Abstract
A pivotal step in the transformation of an endosymbiotic cyanobacterium to a plastid some 1.5 billion years ago was the evolution of a protein import apparatus, the TOC/TIC machinery, in the common ancestor of Archaeplastida. Recently, a putative new TIC member was identified in Arabidopsis thaliana: TIC214. This finding is remarkable for a number of reasons: (1) TIC214 is encoded by ycf1, so it would be the first plastid-encoded protein of this apparatus; (2) ycf1 is unique to the green lineage (Chloroplastida) but entirely lacking in glaucophytes (Glaucophyta) and the red lineage (Rhodophyta) of the Archaeplastida; (3) ycf1 has been shown to be one of the few indispensable plastid genes (aside from the ribosomal machinery), yet it is missing in the grasses; and (4) 30 years of previous TOC/TIC research missed it. These observations prompted us to survey the evolution of ycf1. We found that ycf1 is not only lacking in grasses and some parasitic plants, but also for instance in cranberry (Ericaceae). The encoded YCF proteins are highly variable, both in sequence length and in the predicted number of N-terminal transmembrane domains. The evolution of the TOC/TIC machinery in the green lineage experienced specific modifications, but our analysis does not support YCF1 to be a general green TIC. It remains to be explained how the apparent complete loss of YCF1 can be tolerated by some embryophytes and whether what is observed for YCF1 function in a member of the Brassicaceae is also true for, e.g., algal and noncanonical YCF1 homologs.
Some of the best evidence we have for the monophyly of plastids is how they import proteins (McFadden and van Dooren, 2004; Zimorski et al., 2014). Establishing a machinery that translocates proteins across the two membranes separating the organelle’s stroma from the cytosol was crucial for the transition of the endosymbiotic diazotrophic-like cyanobacterium to a plastid in the heterotrophic host (Cavalier-Smith, 2000; McFadden and van Dooren, 2004; Dagan et al., 2013). It was also a prerequisite for the successful transfer of genetic material from the endosymbiont to the host nucleus through endosymbiotic gene transfer (EGT; Martin et al., 1993). Most of the EGT that stripped the plastid genome of its coding capacity occurred in the common ancestor from which the three archaeplastidal lineages—Glaucophyta, Rhodophyta (red lineage), and Chloroplastida (green lineage)—evolved (Martin et al., 1998; Zimorski et al., 2014).
Two main protein complexes mediate plastid protein import: the translocon on the outer envelope of chloroplasts (TOC) and the translocon on the inner envelope of chloroplasts (TIC). In accordance with the majority of EGT events, the main TOC/TIC components evolved in the common ancestor of Archaeplastida (Timmis et al., 2004; Kalanon and McFadden, 2008). Most proteins of the TIC complex are of cyanobacterial origin. However, all previously identified TOC and TIC components are encoded on nuclear DNA (Gould et al., 2008; Kalanon and McFadden, 2008; Shi and Theg, 2013), and, thus far, only TIC40 is unique to the Chloroplastida.
Recently, Kikuchi et al. (2013) found the plastid-encoded YCF1 to be part of a 1-MD membrane protein complex that included TIC20. This complex contained neither TIC40 nor TIC110 components that were previously identified (Kessler and Blobel, 1996; Chou et al., 2003), but three novel ones, including YCF1 (Kikuchi et al., 2013). YCF1 was the first plastid-encoded protein identified whose presence was shown to be essential for the survival of Chlamydomonas reinhardtii and tobacco (Nicotiana tabacum), but whose function remained elusive (Boudreau et al., 1997; Drescher et al., 2000). YCF1—or in accordance with its newly proposed function, TIC214—would represent the first plastid-encoded protein of the TIC complex. It would also add to the set of TIC components (including TIC40 and a full-length TIC62) that is unique to the green lineage. In the course of evolution, organelles have lost or transferred 1000s of genes to the host nucleus, but they hardly ever gained any. That alone is remarkable and the apparent lack of ycf1 in Glaucophytes and Rhodophytes (whose import apparatus evolved from the same ancestral TOC/TIC as that of Chloroplastida) prompted us to inspect the distribution and sequence diversity of ycf1 in more detail.
YCF1 AROSE VIA PLASTID GENE GAIN EARLY IN THE GREEN LINEAGE
The sequences of YCF1 proteins are highly divergent, with an average sequence identity well below 25%. This hinders the use of more classical BLAST searches and the generation of reliable phylogenetic trees. Hence, we chose to investigate the phylogenetic distribution of ycf1 among the Archaeplastida using Hidden Markov Model (HMM) analyses on protein-coding genes of 558 archaeplastidal plastid genomes and a set of 55 cyanobacterial genomes from all five sections (NCBI; as of October 16, 2014). To test the sensitivity of our approach, we performed identical searches using protein alignments of 12 TIC20 and 16 TIC110 sequences from all major embryophyte (land plant) lineages (Figure 1; Supplemental Data Set 1). Through automatic genome annotations, protein-coding genes are sometimes incorrectly annotated as pseudogenes (that do not result in functional proteins) or are even missed entirely. To accommodate for this possibility, we performed a successive screen on those genomes for which no hits were detected in our first round of identifying YCF1 homologs. A tBLASTn search was performed (a search of translated nucleotide databases using a protein sequence query) and the output then added to an additional HMM search. By doing so, we identified several, yet with respect to sequence coverage and length, problematic YCF1 homologs. For instance, they might represent fragments of once full-length YCF1 proteins. These results are shown separately (Supplemental Figure 1 and Supplemental Data Set 2), as we do not wish to speculate on their possible functionality or correct annotation, and, more importantly, the difference is irrelevant for our conclusion.
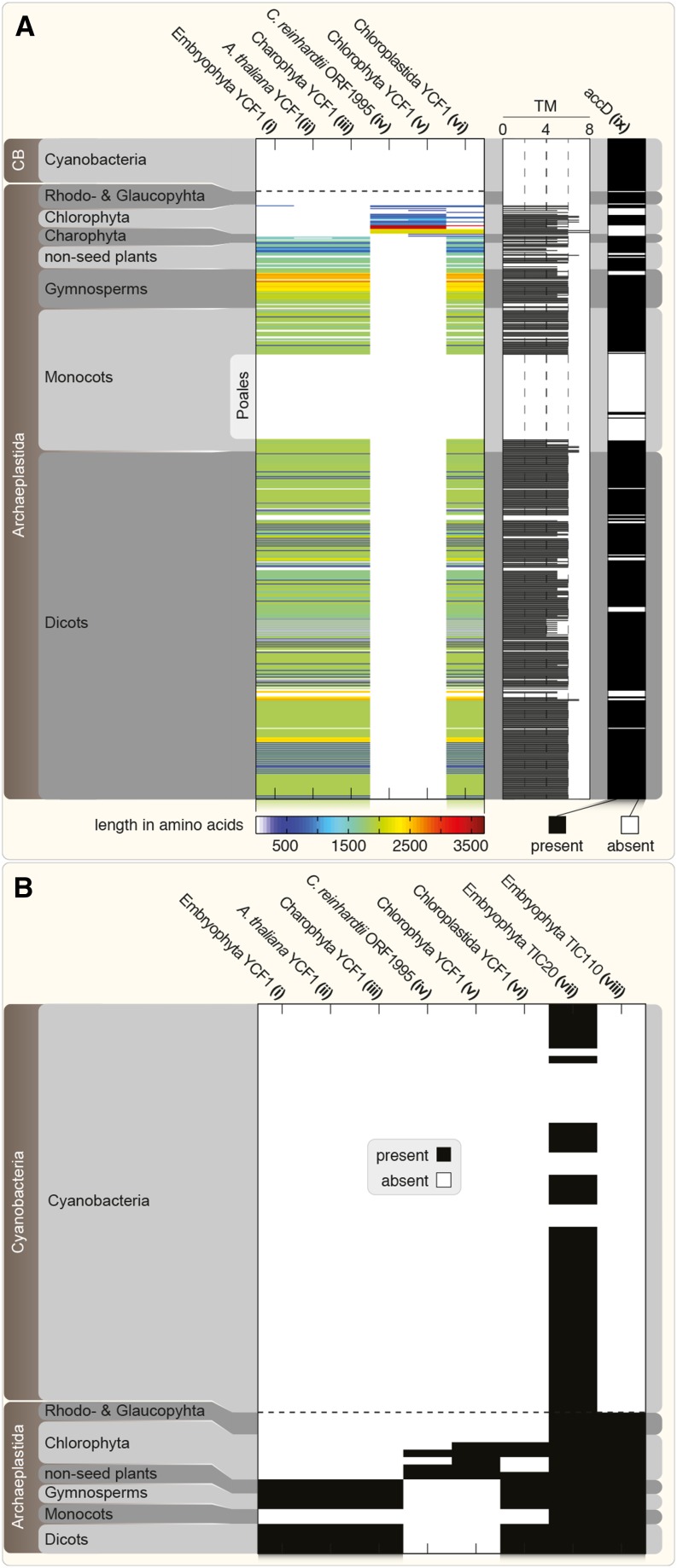
Figure 1.
ycf1 Distribution and Sequence Diversity.
(A) Phylogenetic distribution of YCF1 among 558 archaeplastidal plastid and 55 cyanobacterial genomes. HMM searches were performed based on different alignments or sequence seeds (i to viii). Hits (E-value < 10−5) based on YCF1 alignments (i to vi) among all protein-encoding genes of 558 archaeplastidal plastid and cyanobacterial (CB) genomes are shown on the left, color-coded based on their amino acid length (blue to red; white means absence). Note the absence of YCF1 homologs among cyanobacteria, Rhodophyta, Glaucophyta, and Poales. For all detected YCF1 homologs, the number of TMHMM-predicted transmembrane helices (TM) is shown in the horizontal histogram. For comparison, the results (positive hit, black; no hit, white) of an HMM search against all plastid and cyanobacterial genomes using an alignment of 37 ACCD protein sequences is shown on the right (ix).
(B) The presence or absence (black and white, respectively) of genes encoding TIC20 and TIC110 among all protein-encoding genes of archaeplastidal nuclear and cyanobacterial genomes is shown. The screen is based on HMM searches using embryophyte sequence alignments (vii and viii). In comparison, the data from the YCF1 HMM searches in the archaeplastidal plastid and cyanobacterial genomes are shown in the same fashion. Dashed horizontal lines separate the cyanobacterial and archaeplastidal genome data. HMM and TMHMM results for every species analyzed are individually listed in Supplemental Data Set 1.
A previous comprehensive analysis of the evolution of green plastid genomes suggested ycf1 to represent one of the only few examples, together with ycf2 and matK, of plastid gene gain in the green lineage (Wicke et al., 2011). However, there is also the often overlooked possibility of differential gene loss; the absence of homologs in available cyanobacterial genomes would support the former scenario. We found homologs encoded by cyanobacterial genomes only in the case of TIC20 that we used as a positive control (Figure 1B, vii), but not for YCF1 or TIC110 (Figure 1, i to vi and viii). The universal presence of TIC20 is in line with previous models (Kalanon and McFadden, 2008), and the lack of evidence for differential gene loss indeed supports the scenario that ycf1 was introduced into the plastid genome of the common ancestor of Chloroplastida (Figure 2). However, there is a large degree of ycf1 sequence variability in annotated genomes (particularly among the algae) that points to a more complicated evolutionary history.
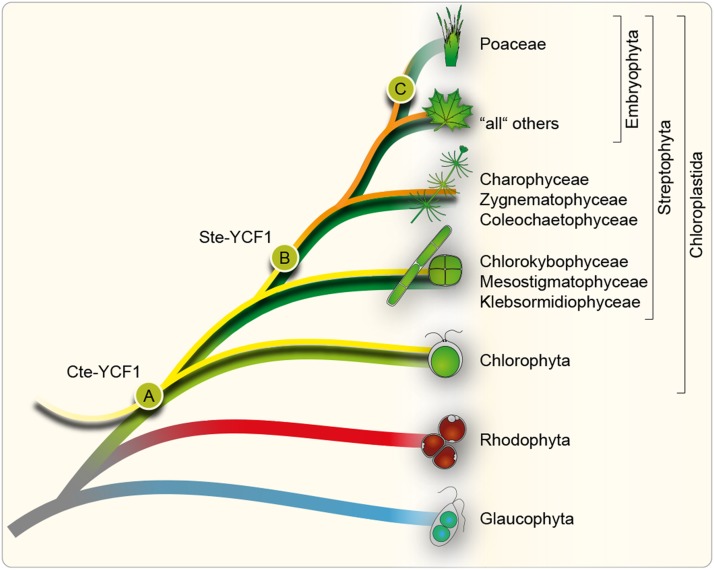
Figure 2.
Cladogram of ycf1 Evolution.
ycf1 was gained in the green lineage (event A), leading to the initial distribution of the ancestral chlorophyte-type (Cte)-YCF1 among all Chloroplastida (yellow trajectory). Within the charophytes, the ancestral “800 amino acid” Cte-YCF1 was transformed into the “1500 amino acid” Streptophyte-type (Ste)-YCF1 (event B), leading to its distribution among Embryophyta and their closest charophyte neighbors (orange trajectory). The vast majority of the embryophytes (“all” others) retained the Ste-YCF1, while it was lost in the Poaceae (event C) and a few other cases mentioned throughout the text. Note that in some chlorophyte algae, the ancestral Cte-YCF1 (initially 800 amino acids in length) also experienced some sequence duplication events that led to much longer versions of YCF1. However, this occurred independently from event B.
YCF1 IS NOT RESTRICTED TO LAND PLANTS, BUT HMM ANALYSIS SUPPORTS A GENERAL SEPARATION BETWEEN STREPTOPHYTE AND CHLOROPHYTE YCF1 SEQUENCES
Using Arabidopsis thaliana YCF1 (ATCG01130; Figure 3, i) as a seed sequence to search for a potentially conserved HMM characteristic for YCF1 returned 466 hits, 462 of which were restricted to embryophytes. Within the angiosperms, no YCF1 homolog was detected among Poales except Typha latifolia (Guisinger et al., 2010; Figure 1A, ii); this observation supports the early loss of ycf1 in the grass lineage (Figure 2). A streptophyte-type YCF1 (Ste-YCF1) was detected only in genomes of charophytes that are currently thought to represent the streptophyte algae most closely related to land plants (these are Charophyceae, Zygnematophyceae, and Coleochaetophyceae; Civaň et al., 2014). A Ste-YCF1 was not detected in genomes of more distantly related taxa, including more distantly related charophytes (Chlorokybophyceae, Mesostigmatophyceae, and Klebsormidiophyceae), let alone rhodophytes, glaucophytes, and cyanobacteria (Figures 1A [ii] and 2; Supplemental Data Set 1). Thus, the approach detected the evolutionarily most distant land plant relative among charophytes using Arabidopsis YCF1 as a seed sequence to build the HMM. To exclude potential false negatives, we performed an HMM search based on an alignment of 25 YCF1 sequences from all major embryophytic lineages. This returned the same pattern, with hits limited to only streptophytes (HMM E-value ≤ 10−5) except for the chlorophyte alga Pseudendoclonium akinetum RF1 (YP_636263.1; Figure 1A, i) and differing in only two hits compared with the At-YCF1 results. Therefore, irrespective of whether we used one sequence or an alignment of 25 to commence our HMM search, the results were the same. This is in contrast to the results of Kikuchi et al. (2013), who found the ycf1 gene present among all Chloroplastida, with the exception of the Poales, but including Chlamydomonas.
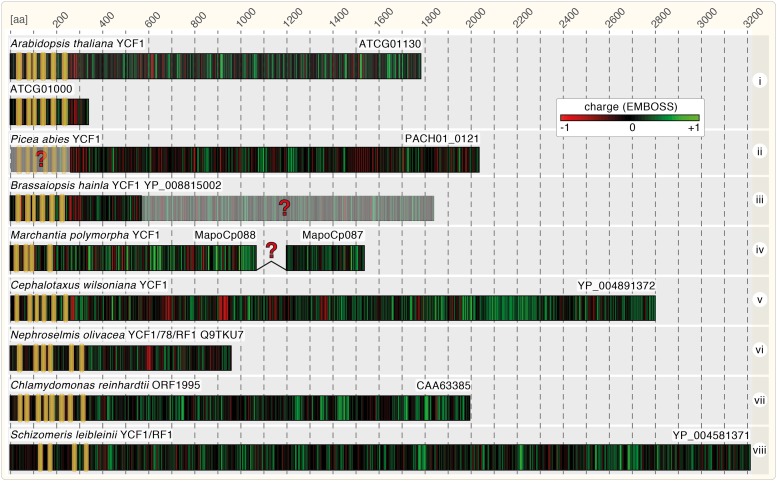
Figure 3.
YCF1 Structural Features and Charge Distribution.
Structure and charge distribution of exemplary YCF1 proteins of eight species (i to viii). Proteins are schematically drawn, but to scale with regard to their amino acid length (scale at the top) based on current genome annotations. Potentially problematic annotations (red question marks) include a possibly incorrect open reading frame annotation (sequence ii), a frameshift (possible sequencing error; sequence iii), and an overlooked potential intron (sequence iv). ATCG01000 is a truncated copy of YCF1 present in the inverted repeat of the Arabidopsis plastid genome. Positive and negative charge distribution is shown in green and red, respectively, based on EMBOSS charge prediction using the standard settings. The positions of TMHMM-predicted TM helices are highlighted by yellow boxes. For each protein the gene ID or NCBI accession is shown at the C terminus of the respective sequence.
The potential ortholog of Ste-YCF1 sequences in Chlamydomonas (Chlorophyte lineage) is Crorf1995 (GenBank X92726.1). Boudreau et al. (1997) were the first to notice structural similarities but were careful to draw a homology between Crorf1995 and the land plant YCF1 and the lack of the latter in the red and glaucophyte lineages. When we used Cr-ORF1995 as a seed sequence (in the same manner as At-YCF1; described above), the search for an HMM returned 23 hits, all limited to the chlorophyte lineage (Figure 1A, iv). Using an alignment of chlorophyte YCF1 sequences (a total of 17, all homologous to Crorf1995 with an E-value < 10−10) returned 28 hits among the chlorophyta and charophyta most closely related to Chlorophyta, but none among other streptophytes. Even when using the only nonstreptophyte protein retrieved above (the chlorophytic P. akinetum RF1; Figure 1A, i) as a seed sequence for an HMM search, we exclusively detected other chlorophyte hits (Supplemental Data Set 1). Only when we used a set of sequences from the alignment of Kikuchi et al. (2013), containing both streptophyte and chlorophyte YCF1 (henceforth named Chloroplastida YCF1), did we detect hits (481) among both Chlorophyta and Streptophyta (Figure 1A, vi). Importantly, the HMM search based on the Chloroplastida YCF1 alignment (Figure 1A, vi) returned hits that are, within the streptophyte lineage, fully consistent with the results obtained by building a streptophyte-only HMM (Figure 1A, i to iii). Yet, within the chlorophyte lineage, there is only a 50% match to the hits returned by using the chlorophyte alignment (Figure 1A, v). Although Cr-ORF1995 is present in the Chloroplastida YCF1 alignment, only 43.5% of the hits match those obtained by just using the Cr-ORF1995 sequence alone (Figure 1A, iv). This means that there is a general separation between streptophyte and chlorophyte YCF1 proteins (that is, Ste-YCF1 and Cte-YCF1; Figure 2). We might then ask how a connection between the algal Cte-YCF1 and the streptophyte Ste-YCF1 sequences was drawn in the first place.
The first chloroplast genome to be sequenced was that of the liverwort Marchantia polymorpha (Ohyama et al., 1986). Four additional embryophyte plastid genomes followed (Shinozaki et al., 1986; Hiratsuka et al., 1989; Wakasugi et al., 1994; Maier et al., 1995) before any algal plastid genome was sequenced. The first algal plastid genomes sequenced were not from green algae, but from the red algae Porphyra purpurea (Reith and Munholland, 1995), the glaucophyte Cyanophora paradoxa (Stirewalt et al., 1995), the heterokontophyte (secondary red) Odontella sinensis (Kowallik et al., 1995), and the euglenophyte Euglena gracilis (secondary green; Hallick et al., 1993). Finally, in 1997, the first plastid genome of a chlorophyte was reported, that of Chlorella vulgaris C-27 (Wakasugi et al., 1997). Wakasugi and colleagues detected a large open reading frame, Cv-ORF819 (NP_045891), for which no homolog was found among published algal plastid genomes, but it shared a few similarities (the N-terminal region encoding transmembrane domains) to the YCF1 found in the land plant plastid genomes available at that time. Wakasugi and colleagues raised concerns with regard to the weak similarity but proposed Cv-ORF819 to represent a potential YCF1 ortholog (Wakasugi et al., 1997); thus, a tenuous bridge from algal chlorophytes to land plant streptophytes was established. With the release of the second chlorophyte plastid genome, that of Nephroselmis olivacea (Turmel et al., 1999), sequence discrepancies between streptophyte and chlorophyte YCF1s were no longer traceable. With the plastid genome data now available, our HMM-based analysis identified Cvorf819 to be homologous to Cte-YCF1s, but its sequence similarity to Ste-YCF1s is somewhat obscure and warrants attention. Therefore, while there is an evolutionary connection between the Cte-YCF1 and Ste-YCF1 proteins, it becomes apparent that in streptophyte evolution, something special happened to YCF1 (Figure 2).
MASSIVE YCF1 SEQUENCE DIVERSITY AND HOW IT MIGHT HAVE EVOLVED
TIC214 is not always 214 kD. It is important to note that YCF1 sequences vary immensely in length. Those chlorophytes (prasinophytes) and charophytes (Mesostigmatophyceae and Chlorokybophyceae; Leliaert et al., 2012) most closely related to the common ancestor of all Chloroplastida possess a Cte-YCF1 of ∼800 to 900 amino acids (Figure 3, vi). Yet in both the streptophyte and chlorophyte trajectories, some lineages encode YCF1 proteins that are >1500 amino acids in length, in some cases above 3000 amino acids. A rather conserved feature, and precisely the one that initially led to the original link of Cte- and Ste-YCF1s (Wakasugi et al., 1997), is the presence of N-terminally encoded transmembrane domains (TMs). For the vast majority of YCF1s, six TM domains are predicted based on TMHMM (Krogh et al., 2001), but the number ranges between four and eight (Figure 1A). In terms of being a TIC-associated membrane protein, the spruces Picea morrisonicola and Picea abies are interesting. Each spruce encodes a single YCF1 of ∼1800 amino acids, but the N-terminal regions encoding the TM domains are missing. As discussed above regarding pseudogenization, we cannot exclude that this is due to a wrong annotation. A region encoding TMs is present upstream of the annotated genes (Figure 3, ii). However, the largest variation is found within the region downstream of the N-terminal TM domains.
Arabidopsis encodes a canonical YCF1 (ATCG01130), but also a very short version of only 343 amino acids (ATCG01000; Figure 3, i) largely lacking the sequence reaching into the stroma (according to Supplemental Figure 17 in Kikuchi et al., 2013). For ATCG01000, this truncation is likely due to the N terminus being encoded on the inverted repeat, but some seed plants, such as Brassaiopsis hainla, might encode only a short YCF1 (574 amino acids; Figure 3, iii). Some basal branching bryophytes even have a split version, maybe due to overlooked introns (Figure 3, iv). The domain of YCF1 reaching into the stroma harbors many highly variable charged motifs arranged in tandem (charge calculations are based on EMBOSS explorer charge prediction; Rice et al., 2000). Both the number and amino acid composition of these motifs vary, which explains why they are difficult to detect and align through classic BLAST searches, especially across distant taxa. This is reminiscent of the coiled regions of intermediate filament proteins, which function as “molecular Velcro” (Rose et al., 2005) and are rapidly evolving (Gould et al., 2011). Thus, what generally unites the streptophyte and chlorophyte YCF1 is not the primary sequence, but the presence of N-terminal TM helices that are followed by a sequence of varying length with repetitive charged motifs.
Whether streptophytic or chlorophytic, previous analyses indicate that ycf1 emerged in the common ancestor of all Chloroplastida (Maul et al., 2002; reviewed in Wicke et al., 2011), apparently as an orphan gene because homologs outside of the green lineage are not known (Figure 2). Ste-YCF1 evolved within the Charophyta and was then vertically inherited by all Embryopyhta. This means that either—although it seems quite unlikely—Cte-YCF1 was lost in the later evolving lineages of charophytes and there was a gain of the Ste-YCF1 or, much more likely, the ancestral Cte-YCF1 (of around 800 amino acids) rapidly evolved into an ∼1500-amino-acid-long protein (Figure 2); maybe through a recombination event of the sequence encoding the repetitive part of the protein as the amount of N-terminal TMs has more less remained unchanged (Figure 1A). For a better phylogenetic resolution, we performed an HMM search based on an alignment of five charophyte YCF1 sequences from charophytes most closely related to land plants (Charophyceae, Zygnematophyceae, and Coleochaetophyceae). This search returned 467 hits that matched the results of the HMM search based on the YCF1 Embryophyta alignment to 100% (Figure 1A, i and iii). Within the charophyte lineage, Ste-YCF1 is only found among these charophytes, but not in the more distantly related ones (Chlorokybophyceae, Mesostigmatophyceae, and Klebsormidiophyceae; Figure 2). This places the origin of Ste-YCF1 at the base of the Charophyceae, which, in accordance with the latest phylogenetic analysis (Civaň et al., 2014), explains their occurrence in the Zygnematophyceae, Coleochaetophyceae, and Embryophytes.
To complicate the matter, the Poales are not the only group within the angiosperms that lack YCF1; the parasitic plant Orobanche purpurea as well as Vaccinium macrocarpon and the genus Erodium have also lost the sequence. One might argue that the loss in parasites such as Orobanche is consistent with the fact that they are no longer phototrophic; however, apicomplexan parasites maintain all essential components of the protein import machinery for their highly streamlined and plastid-derived apicoplast (McFadden, 2011). Furthermore, this does not account for the loss in phototrophs such as Vaccinium, Erodium, and the grasses, and, most importantly, they all need to import proteins from the cytosol. For species for which no nuclear genome sequences are available, we cannot exclude the possibility that ycf1 was transferred from the plastid to the nuclear genome, as it is a frequent event that can occur multiple times independently (Martin et al., 1998; Millen et al., 2001). But should this be the case, it would further complicate YCF1 evolution and diversity; moreover, grass nuclear genomes available do not encode an apparent ycf1 ortholog.
Besides having lost ycf1 from their plastid genome, grasses have replaced the plastid-encoded acetyl-CoA carboxylase D (accD) with a nuclear gene (Konishi et al., 1996). The corresponding protein is posttranslationally targeted to plastids and represents the eukaryotic large homomeric fatty acid (FA) synthase; in dicot plastids, the prokaryotic heteromeric form persists. The fact that grass plastids no longer assemble a heteromeric fatty acid-synthase complex, and at the same time have no need for YCF1, makes it tempting to speculate that YCF1 is involved in the assembly of the ACCase holoenzyme. Among embryophytes, in 80% of the cases where ycf1 was lost from the plastid genome, it co-occurred together with the loss of accD (Delannoy et al., 2011), a trend that we were able to confirm by performing an HMM search using an alignment of 37 ACCD sequences from a broad range of embryophytes (Figure 1A, ix). In case of the above-mentioned O. purpurea, accD is still present in the chloroplast genome, although it does not encode a YCF1 homolog. Erodium and V. macrocarpon, on the other hand, lack ACCD. Whether or not this correlation translates into a functional causality is not known, but is worth investigating.
CONCLUDING REMARKS
Kikuchi et al. (2013) concluded that the complex containing YCF1 “constitutes a general TIC translocon.” While we do not want to question the importance of YCF1 with regard to protein translocation in Arabidopsis, in the light of such sequence variation (some species appear to lack individual TM domains or even the entire gene), it is valid to ask how universal such a claim can be for all Chloroplastida. One could imagine that the emergence of YCF1 is associated with the substitution of phenylalanine at the +1 position of the ancestral N-terminal transit peptide: Based on this amino acid position, glaucophytes and rhodophytes are able to distinguish thousands of plastid proteins from all others translated in the cytosol (Patron and Waller, 2007; Gould, 2008; Bolte et al., 2009). The green lineage Chloroplastida lost this ability, and it has been speculated that the emergence of TOC159 and TOC64 was required to compensate for this remarkable evolutionary drift (Gould et al., 2008). However, these two proteins are needed for the recognition on the outside of the organelle; for TIC214, this scenario would only make sense if it is somehow associated with transit peptide recognition inside the stroma. Also, would not all Chloroplastida then require a functional YCF1? If this protein truly is “a general TIC translocon” (Kikuchi et al., 2013), its sequence diversity is unique in the light of the TOC/TIC machinery and its evolution. Future research needs to determine whether this finding applies to algal and noncanonical Ste- YCF1 homologs and how a complete YCF1 loss is tolerated by some embryophytes. Biochemical data from a broader range of species that also provide a functional explanation for the absence of YCF1 in Poales is direly needed.
Supplemental Data
Supplemental Figure 1. ycf1 distribution based on HMM searches against a data set of all protein data from the plastid and cyanobacterial genomes plus additional tBLASTn hits.
Supplemental Data Set 1. Results for all YCF1 HMM searches against a data set containing all protein data from the plastid and cyanobacterial genomes as shown in Figure 1.
Supplemental Data Set 2. Results for all YCF1 HMM searches against a data set containing all protein data from the plastid and cyanobacterial genomes plus additional tBLASTn hits as shown in Supplemental Figure 1.
Supplementary Material
Acknowledgments
This work is supported by a Grant GO1825/4-1 from the Deutsche Forschungsgemeinschaft to S.B.G.
AUTHOR CONTRIBUTIONS
J.d.V. and F.L.S. performed the computational analysis. S.B.G. and J.S. designed the study. All authors discussed the results and commented on the article that J.d.V., B.B., and S.B.G. wrote.
References
- Bolte K., Bullmann L., Hempel F., Bozarth A., Zauner S., Maier U.G. (2009). Protein targeting into secondary plastids. J. Eukaryot. Microbiol. 56: 9–15. [DOI] [PubMed] [Google Scholar]
- Boudreau E., Turmel M., Goldschmidt-Clermont M., Rochaix J.D., Sivan S., Michaels A., Leu S. (1997). A large open reading frame (orf1995) in the chloroplast DNA of Chlamydomonas reinhardtii encodes an essential protein. Mol. Gen. Genet. 253: 649–653. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. (2000). Membrane heredity and early chloroplast evolution. Trends Plant Sci. 5: 174–182. [DOI] [PubMed] [Google Scholar]
- Chou M.-L., Fitzpatrick L.M., Tu S.-L., Budziszewski G., Potter-Lewis S., Akita M., Levin J.Z., Keegstra K., Li H.-M. (2003). Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J. 22: 2970–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civaň P., Foster P.G., Embley M.T., Séneca A., Cox C.J. (2014). Analyses of charophyte chloroplast genomes help characterize the ancestral chloroplast genome of land plants. Genome Biol. Evol. 6: 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T., et al. (2013). Genomes of Stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol. Evol. 5: 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannoy E., Fujii S., Colas des Francs-Small C., Brundrett M., Small I. (2011). Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol. Biol. Evol. 28: 2077–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher A., Ruf S., Calsa T. Jr., Carrer H., Bock R. (2000). The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 22: 97–104. [DOI] [PubMed] [Google Scholar]
- Gould S.B. (2008). Ariadne’s thread: guiding a protein across five membranes in cryptophytes. J. Phycol. 44: 23–26. [DOI] [PubMed] [Google Scholar]
- Gould S.B., Waller R.F., McFadden G.I. (2008). Plastid evolution. Annu. Rev. Plant Biol. 59: 491–517. [DOI] [PubMed] [Google Scholar]
- Gould S.B., Kraft L.G.K., van Dooren G.G., Goodman C.D., Ford K.L., Cassin A.M., Bacic A., McFadden G.I., Waller R.F. (2011). Ciliate pellicular proteome identifies novel protein families with characteristic repeat motifs that are common to alveolates. Mol. Biol. Evol. 28: 1319–1331. [DOI] [PubMed] [Google Scholar]
- Guisinger M.M., Chumley T.W., Kuehl J.V., Boore J.L., Jansen R.K. (2010). Implications of the plastid genome sequence of typha (typhaceae, poales) for understanding genome evolution in poaceae. J. Mol. Evol. 70: 149–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallick R.B., Hong L., Drager R.G., Favreau M.R., Monfort A., Orsat B., Spielmann A., Stutz E. (1993). Complete sequence of Euglena gracilis chloroplast DNA. Nucleic Acids Res. 21: 3537–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka J., et al. (1989). The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Gen. Genet. 217: 185–194. [DOI] [PubMed] [Google Scholar]
- Kalanon M., McFadden G.I. (2008). The chloroplast protein translocation complexes of Chlamydomonas reinhardtii: a bioinformatic comparison of Toc and Tic components in plants, green algae and red algae. Genetics 179: 95–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler F., Blobel G. (1996). Interaction of the protein import and folding machineries of the chloroplast. Proc. Natl. Acad. Sci. USA 93: 7684–7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S., Bédard J., Hirano M., Hirabayashi Y., Oishi M., Imai M., Takase M., Ide T., Nakai M. (2013). Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339: 571–574. [DOI] [PubMed] [Google Scholar]
- Konishi T., Shinohara K., Yamada K., Sasaki Y. (1996). Acetyl-CoA carboxylase in higher plants: most plants other than gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol. 37: 117–122. [DOI] [PubMed] [Google Scholar]
- Kowallik K.V., Stoebe B., Schaffran I., Kroth–Pancic P., Freier U. (1995). The chloroplast genome of a chlorophyll a+c-containing alga, Odontella sinensis. Plant Mol. Biol. Rep. 13: 336–342. [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305: 567–580. [DOI] [PubMed] [Google Scholar]
- Leliaert F., Smith D.R., Moreau H., Herron M.D., Verbruggen H., Delwiche C.F., De Clerck O. (2012). Phylogeny and molecular evolution of the green algae. Crit. Rev. Plant Sci. 31: 1–46. [Google Scholar]
- Maier R.M., Neckermann K., Igloi G.L., Kössel H. (1995). Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 251: 614–628. [DOI] [PubMed] [Google Scholar]
- Martin W., Brinkmann H., Savonna C., Cerff R. (1993). Evidence for a chimeric nature of nuclear genomes: eubacterial origin of eukaryotic glyceraldehyde-3-phosphate dehydrogenase genes. Proc. Natl. Acad. Sci. USA 90: 8692–8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Stoebe B., Goremykin V., Hapsmann S., Hasegawa M., Kowallik K.V. (1998). Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393: 162–165. [DOI] [PubMed] [Google Scholar]
- Maul J.E., Lilly J.W., Cui L., dePamphilis C.W., Miller W., Harris E.H., Stern D.B. (2002). The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell 14: 2659–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden G.I. (2011). The apicoplast. Protoplasma 248: 641–650. [DOI] [PubMed] [Google Scholar]
- McFadden G.I., van Dooren G.G. (2004). Evolution: red algal genome affirms a common origin of all plastids. Curr. Biol. 14: R514–R516. [DOI] [PubMed] [Google Scholar]
- Millen R.S., et al. (2001). Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13: 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K., Fukuzawa H., Kohchi T., Shirai H., Sano T., Sano S., Umesono K., Shiki Y., Takeuchi M., Chang Z. (1986). Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 322: 572–574. [Google Scholar]
- Patron N.J., Waller R.F. (2007). Transit peptide diversity and divergence: A global analysis of plastid targeting signals. BioEssays 29: 1048–1058. [DOI] [PubMed] [Google Scholar]
- Reith M., Munholland J. (1995). Complete nucleotide sequence of the Porphyra purpurea chloroplast genome. Plant Mol. Biol. Rep. 13: 333–335. [Google Scholar]
- Rice P., Longden I., Bleasby A. (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16: 276–277. [DOI] [PubMed] [Google Scholar]
- Rose A., Schraegle S.J., Stahlberg E.A., Meier I. (2005). Coiled-coil protein composition of 22 proteomes—differences and common themes in subcellular infrastructure and traffic control. BMC Evol. Biol. 5: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.-X., Theg S.M. (2013). The chloroplast protein import system: from algae to trees. Biochim. Biophys. Acta 1833: 314–331. [DOI] [PubMed] [Google Scholar]
- Shinozaki K., et al. (1986). The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 5: 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirewalt V.L., Michalowski C.B., Loffelhardt W., Bohnert H.J., Bryant D.A. (1995). Nucleotide sequence of the cyanelle genome from Cyanophora paradoxa. Plant Mol. Biol. Rep. 13: 327–332. [Google Scholar]
- Timmis J.N., Ayliffe M.A., Huang C.Y., Martin W. (2004). Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5: 123–135. [DOI] [PubMed] [Google Scholar]
- Turmel M., Otis C., Lemieux C. (1999). The complete chloroplast DNA sequence of the green alga Nephroselmis olivacea: insights into the architecture of ancestral chloroplast genomes. Proc. Natl. Acad. Sci. USA 96: 10248–10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi T., et al. (1997). Complete nucleotide sequence of the chloroplast genome from the green alga Chlorella vulgaris: the existence of genes possibly involved in chloroplast division. Proc. Natl. Acad. Sci. USA 94: 5967–5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi T., Tsudzuki J., Ito S., Nakashima K., Tsudzuki T., Sugiura M. (1994). Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc. Natl. Acad. Sci. USA 91: 9794–9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S., Schneeweiss G.M., dePamphilis C.W., Müller K.F., Quandt D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76: 273–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimorski V., Ku C., Martin W.F., Gould S.B. (2014). Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol. 22: 38–48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.