Abstract
Instructive programs guiding cell fate decisions in the developing mouse embryo are controlled by a few so-termed master regulators. Genetic studies demonstrate that the T-box transcription factor Eomesodermin (Eomes) is essential for epithelial to mesenchymal transition (EMT), mesoderm migration and specification of definitive endoderm (DE) during gastrulation1. Here we report that Eomes expression within the primitive streak marks the earliest cardiac mesoderm and promotes formation of cardiovascular progenitors by directly activating the bHLH transcription factor Mesp1 upstream of the core cardiac transcriptional machinery2-4. In marked contrast to Eomes/Nodal signalling interactions that cooperatively regulate anterior-posterior (A-P) axis patterning and allocation of the DE cell lineage1, 5-8, formation of cardiac progenitors requires only low levels of Nodal activity accomplished via a Foxh1/Smad4 independent mechanism. Collectively our experiments demonstrate that Eomes governs discrete context dependent transcriptional programmes that sequentially specify cardiac and DE progenitors during gastrulation.
Keywords: Eomesodermin, Mesp1/2, cardiac specification, definitive endoderm, gastrulation
Much has been learned about the coordinated activities of key regulatory networks of transcription factors and growth factor signalling pathways governing cell fate decisions during gastrulation9. Nascent mesoderm is induced as epiblast cells ingress through the primitive streak (PS) and undergo EMT. Distinct mesodermal sub-populations become allocated according to the timing and order of these cell movements. Thus extra-embryonic mesoderm arises from the posterior PS, while cardiac, paraxial and lateral plate precursors emerge sequentially as the PS elongates towards the distal tip of the embryo. Fate mapping experiments demonstrate that DE progenitors are specified in the most anterior region of the PS (APS), in close proximity to the cardiovascular progenitors10, 11.
Mesoderm formation and patterning along the proximodistal axis of the PS is known to be regulated by dose-dependent Nodal/Smad2/3 activities7. The highest level of Nodal/Smad2/3 signaling is required to specify APS derivatives, namely the DE, node and notochord5, 6, 8. The transcription factor Smad4, and its DNA-binding partner the forkhead transcription factor Foxh1, also play essential roles in APS specification6, 12, 13. Additionally, the T-box transcription factor Eomes acts cooperatively with the Nodal/Smad2/3 pathway to promote delamination of nascent mesoderm and specification of APS fates1.
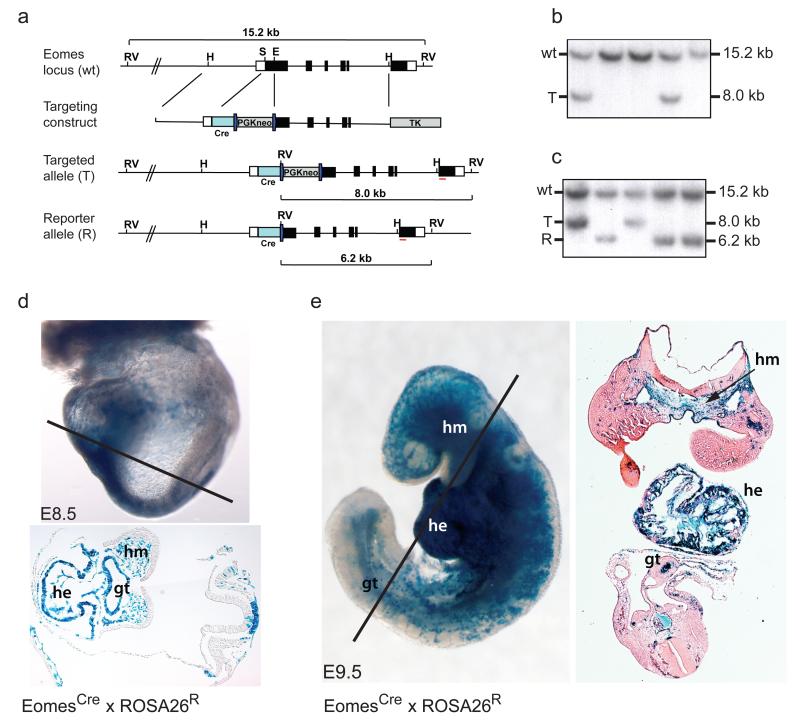
Eomes expression is initiated in the prospective posterior aspect of the epiblast at embryonic day 5.75 (E5.75)14. During gastrulation expression is maintained in the distal PS14, 15, encompassing the same region where cranial, cardiac and paraxial mesodermal sub-cell populations are generated10. Inactivation of Eomes in the epiblast results in a severe block in EMT and arrests development at gastrulation1. To further characterise Eomes functional contributions within the mesodermal cell lineages we generated an EomesCre reporter allele. Cre mRNA expression recapitulates endogenous expression (Supplementary Information, Fig. S1a) allowing derivation of a fate map of Eomes expressing cells in later stage embryos (Fig. 1). EomesCre/+ males were mated to females carrying the ROSA26R reporter strain16 and the resulting embryos stained for LacZ activity (Fig. 1d, e, Supplementary Information, Fig. S1b). Surprisingly, we found in E8.5 and E 9.5 EomesCre; ROSA26R embryos that LacZ expressing cells are mostly absent from the somites, intermediate and lateral plate mesoderm and largely restricted to the head mesenchyme, cardiac mesoderm and vasculature (Fig. 1d, e). As expected1, the DE and gut tube are exclusively comprised of LacZ marked EomesCre+ descendants. At later stages endodermal but not mesodermal components of developing organs derived from the gut tube are LacZ positive (Supplementary Information, Fig. S1c). Eomes expressing cells thus give rise to two discrete progenitor cell populations during gastrulation, namely the prospective cranial and cardiac mesoderm that emerge from the PS at early stages, and APS derivatives giving rise to the DE, node and notochord. In striking contrast Eomes+ cells are excluded from the majority of mesodermal tissues derived from the paraxial and lateral plate mesoderm. These observations strongly suggest that a discrete sub-population of cells within the pre-gastrulation epiblast preferentially ingress and migrate anteriorly as a cohort to form the cardiac crescent and prospective head mesoderm.
Figure 1.
Fate mapping EomesCre expressing cells reveals selective contributions to DE and cardiovascular cell lineages.
(a) Targeting strategy used to generate the EomesCre reporter allele. Cre recombinase coding sequences were inserted into Exon1 of the Eomes locus. RV, EcoRV; H, HpaI; S, SphI; E, EagI; Cre, Cre recombinase. (b) ES clones were screened by Southern blot on EcoRV digested DNA using a 3′ probe (red line in a) to detect wildtype (wt) and targeted (T) alleles. (c) Correctly targeted ES cells were transiently transfected with Cre to excise the PGK-neo selection cassette and generate the reporter allele (R), as shown by Southern blot. (d, e) Fate mapping experiments demonstrate that descendants of EomesCre expressing cells contribute to the myocardium and endocardium of the heart (he), the head mesenchyme (hm), vasculature and the endoderm of the primary gut tube (gt), but rarely colonize other mesoderm tissues formed from paraxial and lateral plate mesoderm. Sections were counterstained with eosin to highlight non-labelled cells.
To directly examine whether Eomes function is required for specification of cardiovascular progenitors we analysed E7.5 embryos carrying an epiblast-specific Eomes deletion (EomesCA/N;Sox2.Cre)1 by whole mount in-situ hybridization (WISH). Embryos lacking Eomes function strongly express mesodermal marker genes, including Brachyury, Fgf8 and Snail1. However in striking contrast expression of early cardiac marker genes, Myl7 (also known as Mlc2a), Wnt2 and Nkx2.5 was absent (Fig. 2a). Moreover we observe severely reduced expression of early vascular marker genes such as Agtrl1, Rasgrp3 and Klhl6 (Fig. 2a).
Figure 2.
Eomes functional loss disrupts specification of cardiovascular progenitors.
(a) Whole-mount in situ hybridisation analysis of cardiac mesoderm (Myl7, Wnt2, Nkx2.5) and vascular (Agtrl1, Rasgrp3, Klhl6) markers in control and EomesN/CA;Sox2.Cre mutant embryos. Eomes mutants entirely lack expression of cardiac marker genes and show significantly reduced expression of vascular markers. In contrast in wild type embryos vascular markers broadly delineate the embryonic and extra-embryonic vasculature at E8.5-9.5. (b) Schematic diagram illustrating the protocol for generation of chimeric embryos. Eomes-null ES cells were introduced into wild type ROSA26LacZ blastocysts. (c, d) Histological sections of two independent LacZ stained E9.5 chimeric embryos were counterstained with Eosin to identify Eomes mutant cell populations (pink). The myocardium and endocardium of the heart (he) and endoderm of the gut tube (gt) are exclusively comprised of LacZ positive wild type cells. Relatively few Eomes null cells colonize the head mesenchyme (hm), whereas the majority of other tissues are comprised of mixed populations of Eomes null and wild-type cells. Scale bar is 500 μm.
To test whether loss of cardiac gene expression reflects a cell autonomous Eomes requirement we examined the developmental potential of Eomes null embryonic stem (ES) cells in the context of chimeric embryos. Eomes null ES cells1 were injected into wild type blastocysts carrying the ROSA26LacZ allele17 and the resulting embryos analysed by X-Gal staining (Fig. 2b). As expected1 at E8.5 and E9.5 Eomes null cells efficiently contribute to the majority of mesodermal tissues but are entirely excluded from the forming gut tube. Strikingly in all cases examined (n=10), the myocardium and endocardium of the developing heart were also exclusively comprised of wild type LacZ+ cells (Fig. 2c, d). Thus we conclude that Eomes plays an essential cell autonomous role during allocation of both the DE and cardiovascular progenitors.
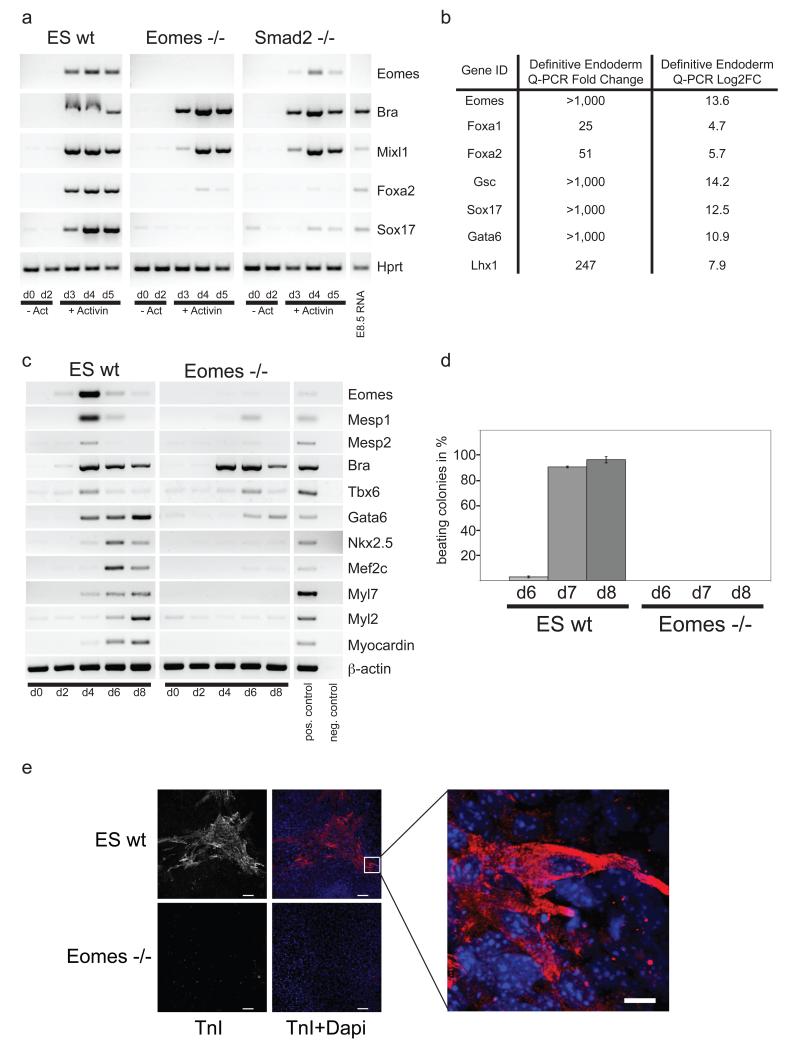
To further evaluate Eomes contributions we exploited culture protocols that promote ES cell differentiation towards either DE or cardiac fates. To elicit DE formation, ES cells harboring an EomesGFP reporter allele18 were cultured in the presence of high doses of ActivinA (50ng/ml, Supplementary Information, Fig. S2a). After 4 days, as judged by GFP expression or staining with an Eomes antibody, >95% of cells were strongly positive (Supplementary Information, Fig. S2b, and data not shown). Next we compared wild type (CCE), Eomes null (6A6)1 and Smad2 null (KT15)7 ES cells and monitored DE marker gene expression by RT-PCR. Eomes expression was detectable 24hrs after ActivinA treatment in wild type cells, severely reduced in Smad2-deficient cells and absent in Eomes null cells (Fig. 3a). In chimera studies Smad2-deficient cells robustly contribute to all mesodermal lineages but are excluded from DE derivatives19 and as shown here only inefficiently up-regulate expression of Eomes or DE markers.
Figure 3.
Eomes null ES cells fail to give rise to cardiomyocytes.
(a) Wild-type, Eomes−/− and Smad2−/− ES cells were cultured in the presence of high-doses of ActivinA. Semi-quantitative RT-PCR analysis shows that Eomes−/− and Smad2−/−cultures strongly express mesoderm marker genes such as Brachyury and Mixl1, but lack expression of the DE marker genes, Foxa2 and Sox17. (b) Quantitative RT-PCR (qRT-PCR) analysis confirms the dramatically reduced DE marker transcript levels in Eomes−/−at day4 of ActivinA-induced differentiation. (c) Wild-type and Eomes mutant embryoid bodies (EBs) were induced to form cardiomyocytes in hanging drop cultures. Semi-quantitative RT-PCR analysis reveals that cardiac-specific transcription factors (Mesp1, Mesp2, Gata6, Nkx2.5, Mef2c and Myocardin), as well as structural proteins (Myl7, Myl2) are significantly down-regulated in Eomes null EBs while expression of the panmesodermal marker Brachyury is unaffected. (d) Clusters of beating cardiomyocytes are readily detectable in wild-type EB outgrowths but absent in Eomes null cultures at day 7. Error bars represent standard error of the mean (s.e.m.) of three independent experiments. (e) At day 8 TroponinI (TnI)-positive cardiomyocytes are detectable in wild type outgrowths but are entirely absent in Eomes mutant cultures. Higher magnification reveals characteristic cross striation of myofibrils. Scale bar, 100μm for the overview and 10μm for the higher magnification image.
Differentiating Eomes null ES cells strongly express PS and mesodermal marker genes, including Brachyury and Mixl1 but expression of DE marker genes including Sox17 and Foxa2 was drastically reduced. Similar conclusions were reached in Q-RT-PCR experiments examining a panel of key DE-associated genes including Gsc, Gata6 and Lhx1 (Fig. 3b).
Next we examined the development of cardiac progenitors via embryoid body (EB) differentiation in hanging drops with high serum-stimulation20 (Supplementary Information, Fig. S2c, d). In wild type cultures Eomes transcripts are detectable by RT-PCR on day 2, peak around day 4 and are down-regulated by day 6 (Fig. 3c). Coincident with highest levels of Eomes expression, early cardiac markers Mesp1 and Myl7 are upregulated beginning at around day 4. In Eomes null EBs the pan-mesodermal marker Brachyury is robustly induced (Fig.3c), but in marked contrast expression of cardiac-specific genes including Mesp1, Myl7 Myl2, Myocardin, Nkx2.5 and Mef2c was entirely absent. As judged by microscopic observation and staining for the cardiac protein TroponinI (TnI), close to 100% of wild type EBs contain clusters of contractile cardiomyocytes after day 7 of differentiation (Fig. 3d, e, Supplementary Information, Movie SM1). Eomes null EBs fail to differentiate into cardiomyocytes by these criteria (Fig. 3d, e, Supplementary Information, Movie SM2). Collectively these results suggest that Eomes acts upstream of the transcriptional hierarchy that specifies cardiac fates during gastrulation.
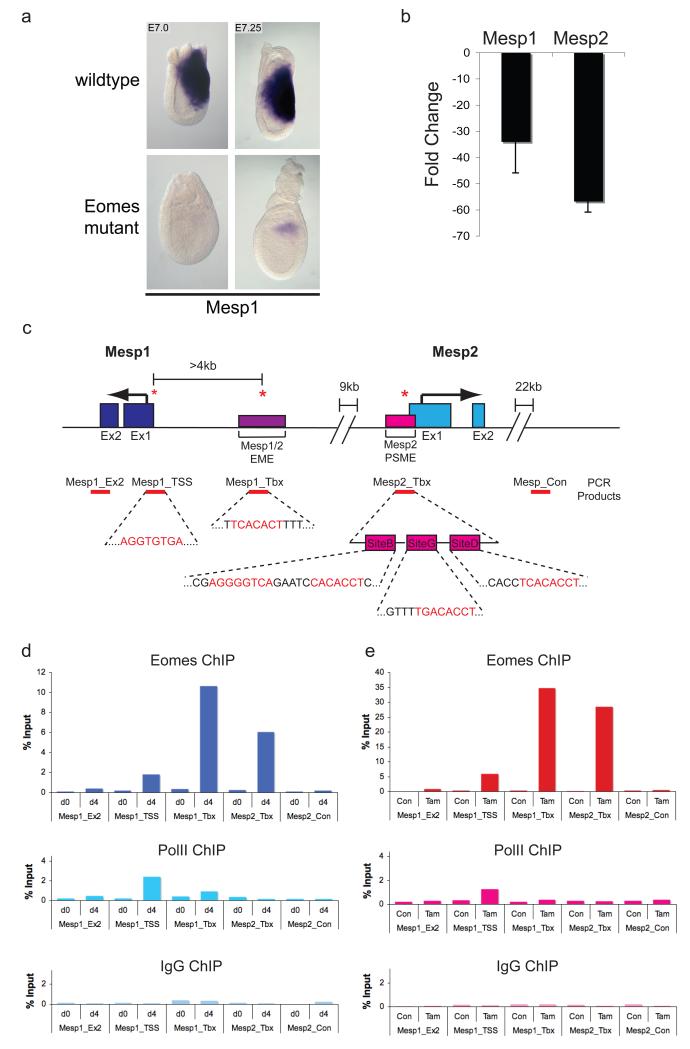
The bHLH transcription factor Mesp1 has been described a master regulator of multipotent cardiovascular progenitor specification21-23. Genetic fate mapping experiments demonstrate that Mesp1 expression marks cardiac progenitors that give rise to the myocardial and endocardial derivatives22. Moreover Mesp1 activity has also been implicated in the early steps of cardiac lineage specification in vitro2-4. Mesp1 is strongly expressed at the onset of gastrulation (E6.5) along the forming PS, marking the prospective cardiac mesoderm and is then rapidly down-regulated starting around E7.522. Mesp1 mutants form cardiac mesoderm but heart morphogenesis is highly compromised leading to cardia bifida21, 22. Mesp1 and Mesp2 are functionally interchangeable in vivo21. Thus premature up-regulated expression of the closely related family member Mesp2 in the loss of function mutants is sufficient to generate cardiac progenitors and partially rescue heart morphogenesis23. Mesp1; Mesp2 double mutant embryos block at gastrulation, and display an EMT phenotype that closely resembles the Eomes loss of function phenotype1, 23. At E7.0 Mesp1 is completely absent in Eomes mutants (Fig. 4a). At slightly later stages (E7.25) a few exceptional Eomes mutants display weak Mesp1 staining. At E7.25 Mesp2 expression is undetectable in WISH experiments24. However Q-RT-PCR analysis demonstrates that Mesp1 and Mesp2 expression are both dramatically down-regulated (Fig. 4b). As shown above differentiating Eomes null ES cells lack the ability to upregulate Mesp1 and Mesp2 (Fig. 3c). Collectively these experiments strongly suggest that Eomes acts upstream of Mesp1/2 to regulate EMT and control allocation of cardiac progenitors.
Figure 4.
Eomes directly binds conserved T-box sites within the Mesp1 locus to activate expression.
(a) Mesp1 robustly expressed in wild-type embryos at E7.0, is absent in EomesN/CA;Sox2Cre mutants. At slightly later stages (E7.25) EomesN/CA;Sox2Cre embryos occasionally show weak expression that likely reflects activity of Tbx6, known to be expressed in E7.5 Eomes mutants1. (b) As judged by qRT-PCR Mesp1 and Mesp2 transcripts are dramatically reduced in E7.25 Eomes mutant embryos. Error bars represent standard deviation (s.d.), n= 5 per genotype. (c) Diagrammatic representation of cis-regulatory elements in the Mesp1/2 locus. The positions of previously identified T-box sites within the Mesp1/2 EME and Mesp2 PSME (nomenclature according to ref26, 27) and a novel putative T-box site identified near the Mesp1 TSS are indicated. The Mesp2 PSME contains 3 binding elements (Sites B, G, D) that contain T-box binding motifs. T-box consensus sequences are indicated in red. Red bars indicate areas amplified by qPCR after ChIP using different antibodies. Ex1, Exon1; Ex2, Exon2; EME, early mesoderm enhancer; PSME, pre-somitic mesoderm enhancer; TSS, transcriptional start site; Tbx, T-box site. (d) ChIP analysis of P19Cl6 cells treated for 4 days with DMSO using antibodies specific for Eomes, RNA-Polymerase II (PolII) or an IgG control. Specific enrichment for genomic loci containing T-box sites (Mesp1_TSS, Mesp1_Tbx, Mesp2_Tbx) was observed using the Eomes-specific antibody. The PolII antibody gave specific enrichment of the Mesp1_TSS, but not other tested regions of the Mesp1/2 locus. Specific binding to T-box sites (indicated by *) in the Mesp1/2 EME, the Mesp2 PSME and the Mesp1 TSS was detectable after 4 days, but not at day 0. d0, day 0; d4, day4 of DMSO differentiation of P19Cl6 cells. (e) Cells expressing a tamoxifen inducible Eomes fusion construct (P19EoER cells), also shows Eomes binding to these T-box sites at day 4 of tamoxifen treatment. Con, non-tamoxifen induced control P19EoER cells; Tam, day 4 tamoxifen treated P19EoER cells. The best representative plots of three independent experiments are shown.
Mesp1 and Mesp2 are closely linked on chromosome 7, and arranged in opposite orientation with the two transcriptional start (TS) sites separated by approximately 17 kb25. The cis-acting regulatory elements responsible for controlling temporally and spatially restricted expression patterns are well characterized25-27. An evolutionary conserved early mesoderm enhancer (EME, Fig. 4c, Supplementary Information, Fig. S3b, d) recapitulates Mesp1 expression in nascent mesoderm25, while regulatory sequences adjacent to the Mesp2 TS site direct mesodermal expression in the PS, presomitic mesoderm and developing somites26. Both elements contain T-box consensus binding sites25-27 (Supplementary Information, Fig. S3). Additionally we have identified a further conserved T-box binding element in close proximity to the Mesp1 TS site (Fig. 4c, Supplementary Information, Fig. S3b, c).
To further investigate Mesp1/2 activation during cardiovascular lineage commitment, we used the P19Cl6 embryonal carcinoma (EC) cell line that efficiently differentiates into beating cardiomyocytes in the presence of 1% DMSO28. Transient Eomes expression seen initially at day 2 is down regulated by day 6, associated with increased Mesp1 expression levels (Supplementary information, Fig. S4a). Activation of Eomes estrogen-receptor fusion protein in the presence of tamoxifen (EomesER, Supplementary Information, Fig. S4b) also efficiently induces strong Mesp1 expression within 24 hours as assayed by Q-PCR (Supplementary Information, Fig. S4g). To directly evaluate Eomes occupancy at the Mesp1/2 locus we performed chromatin immunoprecipitation (ChIP) analysis with day 4 DMSO-treated P19Cl6 cells as well as tamoxifen-treated P19Cl6EomesER (P19EoER) cells. Eomes binding to all three conserved T-box site-containing regions within the Mesp1/2 locus was clearly observed (Fig. 4d, e, Supplementary Information, Fig. S5). The EME, which controls early Mesp1 expression in nascent mesoderm25 gave the strongest signal. Occupancy at other genomic regions, or in uninduced cells was undetectable. The T-box sites adjacent to the Mesp2 TS site are known to be occupied by Tbx6 in pre-somitic mesoderm27. This cis-acting regulatory element, as well as the minimal 220 bp Mesp1 EME T-box element, regulates Mesp2 and Mesp1 expression respectively at later stages during somitogenesis26. Additional T-box family members are also known to be expressed in the early gastrulation stage embryo14, 15, 29-31. However only Brachyury and Eomes are exclusively present in the posterior epiblast and nascent mesoderm overlapping with sites of Mesp1 expression. Specification of early heart progenitors proceeds normally in T mutant embryos. Therefore a strong argument can be made that the earliest Mesp1 expression domain marking the cardiac progenitors is directly activated by Eomes occupancy at these T-box sites.
Genetic studies analyzing double heterozygous mutant embryos demonstrate that Eomes and Nodal function cooperatively in A-P axis patterning and formation of APS derivatives1. We wondered whether dose-dependent Nodal signals could potentially regulate Eomes-dependent Mesp1 activation. To test this possibility first we examined Smad46 and Foxh1 mutant embryos12 at E6.5 and E7.5. Interestingly Mesp1 expression is unaffected by loss of either Smad4 or Foxh1 (Fig. 5a). The lim-homeodomain transcription factor Lhx1 is also required for specification of DE and anterior axial midline tissues32, 33. Similarly we observe that compromised DE development has no noticeable impact on specification of Mesp1+ cardiac progenitors. Thus Lhx1 loss of function embryos display strong expression of both Mesp1 and Eomes (Fig. 5a and data not shown). Conversely it is known that expression of the APS markers Lhx1 and Gsc is unperturbed in Mesp1/2 double mutant embryos23. Lowering Nodal5, 34 or Smad2/37 levels during gastrulation also selectively disturbs DE specification. Next, we manipulated ActivinA concentrations in differentiating ES cells and confirmed that low levels (5ng/ml) are sufficient for robust Mesp1 expression, while conversely maintaining cultures in high ActivinA concentrations (50ng/ml) leads to induction of Sox17 (Supplementary Information, Fig. S6a, b). Collectively, these results strongly suggest that specification of both cardiac and DE progenitors requires Eomes, but these lineages arise independently, dependent on local Nodal/Smad/Foxh1 signalling levels.
Figure 5.
Eomes and dose-dependent Nodal/Smad2/3 signalling levels control cardiac mesoderm and definitive endoderm specification during gastrulation.
(a) Smad4 and Foxh1 are critical Nodal pathway components for transducing high levels of signalling6, 12, 13. Mesp1 is expressed normally in E6.5 and 7.5 Foxh1 null embryos and in embryos lacking Smad4 in the epiblast only (Smad4Δ). Mesp1 expression is also efficiently induced in Lhx1 mutant embryos, which display DE and midline mesoderm defects. However the failure of A-P axis rotation results in induction of Mesp1 throughout the proximal epiblast. (b) Eomes activity regulates formation of both cardiac mesoderm and DE progenitors during gastrulation. Eomes+ epiblast cells confined to the posterior side of the embryo prior to overt streak formation are exposed to low levels of Nodal signalling. Eomes-dependent activation of Mesp1/2 marks the earliest cardiac progenitors induced in the forming PS. Mesp1/2 expression leads to activation of the Nodal antagonist Lefty223 and direct repression of DE genes2. As cells begin to migrate away from the PS Mesp1/2 expression is down-regulated via a negative feedback loop2, 23, 25. In contrast the Eomes expression domain extends distally and overlaps with increased Nodal signalling levels as the PS elongates. Eomes, acting cooperatively with Nodal/Smad4/FoxH1 dependent signals in the APS, induces DE. (c) At later stages from E7.5 onwards Tbx6 expression in the pre-somitic mesoderm activates a second wave of Mesp1/2 expression in the pre-somitic mesoderm via occupancy of the conserved T-box regulatory elements.
Multipotent mesodermal progenitor cells that give rise to diverse tissues of the emerging body plan become progressively allocated as epiblast cells transit the PS. Fate mapping and grafting studies have shown cranial and cardiac mesoderm derive from the earliest wave of cells that exit at the mid streak stage, whereas pre-somitic/paraxial and lateral plate mesoderm emerge at more posterior levels at late streak stages10, 11. Remarkably the present experiments identify a sub-set of proximal posterior epiblast cells already committed to adopt a cardiac fate many hours prior to mesoderm induction and overt PS formation. The early Eomes expression domain marks cardiac progenitors programmed to activate Mesp1, previously identified as the master regulator that acts instructively to specify cardiovascular cell fates. Recent studies demonstrate that Mesp1 initiates global changes in gene expression by directly binding to regulatory sequences at the promoters of many key genes in the core cardiac transcriptional machinery. Mesp1 upregulates expression of Hand2, Myocardin, Nxk2.5 and Gata4, and represses genes governing pluripotency and mesodermal and endodermal cell fates2. Additionally Mesp1 promotes EMT via selective up-regulation of the zinc-finger repressor Snail4, allowing the nascent cardiac progenitors to migrate anteriorly to underlie the developing headfolds where they coalesce to form the cardiac crescent. Acting a few hours later during PS elongation Eomes/Nodal signalling results in specification of DE1 that gives rise to the entire gut endoderm lineage. Eomes was recently shown to be a key player in the transcriptional network upstream of DE specification during human ES cell differentiation35.
How does a single transcription factor Eomes govern allocation of two independent, non-overlapping, multipotent progenitor cell populations as epiblast cells sequentially transit the PS? We suggest that the key parameter controlling cardiac versus DE cell fate is the timing and duration of exposure to Nodal signaling (Fig. 5b). Low levels of Nodal activity in the posterior epiblast are sufficient to activate Eomes and induce cardiac mesoderm formation at early post-implantation stages (Supplementary Information, Fig. S6d). Eomes expression is necessary and sufficient to activate Mesp1/2, promote EMT and migration of a discrete mesodermal sub-population that gives rise to the cardiac crescent. Within this early sub-set Mesp1 directly represses genes required for formation of DE including Foxa2, Gsc and Sox172. Mesp1/2 expression also activates expression of the Nodal antagonist Lefty223 to further insulate cardiac progenitors as they migrate away from the source of Nodal signaling (Supplementary Information, Fig. S6e). Consistent with this idea, up-regulated Nodal signalling in Tgif1/2 double mutant embryos inhibits Mesp1 expression, whereas decreased levels of Nodal activity rescues Mesp1 expression36.
Eomes activation of Mesp1/2 in cardiac progenitors is only transient due to a Mesp1/2 autoregulatory negative feedback loop2, 23, 25. At late streak stages expression of Mesp1/2 is re-activated in pre-somitic mesoderm by Tbx6 occupancy of the conserved T-box sites27 (Fig. 5c). In contrast during streak elongation Eomes expression is maintained by high levels of Nodal/Smad2 signaling downstream of a Wnt3/Lef1 feed-forward loop8. Acting together with Nodal-dependent Smad2/3/4/Foxh1 transcription complexes5-7, 12, 34, Eomes promotes formation of APS progenitors that give rise to the DE, node and notochord. It will be interesting to learn more about developmental regulation of Eomes transcriptional partnerships and cell type specific changes in chromatin architecture governing T-box site occupancy at the Mesp1/2 locus and selection of target genes in the DE cell lineage.
Methods
Generation of the EomesCre reporter allele
The EomesCre allele was generated using the same strategy as previously described for the EGFP knock-in allele18. The targeting vector encompasses a 8.25 kb HpaI fragment of the Eomes locus. Cre coding sequences were introduced into the Exon1 start site followed by a LoxP-flanked neomycin resistance cassette between the SphI (translational start) and EagI sites, resulting in removal of ~500 bp of the endogenous 5′ coding region. The 3′ homology arm was flanked by a pMC1.TK negative selection cassette. Linearised targeting vector was electroporated into CCE ES cells and drug resistant ES cell colonies screened by Southern blot using a 3′ external probe on EcoRV digested DNA (wt allele; 15.2 kb; targeted allele; 8.0 kb). Correctly targeted clones were transfected with pMC1.Cre and resulting subclones screened by Southern blot to detect the reporter allele (6.2 kb). Two independent excised cell clones were used to generate germline chimeras. Offspring were genotyped by PCR using Cre specific primers (Cre-fw: 5′-GCATAACCAGTGAAACAGCATTGCTG-3′, Cre-rev: 5′-GGACATGTTCAGGGATCGCCAGGCG-3′). The strain was maintained on a 129SvEv/C57Bl/6 mixed genetic background.
Mouse strains, genotyping and generation of chimeric embryos
All animal procedures were approved by the Ethical Review Committee of the University of Oxford. The ROSA26R reporter16, ROSA26 gene-trap17, Foxh112, Smad4CA 6, EomesCA/N;Sox2.Cre1 and Lhx132 strains were genotyped as previously described. For generation of chimeric embryos blastocysts recovered from matings of ROSA26 males to CD1 outbred females were injected with 12-14 Eomes null ES cells1 and transferred into E2.5 pseudopregnant foster females.
Whole-mount in situ hybridisation, LacZ staining and histology
Whole-mount in situ hybridisation analysis of E6.5-9.5 embryos dissected and fixed with 4% PFA overnight at 4 °C was performed according to standard protocols37 using probes for Agtrl1,Cre, Eomes, Klhl6, Mesp1, Myl7, Nkx2.5, Rasgrp3, and Wnt2. LacZ staining was performed as described37. For histology, embryos were post-fixed in 4% PFA, dehydrated through an ethanol series, embedded in paraffin, sectioned at 8 μM and Eosin-counterstained.
Definitive endoderm and cardiomyocyte differentiation assays
Wild-type (CCE), Eomes null (6A6) and Smad2 null (KT 15) feeder-depleted ES cells were cultured in DMEM (Invitrogen) with 15% FCS, 1% non-essential amino acids, 0.1 mM β–mercaptoethanol and 1000 U/ml recombinant LIF (Millipore). To induce DE differentiation, ES cells were cultured in serum-free ESGRO Complete clonal grade medium (Millipore) on 0.1% gelatin-coated dishes for a minimum of 2-3 passages before seeding at low density (18,000 cells/ml) in ESGRO Complete basal medium (Millipore) in bacteriological grade dishes, to promote embryoid body (EB) formation. 50 ng/ml Activin A (R&D systems) was added after 48hrs. For titration of ActivinA effects, the medium was changed after 72 hrs and new medium added with 5 or 50 ng/ml ActivinA. For cardiac differentiation, ES cells were re-suspended at 1 × 104 cells/ml in IMDM (Invitrogen) supplemented with 20% fetal calf serum, 1% non-essential amino acids, and 0.1 mM β–mercaptoethanol in hanging drops (10 μl) plated on the inside lids of bacteriological dishes. After 48hrs EBs were transferred in 10 ml medium to 10 cm bacteriological dishes. At day 4 EBs were plated on tissue culture dishes, allowed to adhere and scored at d7. EBs were plated on fibronectin-coated 15 μ-Slide 2×9well (Ibidi) and cultured for additional 96 hrs prior to imaging and immunostaining. The number and percentage of beating EBs was counted in three independent experiments. Live cell imaging was recorded on a Zeiss Observer Z.1 microscope equipped with an Axiocam MRm camera at frame rates of 10/s and 36/s.
P19Cl6 cell culture, differentiation and generation of EomesER expressing sub-clones
P19Cl6 EC cells were routinely cultured in α-MEM (Invitrogen) supplemented with 10% FCS. To induce differentiation, cells were seeded at 3.7×105 cells/6 cm dish in media containing 1% DMSO (Sigma). Cells were electroporated with linearised pCAGEomesER-IRESPuro vector38 and selected in 1 μg/ml puromycin, to generate P19EoER sub-clones. Expression of the EomesER fusion protein was confirmed by Western blot analysis. For EomesER activation 1 μg/ml of 4-Hydroxytamoxifen (Sigma, H7904) was added to the culture media.
RNA isolation, One-step & quantitative RT-PCR analysis
RNA was isolated using the RNeasy Kit (Qiagen) according to the manufacturer’s instructions, using on-column DNase treatment. cDNA was generated using the SuperScriptIII kit (Invitrogen) with oligo-dT primers. Q-RT-PCR was performed using the Quantitect SYBRGreen PCR kit and a Rotar-gene Q (Qiagen) and analysed using the ΔΔCt method, as described previously39. For one-step RT-PCR analysis, the One-step RT-PCR kit (Qiagen) was used with gene specific primers according to manufacturer’s instructions. Primer sequences are provided in Supplementary Information Table S1.
Chromatin Immunoprecipitation
Cells were cross-linked for 10 minutes at RT with 1% (v/v) formaldehyde and quenched with 125 mM glycine. Prepared chromatin was sonicated to 200-500 bp and immunoprecipitated with 15 μg of anti-Eomes (Abcam, ab23345), anti-PolII N-terminal (Santa Cruz, sc-899x) or normal rabbit IgG (Santa Cruz, sc-2027). Eluted DNA was recovered by phenol-chloroform extraction, precipitated with ethanol and resuspended in TE buffer. ChIP material was analysed using a Rotar-Gene Q (Qiagen) and SYBRGreen master mix (Qiagen). The amount of precipitated DNA was compared to the starting input material, as percentage of input. Each ChIP experiment was performed at least three times on separate biological samples. Q-PCR was performed in triplicate. ChIP primer sequences are provided in Supplementary Information Table S2.
Immunofluorescence
ES or P19Cl6 cultures were fixed in 4% PFA for 10 min at RT and permeabilised with 0.2% TritonX/PBS for 20 min before blocking for 1 hr in 10% FCS, 0.3% BSA, 0.3% TritonX in PBS. Primary antibodies used include rabbit anti-Eomes (Abcam, ab23345, 1:100), rat anti-Eomes (eBioscience, 14-4876, 1:100), goat anti-Gata4 (Santa Cruz, sc-1237, 1:100), mouse anti-Troponin I (Abcam, ab19615, 1:200) and mouse anti-smooth muscle actin (Dako, M0851, 1:100). Primary antibodies were applied in blocking solution overnight at 4 °C. Cells were washed with 0.1% TrionX/PBS and incubated in blocking solution for 1 hr at RT with appropriate conjugated secondary antibodies; goat anti-rabbit Alexa Fluor 555; goat anti-rat Alexa Fluor 488; donkey anti goat Alexa Fluor 488, goat anti-mouse Alexa Fluor 488 (all Molecular Probes/Invitrogen, 1:500) and donkey anti-mouse Cy3 (Jackson Immuno Research, 1:2000). Coverslips were mounted with Vectashield mountant containing DAPI (Vector Labs, H-1200). Fluorescent images were captured with a Zeiss epifluorescence microscope or an inverted laser scanning microscope (LSM 510 Meta Duo Live) equipped with a 25x/63x Plan-Apochromate objective.
Western blot analysis
Cell lysates were prepared using RIPA buffer and subjected to SDS-PAGE and transferred onto PVDF membranes. Membranes were blocked with 5% milk in TBS-T, incubated in primary antibodies overnight including rabbit anti-Eomes (Abcam, ab23345, 1:1000), rabbit anti-phospho-Smad2 (Cell Signaling, 3101, 1:1000) and mouse anti-γ-tubulin (Sigma, T6557, 1:3000). Secondary antibodies were donkey anti-rabbit-HRP (Amersham NA934V, 1:2000) and goat anti-mouse (Dako, P0447, 1:2000). Blots were developed by chemiluminescence using ECL plus (Amersham).
Supplementary Material
Acknowledgements
We thank Nicole Hortin, Ahmed Salman and Mihael Pavlovic for technical assistance, Christopher Böhlke and Alexis Hofherr for help with imaging techniques, Sonia Stefanovic for Q-PCR primer optimization, and Hitoshi Niwa and Yumiko Saga for plasmids. This work was supported by the Emmy Noether Programme and SFB850 of the German Research Council (DFG) to S.J.A. and a Programme Grant from the Wellcome Trust to E.J.R.
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Arnold SJ, Hofmann UK, Bikoff EK, Robertson EJ. Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development. 2008;135:501–511. doi: 10.1242/dev.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bondue A, et al. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 3.David R, et al. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat Cell Biol. 2008;10:338–345. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- 4.Lindsley RC, et al. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell. 2008;3:55–68. doi: 10.1016/j.stem.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vincent SD, Dunn NR, Hayashi S, Norris DP, Robertson EJ. Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev. 2003;17:1646–1662. doi: 10.1101/gad.1100503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu GC, Dunn NR, Anderson DC, Oxburgh L, Robertson EJ. Differential requirements for Smad4 in TGFbeta-dependent patterning of the early mouse embryo. Development. 2004;131:3501–3512. doi: 10.1242/dev.01248. [DOI] [PubMed] [Google Scholar]
- 7.Dunn NR, Vincent SD, Oxburgh L, Robertson EJ, Bikoff EK. Combinatorial activities of Smad2 and Smad3 regulate mesoderm formation and patterning in the mouse embryo. Development. 2004;131:1717–1728. doi: 10.1242/dev.01072. [DOI] [PubMed] [Google Scholar]
- 8.Ben-Haim N, et al. The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev Cell. 2006;11:313–323. doi: 10.1016/j.devcel.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- 10.Lawson KA, Meneses JJ, Pedersen RA. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development. 1991;113:891–911. doi: 10.1242/dev.113.3.891. [DOI] [PubMed] [Google Scholar]
- 11.Tam PP, Parameswaran M, Kinder SJ, Weinberger RP. The allocation of epiblast cells to the embryonic heart and other mesodermal lineages: the role of ingression and tissue movement during gastrulation. Development. 1997;124:1631–1642. doi: 10.1242/dev.124.9.1631. [DOI] [PubMed] [Google Scholar]
- 12.Hoodless PA, et al. FoxH1 (Fast) functions to specify the anterior primitive streak in the mouse. Genes Dev. 2001;15:1257–1271. doi: 10.1101/gad.881501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto M, et al. The transcription factor FoxH1 (FAST) mediates Nodal signaling during anterior-posterior patterning and node formation in the mouse. Genes Dev. 2001;15:1242–1256. doi: 10.1101/gad.883901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russ AP, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- 15.Ciruna BG, Rossant J. Expression of the T-box gene Eomesodermin during early mouse development. Mech Dev. 1999;81:199–203. doi: 10.1016/s0925-4773(98)00243-3. [DOI] [PubMed] [Google Scholar]
- 16.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 17.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 18.Arnold SJ, Sugnaseelan J, Groszer M, Srinivas S, Robertson EJ. Generation and analysis of a mouse line harboring GFP in the Eomes/Tbr2 locus. Genesis. 2009;47:775–781. doi: 10.1002/dvg.20562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tremblay KD, Hoodless PA, Bikoff EK, Robertson EJ. Formation of the definitive endoderm in mouse is a Smad2-dependent process. Development. 2000;127:3079–3090. doi: 10.1242/dev.127.14.3079. [DOI] [PubMed] [Google Scholar]
- 20.Kuzmenkin A, et al. Functional characterization of cardiomyocytes derived from murine induced pluripotent stem cells in vitro. FASEB J. 2009;23:4168–4180. doi: 10.1096/fj.08-128546. [DOI] [PubMed] [Google Scholar]
- 21.Saga Y. Genetic rescue of segmentation defect in MesP2-deficient mice by MesP1 gene replacement. Mech Dev. 1998;75:53–66. doi: 10.1016/s0925-4773(98)00077-x. [DOI] [PubMed] [Google Scholar]
- 22.Saga Y, et al. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- 23.Kitajima S, Takagi A, Inoue T, Saga Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development. 2000;127:3215–3226. doi: 10.1242/dev.127.15.3215. [DOI] [PubMed] [Google Scholar]
- 24.Saga Y, Hata N, Koseki H, Taketo MM. Mesp2: a novel mouse gene expressed in the presegmented mesoderm and essential for segmentation initiation. Genes Dev. 1997;11:1827–1839. doi: 10.1101/gad.11.14.1827. [DOI] [PubMed] [Google Scholar]
- 25.Haraguchi S, et al. Transcriptional regulation of Mesp1 and Mesp2 genes: differential usage of enhancers during development. Mech Dev. 2001;108:59–69. doi: 10.1016/s0925-4773(01)00478-6. [DOI] [PubMed] [Google Scholar]
- 26.Oginuma M, Hirata T, Saga Y. Identification of presomitic mesoderm (PSM)-specific Mesp1 enhancer and generation of a PSM-specific Mesp1/Mesp2-null mouse using BAC-based rescue technology. Mech Dev. 2008;125:432–440. doi: 10.1016/j.mod.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Yasuhiko Y, et al. Functional importance of evolutionally conserved Tbx6 binding sites in the presomitic mesoderm-specific enhancer of Mesp2. Development. 2008;135:3511–3519. doi: 10.1242/dev.027144. [DOI] [PubMed] [Google Scholar]
- 28.Habara-Ohkubo A. Differentiation of beating cardiac muscle cells from a derivative of P19 embryonal carcinoma cells. Cell Struct Funct. 1996;21:101–110. doi: 10.1247/csf.21.101. [DOI] [PubMed] [Google Scholar]
- 29.Chapman DL, et al. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dyn. 1996;206:379–390. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 30.Chapman DL, Agulnik I, Hancock S, Silver LM, Papaioannou VE. Tbx6, a mouse T-Box gene implicated in paraxial mesoderm formation at gastrulation. Dev Biol. 1996;180:534–542. doi: 10.1006/dbio.1996.0326. [DOI] [PubMed] [Google Scholar]
- 31.Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- 32.Shawlot W, Behringer RR. Requirement for Lim1 in head-organizer function. Nature. 1995;374:425–430. doi: 10.1038/374425a0. [DOI] [PubMed] [Google Scholar]
- 33.Tam PP, Khoo PL, Wong N, Tsang TE, Behringer RR. Regionalization of cell fates and cell movement in the endoderm of the mouse gastrula and the impact of loss of Lhx1(Lim1) function. Dev Biol. 2004;274:171–187. doi: 10.1016/j.ydbio.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 34.Norris DP, Brennan J, Bikoff EK, Robertson EJ. The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development. 2002;129:3455–3468. doi: 10.1242/dev.129.14.3455. [DOI] [PubMed] [Google Scholar]
- 35.Teo AK, et al. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011 doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powers SE, et al. Tgif1 and Tgif2 regulate Nodal signaling and are required for gastrulation. Development. 2010;137:249–259. doi: 10.1242/dev.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating The Mouse Embryo: A Laboratory Manual. 3rd edn Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2003. [Google Scholar]
- 38.Niwa H, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 39.Costello I, Biondi CA, Taylor JM, Bikoff EK, Robertson EJ. Smad4-dependent pathways control basement membrane deposition and endodermal cell migration at early stages of mouse development. BMC Dev Biol. 2009;9:54. doi: 10.1186/1471-213X-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Badis G, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.