng et al., 2011 | |
| Transcription elongation factor | greA | Van | Down | 1 | Pieper et al., 2006 | Trnascription | |
| Lin | Up | 1 | Feng et al., 2011 | ||||
| Translation, ribosomal structure and biogenesis | 5-methylaminomethyl-2-thiouridylate)-methyltransferase | trmU | Van | Up | 1 | Wang et al., 2010 | tRNA modification |
| 16S rRNA processing protein | rimM | Lin | Down | 1 | Feng et al., 2011 | Ribosome maturation | |
| Acetyltransferase | rimL | Lin | Up | 1 | Feng et al., 2011 | Ribosome modification | |
| Methionine aminopeptidase | map | Lin | Up | 1 | Feng et al., 2011 | Amino-terminal maturation | |
| Ribosomal subunit interface protein | spr2011 | Lin | Up | 1 | Feng et al., 2011 | Ribosome regulation | |
| Ribosomal protein S4 | rpsD | Dap | Up | 1 | Fischer et al., 2011 | Ribosomal subunit protein | |
| Ribosomal protein S10 | rpsJ | Met | Up | 1 | Enany et al., 2014 | Ribosomal subunit protein | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein S13 | rpsM | Met | Up | 1 | Enany et al., 2014 | Ribosomal subunit protein | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein S3 | rpsC | Met | Up | 1 | Enany et al., 2014 | Ribosomal subunit protein | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein L7/L12 | rplL | Met | Up | 1 | Enany et al., 2014 | Ribosomal subunit protein | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Translational initiation factor IF-2 | infB | Van | Down | 1 | Pieper et al., 2006 | Translation | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Essential GTPase | era | Van | Down | 1 | Pieper et al., 2006 | Ribosome maturation | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein L2 | rplB | Van | Down | 1 | Hessling et al., 2013 | Ribosomal subunit protein | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| GTP-binding protein | engA | Van | Down | 1 | Scherl et al., 2006 | Ribosome maturation | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein S18 | rpsR | Lin | Up | 1 | Feng et al., 2011 | Ribosomal subunit protein | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein L3 | rplC | Lin | Up | 1 | Bernardo et al., 2004 | Ribosomal subunit protein | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein L27 | rpmA | Lin | Up | 1 | Bernardo et al., 2004 | Ribosomal subunit protein | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein L22 | rplV | Lin | Up | 1 | Bernardo et al., 2004 | Ribosomal subunit protein | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein S9 | rplI | Lin | Up | 1 | Bernardo et al., 2004 | Ribosomal subunit protein | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein L15 | rplO | Lin | Up | 1 | Bernardo et al., 2004 | Ribosomal subunit protein | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein L13 | rplM | Lin | Up | 1 | Bernardo et al., 2004 | Ribosomal subunit protein | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein L4 | rplD | Lin | Up | 1 | Bernardo et al., 2004 | Ribosomal subunit protein | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein L1 | rplA | Lin | Up | 2 | Bernardo et al., 2004; Feng et al., 2011 | Ribosomal subunit protein | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| 30S ribosomal protein S1 | rpsA | Met | Up | 1 | Enany et al., 2014 | Ribosomal subunit protein | |
| Van | Down | 1 | Drummelsmith et al., 2007 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein L14 | rplN | Met | Up | 1 | Enany et al., 2014 | Ribosomal subunit protein | |
| Van | Down | 1 | Hessling et al., 2013 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein L21 | rplU | Met | Up | 1 | Enany et al., 2014 | Ribosomal subunit protein | |
| Lin | Up | 2 | Bernardo et al., 2004; Feng et al., 2011 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein S6 | rpsF | Met | Up | 1 | Enany et al., 2014 | Ribosomal subunit protein | |
| Lin | Up | 1 | Feng et al., 2011 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein L6 | rplF | Van | Down | 1 | Hessling et al., 2013 | Ribosomal subunit protein | |
| Lin | Up | 1 | Bernardo et al., 2004 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Ribosomal protein S2 | rpsB | Van | Up | 1 | Wang et al., 2010 | Ribosomal subunit protein | |
| Lin | Up(down) | 1(1) | Bernardo et al., 2004; Feng et al., 2011 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Elongation factor Tu | tuf | Van | Down(up) | 1(1) | Drummelsmith et al., 2007; Wang et al., 2010 | Translation | |
| Lin | Up | 2 | Bernardo et al., 2004; Feng et al., 2011 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| 50S ribosomal protein L20 | rplT | Van | Up | 1 | Drummelsmith et al., 2007 | Ribosomal subunit protein | |
| Lin | Down | 1 | Feng et al., 2011 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Elongation factor Ts | tsf | Van | Down | 1 | Pieper et al., 2006 | Translation | |
| Lin | Up | 1 | Feng et al., 2011 | ||||
| Dap | Down | 1 | Fischer et al., 2011 | ||||
| Translational elongation factor G | fusA | Met | Up | 1 | Enany et al., 2014 | Translation | |
| Van | Down(up) | 1(1) | Drummelsmith et al., 2007; Wang et al., 2010 | ||||
| Lin | Up | 1 | Bernardo et al., 2004 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Post-translational modification, protein turnover, chaperones | ATP-dependent Clp protease proteolytic subunit | clpP | Met | Up | 1 | Cordwell et al., 2002 | Protein degradation |
| Preprotein translocase | secY | Van | Up | 1 | Scherl et al., 2006 | Protein translocation | |
| ATP-dependent chaperone protein | clpB | Van | Up | 1 | Hessling et al., 2013 | Protein degradation | |
| Aminopeptidase | pepS | Van | Down | 1 | Drummelsmith et al., 2007 | Protein degradation | |
| Foldase precursor | prsA | Van | Up | 3 | Scherl et al., 2006; Drummelsmith et al., 2007; Hessling et al., 2013 | Chaperone | |
| Chaperone | groS | Van | Up | 1 | Pieper et al., 2006 | Chaperone | |
| Methionine sulfoxide reductase A | msrA | Van | Up | 2 | Scherl et al., 2006; Chen et al., 2013 | Protein modification | |
| Carboxy-terminal processing peptidase | ctpA | Van | Up | 1 | Scherl et al., 2006 | Protein processing | |
| Cell wall-associated serine proteinase precursor | prtA | Lin | Up | 1 | Feng et al., 2011 | Protein degradation | |
| Methionine sulfoxide reductase B | SA1256 | Met | Up | 1 | Cordwell et al., 2002 | Protein modification | |
| Van | Up | 1 | Scherl et al., 2006 | ||||
| Glutamyl-aminopeptidase | pepA | Van | Up | 1 | Wang et al., 2010 | Protein degradation | |
| Lin | Up | 1 | Feng et al., 2011 | ||||
| Chaperone | dnaK | Van | Up(down) | 2(1) | Scherl et al., 2006; Drummelsmith et al., 2007; Wang et al., 2010 | Chaperone | |
| Lin | Up | 1 | Bernardo et al., 2004 | ||||
| Signal peptidase B | spsB | Van | Up | 2 | Scherl et al., 2006; Drummelsmith et al., 2007 | Cleavage of signal peptide | |
| Dap | Down | 1 | Fischer et al., 2011 | ||||
| Peptide methionine sulfoxide reductase regulator | msrR | Van | Up | 1 | Scherl et al., 2006 | Protein modification | |
| Dap | Down | 1 | Fischer et al., 2011 | ||||
| Peptidase | ftsH | Van | Up | 2 | Scherl et al., 2006; Drummelsmith et al., 2007 | Protein degradation | |
| Dap | Down | 1 | Fischer et al., 2011 |
Pieper et al. showed that purine ribonucleotide biosynthesis (PRNBS) pathway enzymes, which are under the control of the PurR regulator, strongly increased in protein abundance in the vancomycin-resistant S. aureus strain VP32 having a vancomycin MIC of 32 μg/ml when compared with the vancomycin-intermediate S. aureus strain HIP5827 (MIC = 8 μg/ml) (Pieper et al., 2006). Notably, among them, several proteins such as amidophosphoribosyltransferase (PurF), phosphoribosylamine-glycine ligase (PurD), phosphoribosylglycinamidine cyclo-ligase (PurM), phosphoribosylaminoimidazole-succinocarboxamide synthase (PurC), adenylosuccinate lyase (PurB), and bifunctional purine biosynthesis protein (PurH), were also changed in protein abundance in cases of other antibiotics such as daptomycin and linezolid (Table 4). Microarray transcription analysis of clinical VISA isolates already showed that among the 35 genes with increased transcription in vancomycin-resistant S. aureus strain VP32 when compared with those of their VISA parent strains HIP5827 and P100, 15 were involved in purine biosynthesis or transport (Mongodin et al., 2003). They hypothesized that increased energy (ATP) is required to generate the thicker cell walls that characterize resistant mutants (Mongodin et al., 2003). However, contrary to these results, other comparative proteomic analyses between vancomycin-susceptible strains and vancomycin-intermediate S. aureus strains did not show similar results (Scherl et al., 2006; Drummelsmith et al., 2007; Chen et al., 2013). Therefore, these results imply that VRSA may more efficiently compensate for a fitness cost of antibiotic resistance such as ATP requirement than VISA.
Table 4.
Differentially expressed proteins identified by the quantitative proteomic approach: proteins involved in amino acid, nucleotide, coenzyme, and inorganic ion metabolisms.
| Biological process | Protein name | Gene | Antibiotics | Regulation | Frequency of difference | References | Protein description |
|---|---|---|---|---|---|---|---|
| Amino acid transport and metabolism | Threonine deaminase | ilvA | Met | Down | 1 | Cordwell et al., 2002 | Threonine metabolism |
| Aspartate kinase | lysC | Van | Up | 1 | Hessling et al., 2013 | Biosynthesis of lysine, methionine, threonine | |
| Aspartate semialdehyde dehydrogenase | Asd | Van | Up | 1 | Hessling et al., 2013 | Biosynthesis of lysine, methionine, threonine | |
| Dihydrodipicolinate synthase | dapA | Van | Up | 1 | Hessling et al., 2013 | Biosynthesis of lysine | |
| Tetrahydrodipicolinate acetyltransferase | dapD | Van | Up | 1 | Hessling et al., 2013 | Biosynthesis of lysine | |
| Diaminopimelate decarboxylase | lysA | Van | Up | 1 | Hessling et al., 2013 | Biosynthesis of lysine | |
| Arginine deiminase | arcA | Van | Down | 1 | Ramos et al., 2015 | Arginine metabolism | |
| 4-Hydroxy-tetrahydrodipicolinate reductase | dapB | Van | Up | 1 | Hessling et al., 2013 | Biosynthesis of lysine | |
| Cystathionine β-lyase | metC | Van | Up | 1 | Wang et al., 2010 | Biosynthesis of methionine | |
| Cystathionine γ-synthase | metB | Van | Up | 1 | Drummelsmith et al., 2007 | Biosynthesis of methionine | |
| Chorismate mutase | aroA | Van | Down | 1 | Drummelsmith et al., 2007 | Biosynthesis of aromatic amino acids | |
| Urease α subunit | ureC | Van | Up | 1 | Drummelsmith et al., 2007 | Urea metabolism | |
| Urease accessory protein | ureE | Van | Up | 2 | Scherl et al., 2006; Drummelsmith et al., 2007 | Urea metabolism | |
| Branched-chain amino acids aminotransferase | ilvE | Van | Down | 2 | Pieper et al., 2006; Scherl et al., 2006 | Biosynthesis of branched-chain amino acids | |
| Aetylornithine aminotransferase 2 | argD | Van | Up | 1 | Pieper et al., 2006 | Biosynthesis of lysine | |
| Amino acid ABC transporter amino acid-binding protein | glnH | Lin | Up | 1 | Feng et al., 2011 | Glutamine transport | |
| Tryptophan synthase subunit α | trpA | Lin | Down | 1 | Feng et al., 2011 | Biosynthesis of aromatic amino acids | |
| Asparagine synthetase | asnA | Lin | Down | 1 | Feng et al., 2011 | Biosynthesis of asparagine | |
| Aminotransferase | aspC | Lin | Up | 1 | Feng et al., 2011 | Biosynthesis of lysine, methionine, threonine | |
| Carbamate kinase | arcC | Van | Down | 2 | Scherl et al., 2006; Ramos et al., 2015 | Allantoin degradation | |
| Lin | Up | 1 | Feng et al., 2011 | ||||
| 1-Pyrroline-5-carboxylate dehydrogenase | rocA | Van | Up | 1 | Scherl et al., 2006 | Proline degradation | |
| Dap | Down | 1 | Fischer et al., 2011 | ||||
| Serine hydroxymethyltransferase | glyA | Van | Down | 1 | Drummelsmith et al., 2007 | Biosynthesis of glycine | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Glutamine synthetase type 1 | glnA | Lin | Down | 1 | Feng et al., 2011 | Nitrogen assimilation | |
| Dap | Down | 1 | Fischer et al., 2011 | ||||
| Nucleotide transport and metabolism | Bifunctional pyrimidine regulatory protein /uracil phosphoribosyltransferase | pyrR | Met | Down | 1 | Cordwell et al., 2002 | Salvage pathways of pyrimidine ribonucleotides |
| Purine nucleoside phosphorylase | deoD | Van | Down | 1 | Pieper et al., 2006 | Guanosine nucleotides de novo biosynthesis | |
| Putative purine biosynthesis protein | purS | Van | Up | 1 | Pieper et al., 2006 | Purine biosynthesis | |
| Adenylosuccinate synthetase | purA | Van | Up | 1 | Pieper et al., 2006 | Purine biosynthesis | |
| Phosphoribosylaminoimidazole carboxylase | purK | Van | Up | 1 | Pieper et al., 2006 | Purine biosynthesis | |
| Phosphoribosylglycinamidine synthase I | purQ | Van | Up | 1 | Pieper et al., 2006 | Purine biosynthesis | |
| Phosphoribosylglycinamidine synthase II | purL | Van | Up | 1 | Pieper et al., 2006 | Purine biosynthesis | |
| Phosphoribosylglycinamide formyltransferase | purN | Van | Up | 1 | Pieper et al., 2006 | Purine biosynthesis | |
| Phosphoribosykaminoimidazole carboxylase, catalytic subunit | purE | Van | Up | 1 | Pieper et al., 2006 | Purine biosynthesis | |
| GMP synthase | guaA | Van | Down | 2 | Scherl et al., 2006; Ramos et al., 2015 | Biosynthesis of guanosine nucleotides | |
| The pur operon repressor | purR | Lin | Up | 1 | Feng et al., 2011 | Purine biosynthesis | |
| Ribose-phosphate pyrophosphokinase | prs | Van | Up | 2 | Scherl et al., 2006; Drummelsmith et al., 2007 | Purine biosynthesis | |
| Lin | Up | 1 | Feng et al., 2011 | ||||
| Amidophosphoribosyltransferase | purF | Van | Up | 1 | Pieper et al., 2006 | Purine biosynthesis | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Phosphoribosylamine-glycine ligase | purD | Van | Up | 1 | Pieper et al., 2006 | Purine biosynthesis | |
| Dap | Down | 1 | Fischer et al., 2011 | ||||
| Phosphoribosylglycinamidine cyclo-ligase | purM | Van | Up | 1 | Pieper et al., 2006 | Purine biosynthesis | |
| Dap | Down | 1 | Fischer et al., 2011 | ||||
| GMP reductase | guaC | Van | Up | 1 | Pieper et al., 2006 | The purine salvage pathway | |
| Dap | Down | 1 | Fischer et al., 2011 | ||||
| Dihydroorotase | pyrC | Van | Down | 1 | Drummelsmith et al., 2007 | Pyrimidine biosynthesis | |
| Dap | Down | 1 | Fischer et al., 2011 | ||||
| Carbamoyl phosphate synthase large subunit | carB | Van | Down | 1 | Scherl et al., 2006 | Pyrimidine biosynthesis | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Phosphoribosylaminoimidazole-succinocarboxamide synthase | purC | Van | Up | 1 | Pieper et al., 2006 | Purine biosynthesis | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Adenylosuccinate lyase | purB | Met | Up | 1 | Enany et al., 2014 | Purine biosynthesis | |
| Van | Up | 2 | Pieper et al., 2006 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Bifunctional purine biosynthesis protein | purH | Van | Up | 1 | Pieper et al., 2006 | Purine biosynthesis | |
| Lin | Down | 1 | Feng et al., 2011 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Uracil phosphoribosyltransferase | upp | Van | Down | 2 | Scherl et al., 2006; Drummelsmith et al., 2007 | Salvage pathways of pyrimidine ribonucleotides | |
| Lin | Down | 1 | Feng et al., 2011 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Coenzyme transport and metabolism | 3-Hydroxy-3-methylglutaryl-CoA synthase | mvaS | Met | Up | 1 | Cordwell et al., 2002 | Isoprenoid biosynthesis |
| Thiamin-biosynthesis protein | thiL | Met | Up | 1 | Cordwell et al., 2002 | Thiamin biosynthesis | |
| δ-aminoevulinic acid dehydratase | hemB | Van | Down | 1 | Pieper et al., 2006 | Heme biosynthesis | |
| Molybdopterin converting factor subunit 2 | moaE | Van | Down | 1 | Pieper et al., 2006 | Molybdenum cofactor biosynthesis | |
| 2-Dehydropantoate 2-reductase | panE | Van | Down | 1 | Drummelsmith et al., 2007 | Pantothenate and coenzyme A biosynthesis | |
| 6-Pyruvoyl tetrahydrobiopterin synthase | ptpS | Van | Up | 1 | Drummelsmith et al., 2007 | Tetrahydrobiopterin biosynthesis | |
| Phosphopantetheine adenylyltransferase | coaD | Van | Up | 1 | Drummelsmith et al., 2007 | Pantothenate and coenzyme A biosynthesis | |
| Coenzyme A disulfide reductase | cdr | Van | Down | 1 | Scherl et al., 2006 | Pantothenate and coenzyme A biosynthesis | |
| Hydroxyethylthiazole kinase | thiM | Lin | Up | 1 | Feng et al., 2011 | Thiamin biosynthesis | |
| 3-Methyl-2-oxobutanoate hydroxymethyltransferase | panB | Met | Up | 1 | Enany et al., 2014 | Pantothenate and coenzyme A biosynthesis | |
| Van | Down(up) | 1(1) | Drummelsmith et al., 2007; Wang et al., 2010 | ||||
| Bifunctional 5,10-methylene-tetrahydrofolate dehydrogenase/5,10-methylene-tetrahydrofolate cyclohydrolase | folD | Van | Up | 1 | Pieper et al., 2006 | N10-formyl-tetrahydrofolate biosynthesis | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| 6,7-Dimethyl-8-ribityllumazine synthase | ribH | Lin | Up | 1 | Feng et al., 2011 | Flavin biosynthesis | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Pyridoxal biosynthesis lyase | pdxS | Lin | Down | 2 | Drummelsmith et al., 2007; Feng et al., 2011 | Pyridoxal 5′-phosphate biosynthesis. | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Inorganic ion transport and metabolism | ABC protein/substrate binding protein subunit—metal ion transport | lmb | Lin | Up | 1 | Feng et al., 2011 | Metal ion transport |
| Non-heme iron-containing ferritin | dpr | Lin | Up | 1 | Feng et al., 2011 | Iron tansport | |
| Iron-compound ABC transporter permease | fatD | Lin | Up | 1 | Feng et al., 2011 | Iron tansport | |
| Lipoprotein similar to streptococcal adhesin | psaA | Lin | Up | 1 | Feng et al., 2011 | Manganese transport; pneumococcal attachment | |
| Dap | Down | 1 | Fischer et al., 2011 |
Abundance changes were also found in proteins such as the single-stranded DNA binding protein (TraM), DNA gyrase subunit B (GyrB), and topoisomerase IV subunit B (ParE), which catalyze or influence the fidelity of DNA replication and repair (Table 5). This result is consistent with the increasing evidence that exposure to antibiotics in bacteria leads to increased mutation rates in the genome, to favor their survivals under antibiotic pressure (Napolitano et al., 2000; Friedberg et al., 2002; Pieper et al., 2006). Expression levels of many enzymes involved in energy metabolisms, including L-lactate dehydrogenase (LdhA), glucose-6-phosphate isomerase (Pgi), succinyl-CoA synthetase (SucCD), phosphoglycerate kinase (Pgk), nitrate reductase alpha chain (NarG), and aconitate hydratase (CitB), were also changed. In fact, comparative proteomic analyses show that proteins involved in energy metabolism, protein synthesis, and envelope biogenesis, most frequently exhibit abundance change in antibiotic-resistant strains (Table 3). In many cases, proteins playing a role in energy metabolism were up-regulated in antibiotic-resistant strains (Table 3). This phenomenon may be explained by a prior hypothesis that increased energy (ATP) is required to generate the thicker cell walls or to pump antibiotics out of the cell using efflux pumps. This study also showed the changes of proteins involved in cell envelope biogenesis, such as D-Ala-D-Ala ligase (Ddl), D-Ala-D-Lac ligase (VanA), peptidoglycan hydrolase (LytM), cell division and cell wall biosynthesis protein (MraZ), putative cell wall transglycosylase (SceD), and glucosamine-fructose-6-phosphate aminotransferase (GlmS) (Pieper et al., 2006).
Table 3.
Differentially expressed proteins identified by the quantitative proteomic approach: proteins involved in energy metabolism.
| Biological process | Protein name | Gene | Antibiotics | Regulation | Frequency of difference | References | Protein description |
|---|---|---|---|---|---|---|---|
| Energy production and conversion | Pyruvate dehydrogenase E1 component beta subunit | pdhB | Met | Up | 1 | Enany et al., 2014 | Acetyl-CoA biosynthetic process from pyruvate |
| D-Lactate dehydrogenase | ldhD | Met | Up | 1 | Enany et al., 2014 | Pyruvate metabolism | |
| Formyltetrahydrofolate synthetase | fhs | Van | Up | 1 | Pieper et al., 2006 | Glyoxylate and dicarboxylate metabolism and one carbon pool by folate | |
| Succinyl-CoA synthetase alpha chain | sucD | Van | Up | 1 | Pieper et al., 2006 | The citric acid cycle | |
| Aconitate hydratase | citB | Van | Up(down) | 1(1) | Pieper et al., 2006; Drummelsmith et al., 2007 | The citric acid cycle | |
| Isocitrate dehydrogenase | citC | Van | Up | 1 | Drummelsmith et al., 2007 | The citric acid cycle | |
| Citrate lyase | citF | Van | Up | 1 | Wang et al., 2010 | Acetyl-CoA metabolic process | |
| ATP synthase γ chain | atpG | Van | Up(down) | 2(1) | Pieper et al., 2006; Scherl et al., 2006; Wang et al., 2010 | ATP formation | |
| Pyruvate carboxylase | pycA | Van | Down | 1 | Pieper et al., 2006 | Anaplerotic reaction | |
| Malate:quinone oxidoreductase 1 | mqo2 | Van | Up | 2 | Scherl et al., 2006; Drummelsmith et al., 2007 | The citric acid cycle | |
| 2-Dehydro-3-deoxyphosphogluconate aldolase | eda | Van | Up | 1 | Wang et al., 2010 | Glycolysis | |
| Glyceraldehyde-3-phosphate dehydrogenase | gapA | Van | Up(down) | 1(1) | Wang et al., 2010; Ramos et al., 2015 | Glycolysis | |
| Dihydrolipoamide succinyltransferase | odhB | Van | Down | 1 | Scherl et al., 2006 | The citric acid cycle and lysine degradation. | |
| Glycerophosphoryl diester phosphodiesterase | glpQ | Lin | Up | 1 | Bernardo et al., 2004 | Glycerol and glycerophosphodiester degradation | |
| Lactate oxidase | lctO | Lin | Up | 1 | Feng et al., 2011 | Lactate oxidation | |
| Flavodoxin/nitric oxide synthase | flaV | Lin | Up | 1 | Feng et al., 2011 | Flavodoxin biosynthesis | |
| Gluconate 5-dehydrogenase | gno | Lin | Up | 1 | Feng et al., 2011 | Gluconate oxidation | |
| Phosphoglycolate phosphatase | gph | Lin | Up | 1 | Feng et al., 2011 | Glyoxylate and dicarboxylate metabolism | |
| Enolase (2-phosphoglycerate dehydratase) | edo | Dap | Up | 1 | Fischer et al., 2011 | Glycolysis | |
| Triose-phosphate isomerase | tpiA | Met | Up | 1 | Enany et al., 2014 | Glycolysis | |
| Van | Down | 1 | Ramos et al., 2015 | ||||
| Alcohol dehydrogenase | adhE | Met | Down(up) | 1(1) | Cordwell et al., 2002; Enany et al., 2014 | Fermentation | |
| Van | Up | 2 | Drummelsmith et al., 2007; Wang et al., 2010 | ||||
| Alcohol dehydrogenase | adhP | Met | Up | 1 | Enany et al., 2014 | Fermentation | |
| Lin | Up | 1 | Feng et al., 2011 | ||||
| 2,3-Bisphosphoglycerate-dependent phosphoglycerate mutase | gpmA | Van | Up(down) | 2(1) | Scherl et al., 2006; Drummelsmith et al., 2007; Chen et al., 2013 | Glycolysis | |
| Lin | Up | 1 | Feng et al., 2011 | ||||
| Nitrate reductase α chain | narG | Van | Down | 1 | Pieper et al., 2006 | Anaerobic respiration | |
| Lin | Down | 1 | Fischer et al., 2011 | ||||
| Phosphoglycerate kinase | pgk | Van | Down | 2 | Pieper et al., 2006; Scherl et al., 2006 | Glycolysis | |
| Lin | Up | 1 | Feng et al., 2011 | ||||
| Phosphopyruvate hydratase | eno | Van | Up | 1 | Scherl et al., 2006 | Glycolysis | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Succinate dehydrogenase flavoprotein subunit | sdhA | Van | Up | 1 | Scherl et al., 2006 | The citric acid cycle | |
| Dap | Down | 1 | Fischer et al., 2011 | ||||
| Pyruvate dehydrogenase α subunit | pdhA | Van | Up | 1 | Wang et al., 2010 | Acetyl-CoA biosynthetic process from pyruvate | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Citrate synthase II | citZ | Van | Up | 1 | Drummelsmith et al., 2007 | The citric acid cycle | |
| Dap | Down | 1 | Fischer et al., 2011 | ||||
| Succinyl-CoA synthetase β chain | sucC | Van | Up | 2 | Pieper et al., 2006; Drummelsmith et al., 2007 | The citric acid cycle | |
| Dap | Down | 1 | Fischer et al., 2011 | ||||
| Aminoethyltransferase | gcvT | Van | Up | 1 | Pieper et al., 2006 | Glycine cleavage | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Glyceraldehyde-3-phosphate dehydrogenase 1 | gapA | Lin | Up | 1 | Feng et al., 2011 | Glycolysis | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| L-Lactate dehydrogenase | ldhA | Met | Up | 1 | Enany et al., 2014 | Fermentation | |
| Van | Down(up) | 2(1) | Pieper et al., 2006; Scherl et al., 2006; Wang et al., 2010 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Fructose-bisphosphate aldolase | fba | Van | Up | 2 | Wang et al., 2010; Ramos et al., 2015 | Glycolysis | |
| Lin | Up | 1 | Feng et al., 2011 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Acetate kinase | ackA | Met | Up | 1 | Enany et al., 2014 | Fermentation | |
| Van | Up(down) | 1(1) | Scherl et al., 2006; Drummelsmith et al., 2007 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Glucose-6-phosphate isomerase | pgi | Van | Down(up) | 1(1) | Pieper et al., 2006; Scherl et al., 2006 | Glycolysis | |
| Lin | Up | 1 | Feng et al., 2011 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Carbohydrate transport and metabolism | ABC transporter, ATP binding protein | stpC | Van | Down | 1 | Drummelsmith et al., 2007 | Carbohydrate transport |
| PTS transport system, fructose-specific IIABC component | fruA | Van | Down | 1 | Drummelsmith et al., 2007 | Fructose transport | |
| ABC transporter, ATP binding protein | vraD | Van | Up | 1 | Drummelsmith et al., 2007 | Bacitracin tansport | |
| Phosphoglycerate mutase 1 | pgm | Van | Up | 1 | Wang et al., 2010 | The breakdown of glycogen and metabolism of galactose and maltose | |
| 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase | gpmI | Van | Up | 1 | Drummelsmith et al., 2007 | Carbohydrate degradation | |
| Glycerol kinase | glpK | Van | Up | 1 | Drummelsmith et al., 2007 | Carbohydrate degradation | |
| Lactose PTS system repressor | fruR | Lin | Up | 1 | Feng et al., 2011 | Lactose transport | |
| Glucosamine-6-phosphate isomerase | nagB | Lin | Up | 1 | Feng et al., 2011 | Glucosamine metabolism | |
| Galactose-6-phosphate isomerase | lacB | Lin | Up | 1 | Feng et al., 2011 | Galactose metabolism | |
| Tagatose-6-phosphate kinase | lacC | Lin | Up | 1 | Feng et al., 2011 | Tagatose metabolism | |
| Tagatose 1,6-diphosphate aldolase | lacD | Lin | Up | 1 | Feng et al., 2011 | Tagatose metabolism | |
| β-N-acetylhexosaminidase | strH | Lin | Up | 1 | Feng et al., 2011 | Hexosamine metabolism | |
| β-galactosidase | bgaA | Lin | Up | 1 | Feng et al., 2011 | Lactose metabolism | |
| PTS system transporter subunit IIB | spr0563 | Lin | Up | 1 | Feng et al., 2011 | Carbohydrate transport | |
| PTS system transporter subunit IIA | spr0562 | Lin | Up | 1 | Feng et al., 2011 | Carbohydrate transport | |
| PTS system transporter subunit IIB | spr0060 | Lin | Up | 1 | Feng et al., 2011 | Carbohydrate transport | |
| Fructokinase | scrK | Lin | Up | 1 | Feng et al., 2011 | Fructose metabolism | |
| Glucokinase | glcK | Van | Down | 1 | Scherl et al., 2006 | Glucose metabolism | |
| Lin | Up | 1 | Feng et al., 2011 | ||||
| Catabolite control protein A | ccpA | Met | Down | 1 | Cordwell et al., 2002 | Carbon catabolite repression | |
| Van | Up | 1 | Wang et al., 2010 | ||||
| Lin | Up | 1 | Feng et al., 2011 |
Similar to prior reports, Drummelsmith et al. showed the high level inductions of cell wall metabolism-related proteins such as MecA, LytM, GlmS, and SceD in the VISA type strain Mu50 when compared with the vancomycin-sensitive strain CMRSA-2 (Drummelsmith et al., 2007). In particular, they selected SceD for further study based on its high level of induction (approximately 16-fold) in VISA, and relative sceD mRNA expression levels were compared between 25 VSSA and VISA clinical isolates by real-time RT-PCR (Drummelsmith et al., 2007). The sceD mRNA was significantly induced in all VISA isolates relative to all VSSA strains, and they suggest that SceD expression level could serve as a molecular diagnostic marker for the rapid detection of VISA (Drummelsmith et al., 2007). Interestingly, SceD was also up-regulated in both daptomycin-resistant (Song et al., 2013) and linezolid-resistant strains (Bernardo et al., 2004), suggesting the importance of this protein in antibiotic resistance. They also identified other proteins involved in cell envelope metabolism as a highly up-regulated protein in VISA; UDP-GlcNAc 1-carboxyvinyltransferase 1 (MurA), bifunctional autolysin (Atl), immunodominant antigen A (IsaA), UDP-glucose/GDP-mannose dehydrogenase (CapO), and UDP-N-acetyltalosamine 2-epimerase (CapG) (Table 6). Among them, IsaA was also up-regulated in VISA at other two studies (Scherl et al., 2006; Chen et al., 2013). In addition, its expression level increased in both methicillin-resistant and daptomycin-resistant strains (Cordwell et al., 2002; Fischer et al., 2011), and decreased in linezolid-resistant strains (Bernardo et al., 2004), suggesting the importance of this protein. The housekeeping protein IsaA is a highly immunogenic, non-covalently cell wall-bound lytic transglycosylase that is co-regulated with a glycylglycine endopeptidase LytM (Stapleton et al., 2007; Lorenz et al., 2011). S. aureus has two putative peptidoglycan hydrolases, IsaA and SceD, and SceD can compensate for the loss of IsaA (Stapleton et al., 2007). The fact that both peptidoglycan hydrolases (IsaA and SceD) are involved in antibiotic resistance strongly indicates the importance of cell wall dynamics in antibiotic resistance mechanism.
Table 6.
Differentially expressed proteins identified by the quantitative proteomic approach: proteins involved in envelope biogenesis.
| Biological process | Protein name | Gene | Antibiotics | Regulation | Frequency of difference | References | Protein description |
|---|---|---|---|---|---|---|---|
| Cell wall, membrane, envelope biogenesis | Acyl carrier protein | acpP | Met | Up | 1 | Enany et al., 2014 | Membrane biosynthesis |
| 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | ispD | Met | Up | 1 | Enany et al., 2014 | Isoprenoid biosynthesis | |
| Capsular polysaccharide synthesis enzyme | cap8H | Van | Up | 1 | Scherl et al., 2006 | Capsular polysaccharide biosynthesis | |
| Isopentenyl-diphosphate delta-isomerase | fni | Van | Down | 1 | Drummelsmith et al., 2007 | Biosynthesis of isoprenoids | |
| Malonyl CoA-ACP transacylase | fabD | Van | Down | 1 | Scherl et al., 2006 | Fatty acid biosynthesis | |
| Teichoic acid biosynthesis protein B | tagB | Van | Up | 1 | Scherl et al., 2006 | Teichoic acid biosynthesis | |
| Capsular polysaccharide synthesis protein Cap5D | capD | Van | Up | 1 | Scherl et al., 2006 | Capsular polysaccharide biosynthesis | |
| Capsular polysaccharide synthesis protein Cap5M | capM | Van | Up | 2 | Scherl et al., 2006; Hessling et al., 2013 | Capsular polysaccharide biosynthesis | |
| Capsular polysaccharide synthesis protein Cap5A | capA | Van | Up | 1 | Scherl et al., 2006 | Capsular polysaccharide biosynthesis | |
| UDP-glucose/GDP-mannose dehydrogenase | capO | Van | Up | 1 | Drummelsmith et al., 2007 | Capsular polysaccharide biosynthesis | |
| D-alanine-d-alanine ligase | ddl | Van | Up | 3 | Pieper et al., 2006; Hessling et al., 2013; Ramos et al., 2015 | Peptidoglycan biosynthesis | |
| D-alanine-d-alanine dipeptidase | ddpX | Van | Up | 1 | Ramos et al., 2015 | Peptidoglycan biosynthesis | |
| D-alanine-d-lactate dipeptidase | vanX | Van | Up | 1 | Wang et al., 2010 | Peptidoglycan biosynthesis | |
| D-alanine-d-lactate ligase | vanB | Van | Up | 1 | Wang et al., 2010 | Peptidoglycan biosynthesis | |
| Surface determinant protein A | isdA | Van | Down | 2 | Scherl et al., 2006; Drummelsmith et al., 2007 | ||
| UDP-N-acetyltalosamine 2-epimerase | capG | Van | Up | 1 | Drummelsmith et al., 2007 | Capsular polysaccharide biosynthesis | |
| Glycosyltransferase | sgtB | Van | Up | 1 | Scherl et al., 2006 | ||
| Penicillin binding protein 2A | mecA | Van | Up | 2 | Scherl et al., 2006; Drummelsmith et al., 2007 | Peptidoglycan biosynthesis | |
| Peptidoglycan hydrolase | lytM | Van | Up | 2 | Pieper et al., 2006; Drummelsmith et al., 2007 | Peptidoglycan degradation | |
| UDP-N-acetylmuramyl tripeptide synthetase | murE | Van | Up | 1 | Scherl et al., 2006 | Peptidoglycan biosynthesis | |
| Enoyl-CoA hydratase | phaB | Lin | Down | 1 | Feng et al., 2011 | Fatty acid β-oxidation | |
| 3-Ketoacyl-ACP reductase | fabG | Lin | Down | 1 | Feng et al., 2011 | Fatty acids biosynthesis | |
| Acetyl-CoA carboxylase biotin carboxyl carrier protein subunit | accB | Lin | Down | 1 | Feng et al., 2011 | Fatty acid biosynthesis | |
| Acetyl-CoA carboxylase subunit α | accA | Lin | Down | 1 | Feng et al., 2011 | Fatty acid biosynthesis | |
| Control of cell shape; membrane-associated protein | mreBH | Dap | Up | 1 | Wecke et al., 2009 | Control of cell shape | |
| Squalene synthase | crtN | Dap | Down | 1 | Fischer et al., 2011 | Isoprenoid biosynthesis | |
| Glucosamine-fructose-6-phosphate aminotransferase | glmS | Van | Up | 2 | Pieper et al., 2006; Drummelsmith et al., 2007 | Peptidoglycan biosynthesis | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| UDP-GlcNAc 1-carboxyvinyltransferase 1 | murA | Van | Up | 1 | Drummelsmith et al., 2007 | Peptidoglycan biosynthesis | |
| Dap | Down | 1 | Fischer et al., 2011 | ||||
| 3-Oxoacyl-ACP synthase II | fabF | Van | Up | 2 | Scherl et al., 2006; Wang et al., 2010 | Fatty acid biosynthesis | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Bifunctional N-acetylglucosamine-1-phosphate uridyltransferase/glucosamine-1-phosphate acetyltransferase | glmU | Lin | Up | 1 | Feng et al., 2011 | Peptidoglycan biosynthesis | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| CHAP (Cysteine, Histidine-dependent Amidohydrolases/Peptidases)-domain amidase | ssaA | Met | Up | 1 | Cordwell et al., 2002 | Peptidoglycan degradation | |
| Van | Up | 2 | Scherl et al., 2006; Drummelsmith et al., 2007 | ||||
| Lin | Down | 1 | Bernardo et al., 2004 | ||||
| Triacylglycerol lipase precursor | lipA | Met | Up | 1 | Enany et al., 2014 | Lipoate biosynthesis | |
| Lin | Up | 1 | Bernardo et al., 2004 | ||||
| Dap | Down | 1 | Fischer et al., 2011 | ||||
| Aminoacyltransferase | femA | Met | Up | 1 | Cordwell et al., 2002 | Peptidoglycan biosynthesis | |
| Van | Up | 2 | Scherl et al., 2006; Hessling et al., 2013 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Penicillin-binding protein 1 | pbpA | Met | Up | 1 | Cordwell et al., 2002 | Peptidoglycan biosynthesis | |
| Van | Up | 1 | Scherl et al., 2006 | ||||
| Dap | Down | 1 | Fischer et al., 2011 | ||||
| Hydroxymyristoyl ACP dehydratase | fabZ | Met | Up | 1 | Enany et al., 2014 | Fatty acid biosynthesis | |
| Van | Down | 1 | Drummelsmith et al., 2007 | ||||
| Lin | Down | 1 | Feng et al., 2011 | ||||
| Dap | Down | 1 | Fischer et al., 2011 |
To identify the resistance mechanisms of hVISA with a vancomycin MIC of ≤2 μg/ml, Chen et al. compared proteomic profiles of six pairs of isogenic hVISA and VSSA strains and unrelated hVISA (n = 24) and VSSA stains (n = 30) (Chen et al., 2013). They identified five proteins up-regulated in the hVISA strains; IsaA, MsrA, Asp32, 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase (GpmA), and AhpC. Consistent with this result, MsrA was up-regulated in a prior study using comparative proteomics (Scherl et al., 2006) and in the DNA microarray study, and the msrA gene was also over-expressed in VISA strains (Cui et al., 2005). MsrA, catalyzing the reversible oxidation-reduction of methionine sulfoxide to methionine, has a key function as a repair enzyme for proteins inactivated by oxidation (Chen et al., 2013). The msrA gene is highly induced by cell wall-active antibiotics, such as oxacillin and vancomycin (Chen et al., 2013). The increased level of MsrA can enhance peptidoglycan biosynthesis which results in cell wall thickening, and gene knockout of the msrA gene weakened vancomycin and β-lactam resistance of S. aureus strains (Cui et al., 2005). In addition, MsrA is involved in virulence in several bacteria (Sasindran et al., 2007). Taken together, these observations suggest the important role of methionine sulfoxide in antibiotic resistance. Although in other studies, the abundance of GpmA, which plays a physiological role in glycolysis, has been reported to be changed in VISA (Table 3), its exact role in antibiotic resistance has not been determined. AhpC, an alkyl hydroperoxide reductase subunit C, plays an important role in oxidative-stress resistance of S. aureus (Cosgrove et al., 2007). Interestingly, it was reported that AhpC is up-regulated in strains resistant to methicillin, vancomycin, and daptomycin antibiotics (Table 7). However, up to now, there is no report investigating the direct role of AhpC in antibiotic resistance. It is noteworthy that several proteins involved in oxidative-stress resistance, such as AhpC, SodA, catalase (KatA), and superoxide dismutase (SodM), show the abundance change of proteins in antibiotic-resistant strains (Table 7), and in most cases, their expression is up-regulated. In spite of these interesting results, the relationship between these proteins and antibiotic resistance was not determined.
Table 7.
Differentially expressed proteins identified by the quantitative proteomic approach: proteins involved in stress response.
| Biological process | Protein name | Gene | Antibiotics | Regulation | Frequency of difference | References | Protein description |
|---|---|---|---|---|---|---|---|
| General stress-related proteins | Cold shock protein | cspA | Met | Up | 1 | Cordwell et al., 2002 | Cold shock tolerance |
| Dps family protein | dps | Van | Down | 1 | Ramos et al., 2015 | Protection of DNA from damage | |
| Two-component regulator protein | vanR | Van | Up | 1 | Ramos et al., 2015 | The VanS/VanR two-component system in response to extracellular glycopeptide antibiotic | |
| Lactoylglutathione lyase | gloA | Van | Up | 1 | Wang et al., 2010 | Methylglyoxal degradation | |
| Cell stress stimulon response regulator | vraR | Van | Up | 2 | Scherl et al., 2006; Drummelsmith et al., 2007 | The two-component regulatory system VraS/VraR involved in the control of the cell wall peptidoglycan biosynthesis | |
| HTH-type transcriptional regulator | sarS | Van | Down | 1 | Drummelsmith et al., 2007 | Transcriptional regulator that controls expression of some virulence factors in a cell density-dependent manner | |
| Accessory gene regulator A | agrA | Van | Down | 2 | Scherl et al., 2006; Drummelsmith et al., 2007 | The regulation of virulence proteins | |
| Signal transduction protein TRAP | traP | Van | Up | 1 | Scherl et al., 2006 | A major regulator of staphylococcal pathogenesis | |
| Thioredoxin reductase | trxB | Van | Down | 1 | Drummelsmith et al., 2007 | Thioredoxin pathway | |
| Competence protein | cglA | Lin | Down | 1 | Feng et al., 2011 | Competence regulation | |
| Competence protein | cglB | Lin | Down | 1 | Feng et al., 2011 | Competence regulation | |
| Phosphate transporter | phoU | Lin | Down | 1 | Feng et al., 2011 | Phosphate starvation | |
| Conserved membrane protein; phage-shock protein A homolog (three-component regulatory system) | liaIH | Dap | Up | 1 | Wecke et al., 2009 | Regulation of membrane permeability | |
| Undecaprenyl pyrophosphate phosphatase | bcrC | Dap | Up | 1 | Wecke et al., 2009 | Bacitracin resistance | |
| Superoxide dismutase | sodM | Met | Up | 2 | Cordwell et al., 2002; Enany et al., 2014 | Resistance to oxidative stress | |
| Van | Down | 1 | Drummelsmith et al., 2007 | ||||
| Competence damage-inducible protein A | cinA | Van | Up | 1 | Pieper et al., 2006 | Competence regulation | |
| Lin | Down | 1 | Feng et al., 2011 | ||||
| Two-component sensor histidine kinase | vraS | Van | Up | 1 | Scherl et al., 2006 | The two-component regulatory system VraS/VraR involved in the control of the cell wall peptidoglycan biosynthesis | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Staphylococcus accessory regulator A | sarA | Van | Up | 1 | Drummelsmith et al., 2007 | Regulation of the virulence factors | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| GTP pyrophosphokinase | relA | Van | Down | 1 | Drummelsmith et al., 2007 | Stringent response | |
| Dap | Up | 2 | Wecke et al., 2009; Fischer et al., 2011 | ||||
| Choline dehydrogenase | betA | Van | Up | 1 | Scherl et al., 2006 | Glycine betaine biosynthesis | |
| Dap | Down | 1 | Fischer et al., 2011 | ||||
| GTP-sensing transcriptional pleiotropic repressor | codY | Lin | Up | 1 | Feng et al., 2011 | Transcription regulation in response to the GTP level | |
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Alkaline shock protein 23 | asp23 | Met | Up | 2 | Cordwell et al., 2002; Enany et al., 2014 | Alkaline pH tolerance | |
| Van | Down | 1 | Hessling et al., 2013 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Catalase | katA | Met | Up | 1 | Cordwell et al., 2002 | Resistance to oxidative stress | |
| Van | Up | 1 | Scherl et al., 2006 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Superoxide dismutase | sodA | Met | Up | 2 | Cordwell et al., 2002; Enany et al., 2014 | Resistance to oxidative stress | |
| Van | Up | 1 | Wang et al., 2010 | ||||
| Lin | Up | 1 | Feng et al., 2011 | ||||
| Cold shock protein | cspB | Met | Up | 1 | Cordwell et al., 2002 | Cold shock tolerance | |
| Van | Down | 1 | Drummelsmith et al., 2007 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Cold shock protein | cspC | Met | Up | 1 | Cordwell et al., 2002 | Cold shock tolerance | |
| Van | Down | 1 | Drummelsmith et al., 2007 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Alkyl hydroperoxide reductase subunit C | ahpC | Met | Up | 1 | Enany et al., 2014 | Resistance to oxidative stress | |
| Van | Up | 2 | Scherl et al., 2006; Chen et al., 2013 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Virulence-related proteins | Secreted virulence factor | esxA | Van | Down | 1 | Drummelsmith et al., 2007 | Pathogenesis |
| Extracellular ECM and plasma binding protein | ssp | Van | Up | 1 | Scherl et al., 2006 | Pathogenesis | |
| Cell surface-associated protein | sdrE | Van | Down | 1 | Hessling et al., 2013 | Pathogenesis | |
| Clumping factor A | clfA | Van | Down | 1 | Hessling et al., 2013 | Pathogenesis | |
| Secretory extracellular matrix and plasma binding protein | empbp | Van | Down | 1 | Hessling et al., 2013 | Pathogenesis | |
| Enterotoxin type I | sei | Van | Down | 1 | Hessling et al., 2013 | Pathogenesis | |
| Cysteine protease precursor | sspB1 | Van | Down | 1 | Hessling et al., 2013 | Pathogenesis | |
| Leukotoxin | lukD | Van | Down | 1 | Hessling et al., 2013 | Pathogenesis | |
| Leukotoxin | lukE | Van | Down | 1 | Hessling et al., 2013 | Pathogenesis | |
| Phospholipase C | hlb | Van | Down | 1 | Hessling et al., 2013 | Pathogenesis | |
| HysA | hysA | Van | Down | 1 | Hessling et al., 2013 | Pathogenesis | |
| γ-hemolysin, component C | hlgC | Van | Down | 1 | Hessling et al., 2013 | Pathogenesis | |
| Lipase | geh | Van | Down | 1 | Hessling et al., 2013 | Pathogenesis | |
| Accessory protein Z | sarZ | Van | Down | 1 | Hessling et al., 2013 | Pathogenesis | |
| α-hemolysin | SAV1163 | Lin | Down | 1 | Bernardo et al., 2004 | Pathogenesis | |
| Respiratory response protein | srrA | Met | Up | 1 | Cordwell et al., 2002 | Pathogenesis | |
| Van | Down | 1 | Scherl et al., 2006 | ||||
| Fibrinogen-binding protein | efb | Met | Up | 1 | Enany et al., 2014 | Pathogenesis | |
| Van | Down | 1 | Hessling et al., 2013 | ||||
| Immunoglobulin G binding protein A | spa | Van | Down | 2 | Pieper et al., 2006; Drummelsmith et al., 2007 | Pathogenesis | |
| Lin | Down | 1 | Bernardo et al., 2004 | ||||
| Bifunctional autolysin | atl | Van | Up | 1 | Drummelsmith et al., 2007 | Pathogenesis; Cell wall biogenesis/degradation | |
| Lin | Down | 1 | Bernardo et al., 2004 | ||||
| Dap | Up | 1 | Fischer et al., 2011 | ||||
| Immunodominant antigen A | isaA | Met | Up | 1 | Cordwell et al., 2002 | Pathogenesis; Cell wall biogenesis/degradation | |
| Van | Up | 3 | Scherl et al., 2006; Drummelsmith et al., 2007; Chen et al., 2013 | ||||
| Lin | Down | 1 | Bernardo et al., 2004 | ||||
| Dap | Up | 1 | Fischer et al., 2011 |
Hassling et al. analyzed proteomic profiles of vancomycin-susceptible S. aureus strain COL under the sublethal vancomycin exposure (4.5 μg/ml) (Hessling et al., 2013). They found the specific increase of proteins involved in the synthesis of lysine which are essential for the synthesis of the peptidoglycan precursor pentapeptide; aspartate kinase (LysC), aspartate semialdehyde dehydrogenase (Asd), dihydrodipicolinate synthase (DapA), 4-hydroxy-tetrahydrodipicolinate reductase (DapB), diaminopimelate decarboxylase (LysA), and tetrahydrodipicolinate acetyltransferase (DapD). An increase of lysine synthesis proteins can lead to an overall increase of peptidoglycan synthesis. Induction of genes involved in lysine synthesis under cell wall stress conditions have been documented before by two transcriptome studies (Kuroda et al., 2003; Sobral et al., 2007). Consistent with the previous report (Scherl et al., 2006), this report also showed that several proteins regulated by the two-component system VraSR increased in amount after vancomycin addition (Hessling et al., 2013). Additionally, they identified two important regulators (the alternative sigma factor σB and the two-component system SaeRS regulating numerous virulence genes) that play a role in vancomycin stress response. The cluster of proteins under positive σB control mainly increased, whereas negatively regulated proteins primarily decreased in amount after vancomycin addition (Hessling et al., 2013). The induction of σB regulon by vancomycin has been found in another report (Chen et al., 2013). Increase of the σB activity has also been observed in strains resistant to teicoplanin (Bischoff and Berger-Bächi, 2001) or methicillin (Cordwell et al., 2002). Hassling et al. also found decreased expression levels of most proteins with a virulence related function (Hessling et al., 2013). However, because the great majority of virulence genes in previous transcriptome studies under cell wall stress in S. aureus have been shown to be up-regulated (Kuroda et al., 2003; Utaida et al., 2003; Sobral et al., 2007), the role of virulence genes in antibiotic resistance needs to be determined.
Lastly, Wang et al. and Ramos et al. performed proteomic analysis of vancomycin-resistant E. faecalis strains (V583, V306, and SU18) under 64 μg/ml vancomycin treatment (Wang et al., 2010; Ramos et al., 2015). Vancomycin induced expression of vancomycin resistance-related proteins such as VanA, VanX, D-Ala-D-Ala dipeptidase (DdpX), VanR, and VanB (Wang et al., 2010; Ramos et al., 2015). Distinctively, Wang et al. found that six proteins (Pgm, Ldh, Gap-2, RpsB, EF2076, and sex pheromone cAD1 precursor lipoprotein) exhibited clear post-translational modifications and vancomycin induced phosphorylation of Ser/Thr in Ldh, Gap-2, and sex pheromone cAD1 precursor lipoprotein (EF3256) (Wang et al., 2010). Ramos et al. showed that metabolism-related proteins, such as TipA, GMP synthase (GuaA), and glyceraldehyde-3-phosphate dehydrogenase (GapB), were down-regulated under vancomycin treatment (Ramos et al., 2015).
Linezolid
There was one study exploring comparative proteomic profiles in linezolid-susceptible S. pneumonia strains and linezolid-resistant S. pneumonia strains, and one study analyzing global proteomes of a linezolid- susceptible S. aureus under linezolid stresses (Bernardo et al., 2004; Feng et al., 2011). Through the comparison between linezolid-susceptible S. pneumonia strains (1974 and R6) with linezolid MICs of 0.5–0.75 μg/ml and linezolid-resistant S. pneumonia strains (1974M2-LZD and R6M2-LZD) with MIC of 32 μg/ml, Feng et al. showed that the proteomic and transcriptomic approaches were poorly correlated with previously known resistance factors (23S rRNA, ribosomal proteins L3 and L4, RNA methyltransferase Cfr, and ABC transporter PatA and PatB), as modulated proteins rarely had significant concomitant changes at the expression level (Feng et al., 2011). They found increased expression of proteins involved in the metabolism and transport of carbohydrates in linezolid-resistant S. pneumoniae strains (Feng et al., 2011). Through inactivation of target genes in the linezolid-resistant strains (1974M2-LZD and R6M2-LZD), they identified two ABC transporter substrate-binding proteins (Spr0083 and Spr1527) and the catabolite control protein A (CcpA) as factors associated with resistance to linezolid (Feng et al., 2011). CcpA is known to function as the global regulator controlling the efficient utilization of sugars through carbon catabolite repression (CCR) in Gram-positive bacteria (Stülke and Hillen, 2000). Inactivation of the ccpA gene in S. aureus affected growth, glucose metabolism, and expression of virulence genes (Seidl et al., 2006). CcpA inactivation was also linked to the down-regulation of glycolytic genes in Bacillus cereus (van der Voort et al., 2008; Feng et al., 2011). Therefore, the increased level of CcpA may cause the increased expression of glycolytic enzymes in linezolid-resistant S. pneumonia strains. In S. aureus, the correlation between antibiotic resistance and CcpA has already been reported, as CcpA inactivation significantly reduced the oxacillin resistance levels in MRSA and the teicoplanin resistance level in a glycopeptide-intermediate-resistant S. aureus strain (Seidl et al., 2006). Table 3 shows the possibility that CcpA may also be involved in methicillin and vancomycin resistance. Together with CcpA, inactivation of two ABC transporters putatively involved in the sugar transport (Spr0083 and Spr1527) also reduced resistance to linezolid of S. pneumonia (Feng et al., 2011). Notably, S. pneumoniae is predicted to be highly dependent on external sugars to fulfill its energy requirements by substrate-level phosphorylation as it lacks functional electron transport chain and tricarboxylic acid cycle (Tettelin et al., 2001; Feng et al., 2011). This process eventually leads to the formation of lactate and acetate by the lactate dehydrogenase and lactate oxidase enzymes and these proteins were also found to be overexpressed in linezolid-resistant S. pneumonia strains (Tettelin et al., 2001; Feng et al., 2011). Therefore, these results imply increased energy requirements associated with resistance mechanism to linezolid in S. pneumonia (Feng et al., 2011). To sustain a fitness cost associated with resistance mechanisms such as the 23S rRNA mutations (Besier et al., 2008), S. pneumonia seems to select an increased metabolism of sugars as a secondary adaptation.
This study also showed that several genes involved in the biosynthesis of fatty acids, including enoyl-CoA hydratase (PhaB), 3-ketoacyl-ACP reductase (FabG), acetyl-CoA carboxylase biotin carboxyl carrier protein subunit (AccB), acetyl-CoA carboxylase subunit alpha (AccA), and hydroxymyristoyl-ACP dehydratase (FabZ), were down-regulated in linezolid-resistant strains (Feng et al., 2011). Whether this is directly related to linezolid resistance remains to be established, but it is intriguing that the cell wall inhibitor penicillin also causes a down-regulation of several genes of this pathway in S. pneumoniae (Rogers et al., 2007; Feng et al., 2011). Interestingly, expression levels of FabZ are changed in all cases of the four antibiotics (Table 6), even though its expression increased in methicillin-resistant strains and decreased in strains resistant to other antibiotics. Many numbers of ribosomal proteins were found to be overexpressed or down-regulated in linezolid-resistant strains, but whether this pattern is due to the mechanism of action of linezolid (which targets the ribosome) remains to be established. Although recent several lines of evidence indicate the presence of functional selective ribosomal subpopulations that exhibit variations in the RNA or the protein components and modulate the translational program in response to environmental changes (Byrgazov et al., 2013), it is difficult to obtain any information from variation patterns of ribosomal proteins in this study.
Bernardo et al. compared the change of proteomic profiles of a linezolid- susceptible S. aureus strain ATCC 29213 (MIC = 2.5 μg/ml) under linezolid stresses (12.5, 25, 50, and 90% of MIC) (Bernardo et al., 2004). They found that linezolid reduced in a dose-dependent manner the secretion of specific virulence factors, including bifunctional autolysin (Atl), immunoglobulin G binding protein A (Spa), and α-hemolysin (SAV1163), CHAP-domain amidase (SsaA), and immunodominant antigen A (IsaA). This result is similar to the proteomic result that analyzes protein profiles of S. aureus under the sublethal vancomycin exposure (Hessling et al., 2013).
Daptomycin
There were one study examining comparative proteomic profiles in daptomycin-susceptible and daptomycin-resistant S. aureus strains, and one study analyzing global proteomes of daptomycin-susceptible B. subtilis under daptomycin stress (Wecke et al., 2009; Fischer et al., 2011). Unlike other three antibiotics (methicillin, vancomycin, and linezolid), specific genetic determinant of the daptomycin-resistant strain was not determined. Probable daptomycin resistance-related proteins (MprF, YycG, RpoB, and RpoC) identified in previous reports (Jones et al., 2008; Baltz, 2009) were not identified in comparative proteomic analyses (Tables 2–8). In 2011, Fisher et al. compared proteomic profiles in the daptomycin-susceptible S. aureus strain 616 with a daptomycin MIC of 0.5 μg/ml and the daptomycin-resistant S. aureus strain 701 with MIC of 2 μg/ml (Fischer et al., 2011). Comparative proteomics and transcriptomic approach revealed a differential abundance of proteins in various functional categories, including cell wall-associated targets and biofilm formation proteins (Fischer et al., 2011). Phenotypically, daptomycin-susceptible strains, and daptomycin-resistant strains showed major differences in their ability to develop bacterial biofilms in the presence of the antibacterial lipid, oleic acid (Fischer et al., 2011). Transcriptomic approach showed different expressions of some important genes, such as the key genes (yycFGHI) affecting cell membrane lipid homeostasis, cell wall metabolism and biofilm formation, and two-component regulation system genes (agr, saeRS, and vraRS) involved in pathogenesis of methicillin-resistant strains (Fischer et al., 2011). However, through proteomic research, only several proteins, including Asp23, 3-oxoacyl-ACP synthase II (FabF), GTP-sensing transcriptional pleiotropic repressor (CodY), and PurH, was identified as proteins involved in daptomycin resistance.
Table 8.
Differentially expressed proteins identified by the quantitative proteomic approach: proteins of unknown function.
| Biological process | Protein name | Gene | Antibiotics | Regulation | Frequency of difference | References | Protein description |
|---|---|---|---|---|---|---|---|
| General function prediction only | Metal-dependent hydrolase | SA1529 | Met | Down | 1 | Cordwell et al., 2002 | Protein degradation |
| Aldehyde dehydrogenase | SAV2122 | Met | Up | 1 | Enany et al., 2014 | Energy metabolism | |
| Putative transaldolase | tal | Van | Down | 1 | Drummelsmith et al., 2007 | Energy metabolism | |
| Putative transcription factor | SA2296 | Van | Up | 1 | Scherl et al., 2006 | Gene expression | |
| Penicillin binding methicillin resistant-related protein | fmtA | Van | Up | 1 | Scherl et al., 2006 | Peptidoglycan biosynthesis | |
| Putative cell wall transglycosylase | sceD | Van | Up | 1 | Pieper et al., 2006 | Peptidoglycan degradation | |
| Lin | Up | 1 | Bernardo et al., 2004 | ||||
| ABC protein/substrate binding protein subunit—Sugar transport | spr0083 | Lin | Up | 1 | Feng et al., 2011 | Carbohydrate transport | |
| ABC protein/substrate binding protein subunit—sugar transport | spr1527 | Lin | Up | 1 | Feng et al., 2011 | Carbohydrate transport | |
| Maltose/maltodextrin-binding protein | SA0207 | Dap | Down | 1 | Fischer et al., 2011 | Carbohydrate transport | |
| Function unknown | Unknown | SA1238 | Met | Up | 1 | Cordwell et al., 2002 | Unknown |
| Unknown | SA1051 | Met | Up | 1 | Cordwell et al., 2002 | Unknown | |
| Unknown | SA0940 | Met | Up | 1 | Cordwell et al., 2002 | Unknown | |
| Unknown | SA1868 | Met | Up | 1 | Cordwell et al., 2002 | Unknown | |
| Unknown | SA1813 | Met | Up | 1 | Cordwell et al., 2002 | Unknown | |
| Unknown | SA2302 | Met | Down | 1 | Cordwell et al., 2002 | Unknown | |
| Unknown | SA0759 | Met | Down | 1 | Cordwell et al., 2002 | Unknown | |
| Unknown | SA1812 | Met | Up | 1 | Cordwell et al., 2002 | Unknown | |
| Unknown | SA0587 | Met | Up | 1 | Cordwell et al., 2002 | Unknown | |
| Unknown | SA0772 | Met | Up | 1 | Cordwell et al., 2002 | Unknown | |
| Unknown | SA0587 | Met | Up | 1 | Cordwell et al., 2002 | Unknown | |
| Unknown | SA1455 | Met | Up | 1 | Cordwell et al., 2002 | Unknown | |
| Unknown | SA0919 | Met | Down | 1 | Cordwell et al., 2002 | Unknown | |
| Unknown | SA1709 | Met | Down | 1 | Cordwell et al., 2002 | Unknown | |
| Unknown | SA0022 | Van | Down | 1 | Scherl et al., 2006 | Unknown | |
| Unknown | SA2113 | Van | Up | 1 | Scherl et al., 2006 | Unknown | |
| Unknown | vraX | Van | Up | 1 | Scherl et al., 2006 | Unknown | |
| Unknown | SA0423 | Lin | Down | 1 | Bernardo et al., 2004 | Unknown | |
| Unknown | SAV0719 | Lin | Down | 1 | Bernardo et al., 2004 | Unknown | |
| Unknown | spr1987 | Lin | Down | 1 | Feng et al., 2011 | Unknown | |
| Unknown | spr0033 | Lin | Up | 1 | Feng et al., 2011 | Unknown | |
| Unknown | spr0125 | Lin | Down | 1 | Feng et al., 2011 | Unknown | |
| Unknown | spr0895 | Lin | Up | 1 | Feng et al., 2011 | Unknown | |
| Unknown | spr0618 | Lin | Up | 1 | Feng et al., 2011 | Unknown | |
| Unknown | spr0997 | Lin | Down | 1 | Feng et al., 2011 | Unknown | |
| Unknown | spr1332 | Lin | Up | 1 | Feng et al., 2011 | Unknown | |
| Unknown | spr1693 | Lin | Up | 1 | Feng et al., 2011 | Unknown | |
| Unknown | spr1726 | Lin | Up | 1 | Feng et al., 2011 | Unknown | |
| Unknown | spr1758 | Lin | Down | 1 | Feng et al., 2011 | Unknown | |
| Unknown | spr2029 | Lin | Down | 1 | Feng et al., 2011 | Unknown | |
| Unknown | spr0174 | Lin | Up | 1 | Feng et al., 2011 | Unknown | |
| Unknown | spr0370 | Lin | Up | 1 | Feng et al., 2011 | Unknown | |
| Unknown | SA0269 | Dap | Down | 1 | Fischer et al., 2011 | Unknown | |
| Unknown | SA0591 | Van | Up | 1 | Scherl et al., 2006 | Unknown | |
| Unknown | SA1528 | Met | Down | 1 | Cordwell et al., 2002 | Unknown | |
| Dap | Down | 1 | Fischer et al., 2011 |
Wecke et al. searched proteins induced by daptomycin, through the proteomic approach of a daptomycin-susceptible B. subtilis strain W168 under daptomycin treatment of sublethal amount (1 μg/ml) (Wecke et al., 2009). They identified LiaI and LiaH proteins exclusively and strongly induced (429-fold) by daptomycin. This result is in good agreement with data analyzing genes induced by daptomycin through transcriptome profiling (Muthaiyan et al., 2008). LiaH is a conserved membrane protein similar to a phage shock protein A (PspA) of E. coli, and its expression is regulated by the cell envelope stress-sensing two-component system LiaRS (Jordan et al., 2006; Hachmann et al., 2009; Wecke et al., 2009). Inactivation of liaH leads to 3-fold increased susceptibility to daptomycin and this susceptibility was further exacerbated in cells additionally lacking the paralogous gene pspA (Hachmann et al., 2009). In E. coli, the pspA gene is induced upon phage infection, osmotic shock, exposure to ethanol, or temperature increase, and functions to help cells manage the impacts of agents impairing cell membrane function (Joly et al., 2010). A recent report showed that deletion of the response regulator LiaR regulating expression of liaIH in daptomycin-resistant E. faecalis reversed resistance to daptomycin, and resulted in hypersusceptibility to daptomycin (Reyes et al., 2015). Therefore, these results indicate that LiaR is a master regulator protecting cell membrane to diverse antimicrobial agents, through regulating expression of various genes such as the liaH gene (Reyes et al., 2015).
Conclusion
Although specific genetic determinants of resistance mechanisms to methicillin, vancomycin, and linezolid were identified through non-proteomic approaches (e.g., van genes in vancomycin resistance) (Table 1), recent comparative proteomic methods provide new opportunities to understand the antibiotic resistance mechanism. In particular, in the case of recently used antibiotics such as daptomycin, specific genetic determinant(s) of antibiotic resistance was not fully determined through non-proteomic approaches. Therefore, quantitative proteomic methods can be a good tool to find an important protein involved in daptomycin resistance. Actually, a proteomic research identified LiaH as a highly induced protein by daptomycin treatment (Muthaiyan et al., 2008) and a subsequent report showed that the expression level of this protein is important to daptomycin-resistant phenotype (Reyes et al., 2015). These results show that quantitative proteomic analysis can be used as an effective tool to find novel resistance mechanisms.
Interestingly, comparative proteomic approaches in methicillin, linezolid, and daptomycin, except for vancomycin, were poorly correlated with known resistance-related factors found by non-proteomic approaches (Table 2). This result may be caused by a lack of comparative proteomic studies in three antibiotics, or imply the existence of novel resistance mechanisms different from previously known resistance mechanisms found by non-proteomic approaches. Through summarizing recent data of comparative proteomic researches of four clinically important antibiotics, we can find proteins of which expression levels are changed only in the resistance mechanism to specific antibiotic, such as LiaH in daptomycin resistance and PrsA in vancomycin resistance. It is necessary to determine whether these proteins affect antibiotic resistance through regulating previously known resistance-related determinants or by a novel mechanism. Another interesting result is that many proteins identified by comparative proteomic analyses seem to be simultaneously involved in resistance mechanism to two or more antibiotics (Tables 2–8). These proteins include cold shock proteins (CspABC), sporulation protein G (SpoVG), alkyl hydroperoxide reductase subunit C (AhpC), L-lactate dehydrogenase (LdhA), triacylglycerol lipase precursor (LipA), superoxide dismutase (SodA), catalase (KatA), elongation factor G (FusA), CHAP-domain amidase (SsaA), two component system (VraSR), penicillin binding methicillin resistant-related protein (FmtA), adenylosuccinate lyase (PurB), glucose-6-phosphate isomerase (Pgi), catabolite control protein A (CcpA), putative cell wall transglycosylase (SceD), immunodominant antigen A (IsaA), bifunctional autolysin (Atl), the σB regulon, and hydroxymyristoyl-ACP dehydratase (FabZ). These proteins can be divided into two groups, proteins involved in bacterial envelope regulation and proteins compensating for a fitness cost of antibiotic resistance. Proteins such as LipA, VraSR, FmtA, SsaA, SceD, IsaA, Atl, and FabZ, are directly or indirectly involved in envelope regulation. In order to modify or thicken the bacterial cell wall for antibiotic resistance, cells require abundant energy, and proteins involved in stress adaptation are necessary to neutralize various damages by antibiotic. To sustain these fitness costs associated with resistance mechanisms, proteins involved in energy metabolism (LdhA, FusA, Pgi, PurB, and CcpA) and stress-related proteins (CspABC, SpoVG, AhpC, SodA, KatA, and the σB regulon) seem to be identified in resistance mechanisms to several antibiotics. Therefore, these proteomic results confirm that antibiotic resistance requires a fitness cost.
Detailed studies on the mechanism by which these proteins affect antibiotic resistance are required. In particular, because these proteins can act as the global factor affecting resistance mechanisms to most antibiotics, it is necessary to examine whether they affect resistance mechanism of other antibiotics having different action modes. These studies will provide important clues for understanding and managing antibiotic resistance.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the National Research Laboratory Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (No. 2011-0027928); the Cooperative Research Program for Agriculture Science and Technology Development (No. PJ01103103) of Rural Development Administration in Republic of Korea; and the National Research Foundation of the Ministry of Education, Republic of Korea (2012R1A1A2044184).
References
- Ament P. W., Jamshed N., Horne J. P. (2002). Linezolid: its role in the treatment of gram-positive, drug-resistant bacterial infections. Am. Fam. Physician. 65, 663–671. Available online at: http://www.aafp.org/afp/2002/0215/p663.html [PubMed] [Google Scholar]
- Baltz R. H. (2009). Daptomycin: mechanisms of action and resistance, and biosynthetic engineering. Curr. Opin. Chem. Biol. 13, 144–151. 10.1016/j.cbpa.2009.02.031 [DOI] [PubMed] [Google Scholar]
- Berger-Bächi B., Barberis-Maino L., Strässle A., Kayser F. H. (1989). FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol. Gen. Genet. 219, 263–269. [DOI] [PubMed] [Google Scholar]
- Bernardo K., Pakulat N., Fleer S., Schnaith A., Utermöhlen O., Krut O., et al. (2004). Subinhibitory concentrations of linezolid reduce Staphylococcus aureus virulence factor expression. Antimicrob. Agents Chemother. 48, 546–555. 10.1128/AAC.48.2.546-555.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besier S., Ludwig A., Zander J., Brade V., Wichelhaus T. A. (2008). Linezolid resistance in Staphylococcus aureus: gene dosage effect, stability, fitness costs, and cross-resistances. Antimicrob. Agents Chemother. 52, 1570–1572. 10.1128/AAC.01098-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff M., Berger-Bächi B. (2001). Teicoplanin stress-selected mutations increasing σB activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 45, 1714–1720. 10.1128/AAC.45.6.1714-1720.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle-Vavra S., Yin S., Daum R. S. (2006). The VraS/VraR two-component regulatory system required for oxacillin resistance in community-acquired methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 262, 163–171. 10.1111/j.1574-6968.2006.00384.x [DOI] [PubMed] [Google Scholar]
- Boyle-Vavra S., Yin S., Jo D. S., Montgomery C. P., Daum R. S. (2013). VraT/YvqF is required for methicillin resistance and activation of the VraSR regulon in Staphylococcus aureus. Antimicrob. Agents Chemother. 57, 83–95. 10.1128/AAC.01651-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buntaran L., Hatta M., Sultan A. R., Dwiyanti R., Sabir M. (2013). Sccmec type II gene is common among clinical isolates of methicillin-resistant Staphylococcus aureus in Jakarta, Indonesia. BMC Res. Notes 6:110. 10.1186/1756-0500-6-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrgazov K., Vesper O., Moll I. (2013). Ribosome heterogeneity: another level of complexity in bacterial translation regulation. Curr. Opin. Microbiol. 16, 133–139. 10.1016/j.mib.2013.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers H. F. (1997). Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Liu Y., Zhao C., Xiao D., Zhang J., Zhang F., et al. (2013). Comparative proteomics-based identification of genes associated with glycopeptide resistance in clinically derived heterogeneous vancomycin-intermediate Staphylococcus aureus strains. PLoS ONE 8:e66880. 10.1371/journal.pone.0066880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordwell S. J., Larsen M. R., Cole R. T., Walsh B. J. (2002). Comparative proteomics of Staphylococcus aureus and the response of methicillin-resistant and methicillin-sensitive strains to Triton X-100. Microbiology. 148, 2765–2781. 10.1099/00221287-148-9-2765 [DOI] [PubMed] [Google Scholar]
- Cosgrove K., Coutts G., Jonsson I. M., Tarkowski A., Kokai-Kun J. F., Mond J. J., et al. (2007). Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J. Bacteriol. 189, 1025–1035. 10.1128/JB.01524-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courvalin P. (2005). Genetics of glycopeptide resistance in gram-positive pathogens. Int. J. Med. Microbiol. 294, 479–486. 10.1016/j.ijmm.2004.10.002 [DOI] [PubMed] [Google Scholar]
- Cuaron J. A., Dulal S., Song Y., Singh A. K., Montelongo C. E., Yu W., et al. (2013). Tea tree oil-induced transcriptional alterations in Staphylococcus aureus. Phytother. Res. 27, 390–396. 10.1002/ptr.4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Lian J. Q., Neoh H. M., Reyes E., Hiramatsu K. (2005). DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 49, 3404–3413. 10.1128/AAC.49.8.3404-3413.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaune A., Poupel O., Mallet A., Coic Y. M., Msadek T., Dubrac S. (2011). Peptidoglycan crosslinking relaxation plays an important role in Staphylococcus aureus WalKR-dependent cell viability. PLoS ONE 6:e17054. 10.1371/journal.pone.0017054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler V., Meier P. S., Heusser R., Berger-Bächi B., McCallum N. (2011). Induction kinetics of the Staphylococcus aureus cell wall stress stimulon in response to different cell wall active antibiotics. BMC Microbiol. 11:16. 10.1186/1471-2180-11-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummelsmith J., Winstall E., Bergeron M. G., Poirier G. G., Ouellette M. (2007). Comparative proteomics analyses reveal a potential biomarker for the detection of vancomycin-intermediate Staphylococcus aureus strains. J. Proteome Res. 6, 4690–4702. 10.1021/pr070521m [DOI] [PubMed] [Google Scholar]
- Enany S., Yoshida Y., Yamamoto T. (2014). Exploring extra-cellular proteins in methicillin susceptible and methicillin resistant Staphylococcus aureus by liquid chromatography-tandem mass spectrometry. World J. Microbiol. Biotechnol. 30, 1269–1283. 10.1007/s11274-013-1550-7 [DOI] [PubMed] [Google Scholar]
- Fan X., Liu Y., Smith D., Konermann L., Siu K. W., Golemi-Kotra D. (2007). Diversity of penicillin-binding proteins. Resistance factor FmtA of Staphylococcus aureus. J. Biol. Chem. 282, 35143–35152. 10.1074/jbc.M706296200 [DOI] [PubMed] [Google Scholar]
- Feng J., Billal D. S., Lupien A., Racine G., Winstall E., Légaré D., et al. (2011). Proteomic and transcriptomic analysis of linezolid resistance in Streptococcus pneumoniae. J. Proteome Res. 10, 4439–4452. 10.1021/pr200221s [DOI] [PubMed] [Google Scholar]
- Fischer A., Yang S. J., Bayer A. S., Vaezzadeh A. R., Herzig S., Stenz L., et al. (2011). Daptomycin resistance mechanisms in clinically derived Staphylococcus aureus strains assessed by a combined transcriptomics and proteomics approach. J. Antimicrob. Chemother. 66, 1696–1711. 10.1093/jac/dkr195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E. C., Wagner R., Radman M. (2002). Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science 296, 1627–1630. 10.1126/science.1070236 [DOI] [PubMed] [Google Scholar]
- Gardete S., Wu S. W., Gill S., Tomasz A. (2006). Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob. Agents Chemother. 50, 3424–3434. 10.1128/AAC.00356-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gov Y., Borovok I., Korem M., Singh V. K., Jayaswal R. K., Wilkinson B. J., et al. (2004). Quorum sensing in Staphylococci is regulated via phosphorylation of three conserved histidine residues. J. Biol. Chem. 279, 14665–14672. 10.1074/jbc.M311106200 [DOI] [PubMed] [Google Scholar]
- Hachmann A. B., Angert E. R., Helmann J. D. (2009). Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob. Agents Chemother. 53, 1598–1609. 10.1128/AAC.01329-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaki H., Inaba Y., Sasaki K., Hiramatsu K. (1998). A novel method of detecting Staphylococcus aureus heterogeneously resistant to vancomycin (hetero-VRSA). Jpn. J. Antibiot. 51, 521–530. [PubMed] [Google Scholar]
- Hao H., Dai M., Wang Y., Huang L., Yuan Z. (2012). Key genetic elements and regulation systems in methicillin-resistant Staphylococcus aureus. Future Microbiol. 7, 1315–1329. 10.2217/fmb.12.107 [DOI] [PubMed] [Google Scholar]
- Healy V. L., Lessard I. A., Roper D. I., Knox J. R., Walsh C. T. (2000). Vancomycin resistance in enterococci: reprogramming of the D-Ala-D-Ala ligases in bacterial peptidoglycan biosynthesis. Chem. Biol. 7, R109–R119. 10.1016/s1074-5521(00)00116-2 [DOI] [PubMed] [Google Scholar]
- Herrmann D. J., Peppard W. J., Ledeboer N. A., Theesfeld M. L., Weigelt J. A., Buechel B. J. (2008). Linezolid for the treatment of drug-resistant infections. Expert Rev. Anti. Infect. Ther. 6, 825–848. 10.1586/14787210.6.6.825 [DOI] [PubMed] [Google Scholar]
- Hessling B., Bonn F., Otto A., Herbst F. A., Rappen G. M., Bernhardt J., et al. (2013). Global proteome analysis of vancomycin stress in Staphylococcus aureus. Int. J. Med. Microbiol. 303, 624–634. 10.1016/j.ijmm.2013.08.014 [DOI] [PubMed] [Google Scholar]
- Hobbs J. K., Miller K., O'Neill A. J., Chopra I. (2008). Consequences of daptomycin-mediated membrane damage in Staphylococcus aureus. J. Antimicrob. Chemother. 62, 1003–1008. 10.1093/jac/dkn321 [DOI] [PubMed] [Google Scholar]
- Howden B. P., Grayson M. L. (2006). Dumb and dumber-the potential waste of a useful antistaphylococcal agent: emerging fusidic acid resistance in Staphylococcus aureus. Clin. Infect. Dis. 42, 394–400. 10.1086/499365 [DOI] [PubMed] [Google Scholar]
- Ippolito J. A., Kanyo Z. F., Wang D., Franceschi F. J., Moore P. B., Steitz T. A., et al. (2008). Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J. Med. Chem. 51, 3353–3356. 10.1021/jm800379d [DOI] [PubMed] [Google Scholar]
- Joly N., Engl C., Jovanovic G., Huvet M., Toni T., Sheng X., et al. (2010). Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 34, 797–827. 10.1111/j.1574-6976.2010.00240.x [DOI] [PubMed] [Google Scholar]
- Jones T., Yeaman M. R., Sakoulas G., Yang S. J., Proctor R. A., Sahl H. G., et al. (2008). Failures in clinical treatment of Staphylococcus aureus Infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 52, 269–278. 10.1128/AAC.00719-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S., Junker A., Helmann J. D., Mascher T. (2006). Regulation of LiaRS-dependent gene expression in bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 188, 5153–5166. 10.1128/JB.00310-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousselin A., Renzoni A., Andrey D. O., Monod A., Lew D. P., Kelley W. L. (2012). The posttranslocational chaperone lipoprotein PrsA is involved in both glycopeptide and oxacillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 56, 3629–3640. 10.1128/AAC.06264-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y., Zhang H. Z., Chambers H. F. (2004). PBP 2a mutations producing very-high-level resistance to β-lactams. Antimicrob. Agents Chemother. 48, 453–459. 10.1128/AAC.48.2.453-459.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koripella R. K., Chen Y., Peisker K., Koh C. S., Selmer M., Sanyal S. (2012). Mechanism of elongation factor-G-mediated fusidic acid resistance and fitness compensation in Staphylococcus aureus. J. Biol. Chem. 287, 30257–30267. 10.1074/jbc.M112.378521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M., Kuroda H., Oshima T., Takeuchi F., Mori H., Hiramatsu K. (2003). Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49, 807–821. 10.1046/j.1365-2958.2003.03599.x [DOI] [PubMed] [Google Scholar]
- Kuroda M., Kuwahara-Arai K., Hiramatsu K. (2000). Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269, 485–490. 10.1006/bbrc.2000.2277 [DOI] [PubMed] [Google Scholar]
- Lee C. R., Cho I. H., Jeong B. C., Lee S. H. (2013). Strategies to minimize antibiotic resistance. Int. J. Environ. Res. Public Health. 10, 4274–4305. 10.3390/ijerph10094274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Bae I. K., Lee S. H. (2012). New definitions of extended-spectrum β-lactamase conferring worldwide emerging antibiotic resistance. Med. Res. Rev. 32, 216–232. 10.1002/med.20210 [DOI] [PubMed] [Google Scholar]
- Levine D. P. (2006). Vancomycin: a history. Clin. Infect. Dis. 42(Suppl. 1), S5–S12. 10.1086/491709 [DOI] [PubMed] [Google Scholar]
- Long K. S., Vester B. (2012). Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob. Agents Chemother. 56, 603–612. 10.1128/AAC.05702-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz U., Lorenz B., Schmitter T., Streker K., Erck C., Wehland J., et al. (2011). Functional antibodies targeting IsaA of Staphylococcus aureus augment host immune response and open new perspectives for antibacterial therapy. Antimicrob. Agents Chemother. 55, 165–173. 10.1128/AAC.01144-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcone G. L., Beltrametti F., Binda E., Carrano L., Foulston L., Hesketh A., et al. (2010). Novel mechanism of glycopeptide resistance in the A40926 producer Nonomuraea sp. ATCC 39727. Antimicrob. Agents Chemother. 54, 2465–2472. 10.1128/AAC.00106-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno K., Sonenshein A. L. (1999). Role of SpoVG in asymmetric septation in Bacillus subtilis. J. Bacteriol. 181, 3392–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAleese F., Wu S. W., Sieradzki K., Dunman P., Murphy E., Projan S., et al. (2006). Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate-S. aureus-type resistance to vancomycin. J. Bacteriol. 188, 1120–1133. 10.1128/JB.188.3.1120-1133.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S., Cuirolo A. X., Plata K. B., Riosa S., Silverman J. A., Rubio A., et al. (2012). VraSR two-component regulatory system contributes to mprF-mediated decreased susceptibility to daptomycin in in vivo-selected clinical strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 56, 92–102. 10.1128/AAC.00432-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R. E., Deshpande L. M., Jones R. N. (2014). Linezolid update: stable in vitro activity following more than a decade of clinical use and summary of associated resistance mechanisms. Drug Resist. Updat. 17, 1–12. 10.1016/j.drup.2014.04.002 [DOI] [PubMed] [Google Scholar]
- Miao V., Coëffet-Legal M. F., Brian P., Brost R., Penn J., Whiting A., et al. (2005). Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology 151, 1507–1523. 10.1099/mic.0.27757-0 [DOI] [PubMed] [Google Scholar]
- Mishra N. N., Yang S. J., Sawa A., Rubio A., Nast C. C., Yeaman M. R., et al. (2009). Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53, 2312–2318. 10.1128/AAC.01682-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D. G., Group P. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 6:e1000097. 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongodin E., Finan J., Climo M. W., Rosato A., Gill S., Archer G. L. (2003). Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 185, 4638–4643. 10.1128/JB.185.15.4638-4643.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthaiyan A., Silverman J. A., Jayaswal R. K., Wilkinson B. J. (2008). Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob. Agents Chemother. 52, 980–990. 10.1128/AAC.01121-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano R., Janel-Bintz R., Wagner J., Fuchs R. P. (2000). All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 19, 6259–6265. 10.1093/emboj/19.22.6259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsom S. W. (2004). MRSA—past, present, future. J. R. Soc. Med. 97, 509–510. 10.1258/jrsm.97.11.509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S. E., Edwards D. H. (2004). Identification of a polar targeting determinant for Bacillus subtilis DivIVA. Mol. Microbiol. 54, 1237–1249. 10.1111/j.1365-2958.2004.04363.x [DOI] [PubMed] [Google Scholar]
- Pieper R., Gatlin-Bunai C. L., Mongodin E. F., Parmar P. P., Huang S. T., Clark D. J., et al. (2006). Comparative proteomic analysis of Staphylococcus aureus strains with differences in resistance to the cell wall-targeting antibiotic vancomycin. Proteomics. 6, 4246–4258. 10.1002/pmic.200500764 [DOI] [PubMed] [Google Scholar]
- Pietiäinen M., François P., Hyyryläinen H. L., Tangomo M., Sass V., Sahl H. G., et al. (2009). Transcriptome analysis of the responses of Staphylococcus aureus to antimicrobial peptides and characterization of the roles of vraDE and vraSR in antimicrobial resistance. BMC Genomics. 10:429. 10.1186/1471-2164-10-429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J., Pogliano N., Silverman J. A. (2012). Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J. Bacteriol. 194, 4494–4504. 10.1128/JB.00011-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhouani H., Pinto L., Poeta P., Igrejas G. (2012). After genomics, what proteomics tools could help us understand the antimicrobial resistance of Escherichia coli? J. Proteomics. 75, 2773–2789. 10.1016/j.jprot.2011.12.035 [DOI] [PubMed] [Google Scholar]
- Ramos S., Chafsey I., Silva N., Hébraud M., Santos H., Capelo-Martinez J. L., et al. (2015). Effect of vancomycin on the proteome of the multiresistant Enterococcus faecium SU18 strain. J. Proteomics. 113, 378–387. 10.1016/j.jprot.2014.10.012 [DOI] [PubMed] [Google Scholar]
- Reyes J., Panesso D., Tran T. T., Mishra N. N., Cruz M. R., Munita J. M., et al. (2015). A liaR deletion restores susceptibility to daptomycin and antimicrobial peptides in multidrug-resistant Enterococcus faecalis. J. Infect. Dis. 211, 1317–1325. 10.1093/infdis/jiu602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers P. D., Liu T. T., Barker K. S., Hilliard G. M., English B. K., Thornton J., et al. (2007). Gene expression profiling of the response of Streptococcus pneumoniae to penicillin. J. Antimicrob. Chemother. 59, 616–626. 10.1093/jac/dkl560 [DOI] [PubMed] [Google Scholar]
- Sasindran S. J., Saikolappan S., Dhandayuthapani S. (2007). Methionine sulfoxide reductases and virulence of bacterial pathogens. Future Microbiol. 2, 619–630. 10.2217/17460913.2.6.619 [DOI] [PubMed] [Google Scholar]
- Sass P., Jansen A., Szekat C., Sass V., Sahl H. G., Bierbaum G. (2008). The lantibiotic mersacidin is a strong inducer of the cell wall stress response of Staphylococcus aureus. BMC Microbiol. 8:186. 10.1186/1471-2180-8-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherl A., François P., Charbonnier Y., Deshusses J. M., Koessler T., Huyghe A., et al. (2006). Exploring glycopeptide-resistance in Staphylococcus aureus: a combined proteomics and transcriptomics approach for the identification of resistance-related markers. BMC Genomics. 7:296. 10.1186/1471-2164-7-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulthess B., Bloes D. A., Berger-Bachi B. (2012). Opposing roles of σB and σB-controlled SpoVG in the global regulation of esxA in Staphylococcus aureus. BMC Microbiol. 12:17. 10.1186/1471-2180-12-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulthess B., Meier S., Homerova D., Goerke C., Wolz C., Kormanec J., et al. (2009). Functional characterization of the σB-dependent yabJ-spoVG operon in Staphylococcus aureus: role in methicillin and glycopeptide resistance. Antimicrob. Agents Chemother. 53, 1832–1839. 10.1128/AAC.01255-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher A., Trittler R., Bohnert J. A., Kümmerer K., Pages J. M., Kern W. V. (2007). Intracellular accumulation of linezolid in Escherichia coli, Citrobacter freundii and Enterobacter aerogenes: role of enhanced efflux pump activity and inactivation. J. Antimicrob. Chemother. 59, 1261–1264. 10.1093/jac/dkl380 [DOI] [PubMed] [Google Scholar]
- Seidl K., Stucki M., Ruegg M., Goerke C., Wolz C., Harris L., et al. (2006). Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 50, 1183–1194. 10.1128/AAC.50.4.1183-1194.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobral R. G., Jones A. E., Des Etages S. G., Dougherty T. J., Peitzsch R. M., Gaasterland T., et al. (2007). Extensive and genome-wide changes in the transcription profile of Staphylococcus aureus induced by modulating the transcription of the cell wall synthesis gene murF. J. Bacteriol. 189, 2376–2391. 10.1128/JB.01439-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Rubio A., Jayaswal R. K., Silverman J. A., Wilkinson B. J. (2013). Additional routes to Staphylococcus aureus daptomycin resistance as revealed by comparative genome sequencing, transcriptional profiling, and phenotypic studies. PLoS ONE 8:e58469 10.1371/journal.pone.0058469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton M. R., Horsburgh M. J., Hayhurst E. J., Wright L., Jonsson I. M., Tarkowski A., et al. (2007). Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J. Bacteriol. 189, 7316–7325. 10.1128/JB.00734-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stülke J., Hillen W. (2000). Regulation of carbon catabolism in Bacillus species. Annu. Rev. Microbiol. 54, 849–880. 10.1146/annurev.micro.54.1.849 [DOI] [PubMed] [Google Scholar]
- Swaney S. M., Aoki H., Ganoza M. C., Shinabarger D. L. (1998). The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob. Agents Chemother. 42, 3251–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H., Nelson K. E., Paulsen I. T., Eisen J. A., Read T. D., Peterson S., et al. (2001). Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293, 498–506. 10.1126/science.1061217 [DOI] [PubMed] [Google Scholar]
- Utaida S., Dunman P. M., Macapagal D., Murphy E., Projan S. J., Singh V. K., et al. (2003). Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149, 2719–2732. 10.1099/mic.0.26426-0 [DOI] [PubMed] [Google Scholar]
- van der Voort M., Kuipers O. P., Buist G., de Vos W. M., Abee T. (2008). Assessment of CcpA-mediated catabolite control of gene expression in Bacillus cereus ATCC 14579. BMC Microbiol. 8:62. 10.1186/1471-2180-8-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., He X., Jiang Z., Wang J., Chen X., Liu D., et al. (2010). Proteomic analysis of the Enterococcus faecalis V583 strain and clinical isolate V309 under vancomycin treatment. J. Proteome Res. 9, 1772–1785. 10.1021/pr901216e [DOI] [PubMed] [Google Scholar]
- Wecke T., Zuhlke D., Mäder U., Jordan S., Voigt B., Pelzer S., et al. (2009). Daptomycin versus Friulimicin B: in-depth profiling of Bacillus subtilis cell envelope stress responses. Antimicrob. Agents Chemother. 53, 1619–1623. 10.1128/AAC.01046-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. N., Schluenzen F., Harms J. M., Starosta A. L., Connell S. R., Fucini P. (2008). The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc. Natl. Acad. Sci. U.S.A. 105, 13339–13344. 10.1073/pnas.0804276105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M. E., Hoch J. A. (2008). Essentiality, bypass, and targeting of the YycFG (VicRK) two-component regulatory system in gram-positive bacteria. J. Bacteriol. 190, 2645–2648. 10.1128/JB.01682-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford N. (1998). Glycopeptide-resistant enterococci: a decade of experience. J. Med. Microbiol. 47, 849–862. [DOI] [PubMed] [Google Scholar]
- Yin S., Daum R. S., Boyle-Vavra S. (2006). VraSR two-component regulatory system and its role in induction of pbp2 and vraSR expression by cell wall antimicrobials in Staphylococcus aureus. Antimicrob. Agents Chemother. 50, 336–343. 10.1128/AAC.50.1.336-343.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]