Abstract
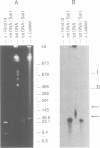
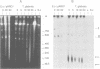
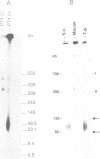
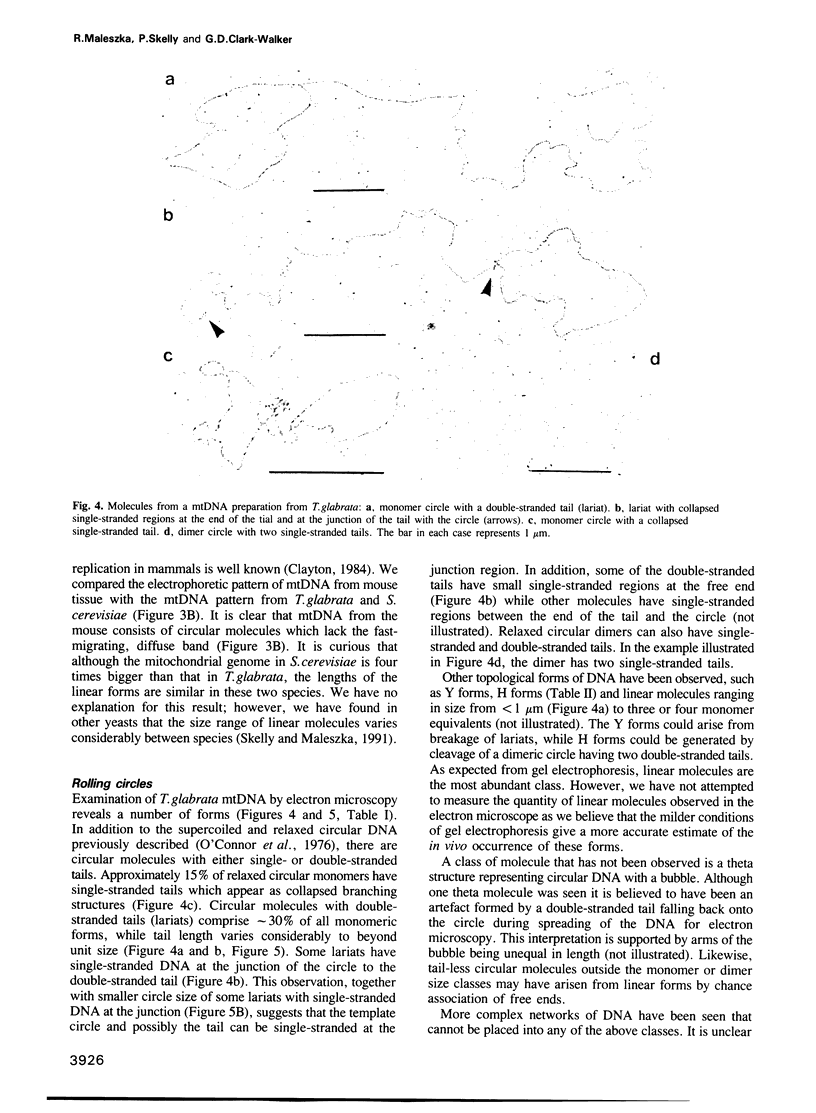
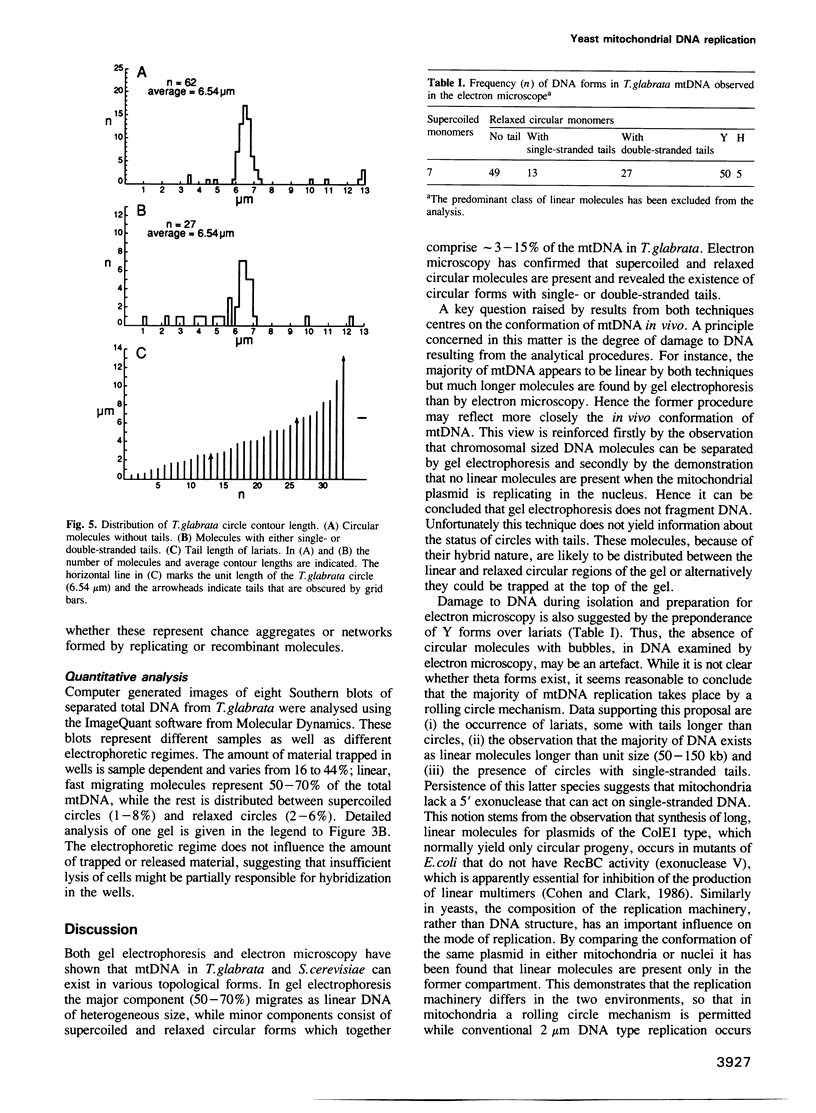
The conformation of mitochondrial DNA (mtDNA) from yeasts has been examined by pulsed field gel electrophoresis and electron microscopy. The majority of mtDNA from Candida (Torulopsis) glabrata (mtDNA unit size, 19 kb) exists as linear molecules ranging in size from 50 to 150 kb or 2-7 genome units. A small proportion of mtDNA is present as supercoiled or relaxed circular molecules. Additional components, detected by electron microscopy, are circular molecules with either single- or double-stranded tails (lariats). The presence of lariats, together with the observation that the majority of mtDNA is linear and 2-7 genome units in length, suggests that replication occurs by a rolling circle mechanism. Replication of mtDNA in other yeasts is thought to occur by the same mechanism. For Saccharomyces cerevisiae, the majority of mtDNA is linear and of heterogeneous length. Furthermore, linear DNA is the chief component of a plasmid, pMK2, when it is located in the mitochondrion of baker's yeast, although only circular DNA is detected when this plasmid occurs in the nucleus. The implications of long linear mtDNA for hypotheses concerning the ploidy paradox and the mechanism of the petite mutation are discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beverley S. M. Characterization of the 'unusual' mobility of large circular DNAs in pulsed field-gradient electrophoresis. Nucleic Acids Res. 1988 Feb 11;16(3):925–939. doi: 10.1093/nar/16.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blok J., Loman H. The effects of gamma-radiation in DNA. Curr Top Radiat Res Q. 1973 Dec;9(2):165–245. [PubMed] [Google Scholar]
- Butow R. A., Fox T. D. Organelle transformation: shoot first, ask questions later. Trends Biochem Sci. 1990 Dec;15(12):465–468. doi: 10.1016/0968-0004(90)90299-q. [DOI] [PubMed] [Google Scholar]
- Christiansen G., Christiansen C. Comparison of the fine structure of mitochondrial DNA from Saccharomyces cerevisiae and S. carlsbergensis: electron microscopy of partially denatured molecules. Nucleic Acids Res. 1976 Feb;3(2):465–476. doi: 10.1093/nar/3.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker G. D. In vivo rearrangement of mitochondrial DNA in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8847–8851. doi: 10.1073/pnas.86.22.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker G. D. Isolation of circular DNA from a mitochondrial fraction from yeast. Proc Natl Acad Sci U S A. 1972 Feb;69(2):388–392. doi: 10.1073/pnas.69.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker G. D., McArthur C. R., Sriprakash K. S. Location of transcriptional control signals and transfer RNA sequences in Torulopsis glabrata mitochondrial DNA. EMBO J. 1985 Feb;4(2):465–473. doi: 10.1002/j.1460-2075.1985.tb03652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker G. D. Size distribution of circular DNA from petite-mutant yeast lacking rho DNA. Eur J Biochem. 1973 Jan 15;32(2):263–267. doi: 10.1111/j.1432-1033.1973.tb02606.x. [DOI] [PubMed] [Google Scholar]
- Clayton D. A. Transcription of the mammalian mitochondrial genome. Annu Rev Biochem. 1984;53:573–594. doi: 10.1146/annurev.bi.53.070184.003041. [DOI] [PubMed] [Google Scholar]
- Cohen A., Clark A. J. Synthesis of linear plasmid multimers in Escherichia coli K-12. J Bacteriol. 1986 Jul;167(1):327–335. doi: 10.1128/jb.167.1.327-335.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R. J., Oakley K. M., Clark-Walker G. D. Elevated levels of petite formation in strains of Saccharomyces cerevisiae restored to respiratory competence. I. Association of both high and moderate frequencies of petite mutant formation with the presence of aberrant mitochondrial DNA. Genetics. 1985 Nov;111(3):389–402. doi: 10.1093/genetics/111.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fox T. D., Sanford J. C., McMullin T. W. Plasmids can stably transform yeast mitochondria lacking endogenous mtDNA. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7288–7292. doi: 10.1073/pnas.85.19.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeben P., Clark-Walker G. D. An approach to yeast classification by mapping mitochondrial DNA from Dekkera/Brettanomyces and Eeniella genera. Curr Genet. 1986;10(5):371–379. doi: 10.1007/BF00418409. [DOI] [PubMed] [Google Scholar]
- Hollenberg C. P., Borst P., van Bruggen E. F. Mitochondrial DNA. V. A 25 micron closed circular duplex DNA molecule in wild-type yeast mitochondria. Stucture and genetic complexity. Biochim Biophys Acta. 1970 May 21;209(1):1–15. [PubMed] [Google Scholar]
- Locker J., Rabinowitz M., Getz G. S. Electron microscopic and renaturation kinetic analysis of mitochondrial DNA of cytoplasmic petite mutants of Saccharomyces cerevisiae. J Mol Biol. 1974 Sep 15;88(2):489–507. doi: 10.1016/0022-2836(74)90497-5. [DOI] [PubMed] [Google Scholar]
- Maleszka R., Clark-Walker G. D. A petite positive strain of Kluyveromyces lactis has a 300 kb deletion in the rDNA cluster. Curr Genet. 1989 Dec;16(5-6):429–432. doi: 10.1007/BF00340722. [DOI] [PubMed] [Google Scholar]
- Maleszka R., Skrzypek M. Assignment of cloned genes to electrophoretically separated chromosomes of the yeast Pachysolen tannophilus. FEMS Microbiol Lett. 1990 May;57(1-2):79–82. doi: 10.1016/0378-1097(90)90416-n. [DOI] [PubMed] [Google Scholar]
- Maniloff J. Anomalous values of Mycoplasma genomes sizes determined by pulse-field gel electrophoresis. Nucleic Acids Res. 1989 Feb 11;17(3):1268–1268. doi: 10.1093/nar/17.3.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur A. J., Monteith J. L. Air movement and heat loss from sheep. II. Thermal insulation of fleece in wind. Proc R Soc Lond B Biol Sci. 1980 Aug 13;209(1175):209–217. doi: 10.1098/rspb.1980.0091. [DOI] [PubMed] [Google Scholar]
- O'Connor R. M., McArthur C. R., Clark-Walker G. D. Respiratory-deficient mutants of Torulopsis glabrata, a yeast with circular mitochondrial deoxyribonucleic acid of 6 mu m. J Bacteriol. 1976 May;126(2):959–968. doi: 10.1128/jb.126.2.959-968.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D., Byers B., Fangman W. L. Size and structure of yeast chromosomal DNA. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3072–3076. doi: 10.1073/pnas.70.11.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Simske J. S., Scherer S. Pulsed-field gel electrophoresis of circular DNA. Nucleic Acids Res. 1989 Jun 12;17(11):4359–4365. doi: 10.1093/nar/17.11.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelly P. J., Maleszka R. Distribution of mitochondrial intron sequences among 21 yeast species. Curr Genet. 1991 Feb;19(2):89–94. doi: 10.1007/BF00326288. [DOI] [PubMed] [Google Scholar]
- Skelly P. J., Maleszka R. Isolation of mitochondrial DNA using pulsed field gel electrophoresis. Nucleic Acids Res. 1989 Sep 25;17(18):7537–7537. doi: 10.1093/nar/17.18.7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypek M., Borsuk P., Maleszka R. Cloning and sequencing of the ornithine carbamoyltransferase gene from Pachysolen tannophilus. Yeast. 1990 Mar-Apr;6(2):141–148. doi: 10.1002/yea.320060208. [DOI] [PubMed] [Google Scholar]
- Thorsness P. E., Fox T. D. Escape of DNA from mitochondria to the nucleus in Saccharomyces cerevisiae. Nature. 1990 Jul 26;346(6282):376–379. doi: 10.1038/346376a0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Young I. G., Anderson S. The genetic code in bovine mitochondria: sequence of genes for the cytochrome oxidase subunit II and two tRNAs. Gene. 1980 Dec;12(3-4):257–265. doi: 10.1016/0378-1119(80)90108-0. [DOI] [PubMed] [Google Scholar]
- van der Bliek A. M., Lincke C. R., Borst P. Circular DNA of 3T6R50 double minute chromosomes. Nucleic Acids Res. 1988 Jun 10;16(11):4841–4851. doi: 10.1093/nar/16.11.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]