Abstract
Photosensitivity in humans can result from defects in repair of light-induced DNA lesions, from photoactivation of chemicals (including certain medications) with sunlight to produce toxic mediators, and by immune reactions to sunlight exposures. Deficiencies in DNA repair and the processing of damaged DNA during replication and transcription may result in mutations and genomic instability. We will review current understanding of photosensitivity to short wavelength ultraviolet light (UV) due to genetic defects in particular DNA repair pathways; deficiencies in some are characterized by an extremely high incidence of cancer in sun-exposed tissues, while in others no cancers have been reported.
Keywords: Photosensitivity, DNA repair, skin cancer, global genomic repair, transcription-coupled repair
1. Introduction
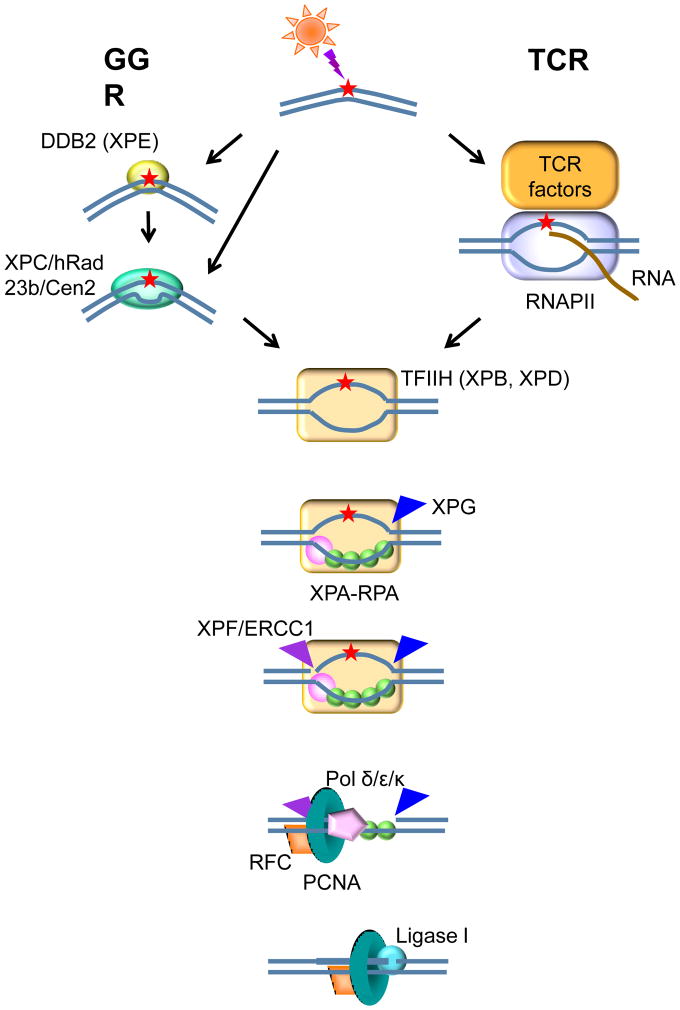
Light emitted by the sun is essential for life on Earth. In particular, it is necessary for photosynthesis in plants and for the production of vitamin D in mammals. However, sunlight can also be harmful; in humans it causes aging of the skin and cancer, in addition to photodermatoses. Cellular biomolecules, particularly nucleic acids, sustain light-induced damage; the types of lesions depend upon the wavelengths to which the cells are exposed. Various strategies have evolved to reverse or remove the photoproducts in DNA that can interfere with vital functions such as replication and transcription. Essentially all living organisms (with the exception of placental mammals) produce highly effective photolyases that can directly reverse the principal sunlight-induced DNA lesions by the process known as photoreactivation. Otherwise, these photoproducts can be recognized and removed by the ubiquitous pathway of nucleotide excision repair (NER). NER operates in two modes, throughout the genome as global genomic repair (GGR), and on the transcribed strands of active genes as transcription-coupled repair (TCR) (reviewed in [1], figure 1).
Figure 1.
The nucleotide excision repair pathway.
GGR is initiated when the lesions are recognized by XPE (DDB2) in complex with DDB1 and Cul4, and/or by the XPC-hRAD23b-Cen2 complex, to open up the DNA at the site of the lesion. The TFIIH complex binds and extends the bubble 5′ and 3′ from the lesion using its component helicases, XPB and XPD, and then XPA and RPA verify the damaged strand, so that the structure-specific endonucleases XPF-ERCC1 and XPG can incise the strand containing the lesion on both sides; DNA polymerases δ,ε and κ [60] in association with PCNA and RFC carry out repair replication using the undamaged complementary strand as template, and ligase I seals the repair patch to the contiguous DNA. Lesions encountered by translocating RNA polymerases can be sensitively detected and repaired through the TCR subpathway of NER, in which there is a hand-off to TFIIH without requiring either XPE or XPC; instead TCR requires CSA, CSB, UVSSA, USP7, XAB2, HMGN1, p300, NEDD4, SPT16 and TFIIS in humans.
UVC (100 to 280 nm) radiation from the sun is completely screened out by dioxygen and ozone in the upper atmosphere. UVB (280 to 315 nm) is greatly attenuated by the ozone layer, but the small fraction of these wavelengths that reach the earth’s surface can affect all life forms. They induce damage to DNA and RNA by direct excitation, with a maximum quantum yield at ~ 260 nm. The primary products caused by these wavelengths are cyclobutane pyrimidine dimers (CPD) and (6–4) pyrimidine-pyrimidone photoproducts (6-4PP), which can form between adjacent pyrimidines in the DNA. Mutations in genes that code for factors involved in the repair of these photoproducts result in enhanced photosensitivity, manifested as acute sunburn, pigmentation anomalies, dryness and atrophy of the skin, and in some patients, a high incidence of cancer in sun-exposed areas.
Although UVA, in particular 340–400 nm radiation, can cause acute reactions in photosensitive patients, it was generally assumed that these wavelengths do not directly affect DNA; however recent studies using powerful analytical tools and effective filters have shown that CPD but not 6-4PP are generated by UVA [2]. Photosensitized reactions (e.g. with tryptophan in proteins) can result in the formation of reactive oxygen species (ROS) leading to oxidation of DNA bases, for example, 8-hydroxyguanine (8-oxo-7,8-dihydroguanine, 8-oxoG) [3, 4]. There are currently no human syndromes for which sunlight sensitivity is caused by deficient repair of oxidized DNA bases. However, inhibition of the various steps in the base excision repair (BER) pathway causes cellular hypersensitivity to UVA [5], and laboratory mice with engineered mutations in genes coding for enzymes that initiate BER are somewhat prone to skin cancer upon exposure to UVB [6], but they are not generally sun-sensitive [7]. NER-deficient human cell lines have been reported to be hypersensitive to ROS [8, 9]; however, mutation spectra analyses in skin tumors from patients defective in NER reveal that essentially all the mutations had been caused by CPD or 6-4PP [10].
Pathologies associated with visible light and radiations in the longer wavelength regions have been reported, albeit rarely; the roles of these wavelengths in skin disease, if any, are poorly understood. By far, most of the hereditary photosensitive disorders involving DNA repair are due to NER deficiencies; exceptions for which the defects are known include Bloom’s and Rothmund-Thompson syndromes and ataxia telangiectasia, as described below and listed in Table I.
Table I.
Mutations that cause photosensitivity in humans
| Pathway | Subpathway | Protein | HUGO, other nomenclature | Disease(s) | Photosensitivity | Cancer predisposition |
|---|---|---|---|---|---|---|
| NER | GGR + TCR | XPA | XP | +++ | +++ | |
| XPB | ERCC3 | XP, XP/CS, COFS | +++ | +++ | ||
| GGR | XPC | XP | +++ | +++ | ||
| GGR + TCR | XPD | ERCC2 | XP, TTD, XP/CS, XP/TTD, COFS | + to +++ | − to +++ | |
| GGR | XPE | DDB2 | XP | + | + | |
| GGR + TCR | XPF | ERCC4 | XP, XFE, XP/CS, FA | + | + | |
| ERCC1 | XFE, COFS, CS | |||||
| XPG | ERCC5 | XP, XP/CS | + to +++ | + to +++ | ||
| TTDA | TTD | + to +++ | − | |||
| TCR | CSA | ERCC8 | CS, UVSS | +++ | − | |
| CSB | ERCC6 | CS, UVSS, DSC, COFS | +++ | − | ||
| UVSSA | UVSS | +++ | − | |||
| TLS | pol η | XP | +++ | +++ | ||
| DNA damage response | ATM | AT | telangiectasia | |||
| RecQ- like DNA helicases | WRN | RecQL2 | Werner’s syndrome | − | ++ | |
| BLM | RecQL3 | Bloom’s syndrome | telangiectasia | ++ | ||
| RTS | RecQL4 | Rothmund-Thomson syndrome | 30% of patients | ++ | ||
| RAPADILINO | − | ++ |
Abbreviations: XP, xeroderma pigmentosum; CS, Cockayne syndrome; COFS, cerebro-oculo-facial-skeletal syndrome; TTD, trichothiodystrophy; XFE, XPF-ERCC1 progeroid syndrome; FA, Fanconi anemia; UVSS, UV-sensitive syndrome; AT, ataxia telangiectasia.
2. DNA repair and photosensitivity
2.1. Photosensitivity and defects in DNA helicases
Five homologs of the recQ gene from Escherichia coli, which codes for a DNA helicase with 3′- 5′ directional specificity involved in recombination and repair of DNA breaks, have been identified in humans. In addition to Bloom’s and Rothmund-Thomson syndromes, Werner’s syndrome is also due to mutations in a RecQ-like helicase, the WRN (RecQL2) gene; although patients exhibit premature aging and early onset sarcomas and mesenchymal tumors, photosensitivity has not been reported. Mutations in the other two human RecQ-like helicases, RECQL1 and RECQL5, have not yet been genetically linked to a disease [11].
2.1.1
Bloom’s syndrome is a rare chromosome breakage disease primarily seen among Ashkenazi Jews. It presents with failure to thrive, stunted growth, small and narrow facies, sun-sensitive facial telangiectasias, immunodeficiency, and increased risk of malignancies. Mutations in the BLM gene, which codes for the RecQL3 (BLM) DNA helicase, are associated with the syndrome. Cells from Bloom’s syndrome patients exhibit high frequencies of sister chromatid exchanges, chromosome aberrations and rearrangements, reflecting the high mutation rate associated with the loss of BLM [12].
2.1.2
Rothmund-Thomson syndrome (RTS), or poikiloderma congenitale, is a rare autosomal recessive disorder attributed to mutations in the RECQL4 helicase gene. Key features include early photosensitivity and poikilodermatous (abnormal pigmentation) skin changes, juvenile cataracts, skeletal dysplasias, and a predisposition to osteosarcoma and skin cancer. The acute phase of the disease appears in early infancy as red patches on the cheeks, spreading later to other areas of the face, extremities or buttocks. Roughly 30% of the patients are photosensitive [13]. Individuals with mutations in RECQL4 can also develop RAPADILINO, a disease very similar to RTS, but without photosensitivity and poikiloderma [14].
2.2. Photosensitivity due to defects in DNA damage response pathways
2.2.1
Ataxia telangiectasia (AT), also known as Louis-Bar syndrome, results in ataxia (lack of muscle control), immune deficiency, elevated cancer incidence, and premature aging. This autosomal recessive disease is caused by mutations in the ATM gene, a key factor in the cellular response to DNA damage, particularly double-strand breaks. Moreover, ATM regulates telomere length, and this might correlate with the progeria observed in most patients. Although AT patients are not photosensitive, telangiectasia due to broken venous capillaries usually appears in childhood several years following ataxia, and this is more evident in sun-exposed areas of the skin, although sun-protected areas such as the flexural surfaces of the extremities and the chest are also typically affected. Other common cutaneous findings that contribute to the progeroid appearance include atrophy of subcutaneous fat and graying hairs.
2.3. Photosensitivity and defects in NER
The spectrum of human disorders resulting from mutations in NER proteins has been presented in a number of recent reviews, including [15–18]. Here we will summarize the major characteristics of each of these diseases.
2.3.1
Xeroderma pigmentosum (XP) is defined by the atrophic, dry, parchment-like texture of the skin, and by high incidence (up to 4000-fold more prevalent than in unaffected individuals) and early development of tumors of the skin and sun-exposed areas, such as the eyes and the tip of the tongue. The disease comprises seven complementation groups, A to G, with defects in various steps of the NER pathway (figure 1), and an eighth group, XP variant (XPV), with normal repair of photoproducts but a defective DNA polymerase, pol η, which has a role in DNA translesion synthesis (TLS) over UV-induced photoproducts. Pol η appears to have evolved to deal specifically with pyrimidine dimers, since it copies DNA containing them with remarkably high fidelity, while it is exceedingly error prone when replicating undamaged DNA. XPV patients are also highly prone to skin cancer; lacking pol η, TLS is carried out by other polymerases (of which there are several in humans) that introduce mutations that can lead to malignancy. Approximately 20% of XP patients develop neurological and cognitive dysfunction due to neuronal loss, manifested as mental retardation, spasticity and microcephaly. The frequency of mental problems varies among the complementation groups: more than 50% of XPD patients exhibit neurological symptoms [19, 20], while such manifestations are very rare in XPC patients. Cells from XPC patients are proficient in TCR of sun-induced photoproducts and bulky DNA lesions; thus one can speculate that TCR is more important than GGR for maintaining genomic integrity in neuronal tissues. This idea is consistent with the evidence for efficient TCR but attenuated GGR in wild type neurons [21]. Tumors in the internal organs of XP patients may occur at a higher frequency than in the overall population, although there is still active debate about whether these cancers are directly due to the NER deficiency [22].
Certain mutations in XP genes may result in combined phenotypes, for example XP/Cockayne syndrome (section 2.3.2) or XP/trychothiodystrophy (section 2.3.6), or in different disorders such as XFE progeroid syndrome (section 2.3.5) [23], in severe and progressive neurodegeneration [24], or in Cockayne syndrome (section 2.3.2) [25]. Some mutations in XPF (ERCC4) result in Fanconi anemia (FA) alone [26] or in combination with XP and Cockayne syndrome [25], and individuals with these mutations belong to the FancQ complementation group of FA [26]. The FA pathway, which involves elements of homologous recombination, NER and translesion synthesis, regulates the replication-dependent removal of DNA interstrand crosslinks. FA patients exhibit defective repair of DNA interstrand crosslinks and their cells are hypersensitive to crosslinking reagents such as mitomycin C and melphalan, but typically not to UVC; however, FA patients defective in XPF might also be sun-sensitive, depending upon whether NER is affected by the particular mutations.
2.3.2
Cockayne syndrome (CS) is a complex disease with a multitude of symptoms. There are two principal complementation groups of CS, CS-A and CS-B, with mutations in the CSA (ERCC8) and CSB (ERCC6) genes, although mutations in XPF (ERCC4) or ERCC1 have been recently found to be causative of CS [25]; in very rare cases, patients with certain mutations in the XPB, XPD, or XPG genes exhibit a combination of symptoms of CS and XP, and mutations in XPF may result in a combined XP-CS-FA phenotype (see above). A handful of CS patients remain unassigned to any of these genes [25]. CS patients present three major characteristics: microcephaly, stunted growth, and progressive neurological dysfunction due to leukodystrophy (progressive degeneration of the white matter); in addition, three to five of the following minor criteria have been recommended for a positive diagnosis: photosensitivity, demyelination of the peripheral nerves, pigmentary retinopathy and/or cataracts, sensorineural hearing loss, cachectic dwarfism with stooped posture, and progeria with shortened lifespan. These are usually accompanied by numerous additional problems, including gait defects, contractures, spasticity, tremors, dental caries, basal ganglia calcifications, hypertension, osteoporosis, and others. Curiously, there have been no reports of cancers in any of the nearly 1000 patients diagnosed with CS to date. Cells from CS patients are defective in the TCR subpathway of NER but they are proficient in the global repair of bulky DNA adducts and UVC-induced photoproducts; moreover, we have recently determined that CSB is required for TCR of 8-oxoG, in cells treated with potassium bromate [27]. Although the vast majority of CS patients display photosensitivity, the developmental and neurological features of the disease might not be related to the TCR defect, as discussed below [28].
2.3.3
Cerebro oculo-facial-skeletal syndrome (COFS) shares some of the traits typical of CS, with additional features such as severe hypotonia, impaired reflexes, poor vision, and distinctive facial features: small eyes, low-set ears, microcephaly and small jaw. The limbs, skull, heart and kidneys may also be abnormal. Mutations in CSB, ERCC1, XPD or XPG can cause COFS. The severity of the symptoms leads to lethality during infancy or early childhood; the patients and/or their cells are usually photosensitive [29].
2.3.4
De Sanctis-Cacchione (DSC) syndrome was first named “xerodermic idiocy”, because the patients exhibited symptoms of xeroderma pigmentosum combined with mental deficiency, progressive neurologic deterioration, dwarfism and gonadal hypoplasia. The disease has been associated with mutations in XP genes, most often XPA [30, 31]. However, two siblings diagnosed with DSC exhibited more severe neurological symptoms and less striking cutaneous manifestations than those typically seen in XP patients; biochemical and complementation assays revealed that these individuals carried mutations in the CSB gene [30].
2.3.5
XFE progeroid syndrome (XFEPS) is characterized by aged and bird-like facies, lack of subcutaneous fat, dwarfism, cachexia and microcephaly. Additional features include sun-sensitivity from birth, learning disabilities, hearing loss, and visual impairment [32]. The causal mutations are in the XPF or ERCC1 genes, whose products form the XPF-ERCC1 endonuclease complex.
2.3.6
Tricothiodystrophy (TTD) comprises six complementation groups, of which only three involve sun sensitivity and are the result of mutations in genes involved in the NER pathway, namely XPB (ERCC3), XPD (ERCC2) and TTDA; the proteins encoded by these genes are subunits of the transcription factor TFIIH. As mentioned in section 2.3.1, some mutations in XPD result in combined XP/TTD phenotypes. TTD patients present with intellectual impairment and other features in common with CS. However, the most notable and unique feature of TTD is the presence of brittle hair and nails, which are used for positive diagnoses based upon the tiger-tail-like pattern of the hair under polarized light. (This reflects the lack of sulphur-containing proteins that normally account for the strength and flexibility of hair.) The acronyms PIBIDS, IBIDS, BIDS and PBIDS have been used to classify patients according to their individual set of symptoms: Photosensitivity, Ichthyosis, Brittle hair, Intellectual impairment, Decreased fertility, and Short stature; patients may also exhibit microcephaly, osteoporosis, skeletal abnormalities, nail abnormalities, premature aging, proneness to infections, dental caries, hearing loss and cataracts. Mutations in the gene coding for M-phase-specific PLK1-interacting protein, MPLKIP (TTDN1) are responsible for the non-photosensitive BIDS; the genetic bases for the other two non-photosensitive complementation groups have not yet been identified [33].
2.3.7
UV-sensitive syndrome (UVSS) was first described by Itoh and colleagues [34]. Individuals with UVSS are sun-sensitive and they present pigmentation anomalies in sun-exposed areas of the skin; however, just as with CS, no tumors have yet been reported in their skin or internal organs. In striking contrast with CS and other DNA repair-deficient diseases, no pathologies other than sunburn and freckles have been associated with UVSS. Although it is likely that there are hundreds or thousands of people with UVSS worldwide, their clinical features are so mild that they may easily elude diagnosis as victims of a genetic disease. To date, only eight patients have been characterized. There are three complementation groups of UVSS, with mutations in the CSA, CSB, or UVSSA genes.
The responses of UVSS and CS cells to treatment with UV light (254 nm) are identical: defective survival, impaired recovery of RNA synthesis, accumulation of p53 correlated with apoptotic response at low UV doses, and proficient global repair of photoproducts; as these observations suggest, cells from both syndromes have been shown to be deficient in TCR of photoproducts ([35] and references therein). As we have shown for CSB, UVSSA is also required for preferential repair of the oxidized base lesion, 8-oxoG [27]. These findings are provocative, because although CS cells are hypersensitive to treatment with agents that primarily induce oxidative DNA lesions in addition to single and/or double strand breaks, UVSS cells exhibit normal resistance to treatment with oxidants [36, 37]. Moreover, using a host cell reactivation (HCR) assay in which undamaged cells were transfected with plasmids treated prior to transfection to induce the oxidized bases, 8-oxoG or thymine glycol, we have demonstrated that expression of the plasmid-coded lacZ reporter gene is defective in CS cells but not in UVSS cells [36]. A possible explanation for these puzzling results is that the CS defect not only eliminates TCR but that it affects other cellular functions such as transcriptional bypass of oxidative damage [38–41], processing of oxidative damage in mitochondria [42–45] or defective neurogenesis [46, 47]
2.4. Other diseases associated with defective DNA repair
It is important to appreciate that not all cases of unusually high incidence of skin cancer can be ascribed to DNA repair defects. There is at least one example in which a patient has normal NER and no sun sensitivity, but presents numerous skin cancers [48]. However, epidermal cells from this patient exhibited reduced HCR of UV-irradiated reporter plasmids, while HCR of abasic site-containing plasmids was normal. The HCR of UV-irradiated plasmids was complemented by UVDE (an endonuclease from Schizosaccharomyces pombe that incises DNA containing CPD, 6-4PP, and AP sites) but not by any of the XP-related proteins. It has been suggested that some novel UV-photoproducts that are recognized by UVDE but not by XP factors could be responsible, but if so, the cause for the lack of sun sensitivity remains elusive.
A syndrome that presents with short stature, hearing loss, premature aging, telangiectasia, neurodegeneration, and photosensitivity, resulting from a homozygous missense (p.Ser228Ile) sequence alteration of the proliferating cell nuclear antigen (PCNA) has been recently described. PCNA is a highly conserved sliding clamp essential for DNA polymerase recruitment and processivity in DNA replication and repair synthesis. Because of this fundamental role, mutations in PCNA that profoundly impair its function would seemingly be incompatible with life. Interestingly, while the p.Ser228Ile alteration appeared to have no effect on protein levels or DNA replication, UV irradiated cells from the patient displayed substantial reductions in both RNA synthesis recovery and survival [49].
Hyperpigmentation has been reported in Fanconi anemia patients with mutations in most genes involved in the FA DNA repair pathway that resolves DNA interstrand crosslinks, but sun sensitivity was found only in the XP-CS-FA patient mutated in XPF described above. Skin photosensitivity was found associated with mutations in LIG4, DNA-PKCS, Ku70 and Ku80, proteins that interact with FA factors [50].
3. Discussion
The TFIIH multifactor complex is essential for both transcription initiation and for NER, so that mutations in any of the proteins in the complex might result in deficiencies in either or both transcription and DNA repair. TFIIH consists of a core sub-complex containing XPB, p62, p52, p44, p34 and TTDA, a cdk-activating kinase (CAK) sub-complex with cdk7, MAT1 and cyclin H, and the XPD helicase that also connects the core and CAK sub-complexes. In addition to these 10 factors, the NER-associated structure-specific endonuclease XPG interacts with and stabilizes TFIIH. The activities ascribed to TFIIH include an ATPase and helicase (XPB, XPD), a protein kinase (cdk7), and an E3 ubiquitin ligase (p44). Reduced concentrations of the complex, increased instability and abnormal function of TFIIH have been reported in all TTD patients tested, suggesting that this may be a “transcription disease” caused by abnormal architecture of the complex; in contrast, mutations in XPD and XPB in XP patients affect the ATPase and/or helicase activities of TFIIH that are specifically necessary for NER [51], although the XPB helicase is required for transcription initiation as well. The mutations in XPG, XPD and XPB that result in XP/CS phenotypes may arise from abnormalities in transcription by RNAPII in addition to defective NER (reviewed in [28]), or by RNAPI with consequent ribosomal stress [52]; defects in other roles of TFIIH, such as phosphorylation of nuclear hormone receptors have also been suggested [53].
The TCR deficiency in CS or UVSS patients is responsible for their sun-sensitivity and pigmentation anomalies. The striking fact, as mentioned earlier, is that the known patients do not present any type of cancer; with the caveat that there are very few patients and, for CS at least, they rarely live to the age at which cancer incidence is maximal in normal human populations. The prevailing explanation is that TCR deficiency results in persistent transcription blockage, leading to cell death by apoptosis, or through the compounded problem of replication fork encounters with the immobilized transcription complexes. Thus, in the absence of TCR there would be few progeny in which mutations could be fixed, while the fully-functional global NER pathway clears the genome of lesions in the surviving cells, to minimize mutagenesis [54].
Although many of the factors involved in TCR have been characterized, and various mechanisms have been suggested, the lack of an in vitro TCR assay has hindered a detailed biochemical understanding of this important pathway. Efforts in several research groups have defined protein complexes plausibly involved in the various steps of TCR in humans: recognition is carried out by a blocked RNAPII, then RNAPII, CSB and XAB2 are required for assembly of the pre-incision complex, while CSA as the component of an E3 ubiquitin ligase complex is required for later events. Chromatin remodeling factors, such as HMGN1 and p300, might be needed for displacing nucleosomes that have become rapidly re-established behind the translocating transcription complex, so that the polymerase can regress, to reveal the lesion to the repair enzymes. Following recognition of the lesion, the TCR pathway requires the common NER factors for incision, removal of the damaged oligonucleotide, repair synthesis and ligation (figure 1). Additional factors continue to be identified: Examples include NEDD4, that is implicated in DNA damage-induced ubiquitination of the RNA polymerase II subunit RPB1 to mark it for proteosomal degradation; UVSSA and USP7 that bind to RNAPII upon transcription arrest and deubiquitinate CSB to prevent its degradation; and the SPT16 FACT chromatin remodeler (reviewed in [1]).
Photoproducts in DNA might also be recognized by repair pathways other than NER. For example, mismatch repair (MMR) complexes can bind bulky lesions like CPD and 6-4 PP; however, the role of MMR appears to be more related to suppression of mutagenesis and apoptosis, than to repair of these lesions. It is also notable that MMR-deficient individuals are not sun-sensitive and do not exhibit a heightened risk of developing skin cancer, although they may be highly susceptible to certain internal cancers. Ultraviolet radiation may result in complex DNA lesions and structures, including double-strand breaks and interstrand crosslinks, such as at sites of clustered oxidized bases [55], or as a consequence of the processing of DNA or RNA polymerases blocked by photoproducts [56]. Such lesions are repaired through specialized mechanisms that require homologous recombination or non-homologous end joining; photosensitivity has not been reported in individuals or laboratory animals with defects in those pathways, with the exception of the individual with a mutation in XPF who exhibits a combined XP-CS-FA phenotype and is sun sensitive (see above).
The spectrum of cancer susceptibility in human populations ranges from extremely high in individuals with mutations in certain DNA repair pathways, to none in rare groups such as CS and UVSS patients, as discussed above. DNA repair proficient individuals carrying polymorphisms in certain NER-related genes may exhibit a moderately higher incidence of gastric cancers [57], prostate cancer [58], head and neck squamous cell carcinomas [59], and other cancers, illustrating the importance of a balanced, fine-tuned NER system dedicated to the elimination of DNA lesions before the essential process of replication can fix mutations in the genome.
The sun-sensitive diseases provide a glimpse into the complex modes for processing genomic damage in cells when they are exposed to photons. The skin, which is the most spatially extensive organ and the main target of sunlight, suffers from a variety of inflammatory, pigmentation related, premature aging and neoplastic pathologies. The correlations between defective biochemical pathways and consequent genetic diseases have yielded important mechanistic insights into the etiology of number of hereditary photodermatoses. The derived understanding of the DNA repair pathways can serve as a foundation for the design of innovative therapeutic approaches for cancer and other degenerative diseases. However, the number of photodermatoses, both hereditary and acquired, for which the molecular pathophysiology is still unknown, offers further challenges and opportunities to fully elucidate the photobiology of skin.
Highlights.
High sensitivity to sunlight can be caused by defects in certain DNA repair pathways
Some DNA repair defects result in high incidence and early appearance of skin tumors
Neurodegeneration and growth abnormalities characterize some DNA repair syndromes
Acknowledgments
Supported by National Institute of Environmental Health Sciences grant ES018834. We apologize to those whose work was inadvertently not cited.
Footnotes
Conflict if interest statement
The manuscript contents have not been previously published nor are they under consideration elsewhere. The authors state no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Spivak G, Ganesan AK. The complex choreography of transcription-coupled repair. DNA Repair (Amst) 2014 doi: 10.1016/j.dnarep.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 2.Cadet J, et al. Photoinduced damage to cellular DNA: direct and photosensitized reactions. Photochem Photobiol. 2012;88(5):1048–65. doi: 10.1111/j.1751-1097.2012.01200.x. [DOI] [PubMed] [Google Scholar]
- 3.Kvam E, Tyrrell RM. Induction of oxidative DNA base damage in human skin cells by UV and near visible radiation. Carcinogenesis. 1997;18(12):2379–84. doi: 10.1093/carcin/18.12.2379. [DOI] [PubMed] [Google Scholar]
- 4.Cadet J, Sage E, Douki T. Ultraviolet radiation-mediated damage to cellular DNA. Mutat Res. 2005;571(1–2):3–17. doi: 10.1016/j.mrfmmm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 5.Kim KJ, et al. Modulation of base excision repair alters cellular sensitivity to UVA1 but not to UVB1. Photochem Photobiol. 2002;75(5):507–12. doi: 10.1562/0031-8655(2002)075<0507:mobera>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 6.Kunisada M, et al. 8-Oxoguanine formation induced by chronic UVB exposure makes Ogg1 knockout mice susceptible to skin carcinogenesis. Cancer Res. 2005;65(14):6006–10. doi: 10.1158/0008-5472.CAN-05-0724. [DOI] [PubMed] [Google Scholar]
- 7.Vens C, et al. The role of DNA polymerase beta in determining sensitivity to ionizing radiation in human tumor cells. Nucleic Acids Res. 2002;30(13):2995–3004. doi: 10.1093/nar/gkf403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D’Errico M, et al. New functions of XPC in the protection of human skin cells from oxidative damage. EMBO J. 2006;25(18):4305–15. doi: 10.1038/sj.emboj.7601277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rünger T, Epe B, Moller K. Repair of ultraviolet B and singlet oxygen-induced DNA damage in xeroderma pigmentosum cells. J Invest Dermatol. 1995;104(1):68–73. doi: 10.1111/1523-1747.ep12613504. [DOI] [PubMed] [Google Scholar]
- 10.Dumaz N, et al. Specific UV-induced mutation spectrum in the p53 gene of skin tumors from DNA-repair-deficient xeroderma pigmentosum patients. Proc Natl Acad Sci U S A. 1993;90(22):10529–33. doi: 10.1073/pnas.90.22.10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brosh RM, Jr, Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 2007;35(22):7527–44. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arora H, et al. Bloom syndrome. Int J Dermatol. 2014 doi: 10.1111/ijd.12408. [DOI] [PubMed] [Google Scholar]
- 13.Hsu S, George S. Rothmund-Thompson syndrome. emedicine 2007 [Google Scholar]
- 14.Siitonen HA, et al. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum Mol Genet. 2003;12(21):2837–44. doi: 10.1093/hmg/ddg306. [DOI] [PubMed] [Google Scholar]
- 15.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10(11):756–68. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 16.Menck CF, Munford V. DNA repair diseases: What do they tell us about cancer and aging? Genet Mol Biol. 2014;37(1 Suppl):220–233. doi: 10.1590/s1415-47572014000200008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamileri I, Karakasilioti I, Garinis GA. Nucleotide excision repair: new tricks with old bricks. Trends Genet. 2012;28(11):566–73. doi: 10.1016/j.tig.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Nouspikel T. DNA repair in mammalian cells : Nucleotide excision repair: variations on versatility. Cell Mol Life Sci. 2009;66(6):994–1009. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor EM, et al. Xeroderma pigmentosum and trichothiodystrophy are associated with different mutations in the XPD (ERCC2) repair/transcription gene. Proc Natl Acad Sci U S A. 1997;94(16):8658–63. doi: 10.1073/pnas.94.16.8658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradford PT, et al. Cancer and neurologic degeneration in xeroderma pigmentosum: long term follow-up characterises the role of DNA repair. J Med Genet. 2011;48(3):168–76. doi: 10.1136/jmg.2010.083022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nouspikel T. DNA repair in differentiated cells: some new answers to old questions. Neuroscience. 2007;145(4):1213–21. doi: 10.1016/j.neuroscience.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 22.Giglia G, et al. Molecular analysis of glioma and skin-tumour alterations in a xeroderma-pigmentosum child. Int J Cancer. 1999;81(3):345–50. doi: 10.1002/(sici)1097-0215(19990505)81:3<345::aid-ijc6>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 23.Niedernhofer LJ, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444(7122):1038–43. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 24.Imoto K, et al. Patients with defects in the interacting nucleotide excision repair proteins ERCC1 or XPF show xeroderma pigmentosum with late onset severe neurological degeneration. Journal of Investigative Dermatology. 2007;127:S92–S92. [Google Scholar]
- 25.Kashiyama K, et al. Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. Am J Hum Genet. 2013;92(5):807–19. doi: 10.1016/j.ajhg.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bogliolo M, et al. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am J Hum Genet. 2013;92(5):800–6. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo J, Hanawalt PC, Spivak G. Comet-FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoGuanine in human cells. Nucleic Acids Res. 2013;41(16):7700–12. doi: 10.1093/nar/gkt524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brooks PJ. Blinded by the UV light: how the focus on transcription-coupled NER has distracted from understanding the mechanisms of Cockayne syndrome neurologic disease. DNA Repair (Amst) 2013;12(8):656–71. doi: 10.1016/j.dnarep.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzumura H, Arisaka O. Cerebro-oculo-facio-skeletal syndrome. Adv Exp Med Biol. 2010;685:210–4. doi: 10.1007/978-1-4419-6448-9_19. [DOI] [PubMed] [Google Scholar]
- 30.Itoh T, Cleaver JE, Yamaizumi M. Cockayne syndrome complementation group B associated with xeroderma pigmentosum phenotype. Hum Genet. 1996;97(2):176–9. doi: 10.1007/BF02265261. [DOI] [PubMed] [Google Scholar]
- 31.Colella S, et al. Identical mutations in the CSB gene associated with either Cockayne syndrome or the DeSanctis-cacchione variant of xeroderma pigmentosum. Hum Mol Genet. 2000;9(8):1171–1175. doi: 10.1093/hmg/9.8.1171. [DOI] [PubMed] [Google Scholar]
- 32.Gregg SQ, Robinson AR, Niedernhofer LJ. Physiological consequences of defects in ERCC1-XPF DNA repair endonuclease. DNA Repair (Amst) 2011;10(7):781–91. doi: 10.1016/j.dnarep.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakabayashi K, et al. Identification of C7orf11 (TTDN1) gene mutations and genetic heterogeneity in nonphotosensitive trichothiodystrophy. Am J Hum Genet. 2005;76(3):510–6. doi: 10.1086/428141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itoh T, et al. UVs syndrome, a new general category of photosensitive disorder with defective DNA repair, is distinct from xeroderma pigmentosum variant and rodent complementation group I. Am J Hum Genet. 1995;56(6):1267–1276. [PMC free article] [PubMed] [Google Scholar]
- 35.Spivak G. UV-sensitive syndrome. Mutat Res. 2005;577:162–169. doi: 10.1016/j.mrfmmm.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 36.Spivak G, Hanawalt PC. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome fibroblasts. DNA Repair (Amst) 2006;5(1):13–22. doi: 10.1016/j.dnarep.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 37.Nardo T, et al. A UV-sensitive syndrome patient with a specific CSA mutation reveals separable roles for CSA in response to UV and oxidative DNA damage. Proc Natl Acad Sci U S A. 2009;106(15):6209–6214. doi: 10.1073/pnas.0902113106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larsen E, et al. Transcription activities at 8-oxoG lesions in DNA. DNA Repair. 2004;3(11):1457–68. doi: 10.1016/j.dnarep.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Pastoriza Gallego M, Armier J, Sarasin A. Transcription through 8-oxoguanine in DNA repair-deficient and DNA repair-proficient mouse embryonic fibroblasts is dependent upon promoter strength and sequence context. 2005 doi: 10.1093/mutage/gem024. submitted. [DOI] [PubMed] [Google Scholar]
- 40.Selby CP, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci U S A. 1997;94(21):11205–9. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charlet-Berguerand N, et al. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 2006;25(23):5481–91. doi: 10.1038/sj.emboj.7601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevnsner T, et al. Mitochondrial repair of 8-oxoguanine is deficient in Cockayne syndrome group B. Oncogene. 2002;21(57):8675–82. doi: 10.1038/sj.onc.1205994. [DOI] [PubMed] [Google Scholar]
- 43.Kamenisch Y, et al. Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J Exp Med. 2010;207(2):379–90. doi: 10.1084/jem.20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D’Errico M, et al. The role of CSA and CSB protein in the oxidative stress response. Mech Ageing Dev. 2013;134(5–6):261–9. doi: 10.1016/j.mad.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 45.Cleaver JE, et al. Mitochondrial reactive oxygen species are scavenged by Cockayne syndrome B protein in human fibroblasts without nuclear DNA damage. Proc Natl Acad Sci U S A. 2014;111(37):13487–92. doi: 10.1073/pnas.1414135111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ciaffardini F, et al. The cockayne syndrome B protein is essential for neuronal differentiation and neuritogenesis. Cell Death Dis. 5:e1268. doi: 10.1038/cddis.2014.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, et al. Dysregulation of gene expression as a cause of Cockayne syndrome neurological disease. Proc Natl Acad Sci U S A. 2014;111(40):14454–9. doi: 10.1073/pnas.1412569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hashimoto S, et al. A new disorder in UV-induced skin cancer with defective DNA repair distinct from xeroderma pigmentosum or Cockayne syndrome. J Invest Dermatol. 2008;128(3):694–701. doi: 10.1038/sj.jid.5701056. [DOI] [PubMed] [Google Scholar]
- 49.Baple EL, et al. Hypomorphic PCNA mutation underlies a human DNA repair disorder. J Clin Invest. 124(7):3137–46. doi: 10.1172/JCI74593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493(7432):356–63. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lehmann AR. XPD structure reveals its secrets. DNA Repair (Amst) 2008;7(11):1912–5. doi: 10.1016/j.dnarep.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 52.Assfalg R, et al. TFIIH is an elongation factor of RNA polymerase I. Nucleic Acids Res. 2012;40(2):650–9. doi: 10.1093/nar/gkr746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ito S, et al. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/CS patients. Mol Cell. 2007;26(2):231–43. doi: 10.1016/j.molcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 54.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–70. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 55.Greinert R, et al. UVA-induced DNA double-strand breaks result from the repair of clustered oxidative DNA damages. Nucleic Acids Res. 2012;40(20):10263–73. doi: 10.1093/nar/gks824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garinis GA, et al. Transcriptome analysis reveals cyclobutane pyrimidine dimers as a major source of UV-induced DNA breaks. EMBO J. 2005;24(22):3952–62. doi: 10.1038/sj.emboj.7600849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J, et al. Nucleotide excision repair related gene polymorphisms and genetic susceptibility, chemotherapeutic sensitivity and prognosis of gastric cancer. Mutat Res Fundam Mol Mech Mutagen. 2014;765C:11–21. doi: 10.1016/j.mrfmmm.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 58.Lockett KL, Snowhite IV, Hu JJ. Nucleotide-excision repair and prostate cancer risk. Cancer Lett. 2005;220(2):125–35. doi: 10.1016/j.canlet.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 59.Khlifi R, Rebai A, Hamza-Chaffai A. Polymorphisms in human DNA repair genes and head and neck squamous cell carcinoma. J Genet. 2012;91(3):375–84. doi: 10.1007/s12041-012-0193-z. [DOI] [PubMed] [Google Scholar]
- 60.Ogi T, et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell. 2010;37(5):714–27. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]