Mutant characterization and complementation analysis reveal that the GTPase activation activity of Arabidopsis RanGAP is needed for its mitotic role as well as sporophyte and gametophyte development.
Abstract
The Ran GTPase activating protein (RanGAP) is important to Ran signaling involved in nucleocytoplasmic transport, spindle organization, and postmitotic nuclear assembly. Unlike vertebrate and yeast RanGAP, plant RanGAP has an N-terminal WPP domain, required for nuclear envelope association and several mitotic locations of Arabidopsis thaliana RanGAP1. A double null mutant of the two Arabidopsis RanGAP homologs is gametophyte lethal. Here, we created a series of mutants with various reductions in RanGAP levels by combining a RanGAP1 null allele with different RanGAP2 alleles. As RanGAP level decreases, the severity of developmental phenotypes increases, but nuclear import is unaffected. To dissect whether the GAP activity and/or the subcellular localization of RanGAP are responsible for the observed phenotypes, this series of rangap mutants were transformed with RanGAP1 variants carrying point mutations abolishing the GAP activity and/or the WPP-dependent subcellular localization. The data show that plant development is differentially affected by RanGAP mutant allele combinations of increasing severity and requires the GAP activity of RanGAP, while the subcellular positioning of RanGAP is dispensable. In addition, our results indicate that nucleocytoplasmic trafficking can tolerate both partial depletion of RanGAP and delocalization of RanGAP from the nuclear envelope.
INTRODUCTION
Ran is a conserved small signaling GTPase of the Ras superfamily, involved in nucleocytoplasmic transport of RNAs and proteins (Avis and Clarke, 1996). Ran is also involved in a number of mitotic processes, such as the regulation of DNA synthesis, spindle assembly, centrosome duplication, chromosome alignment, segregation and decondensation, and nuclear envelope reformation (Ren et al., 1994; Avis and Clarke, 1996; Hughes et al., 1998; Sazer and Dasso, 2000; Gruss and Vernos, 2004; Ciciarello et al., 2007). As a molecular switch, Ran can be either GTP- or GDP-bound, depending on its activating protein (RanGAP) or guanine nucleotide exchange factor (RanGEF). Similar to other small GTPases, Ran has a very low intrinsic GTPase activity and requires stimulation by RanGAP and its accessory factor RanBP1 (Bischoff and Ponstingl, 1991; Bischoff et al., 1994; Seewald et al., 2003). RanGTP and RanGDP have different cellular functions, and the abundance of each form is regulated by the spatial sequestration of RanGAP, RanBP1, and RanGEF (Hopper et al., 1990; Matunis et al., 1996; Izaurralde et al., 1997; Feng et al., 1999).
RanGAP proteins across species share two conserved domains: a leucine-rich repeat (LRR) domain and a C-terminal acidic domain. The LRR domain is the site of Ran interaction and activation (Haberland and Gerke, 1999). While yeast RanGAP (Rna1p) is a cytoplasmic protein, both mammalian and plant RanGAPs contain localization domains (Matunis et al., 1998; Joseph et al., 2002, 2004; Jeong et al., 2005). Plant RanGAPs contain a unique WPP domain (named after a highly conserved Trp-Pro-Pro motif) necessary and sufficient for their subcellular localization. It interacts with two nuclear envelope (NE)-associated coiled-coil protein families, the WIPs (WPP domain-interacting proteins) and the WITs (WPP-domain-interacting tail-anchored proteins) (Rose and Meier, 2001; Xu et al., 2007; Zhao et al., 2008), which are required for RanGAP NE localization. Arabidopsis thaliana RanGAP1 complements the temperature-sensitive Saccharomyces cerevisiae Rna1p mutant rna1-1, indicating that its GTPase activation (GAP) activity is conserved (Ach and Gruissem, 1994; Merkle et al., 1994; Pay et al., 2002). Arabidopsis contains two homologous copies of RanGAP, RanGAP1 and RanGAP2, which share 60% amino acid identity with each other and ∼20% identity with either S. cerevisiae Rna1p or human RanGAP (Rose and Meier, 2001). While single null mutants of either RanGAP1 or RanGAP2 display no visible mutant phenotype, a double null mutant is female gametophyte lethal (Rodrigo-Peiris et al., 2011).
Arabidopsis RanGAP1 relocates during plant cell division in a WPP domain-dependent manner. It associates with the preprophase band (PPB), cortical division zone (CDZ), kinetochores, spindle midzone, outward-growing rim of the phragmoplast, and forming cell plate (Rose and Meier, 2001; Pay et al., 2002; Jeong et al., 2005; Xu et al., 2008). RanGAP1 is one of the few positive markers of the plant CDZ and is therefore possibly involved in constituting a memory that guides the cell plate to the former PPB position (Müller et al., 2006; Azimzadeh et al., 2008; Xu et al., 2008; Van Damme, 2009; Wright et al., 2009; Lipka et al., 2014). This is consistent with data indicating that inducible silencing of plant RanGAP leads to incomplete and irregularly positioned cell walls (Xu et al., 2008).
Studying RanGAP in plants has been challenging, mainly due to the difficulty of separating its potential role in cell division from its vital function in nucleocytoplasmic transport. To address this issue, we generated a series of mutants with progressively lower RanGAP expression. Crucially, these mutants display several phenotypes of varying severity, but no defects in protein nuclear transport. Using these mutants, we determined the contribution of NE localization, GAP activity, and mitotic targeting of RanGAP for its functions in plant cell division and development. Our results suggest that all analyzed functions of RanGAP in plant development require GAP activity, but not subcellular targeting.
RESULTS
RanGAP Mutant Allele Combinations of Increasing Phenotypic Severity Reveal Diverse Developmental Functions
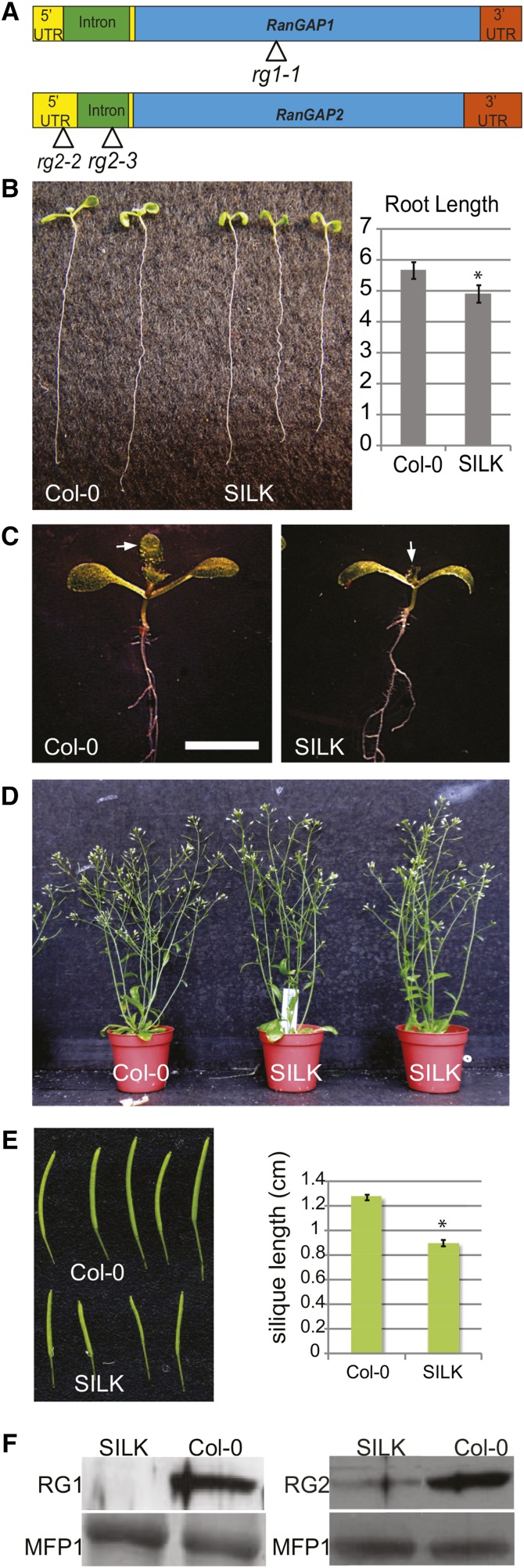
The absence of observable aberrant vegetative phenotypes in the single null mutants, combined with the female gametophytic lethality of the double null mutant, reveal that RanGAP1 and RanGAP2 act redundantly. To dissect the roles of RanGAP in sporophyte development using complementation, we generated combinations of null (rg1-1 and rg2-3; Xu et al., 2008) and knockdown (rg2-2; see below) alleles (Figure 1A), which reduced the levels of RanGAP to a point where functional defects could be observed in viable plants. The inducible RNA interference lines described previously (Xu et al., 2008) could not be used for this purpose because the complementing constructs would be silenced in these genetic backgrounds. A cross between a RanGAP1 (RG1) null mutant (rg1-1) and the RanGAP2 (RG2) knockdown mutant (rg2-2) (Figure 1A) yielded viable rg1-1/rg1-1;rg2-2/rg2-2 plants in the F2 generation (genotype named SILK for short silique knockdown; see below). SILK seedlings were mildly delayed in shoot and root development (Figures 1B and 1C), and mature plants differed only slightly in size or shape from Columbia-0 (Col-0) control plants (Figure 1D). However, SILK siliques were significantly shorter than Col-0 siliques, with an average length decrease of 35% (n ≥ 60, P < 0.0001) (Figure 1E).
Figure 1.
RanGAP Knockdown rg1-1/rg1-1 rg2-2/rg2-2 (SILK) Plants Shows Mild Defects in Growth and Development.
(A) Gene models of Arabidopsis RanGAP1 and RanGAP2 with positional indications of T-DNA insertion lines analyzed. UTR, intron, and exon domains are drawn to scale. UTR, yellow bars upstream and downstream of the first intron.
(B) Initial development of SILK (right) seedlings is delayed when compared with Col-0 seedlings (left). SILK cotyledons are smaller and roots are around 10% shorter than Col-0 (n = 30, Student’s t test P < 0.0001).
(C) Shoot development is delayed in SILK (right) compared with Col-0 (left), represented by smaller cotyledons and delayed development of the first pair of true leaves (marked with an arrow) in the mutant line. Bar = 5 mm.
(D) Mature SILK plants have comparable height and are largely indistinguishable from Col-0.
(E) SILK siliques are around 35% shorter than Col-0 siliques. Mean values and se are shown (n = 60, Student’s t test P < 0.0001).
(F) Immunoblot analysis confirms the rg1-1 and rg2-2 alleles as null and knockdown alleles, respectively. Total protein extracts from 8-d-old Arabidopsis seedlings were incubated with anti-RanGAP1 (left) and anti-RanGAP2 antibodies (right). MFP1 served as a loading control.
Silique dissection and ovule quantification demonstrated that there were 36% fewer ovules per silique in SILK than in Col-0 (Supplemental Figures 1A and 1B). Moreover, of all SILK ovules, 35% were aborted, while Col-0 siliques contained on average only 0.6% aborted ovules. Thus, there is a strong positive correlation between decreased silique length and fewer ovules per silique in SILK (R2 = 0.916, P value = 6.97E-33; Supplemental Figure 1C). Variable integument defects in SILK ovules and female gametophytic arrests at different stages of development were observed (Supplemental Figure 1D), which explains the reduction of the seed set in SILK. These data suggest a role for plant RanGAP in mitotic cell division during female gametophyte development. The rg2-2 allele leads to a substantial reduction in RanGAP2 protein level (Figure 1F). Native promoter-driven RanGAP1 complemented the SILK silique phenotype, implying that the rg2-2 phenotype is caused by a decrease in RanGAP protein (see below).
Plants homozygous null for RanGAP1 and heterozygous null for RanGAP2 (rg1-1/rg1-1 RG2/rg2-3) do not transmit the rg1-1 rg2-3 genotype through the female gametophyte (Rodrigo-Peiris et al., 2011). The rg1-1/rg1-1 RG2/rg2-3 genotype was thus named female gametophyte lethal parent (FELT). To reduce the level of RanGAP beyond SILK, FELT pollen was crossed to SILK stigmas to create sibling populations of rg1-1/rg1-1 RG2/rg2-2 and rg1-1/rg1-1 rg2-2/rg2-3 plants. Approximately half the progeny of this cross had severe pleiotropic developmental defects, while the other half resembled the wild type (Figure 2A). The developmentally abnormal seedlings were genotyped as rg1-1/rg1-1 rg2-2/rg2-3 and named transheterozygote with extreme defects (TWEED), while those that were wild type-like were rg1-1/rg1-1 RG2/rg2-2, referred to here as TWEED siblings. Complementation of this mutant with a native promoter-driven genomic RanGAP1 construct implies that these phenotypes are due to the rg2-2 rg2-3 allele combination (Supplemental Figure 2).
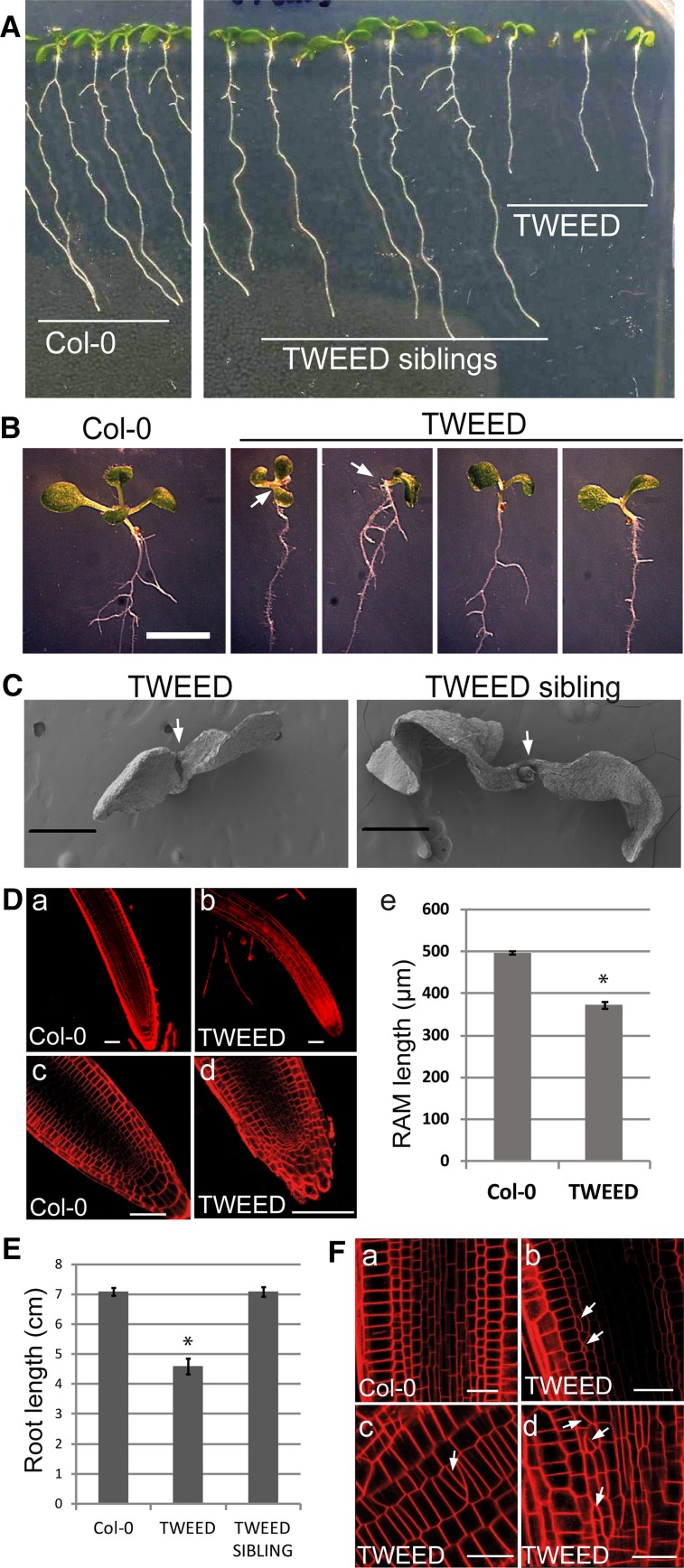
Figure 2.
Seedling Phenotypes of the RanGAP Knockdown rg1-1/rg1-1 rg2-2/rg2-3 (TWEED) Line.
(A) Seven-day-old seedlings of Col-0 (left) and segregating rg1-1/rg1-1 RG2/rg2-3 (TWEED sibling, middle) and rg1-1/rg1-1 rg2-2/rg2-3 (TWEED, right). The TWEED seedlings have less expanded cotyledons and shorter roots.
(B) A close-up view of the seedling phenotypes of the TWEED mutant. Compared with their wild type-like siblings, TWEED seedlings exhibit delayed growth and cotyledon deformations. Bar = 5 mm.
(C) Scanning electron micrographs of 2-d-old seedlings showing the smaller size, decreased cotyledon expansion, and delayed shoot apex development of TWEED seedlings. The first true leaves are visible in TWEED siblings but not in TWEED seedlings themselves. Bars = 50 μm.
(D) FM4-64-stained roots of 7-d-old Col-0 and TWEED seedlings. Compared with Col-0 (a and c), TWEED roots (b and d) have root hairs closer to the root tip and smaller apical meristems. Quantification of the RAM size (E) shows that TWEED seedlings have on average 25% shorter RAM than Col-0. n = 40, error bars represent se, Student’s t test *P < 0.0001. Bars = 50 μm.
(E) The main root length of TWEED seedlings is around 35% shorter than either Col-0 wild-type or TWEED siblings. Measurements performed on 10-d-old seedlings. n = 30, error bars represent se, Student’s t test *P < 0.0001.
(F) TWEED roots (b to d) display disorganized cell files, aberrantly divided cells, and oblique cell walls (white arrows) not observed for Col-0 wild type (A). Bars = 20 μm.
Compared with TWEED sibling plants, TWEED plants showed postgermination abnormalities with a high degree of variability between individuals. These include reduced expansion of the cotyledons and cotyledon deformations (Figure 2B) and a delay in initiating the first pair of true leaves (Figure 2C). The meristematic zone was 25% shorter in TWEED, compared with control Col-0 seedlings, and root hairs developed unusually close to the root tip (Figures 2Da to 2De). The primary root of TWEED seedlings was on average 35% shorter than TWEED siblings or control Col-0 plants (Figure 2E). We also observed irregular cell shapes, obliquely placed cell walls, and disorganized cell files in the root meristems (Figure 2F), resembling the inducible RanGAP silencing phenotype described previously (Xu et al., 2008).
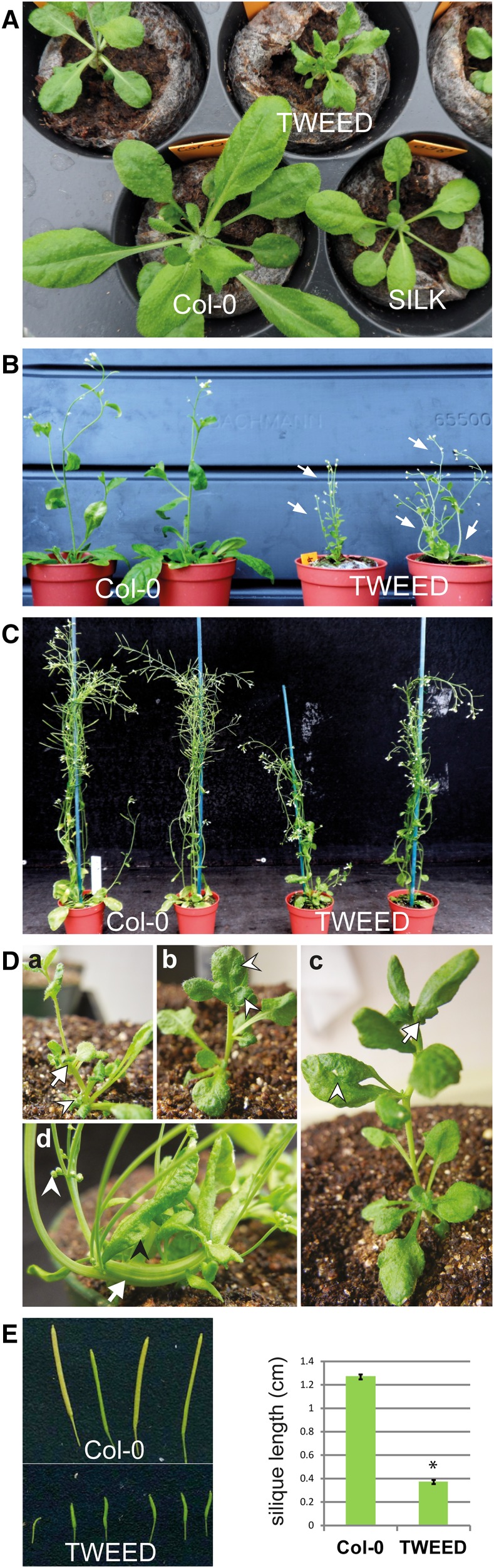
TWEED plants produced smaller rosette leaves and shorter reproductive shoots (Figure 3A). Mature plants were short and bushy, sometimes with multiple rosettes (Figures 3B, 3C, and 3Da). Seedlings often showed abnormal phyllotaxy and fused leaves (Figures 3B, 3Db, and 3Dc). Plants developed slower than control plants and had multiple, bent, and frequently fasciated stems (Figures 3B, 3Dc, and 3Dd) but eventually reached nearly the same height (Figure 3C). The siliques of TWEED plants were on average one-third the size of those of control Col-0 plants, consisting predominantly of aborted female gametophytes and rarely containing fertile seeds (Figure 3E). A mild reduction in the fertility of rg1-1 rg2-3 pollen was observed in comparison to rg1-1 rg2-2 pollen developed on TWEED plants. When TWEED pollen was used to pollinate wild-type (Col-0) flowers, the rg2-3 null allele showed only 77.5% genetic transmission [TE = (107:138) × 100% = 77.5%; Howden et al., 1998] through the pollen, compared with 100% genetic transmission of the competing knockdown rg2-2 allele (Table 1). Together, these data demonstrate that a severe reduction of RanGAP levels leads to significant aberrations in sporophyte development.
Figure 3.
Severe RanGAP Knockdown Mutant rg1-1/rg1-1 rg2-2/rg2-3 (TWEED) Shows Prominent Developmental Phenotypes.
(A) TWEED rosettes (21 d old) are drastically smaller than those of SILK (right bottom) or Col-0 plants (left bottom).
(B) Developing 30-d-old TWEED plants are smaller, shorter, and thinner than Col-0. Multiple rosettes and shoots are common in the mutant line.
(C) Mature TWEED plants (60 d old) grow almost to the height of Col-0 but develop slower and have smaller rosettes.
(D) Representative TWEED phenotypes at several developmental stages. (a) An example of the rosette duplication phenotype in a 3-week-old sibling of the plant highlighted in (b to d). At 24 d of age (b), the plant exhibits abnormal phyllotaxy and leaf fusion (arrowheads). At 30 d of age (c), leaf fusion (arrowhead), as well as stem fasciation and bending (arrow), are present. At 41 d of age (d), the plant shows a lack of apical dominance, slumped appearance, stem fasciation (white arrow), aborted flowers (white arrowhead), and abnormal leaf morphology (black arrowhead).
(E) A 70% decrease in silique length is visible in TWEED (bottom) when compared with Col-0 (top). Mean values and se are shown in the graph (n = 60, Student’s t test *P < 0.0001).
Table 1. Transmission of the rg1-1 rg2-3 Genotype in Comparison to rg1-1 rg2-2 through the Male Gametophyte.
| Cross | RG1/rg1-1; RG2/rg2-2 | RG1/rg1-1; RG2/rg2-3 | No. of Progeny | TE Male | |
|---|---|---|---|---|---|
| Female | Male | ||||
| RG1/RG1; RG2/RG2 | rg1-1/rg1-1; rg2-2/rg2-3 | 138 | 107 | 245 | 77.5 |
TE, transmission efficiency. TE was calculated according to Howden et al. (1998). TE = progeny containing rg1-1 rg2-3 /progeny lacking rg1-1 rg2-3 × 100%.
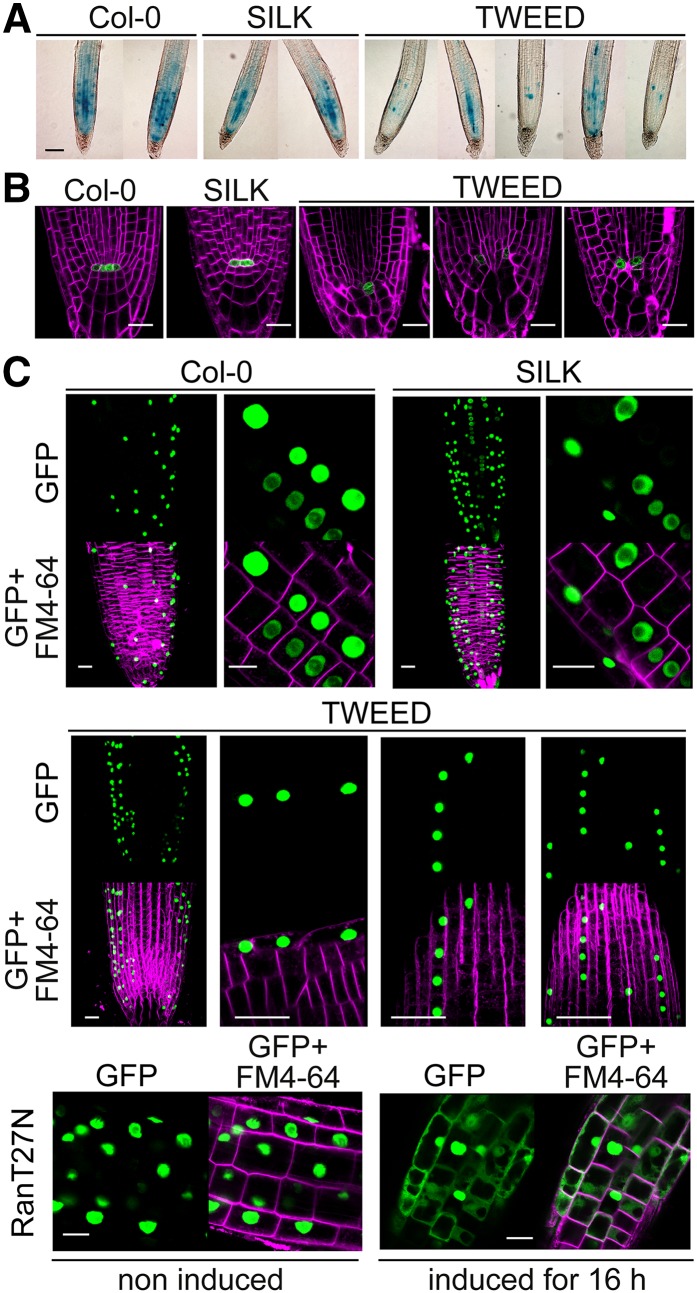
To test whether the short root phenotype of TWEED seedlings might be caused by a decreased rate of cell division, we compared the expression of PromCYCB1:1:GUS (Colón-Carmona et al., 1999) in Col-0, SILK, and TWEED. Compared with wild-type and SILK seedlings, CYCB1:1 expression in the root apical meristem of TWEED seedlings was variable, but overall strongly reduced (Figure 4A), in agreement with the reduced meristem size in TWEED seedlings (Figure 2D). To determine whether disorganized cell files and reduced meristem size in TWEED roots were linked to defects in stem cell maintenance around the quiescent center (QC), we used the QC marker WOX5:GFP (Sarkar et al., 2007). In Col-0 wild-type and SILK seedlings (Figure 4B), WOX5:GFP labeled two to four QC cells, surrounded by well organized stem cell initials. In the TWEED mutant, however, WOX5:GFP-labeled cells were positioned aberrantly when compared with Col-0 (Figure 4B). Also, the columella of TWEED seedlings was not arranged in the distinctive tiers observed in the wild type (Figure 4B). This indicates that the mispatterning in the TWEED root apical meristem (RAM) could be a result of at least partial loss of cell identity or a consequence of aberrant divisions surrounding the QC.
Figure 4.
The Severe RanGAP Knockdown Mutant TWEED Is Defective in Cell Division and Meristem Cell Identity but Shows No Aberrations in Nucleocytoplasmic Transport.
(A) Expression of PromCyclinB1;1:GUS in 7-d-old Col-0, SILK, and TWEED seedlings. While staining was variable within the TWEED line, TWEED seedlings showed a substantial reduction, and SILK seedlings showed only a slight reduction in GUS staining when compared with Col-0. Seedlings were stained for 4 h. Bar = 100 μm.
(B) WOX5:GFP localization in 7-d-old Col-0, SILK, and TWEED roots stained with FM4-64. TWEED mutant seedlings display weaker WOX5:GFP signal and mispositioned WOX5:GFP expressing cells when compared with Col-0 and SILK mutant seedlings. Bar = 20 μm.
(C) N7-GFP localization in Col-0, SILK, TWEED and inducible Ran1T27N seedlings. N7-GFP is localized to the nucleus in Col-0, SILK, TWEED, and uninduced Ran1T27N seedlings. By contrast, the fluorescent fusion protein accumulated in the cytoplasm in estradiol-induced Ran1T27N seedlings. Bar = 20 μm.
Arabidopsis Nuclear Protein Import Robustly Tolerates RanGAP Depletion and Loss of RanGAP Concentration at the NE
Considering the well described role of RanGAP in yeast and animal protein nucleocytoplasmic trafficking, the simplest explanation for the above-described developmental defects would be that they are downstream of impaired protein nuclear shuttling. To address this scenario, nuclear import was tested in the SILK and TWEED genotypes using a nuclear-localized fluorescent marker (GFP-tagged N7) (Cutler and Somerville, 2005). Inducible expression of a dominant-negative Ran mutant (GDP-locked Ran1T27N) resulted in a significant cytoplasmic accumulation of this marker (Figure 4C), demonstrating that the assay is sufficiently robust to detect nuclear import defects. By contrast, no reduction in N7-GFP nuclear import was observed in wild-type, SILK, or TWEED plants. Thus, nuclear import of proteins is not visibly affected in plants with dramatically decreased RanGAP levels.
wifi (wip1-1 wip2-1 wip3-1 wit1-1 wit2-1) is a quintuple null mutant in all five WIP and WIT genes, which encode proteins required for concentrating RanGAP1 at the NE (Zhou and Meier, 2014). To compare the effect of reducing overall RanGAP protein abundance with the effect of reducing the amount of NE-associated RanGAP, 35S:RanGAP1-GFP was expressed in wifi. As anticipated, RanGAP1-GFP does not accumulate at the NE in wifi root meristem cells (Supplemental Figure 3Aa). By contrast, RanGAP1 was still associated with the PPB, kinetochores, phragmoplast, and CDZ (Supplemental Figures 3Aa to 3Af). Thus, wifi can be used to specifically investigate the effect of delocalization of RanGAP from the NE.
We tested nucleocytoplasmic trafficking by transforming wifi with three different transformable fluorescent markers, GFP-NLS-GFP-NES (nucleus and cytoplasm), GFP-NLS(-)-GFP-NES (cytoplasm), and GFP-NLS-GFP-NES(-) (nucleus). None of the fusion proteins changed their distribution pattern in wifi when compared with Col-0 wild type (Supplemental Figures 3B to 3D). Thus, protein nucleocytoplasmic trafficking is not visibly impaired in cells in the absence of RanGAP NE association. The fact that both wifi seedlings and soil-grown plants are phenotypically indistinguishable from Col-0 (Zhou and Meier, 2014) indicates that the observed concentration of RanGAP at the NE is neither required for efficient protein nucleocytoplasmic transport nor for the developmental roles revealed by the mutants described above.
Together, these data show that plants with the rg2-2 rg2-3 allele combination have a variety of developmental defects, while protein nuclear import remains unaffected. Delocalizing RanGAP from the NE, but not its mitotic sites, does not lead to developmental defects and is also tolerated for nuclear import. This suggests that a function of plant RanGAP exists that (1) is important for several aspects of growth and development, (2) does not require concentration of RanGAP at the NE, and (3) is more sensitive to RanGAP depletion than is nucleocytoplasmic trafficking.
Single Amino Acid Substitutions Disrupt Plant RanGAP Activity
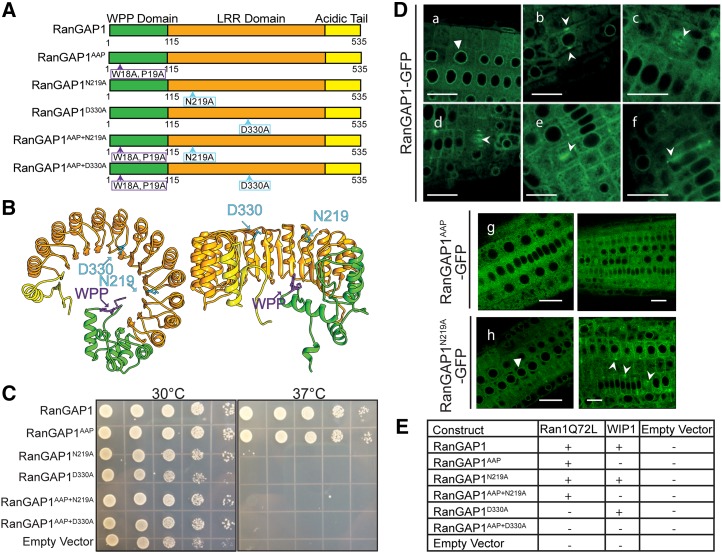
The findings described above prompt the question whether the developmental activity of plant RanGAP is linked to its interaction with Ran and/or to its accumulation at specific sites during cell division. To address this question, we introduced RanGAP1 protein mutants that are defective in GAP activity and/or subcellular targeting (RanGAP1AAP) into the SILK, FELT, and double null mutant genetic backgrounds (Figure 5A).
Figure 5.
Two Point Mutations in the LRR Domain of RanGAP1 Abolish the GAP Activity of the Protein but Not Its Localization and Protein-Protein Interactions.
(A) Schematic representation of mutations and deletions of RanGAP1. The wild-type protein contains the N-terminal WPP domain, the middle LRR domain, and the C-terminal acidic tail. The variant that loses nuclear envelope and mitotic targeting but retains the GAP activity has two point mutations in the WPP domain (WPP/AAP). Two point mutations in the LRR domain (N219A and D330A) block the GAP activity. RanGAP1 variants containing both the mutations in the targeting domain and one of the LRR domain mutations were generated as well.
(B) Schematic 3D representation of RanGAP1. The mutations abolishing the GAP activity are marked and positioned at the top loops of the LRR domain crescent, where Ran protein might interact with RanGAP1 (left, side view; right, top view).
(C) Yeast complementation. The RanGAP1 variants shown in (A) were transformed into the S. cerevisiae rna1-1 temperature-sensitive mutant, and the yeast was grown on selective media at the permissive (30°C) and restrictive (37°C) temperatures. RanGAP1 and RanGAP1AAP complement the temperature sensitivity. However, RanGAP1 variants containing the N219A or D330A mutations (RanGAP1N219A, RanGAP1D330A, RanGAP1AAP+N219A, and RanGAP1AAP+D330A), as well as the empty vector control, were unable to rescue the phenotype.
(D) Point mutations affecting RanGAP1 GAP activity and/or subcellular localization do not interfere with proper protein folding. RanGAP1-GFP (a to f) and RanGAP1N219A-GFP (h) localize in the same manner, while RanGAP1AAP-GFP is exclusively cytoplasmic (g). Triangles indicate nuclear envelope localization (a and h). Arrowheads point to mitotic locations of the fusion proteins. Bars = 20 μm.
(E) Yeast two-hybrid assay. The RanGAP1 variants shown in (A) were transformed into S. cerevisiae and tested for interaction with a GTP-locked form of Ran (Ran1Q72L) or with WIP1. RanGAP1, RanGAP1AAP, RanGAP1N219A, and RanGAP1AAP+N219A all interacted with Ran1Q72L, while those variants containing the D330A mutation did not. RanGAP1 variants with an intact WPP domain interacted with WIP1, while those carrying the AAP mutation did not. Original colony streaks for this table are shown in Supplemental Figure 7. Positive interactions are marked with a “+,” negative interactions are marked with a “−,” and a blank rectangle indicates the combination was not tested.
To identify residues required for Ran binding and activation, we selected and mutagenized (substituted to alanines) 12 amino acids that are conserved in plant, yeast, and human RanGAPs (Supplemental Figure 4) but not in other LRR proteins. This strategy was previously applied to human and yeast RanGAPs (Haberland and Gerke, 1999). The 12 mutagenized RanGAP1 variants were tested for their ability to rescue the temperature-sensitive S. cerevisiae yeast strain rna1-1 (Corbett et al., 1995). As was shown previously, RanGAP1 restored growth of rna1-1 under restrictive conditions (37°C) (Ach and Gruissem, 1994; Joseph et al., 2002; Xu et al., 2007). Similarly, the RanGAP1AAP variant, which has disrupted NE association and targeting to all mitotic sites (Rose and Meier, 2001; Zhao et al., 2008), rescued the growth phenotype under restrictive conditions. Of the 12 variants tested, RanGAP1N219A and RanGAP1D330A were the only protein variants that did not complement the yeast mutant (Figure 5C). Consistently, RanGAP1AAP+N219A and RanGAP1AAP+D330A also did not rescue the growth phenotype, although both protein variants are expressed in yeast (see below). These results show that Asn-219 and Asp-330 are essential for Arabidopsis RanGAP1 to functionally replace Rna1p in yeast. When mapped onto the predicted 3D structure of the LRR domain of RanGAP1 (Zhang, 2008; Roy et al., 2010), the residues Asn-219 and Asp-330 are conserved and exposed to the putative contact site between RanGAP and Ran (Figure 5B; Supplemental Figure 5) (Hillig et al., 1999). Thus, RanGAP1N219A, RanGAP1D330A, RanGAP1AAP+N219A, and RanGAP1AAP+D330A, combined with wild-type RanGAP1 and RanGAP1AAP, made up a RanGAP1 variant series that was used throughout our following experiments (Figure 5A).
To verify that the RanGAP1 variants fold and localize properly in vivo, we visualized their localization using confocal microscopy and tested their ability to interact with WIP1 and Ran1 in a yeast two-hybrid assay. In agreement with previous reports, RanGAP1 associated with the NE in interphase cells (Figure 5Da). At the onset of mitosis, it was located at the PPB, kinetochores, phragmoplast, and the cell plate and marked the CDZ throughout cell division (Figures 5Db to 5Df). The AAP mutant, on the other hand, was diffused throughout the cytoplasm and did not label any mitotic structures (Figure 5Dg). Both RanGAP1N219A and RanGAP1D330A exhibited the same subcellular localization patterns in interphase (Supplemental Figure 6) and in mitosis as wild-type RanGAP1 when expressed in Arabidopsis Col-0 (RanGAP1N219A shown in Figure 5Dh). As anticipated, adding the AAP mutation to the RanGAP1N219A and RanGAP1D330A mutants abolished proper targeting of the fusion proteins (Supplemental Figure 6).
As shown previously, RanGAP1 wild-type protein fused to the GAL4 DNA binding domain (BD) interacts with WIP1 fused to the GAL4 transcription activation domain (AD), while RanGAP1AAP-BD does not (Xu et al., 2007). As expected, RanGAP1N219A and RanGAP1D330A-BD both interacted with WIP1-AD, while RanGAP1AAP+N219A and RanGAP1AAP+D330A-BD did not (Figure 5E; Supplemental Figure 7). These results, together with the subcellular localization of RanGAP1 and its variants, indicate that the point mutations in the LRR domain do not disturb the proper folding and targeting of the protein and shows that the protein variants that do not rescue rna1-1 are expressed in yeast. We also tested in a yeast two-hybrid experiment whether RanGAP1 variants can bind Ran. The RanGAP1 mutants were fused to GAL4 AD and a GTP-locked variant of Arabidopsis Ran (Ran1Q72L) to GAL4 BD. All variants except RanGAP1D330A and RanGAP1AAP+D330A interacted with Ran1Q72L (Figure 5E; Supplemental Figure 7). Since neither RanGAP1N219A nor RanGAP1D330A complemented the yeast rna1-1 mutation, we conclude that either RanGAP1N219A binds Arabidopsis Ran1 but not yeast Ran or that its defect in yeast is not in Ran binding but rather in Ran activation. We then tested the ability of these six variants to rescue the SILK, FELT, and TWEED phenotypes.
The GTPase Activation Function of RanGAP1 Is Necessary for Silique Development
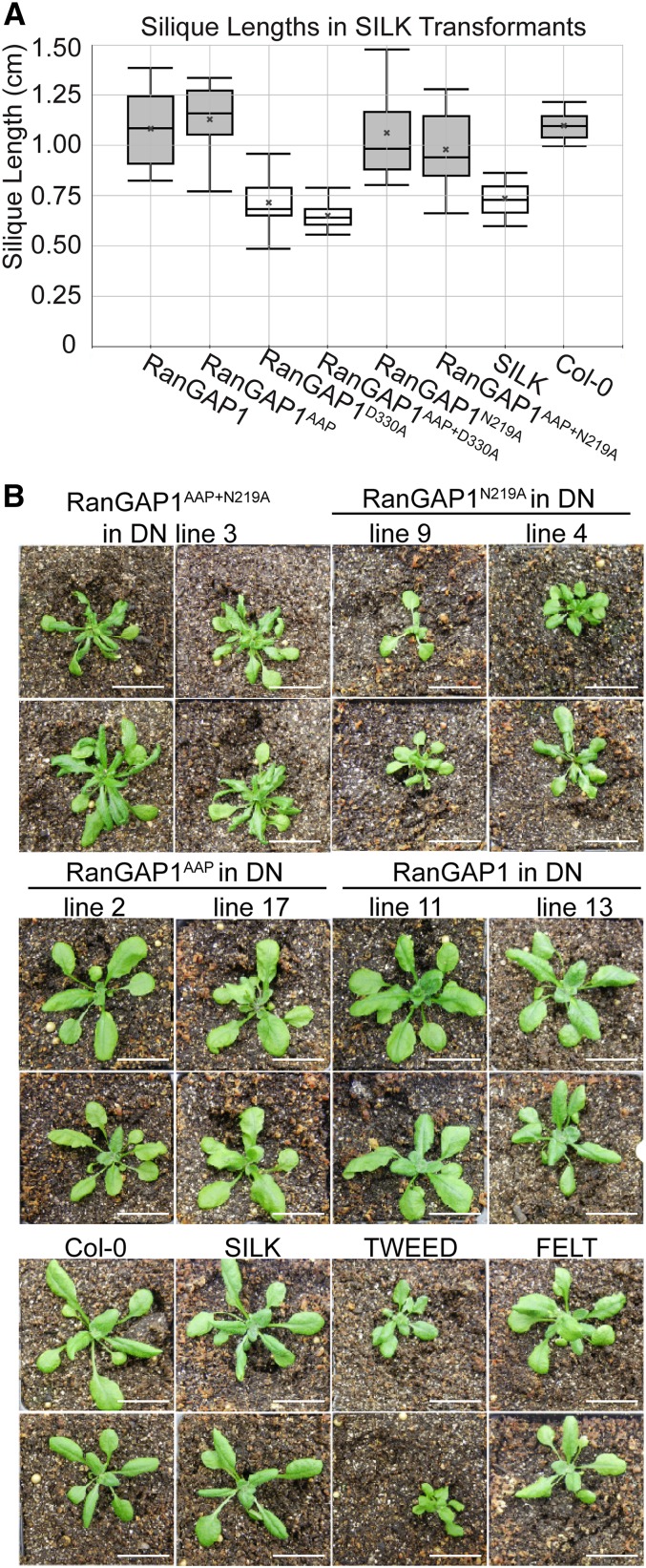
First, SILK mutant plants were transformed with RanGAP1 native promoter-driven and C-terminally GFP-tagged versions of the RanGAP1 variant series shown in Figure 5A. Independent transformants from Agrobacterium tumefaciens-mediated floral dip were analyzed by confocal microscopy to confirm expression and proper localization of GFP fusion proteins. Fusion protein expression was further compared between independent transformants by immunoblot analysis (Supplemental Figure 8A). RanGAP1 and RanGAP1AAP rescued the SILK silique length phenotype to wild-type-like levels, while SILK plants expressing RanGAP1N219A or RanGAP1AAP+N219A had siliques approaching the wild-type length. By contrast, RanGAP1D330A and RanGAP1AAP+D330A did not rescue the SILK silique length defect (Figure 6A). One-way ANOVA followed by Tukey’s honestly significant difference post-hoc test confirmed that the siliques of untransformed SILK plants and SILK plants containing the RanGAP1D330A and RanGAP1AAP+D330A transgenes are statistically significantly different from Col-0 wild type as well as from SILK plants containing the RanGAP1, RanGAP1AAP, RanGAP1N219A, and RanGAP1AAP+N219A transgenes (Supplemental Table 1). To determine whether differences in silique length could be explained by differential expression of transgenic RanGAP1 protein, GFP fusion protein expression was compared between high and low expressing lines (determined from data in Supplemental Figure 8A) of the different RanGAP1 variants by immunoblot analysis. The range of expression levels was similar between constructs (Supplemental Figure 8B), suggesting that the inability of D330A-containing RanGAP1 variants to rescue the SILK silique length phenotype cannot be explained by transgenic protein expression level alone. These data therefore indicate that the GAP activity of RanGAP1 is necessary for its role in silique development but that its subcellular localization is dispensable for this role.
Figure 6.
The GAP Activity of RanGAP1 Is Necessary for Its Role in Silique Elongation and Sporophyte Development.
(A) Silique development depends on the GAP activity of RanGAP1. The silique length for RanGAP1 and its mutant variants in the SILK background. Box represents the 1st and 3rd quartiles of the data set, middle line represents the mean, and whiskers represent the 95th and 5th percentiles. The mean of the data set is represented by a gray X. Boxes in light gray are statistically different from those in dark gray, according to ANOVA followed by the Tukey’s honestly significant difference post-hoc test (see Supplemental Table 1 for test results).
(B) Images of rescued T5 rg1-1/rg1-1 rg2-3/rg2-3 (double null mutant) plants containing various RanGAP1 variant constructs and mutant and wild-type controls grown in parallel. Each line represents an independent transformant. Two plants per line are shown, except for RanGAP1AAP+N219A. The second double null mutant individual identified containing the RanGAP1AAP+N219A variant was sterile and is pictured in Supplemental Figure 10. DN, double null mutant. Bars = 2 cm.
The GTPase Activation Function of RanGAP Is Necessary for Female Gametophyte Development
FELT mutant plants were similarly transformed with the RanGAP1 variant series. As previously shown, the rg1-1 rg2-3 allele combination cannot be passed through the female parent (Rodrigo-Peiris et al., 2011). Basta resistance is linked to the rg2-3 T-DNA. Thus, half the progeny of the FELT (rg1-1/rg1-1 RG2/rg2-3) line are Basta resistant (rg1-1 RG2 female and rg1-1 rg2-3 male gametes) and half are Basta susceptible (rg1-1 RG2 female and rg1-1 RG2 male gametes). However, if a transgenic RanGAP variant is expressed in FELT that allows passage of the rg1-1 rg2-3 genotype through the female gametophyte, then the ratio of Basta resistant to susceptible progeny will be different from 1:1 (Supplemental Figure 9).
Four to five independent FELT transformants for each construct were chosen and populations of T3 seedlings were assayed for Basta resistance. The observed segregation ratios were compared with the expected ratios based on the T2 parent’s genotype (Table 2; Supplemental Figure 9). RanGAP1, RanGAP1AAP, and RanGAP1N219A rescued the female gametophyte lethality (Fisher’s exact test, p value < 0.05, double null progeny identified; Table 2). RanGAP1D330A and RanGAP1AAP+D330A did not rescue the female gametophyte lethality (P > 0.05, no double null progeny identified). The ability of RanGAP1AAP+N219A to rescue the female gametophyte lethality varied depending on the line (Table 2). From the Basta-resistant progeny of two lines per construct, 30 plants were selected and genotyped for the presence or absence of a RanGAP2 wild-type allele. Double homozygous null (rg1-1/rg1-1 rg2-3/rg2-3) individuals were identified in the progeny of FELT plants containing the RanGAP1, RanGAP1AAP, RanGAP1N219A, and RanGAP1AAP+N219A transgenes but not in the progeny of FELT plants containing the RanGAP1D330A or RanGAP1AAP+D330A transgenes. As with SILK, while protein expression varied between independent transformants of the same construct (Supplemental Figure 8C), high and low expressing lines of different constructs had similar expression levels (Supplemental Figure 8D); therefore, this rescue difference cannot be explained by expression level alone. This suggests that the ability of RanGAP1 to effectively bind and/or activate Ran is necessary for its function in female gametogenesis but that RanGAP targeting to specific mitotic locations is dispensable. In addition, the data imply that RanGAP1N219A retains only partial activity in planta, in agreement with the Ran1 Q72L yeast two-hybrid data (Figure 5E).
Table 2. RanGAP GAP Activity Is Required for Female Gametophyte Viability.
| Construct | Line | R | S | R:S Ratio | T2 Genotype | P Value, 1:1 | Conclusion |
|---|---|---|---|---|---|---|---|
| RanGAP1 | 4.1* | 153 | 44 | 3.477 | TT | 1.191E-08 | Rescues |
| 8.4 | 156 | 49 | 3.184 | TT | 4.496E-08 | Rescues | |
| 11.5* | 90 | 53 | 1.698 | Tt | 0.03216 | Rescues | |
| 12.4 | 148 | 65 | 2.277 | Tt | 4.982E-05 | Rescues | |
| 13.1 | 104 | 46 | 2.261 | Tt | 0.0009467 | Rescues | |
| RanGAP1AAP | 2.2* | 242 | 70 | 3.457 | TT | 9.032E-13 | Rescues |
| 3.4 | 90 | 43 | 2.093 | Tt | 0.004192 | Rescues | |
| 12.1 | 339 | 208 | 1.630 | Tt | 7.486E-05 | Rescues | |
| 17.1* | 708 | 398 | 1.779 | Tt | 3.458E-11 | Rescues | |
| 1.2 | 644 | 337 | 1.911 | Tt | 2.433E-12 | Rescues | |
| 3.1 | 584 | 7 | 83.429 | TT | 7.050E-99 | Rescues | |
| RanGAP1N219A | 4.1* | 359 | 8 | 44.875 | Tt | 4.067E-57 | Rescues |
| 5.6 | 526 | 273 | 1.927 | Tt | 1.599E-10 | Rescues | |
| 9.1* | 36 | 1 | 36.000 | TT | 2.526E-06 | Rescues | |
| 3.1* | 104 | 64 | 1.625 | Tt | 0.03664 | Rescues | |
| RanGAP1AAP+N219A | 9.1 | 105 | 98 | 1.071 | Tt | 0.7664 | dnr |
| 11.2* | 121 | 55 | 2.200 | Tt | 0.0004951 | Rescues | |
| RanGAP1D330A | 1.2 | 139 | 116 | 1.198 | Tt | 0.3304 | dnr |
| 2.1 | 171 | 149 | 1.148 | Tt | 0.4289 | dnr | |
| 4.2 | 46 | 34 | 1.353 | Tt | 0.4280 | dnr | |
| 8.1 | 73 | 67 | 1.090 | Tt | 0.8111 | dnr | |
| 10.1* | 54 | 50 | 1.080 | Tt | 0.8897 | dnr | |
| RanGAP1AAP+D330A | 3.2 | 239 | 204 | 1.172 | Tt | 0.2535 | dnr |
| 7.1 | 88 | 89 | 0.989 | Tt | 1.000 | dnr | |
| 10.2 | 162 | 152 | 1.066 | Tt | 0.7495 | dnr | |
| 11.2 | 170 | 150 | 1.133 | Tt | 0.4766 | dnr | |
| 12.1* | 266 | 260 | 1.023 | TT | 0.9019 | dnr |
R, BASTA resistant; S, BASTA susceptible; *, population tested for double null mutant individuals; dnr, does not rescue; TT, homozygous for transgene insertion; Tt, heterozygous for transgene insertion. P values were generated using Fisher’s exact test with a null hypothesis of a 1:1 R:S ratio.
The GTPase Activation Function of Arabidopsis RanGAP Is Necessary for Sporophyte Development
To address whether our RanGAP constructs have the ability to rescue sporophyte developmental phenotypes, we used the progeny of the double null mutants identified in the previous experiment. In 10-d-old sibling seedlings of the plants shown in Figure 6B, GFP fusion protein expression was similar to that seen in the transformed FELT progenitors of the double null mutant lines (Supplemental Figures 8C to 8E). We found that double null plants containing the RanGAP1 and RanGAP1AAP transgenes developed normally, while those containing the RanGAP1AAP+N219A or RanGAP1N219A variant resembled TWEED plants (Figure 6B; Supplemental Figure 10). As in previous experiments, these data could not be explained by differences in transgenic protein expression (Supplemental Figure 8E). These data indicate that N219A-containing RanGAP1 variants are only partially functional for sporophyte development, as double null mutants containing these variants do not resemble wild-type-like FELT (which contains one fully functional copy of RanGAP). This is in line with the observation of a noticeable, but not statistically significant, reduction in silique length in SILK plants with the RanGAP1N219A and RanGAP1AAP+N219A transgenes (Figure 6A).
DISCUSSION
RanGAP Mutant Developmental Phenotypes Are Independent of Nucleocytoplasmic Transport
RanGAP has been shown to be important for a number of cellular processes, such as nucleocytoplasmic transport and cell division in animals and yeast (Di Fiore et al., 2004). However, little is known about the functions of RanGAP in plants. A combination of a null mutant of RanGAP1 (rg1-1) with a knockdown mutant of RanGAP2 (rg2-2) yielded a plant with mild developmental phenotypes that developed shorter siliques (SILK mutant). This suggests that silique development is especially sensitive to RanGAP level perturbation. A further decrease in RanGAP (TWEED mutant) leads to more drastic and pleiotropic phenotypes. Our results suggest that these defects are more likely due to decreased mitotic activity and meristematic maintenance, while nucleocytoplasmic transport is less susceptible to decreased levels of RanGAP in plants.
Phenotypes similar to those of the TWEED mutant were reported for Arabidopsis plants overexpressing rice (Oryza sativa) RAN1 and wheat (Triticum aestivum) RAN1 (Wang et al., 2006; Xu and Cai, 2014), including smaller, bushier plants with shorter roots, fewer lateral roots, reduced apical dominance, and delayed development. However, the authors conclude that these phenotypes are due to an increase in mitotic index, whereas fewer mitotic events occur in the TWEED mutant. Nevertheless, our results, together with the previously published RAN1 overexpression data, suggest that the Ran cycle in plants is important for cell division control. Interestingly, the TWEED mutant exhibits additional phenotypes not reported for RAN1 overexpression lines, namely, disorganized root meristem files and altered division plane orientations.
Several loss-of-function mutants also phenocopy TWEED phenotypes. Stem fasciation has long been tied to mutants in shoot apical meristem identity genes such as clavata, fasciata, and shoot meristemless (Ottoline Leyser and Furner, 1992; Clark et al., 1996). The root phenotypes resemble mutants with perturbed meristem maintenance and cell specification. Dramatic alterations in root development were reported for mutants in genes controlling root tissue patterning, such as SHORT-ROOT, SCARECROW (SCR) (Benfey et al., 1993; Scheres et al., 1995), and PLETHORA (Aida et al., 2004). Roots of TWEED phenocopy plt1 plt2 double mutants, implying that RanGAP might also be essential for QC specification and stem cell activity. Moreover, mutations in Arabidopsis MERISTEM-DEFECTIVE show striking resemblance to TWEED root phenotypes as well. It has been reported that mdf mutants are defective in meristem maintenance and cell specification. In addition, they have strongly decreased and variable mitotic activity, similar to TWEED. Additionally, RAM identity and patterning are tightly linked to cell division regulation. For instance, RETINOBLASTOMA-RELATED interacts with SCR to regulate quiescence in the QC by repressing an asymmetric cell division that generates short-term stem cells (Cruz-Ramírez et al., 2013). The cell cycle switch gene CCS52A (an activator of the anaphase promoting complex/cyclosome [APC/C]) regulates the identity of the QC by repressing mitotic activity in these cells (Vanstraelen et al., 2009). This repression is also achieved by WOX5, which inhibits expression of CYCD3;3 (a proliferation-boosting cyclin) in the QC, thereby maintaining its quiescence (Forzani et al., 2014). The TWEED mutant exhibits a reduction in the population of dividing cells as well, which might be a consequence of lost mitotic identity in RAM, rather than nuclear traffic impairment.
LRR Domain Loop Region Amino Acids Contribute to GAP Activity Differently in Yeast and Plants
All known eukaryotic RanGAP proteins share a conserved 330- to 350-residue LRR domain, followed by an acidic region (Hillig et al., 1999; Rose and Meier, 2001). The LRR domain of yeast Rna1p has a crescent or horseshoe shape with an interior parallel β-sheet and an exterior array of α-helices (Hillig et al., 1999; Rose and Meier, 2001). This type of structure is often involved in the formation of protein-protein interactions (Gay et al., 1991; Kobe and Deisenhofer, 1995; Kobe and Kajava, 2001). Unlike other activating proteins of small GTPases, RanGAP does not act through an arginine finger (Seewald et al., 2002). In yeast, the solvent-exposed β-α loop regions of the LRR crescent constitute the Ran binding interface (Hillig et al., 1999). Because Ran proteins and the LRR structure of RanGAPs are conserved, we postulated that the residues involved in the binding or catalysis of RanGTP in Arabidopsis would be invariant among plant, animal, and yeast sequences. In yeast and animals, these include solvent-accessible residues located in the loop regions connecting the LRR domain’s β-strands with its α-helices (Figure 5B). For instance, it has been postulated that charged amino acids in the β-α loop regions of the LRR domain (like Arg-91 in human RanGAP) are involved in the RanGAP-stimulated GTP hydrolysis of Ran (Haberland and Gerke, 1999). In the case of Arabidopsis RanGAP1, while mutation of a residue analogous to Arg-91 of hsRanGAP led to no functional defect, we found that the mutation of negatively charged aspartic acid (Asp-330) and uncharged asparagine (Asn-219) to alanine abolished the ability of the Arabidopsis protein to rescue the rna1-1 temperature-sensitive phenotype. In plants, however, only Asp-330 was required for full activity. This result was corroborated by yeast two-hybrid data with a GTP-locked form of Arabidopsis Ran (Ran1Q72L). These data suggest a difference in the interaction of Arabidopsis RanGAP with yeast and Arabidopsis Ran. The mutation analogous to Arabidopsis RanGAP1D330A in tobacco (Nicotiana benthamiana) RanGAP2 also leads to a defect in the GAP activity of the protein, suggesting that this residue is crucial for RanGAP function across species (Tameling et al., 2010).
The GAP Activity of RanGAP Is Necessary for Its Function in Arabidopsis Development
We have shown here that in the wifi mutant, which eliminates RanGAP1 targeting to the NE, RanGAP1 is still associated with all mitotic sites. This suggests that mitotic targeting is likely dependent on an as yet unidentified RanGAP1 interacting partner. This is in striking contrast to mammalian cells, where RanBP2 targets RanGAP1 to the NE, kinetochores, and spindle (Joseph et al., 2002). We used the structural information described above to design mutant complementation experiments, in which RanGAP1 protein variants defective in targeting and GAP activity were used to dissect functional requirements for Arabidopsis development. To test whether proper targeting is necessary, the AAP mutation was used, which retains the enzymatic activity of RanGAP1, but is unable to associate with the NE and its mitotic locations. The expression of the RanGAP1AAP variant rescued the short silique phenotype of SILK and the female gametophyte lethality of FELT, indicating that both silique development and female gametogenesis require active RanGAP1 protein but that its subcellular targeting is not crucial for these functions.
However, these data leave some questions unanswered: If the subcellular targeting of RanGAP is dispensable for its function in plant development, then what were the selective pressures that allowed the WPP domain to evolve? One potential explanation could be that under favorable conditions, cytoplasmic RanGAP is sufficient to maintain a Ran gradient at the NE and/or mitotic sites but that under some unknown unfavorable conditions, the ability of the cell to bring RanGAP to specific subcellular locations is vital for plant survival. This idea hinges on the GAP activity being necessary for RanGAP function, which we have shown extensively here. The mathematical model for nucleocytoplasmic transport proposed by Görlich et al. (2003) suggests that, in animals, RanGAP is limiting for RanGTP concentration and that RanGAP and RanGTP concentrations are negatively correlated in a linear fashion. If this is the case in plants, then there may be a minimal cytosolic RanGAP concentration necessary for the appropriate RanGTP gradient to be maintained in RanGAP’s many locations. If that threshold were to be crossed, it is possible that site-specific targeting of RanGAP might become necessary for its function. In light of these results, it would be interesting to carry out a reciprocal experiment testing whether Rna1p could rescue the various RanGAP1 mutant phenotypes in Arabidopsis.
The two “no GAP” variants of RanGAP1 (RanGAP1D330A and RanGAP1N219A) yielded different results. The functionality difference of the RanGAP1N219A point mutation between the yeast and plant systems may be due to a decreased requirement for the Asn-219 residue in planta. Indeed, in the predicted secondary structure, Asn-219 faces into the protein rather than out into the solute (Supplemental Figure 5), which, together with our yeast two-hybrid data, suggests that it is not as vital for the RanGAP-Ran interaction as Asp-330.
Taken together, our data point to the existence of a gradient of sensitivity toward decreasing RanGAP protein levels; while nucleocytoplasmic trafficking appears relatively resistant to RanGAP depletion, other functions seem more susceptible and thus result in cellular and developmental perturbations. This potential dual function of RanGAP at the NE and during mitosis was also reported for other NE-associated proteins. For instance, Rae1, an mRNA export factor associated with the nuclear pore complex, is also involved in spindle formation and chromosome segregation (Lee et al., 2009). Similarly, NUA (the Arabidopsis homolog of Tpr/Mlp1/Mlp2/Megator) (Jacob et al., 2007; Xu et al., 2007) is a nuclear pore complex protein, which is a part of a spindle assembly checkpoint (Ding et al., 2012). Further work is required to shed more light on these developmental functions of RanGAP1 and to determine other molecular players involved in these processes alongside RanGAP.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana (ecotype Col-0) plants were germinated under long-day conditions (16 h light/8 h darkness) at 22°C on Murashige and Skoog (MS) medium. Plants were transformed by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on hygromycin-containing MS solid medium. T-DNA insertion mutants rg1-1 (SALK_058630) and rg2-3 (FLAG_184A06, in the Ws-4 background) were described previously (Xu et al., 2008; Rodrigo-Peiris et al., 2011). The mutant rg2-2 (SALK_006398C) in the Col-0 background was acquired from the ABRC. The insert in rg2-2 was confirmed to reside in an intron ∼370 bp upstream of the start codon of RanGAP2. rg1-1/rg1-1 rg2-2/rg2-2 (SILK) was crossed as the female parent with pollen from rg1-1/ rg1-1 RG2/rg2-3 (FELT) to obtain a transheterozygote rg1-1/rg1-1 rg2-2/rg2-3 (TWEED) and the cosegregating rg1-1/rg1-1 RG2/rg2-2 (TWEED sibling) plants. Thus, both rg1-1/rg1-1 rg2-2/rg2-3 and rg1-1/rg1-1 RG2/rg2-3 are Col-0/Ws-4 hybrids. All genotyping primer sequences are summarized in Supplemental Table 2 (Howden et al., 1998). Primer combinations of RG1-F/LBa1 and RG2-F/fLB were used for genotyping rg1-1 and rg2-3, respectively. For screening of the rg2-2 allele, RG2-R and LBa1 primers were used. The N7 marker line (ABRC number CS84731) contains a fusion protein of GFP and the C terminus of an ankyrin-repeat containing transcription-factor-like protein (GenBank accession number CAA16704).
Plasmid Constructs
All full-length open reading frames of the proteins of interest were recombined into the pDONR221 entry vector (Invitrogen) by a BP reaction. The multisite LR Gateway reaction resulted in translational fusions between RanGAP1 (and its variants) and GFP, driven by the native RanGAP1 promoter. The expression clones were generated in the pH7m34GW destination vector. For the yeast two-hybrid assay, all coding sequences were recombined into the pDEST22 and pDEST32 vectors (Invitrogen) by an LR reaction. For the yeast complementation assay, the yeast expression vector pYL435 was used. All coding sequences were recombined into pYL435 via a LR reaction. A Stratagene QuikChange site-directed mutagenesis kit was used to create point mutations in the LRR domain of RanGAP1 and in Ran1. Thr-27 in Arabidopsis Ran1 (corresponding to Thr-24 in human Ran) was mutated to Asn to construct a nucleotide-free/GDP-locked form designated Ran1T27N, similar to human RanT24N (Klebe et al., 1995). Similarly, Gln-72 of Arabidopsis Ran1 (corresponding to Gln-69 in human Ran) was mutated to Leu to obtain the Ran1Q72L mutant, which resembles the human RanQ69L mutant that fails to hydrolyze GTP and thereby adopts a GTP-locked conformation (Bischoff et al., 1994). HA-Ran1 (control), HA-Ran1T27N, and HA-Ran1Q72L were then recombined into the destination vector pMDC7 for estrogen-inducible expression in plants (Curtis and Grossniklaus, 2003).
Immunoblot Analyses
For immunoblot analysis to compare RanGAP2 expression levels between the wild type (Col-0) and rg1-1/rg1-1 rg2-2/rg2-2, tissues were harvested from seedlings 8 d postgermination. Anti-RanGAP1 and Anti-RanGAP2 antibodies were described previously (Jeong et al., 2005). Anti-MFP1 antibody (OSU91), described previously (Jeong et al., 2003), was used to detect the expression of endogenous MAR binding filament-like protein 1 (MFP1) as a loading control. For GFP fusion protein expression analysis, whole seedlings were harvested at 10 d of age. Immunoblotting was performed with an anti-GFP antibody (1:2000; Roche Biosciences). As a loading control, an SDS-PAGE gel was run in parallel with one-tenth of the protein that was loaded into the immunoblot and stained with Coomassie Brilliant Blue.
GUS Staining
Histochemical staining for GUS activity was performed as described (Jefferson et al., 1987). Briefly, tissues were fixed for 0.5 h in 90% acetone at 4°C, rinsed with 0.2 M Na2HPO4, stained with 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid in 50 mM potassium phosphate buffer (pH 7.0) for 4 h, and then cleared with 70% ethanol.
Ovule Clearing
Dissected pistils were fixed in absolute ethanol:acetic acid 9:1 (v/v) at 4°C overnight and washed with 90% ethanol for 1 h at room temperature. Pistils were subsequently washed with 70% ethanol for 1 h at room temperature and cleared with a clearing solution (8 g chloral hydrate, 1 mL glycerol, and 2 mL water) overnight at room temperature.
Estradiol Induction
Seedlings were germinated on half-strength MS plates for 4 to 7 d and then transferred to liquid half-strength MS medium containing β-estradiol (10 μM; Sigma; for inducible expression), while ethanol (solvent) was used for control (i.e., uninduced) treatments.
Root Meristem Size Analysis
RAM size was calculated as the distance between the QC and the transition zone (indicating the position of the first elongating cortical cell). To calculate the RAM, measurements were performed on 7-d-old seedlings stained with propidium iodide or FM4-64.
Microscopy
Fluorescence was analyzed with confocal microscopes Eclipse C90i (Nikon) and FluoView FV1000 (Olympus). The former is equipped with a plan Apochromat VC box H case (numerical aperture 1.4), and the latter is equipped with a 63× water corrected objective (numerical aperture of 1.2) to scan cells. The GFP fluorescence was imaged with a 488-nm laser. The FM4-64 and propidium iodide-stained seedlings were imaged with a 543-nm laser.
Yeast Transformation
For the yeast two-hybrid assay, plasmids encoding the baits (pDEST32) and preys (pDEST22) were transformed into the yeast strain PJ69-4A (MATa; trp1-901, leu2-3,112, ura3-52, his3-200, gal4gal452, his3-200, gal4gal4 he) by the LiAc method (Gietz et al., 1992). Cotransformed yeast cells were selected on synthetic dextrose (SD) plates without Leu (pDEST32) and without Trp (pDEST22). Interactions between proteins were scored by transferring the colonies on the SD medium lacking Leu, Trp, His, and Ade and incubated at 28°C for 2 to 3 d. The yeast PSY714 (rna1-1, ura3-52, leu2Δ1, trp1, gal+, MATa) temperature-sensitive mutant strain, used for the yeast complementation assay, was a generous gift from Anita Hopper (Corbett et al., 1995). For the rna1-1 yeast strain transformation, the same method was used and transformed cells were plated on SD plates without Ura. After 2 to 3 d, the colonies were streaked on fresh SD-Ura plates in duplicate; one plate was incubated at 28°C, while the other was incubated at 37°C for 2 to 3 d.
TWEED Complementation
A native RanGAP1 promoter-driven RanGAP1 construct was created by first cloning a 1.2-kb fragment containing the 0.73-kb annotated RanGAP1 promoter, 5′ untranslated region (UTR), and 5′ intron into the pMDC162 GUS fusion vector. A 2.2-kb RanGAP1 fragment containing the coding region, 3′UTR, and 3′ intergenic region was then placed between the promoter fragment and the GUS gene by traditional ligation. This construct was then introduced into SILK plants via Agrobacterium tumefaciens-mediated floral dip. These transgenic plants were used as the female parent in a cross with FELT pollen to obtain transgenic TWEED progeny.
SILK Complementation Experiment
Three to five independent transformants were generated for each RanGAP1 variant via Agrobacterium-mediated floral dip. Seeds were collected from dipped plants and germinated on MS media with 1% sucrose, 30 μg/mL hygromycin, and 100 μg/mL carbenicillin. Resistant seedlings were selected and propagated to create independently transformed lines. The progeny of these plants (T2 generation) were grown on MS media with 1% sucrose and 30 μg/mL hygromycin. At least three resistant seedlings were selected and moved to soil. At least 10 mature (yellowing or older) siliques were randomly selected from each of the T2 individuals. Siliques were imaged using a Canoscan LiDE 200 flatbed scanner and measured using ImageJ on the line segment setting. The siliques from each plant were averaged, and this was treated as the data set for each construct. From these averages, the mean, median, and 95th, 75th, 25th, and 5th percentiles for the construct were calculated to generate a box-and-whisker plot. One-way ANOVA followed by Tukey’s honest significant difference post-hoc test was used to determine whether the silique lengths from SILK plants transformed with different constructs were statistically significantly different from each other (Supplemental Table 1). Each independent line was imaged by confocal microscopy to confirm GFP fusion protein expression and localization. Protein expression of independent lines of the same construct, as well as high and low expressing lines of the same construct, were compared via immunoblot (Supplemental Figures 8A and 8B).
FELT Complementation Experiment
The progeny of transformed FELT plants were germinated on MS media with 1% sucrose, 30 μg/mL hygromycin, and 100 μg/mL carbenicillin. Three to five seedlings that were resistant to the hygromycin and that contained at least one rg2-3 allele were selected and propagated. The progeny of these plants (T2 generation) were grown on MS media with 1% sucrose, 30 μg/mL hygromycin, and 10 μg/mL glufosinate ammonium (Basta) and genotyped for rg2-3. T3 generation seeds were plated on MS medium with 30 μg/mL hygromycin to determine whether the transgene was heterozygous or homozygous in the T2 generation. In parallel, T3 seeds were sown on soil and grown under long-day conditions until their first true leaves emerged. These populations of T3 seedlings were sprayed with 120 mg/L Basta three times over a 7-d period and were analyzed for color 2 d after the third spraying. Those whose cotyledons and/or leaves had yellowed were judged susceptible, while those that remained green were judged resistant. These populations were totaled and compared with the overall population numbers to generate segregation ratios for the rg2-3 allele. These were then compared with expected segregation ratios (Supplemental Figure 9) to determine whether the RanGAP1 variant rescued the female gametophyte lethality. Significance from the expected ratio given the null hypothesis that the transgene does not rescue was calculated using Fisher’s exact test. To determine whether any double null mutant progeny resulted from these FELT lines, we randomly chose 30 Basta-resistant plants and genotyped them for the RanGAP2 wild-type and rg2-3 alleles. Those plants that contained only the rg2-3 allele were considered double nulls and analyzed for phenotype. Each independent line was imaged by confocal microscopy to confirm GFP fusion protein expression and localization. Protein expression of independent lines of the same construct, as well as high and low expressing lines of the same construct, were compared via immunoblot (Supplemental Figures 8C and 8D).
Double Null Mutant Complementation Experiment
For the plants shown in Figure 6B, T5 progeny of double null individuals identified in the FELT complementation experiment above were cold treated at 4°C for 3 d and grown on MS media with 0.5% sucrose for 10 d with constant light. They were then transferred to soil and grown under long-day conditions. They were analyzed at 30 d of age. Independent lines were analyzed using siblings of the plants shown in Figure 6 at 10 d of age (Supplemental Figure 8E).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: RanGAP1, At3g63130 and GI825488; RanGAP2, At5g19320 and GI832052; WIP1, At4g26455 and GI7922386; WIP2, At5g56210 and GI835720; WIP3, At1g08290 and GI837349; WIT1, At5g11390 and GI831010; WIT2, At1g68910 and GI843224.
Supplemental Data
Supplemental Figure 1. SILK siliques exhibit developmental abnormalities.
Supplemental Figure 2. TWEED seedling developmental phenotypes are rescued by a native promoter-driven RanGAP1 genomic construct.
Supplemental Figure 3. Delocalization of RanGAP1 from the NE does not lead to nucleocytoplasmic transport defects.
Supplemental Figure 4. Multiple sequence alignment of the LRR domain of Arabidopsis RanGAP1, N. benthamiana RanGAP2, H. sapiens RanGAP1, S. cerevisiae Rna1, and S. pombe Rna1p proteins.
Supplemental Figure 5. A close-up of the 3D representation of RanGAP1.
Supplemental Figure 6. Subcellular localization of RanGAP1 variant constructs in interphase cells.
Supplemental Figure 7. Representative yeast two-hybrid assay results (the summary shown in Figure 5E).
Supplemental Figure 8. Protein expression in SILK, FELT, and double null mutant plants containing RanGAP1 native promoter-driven, C-terminally GFP-tagged versions of the RanGAP1 variant series.
Supplemental Figure 9. Punnett squares representing expected segregation ratios of the rg2-3 allele in the T3 progeny of T2 FELT plants containing RanGAP variant transgenes.
Supplemental Figure 10. Images of a sterile T4 double null mutant plant originating from RanGAP1AAP+N219A in FELT line 11.
Supplemental Table 1. Results from one-way ANOVA and Tukey’s honestly significant difference post-hoc test.
Supplemental Table 2. List of genotyping primers used in this study.
Acknowledgments
We thank Gregory Booton, Patrice Hamel, and Norman Groves for fruitful discussions. This work was supported by grants from the National Science Foundation (NSF-MCB 1243844) and by The Back to Belgium Grant of the Belgian Science Policy Office (selection year 2012). A.H.N.G. is a recipient of a predoctoral fellowship from the Center for RNA Biology at The Ohio State University.
AUTHOR CONTRIBUTIONS
I.M. designed the research and wrote the article. J.B. and A.H.N.G. designed the research, performed experiments, and wrote the article. T.R.-P., X.Z., and B.T. performed experiments and analyzed data. D.V.D. analyzed data.
Glossary
- NE
nuclear envelope
- PPB
preprophase band
- CDZ
cortical division zone
- Col-0
Columbia-0
- QC
quiescent center
- RAM
root apical meristem
- MS
Murashige and Skoog
- SD
synthetic dextrose
- UTR
untranslated region
Footnotes
Articles can be viewed online without a subscription.
References
- Ach R.A., Gruissem W. (1994). A small nuclear GTP-binding protein from tomato suppresses a Schizosaccharomyces pombe cell-cycle mutant. Proc. Natl. Acad. Sci. USA 91: 5863–5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.-S., Amasino R., Scheres B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. [DOI] [PubMed] [Google Scholar]
- Avis J.M., Clarke P.R. (1996). Ran, a GTPase involved in nuclear processes: its regulators and effectors. J. Cell Sci. 109: 2423–2427. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J., Nacry P., Christodoulidou A., Drevensek S., Camilleri C., Amiour N., Parcy F., Pastuglia M., Bouchez D. (2008). Arabidopsis TONNEAU1 proteins are essential for preprophase band formation and interact with centrin. Plant Cell 20: 2146–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey P.N., Linstead P.J., Roberts K., Schiefelbein J.W., Hauser M.T., Aeschbacher R.A. (1993). Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119: 57–70. [DOI] [PubMed] [Google Scholar]
- Bischoff F.R., Klebe C., Kretschmer J., Wittinghofer A., Ponstingl H. (1994). RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. USA 91: 2587–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F.R., Ponstingl H. (1991). Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354: 80–82. [DOI] [PubMed] [Google Scholar]
- Ciciarello M., Mangiacasale R., Lavia P. (2007). Spatial control of mitosis by the GTPase Ran. Cell. Mol. Life Sci. 64: 1891–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.E., Jacobsen S.E., Levin J.Z., Meyerowitz E.M. (1996). The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122: 1567–1575. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Colón-Carmona A., You R., Haimovitch-Gal T., Doerner P. (1999). Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20: 503–508. [DOI] [PubMed] [Google Scholar]
- Corbett A.H., Koepp D.M., Schlenstedt G., Lee M.S., Hopper A.K., Silver P.A. (1995). Rna1p, a Ran/TC4 GTPase activating protein, is required for nuclear import. J. Cell Biol. 130: 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A., Díaz-Triviño S., Wachsman G., Du Y., Arteága-Vázquez M., Zhang H., Benjamins R., Blilou I., Neef A.B., Chandler V., Scheres B. (2013). A SCARECROW-RETINOBLASTOMA protein network controls protective quiescence in the Arabidopsis root stem cell organizer. PLoS Biol. 11: e1001724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Somerville C.R. (2005). Imaging plant cell death: GFP-Nit1 aggregation marks an early step of wound and herbicide induced cell death. BMC Plant Biol. 5: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore B., Ciciarello M., Lavia P. (2004). Mitotic functions of the Ran GTPase network: the importance of being in the right place at the right time. Cell Cycle 3: 305–313. [PubMed] [Google Scholar]
- Ding D., Muthuswamy S., Meier I. (2012). Functional interaction between the Arabidopsis orthologs of spindle assembly checkpoint proteins MAD1 and MAD2 and the nucleoporin NUA. Plant Mol. Biol. 79: 203–216. [DOI] [PubMed] [Google Scholar]
- Feng W., Benko A.L., Lee J.H., Stanford D.R., Hopper A.K. (1999). Antagonistic effects of NES and NLS motifs determine S. cerevisiae Rna1p subcellular distribution. J. Cell Sci. 112: 339–347. [DOI] [PubMed] [Google Scholar]
- Forzani C., Aichinger E., Sornay E., Willemsen V., Laux T., Dewitte W., Murray J.A. (2014). WOX5 suppresses CYCLIN D activity to establish quiescence at the center of the root stem cell niche. Curr. Biol. 24: 1939–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay N.J., Packman L.C., Weldon M.A., Barna J.C.J. (1991). A leucine-rich repeat peptide derived from the Drosophila Toll receptor forms extended filaments with a β-sheet structure. FEBS Lett. 291: 87–91. [DOI] [PubMed] [Google Scholar]
- Gietz D., St Jean A., Woods R.A., Schiestl R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Seewald M.J., Ribbeck K. (2003). Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J. 22: 1088–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss O.J., Vernos I. (2004). The mechanism of spindle assembly: functions of Ran and its target TPX2. J. Cell Biol. 166: 949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland J., Gerke V. (1999). Conserved charged residues in the leucine-rich repeat domain of the Ran GTPase activating protein are required for Ran binding and GTPase activation. Biochem. J. 343: 653–662. [PMC free article] [PubMed] [Google Scholar]
- Hillig R.C., Renault L., Vetter I.R., Drell T. IV, Wittinghofer A., Becker J. (1999). The crystal structure of rna1p: a new fold for a GTPase-activating protein. Mol. Cell 3: 781–791. [DOI] [PubMed] [Google Scholar]
- Hopper A.K., Traglia H.M., Dunst R.W. (1990). The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J. Cell Biol. 111: 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden R., Park S.K., Moore J.M., Orme J., Grossniklaus U., Twell D. (1998). Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149: 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M., Zhang C., Avis J.M., Hutchison C.J., Clarke P.R. (1998). The role of the ran GTPase in nuclear assembly and DNA replication: characterisation of the effects of Ran mutants. J. Cell Sci. 111: 3017–3026. [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Kutay U., von Kobbe C., Mattaj I.W., Görlich D. (1997). The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 16: 6535–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y., Mongkolsiriwatana C., Veley K.M., Kim S.Y., Michaels S.D. (2007). The nuclear pore protein AtTPR is required for RNA homeostasis, flowering time, and auxin signaling. Plant Physiol. 144: 1383–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S.Y., Rose A., Joseph J., Dasso M., Meier I. (2005). Plant-specific mitotic targeting of RanGAP requires a functional WPP domain. Plant J. 42: 270–282. [DOI] [PubMed] [Google Scholar]
- Jeong S.Y., Rose A., Meier I. (2003). MFP1 is a thylakoid-associated, nucleoid-binding protein with a coiled-coil structure. Nucleic Acids Res. 31: 5175–5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J., Liu S.T., Jablonski S.A., Yen T.J., Dasso M. (2004). The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14: 611–617. [DOI] [PubMed] [Google Scholar]
- Joseph J., Tan S.H., Karpova T.S., McNally J.G., Dasso M. (2002). SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156: 595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe C., Bischoff F.R., Ponstingl H., Wittinghofer A. (1995). Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry 34: 639–647. [DOI] [PubMed] [Google Scholar]
- Kobe B., Deisenhofer J. (1995). Proteins with leucine-rich repeats. Curr. Opin. Struct. Biol. 5: 409–416. [DOI] [PubMed] [Google Scholar]
- Kobe B., Kajava A.V. (2001). The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11: 725–732. [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Lee H.S., Wi S.J., Park K.Y., Schmit A.C., Pai H.S. (2009). Dual functions of Nicotiana benthamiana Rae1 in interphase and mitosis. Plant J. 59: 278–291. [DOI] [PubMed] [Google Scholar]
- Lipka E., Gadeyne A., Stöckle D., Zimmermann S., De Jaeger G., Ehrhardt D.W., Kirik V., Van Damme D., Müller S. (2014). The phragmoplast-orienting kinesin-12 class proteins translate the positional information of the preprophase band to establish the cortical division zone in Arabidopsis thaliana. Plant Cell 26: 2617–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis M.J., Coutavas E., Blobel G. (1996). A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135: 1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis M.J., Wu J., Blobel G. (1998). SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140: 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle T., Haizel T., Matsumoto T., Harter K., Dallmann G., Nagy F. (1994). Phenotype of the fission yeast cell cycle regulatory mutant pim1-46 is suppressed by a tobacco cDNA encoding a small, Ran-like GTP-binding protein. Plant J. 6: 555–565. [DOI] [PubMed] [Google Scholar]
- Müller S., Han S., Smith L.G. (2006). Two kinesins are involved in the spatial control of cytokinesis in Arabidopsis thaliana. Curr. Biol. 16: 888–894. [DOI] [PubMed] [Google Scholar]
- Ottoline Leyser H.M., Furner I.J. (1992). Characterisation of three shoot apical meristem mutants of Arabidopsis thaliana. Development 116: 397–403. [Google Scholar]
- Pay A., Resch K., Frohnmeyer H., Fejes E., Nagy F., Nick P. (2002). Plant RanGAPs are localized at the nuclear envelope in interphase and associated with microtubules in mitotic cells. Plant J. 30: 699–709. [DOI] [PubMed] [Google Scholar]
- Ren M., Coutavas E., D’Eustachio P., Rush M.G. (1994). Effects of mutant Ran/TC4 proteins on cell cycle progression. Mol. Cell. Biol. 14: 4216–4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Peiris T., Xu X.M., Zhao Q., Wang H.J., Meier I. (2011). RanGAP is required for post-meiotic mitosis in female gametophyte development in Arabidopsis thaliana. J. Exp. Bot. 62: 2705–2714. [DOI] [PubMed] [Google Scholar]
- Rose A., Meier I. (2001). A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc. Natl. Acad. Sci. USA 98: 15377–15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Kucukural A., Zhang Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A.K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814. [DOI] [PubMed] [Google Scholar]
- Sazer S., Dasso M. (2000). The ran decathlon: multiple roles of Ran. J. Cell Sci. 113: 1111–1118. [DOI] [PubMed] [Google Scholar]
- Scheres B., Di Laurenzio L., Willemsen V., Hauser M.T., Janmaat K., Weisbeek P., Benfey P.N. (1995). Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121: 53–62. [Google Scholar]
- Seewald M.J., Körner C., Wittinghofer A., Vetter I.R. (2002). RanGAP mediates GTP hydrolysis without an arginine finger. Nature 415: 662–666. [DOI] [PubMed] [Google Scholar]
- Seewald M.J., Kraemer A., Farkasovsky M., Körner C., Wittinghofer A., Vetter I.R. (2003). Biochemical characterization of the Ran-RanBP1-RanGAP system: are RanBP proteins and the acidic tail of RanGAP required for the Ran-RanGAP GTPase reaction? Mol. Cell. Biol. 23: 8124–8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling W.I.L., Nooijen C., Ludwig N., Boter M., Slootweg E., Goverse A., Shirasu K., Joosten M.H.A.J. (2010). RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell 22: 4176–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D. (2009). Division plane determination during plant somatic cytokinesis. Curr. Opin. Plant Biol. 12: 745–751. [DOI] [PubMed] [Google Scholar]
- Vanstraelen M., Baloban M., Da Ines O., Cultrone A., Lammens T., Boudolf V., Brown S.C., De Veylder L., Mergaert P., Kondorosi E. (2009). APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 106: 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xu Y., Han Y., Bao S., Du J., Yuan M., Xu Z., Chong K. (2006). Overexpression of RAN1 in rice and Arabidopsis alters primordial meristem, mitotic progress, and sensitivity to auxin. Plant Physiol. 140: 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A.J., Gallagher K., Smith L.G. (2009). discordia1 and alternative discordia1 function redundantly at the cortical division site to promote preprophase band formation and orient division planes in maize. Plant Cell 21: 234–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Cai W. (2014). RAN1 is involved in plant cold resistance and development in rice (Oryza sativa). J. Exp. Bot. 65: 3277–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.M., Meulia T., Meier I. (2007). Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr. Biol. 17: 1157–1163. [DOI] [PubMed] [Google Scholar]
- Xu X.M., Zhao Q., Rodrigo-Peiris T., Brkljacic J., He C.S., Müller S., Meier I. (2008). RanGAP1 is a continuous marker of the Arabidopsis cell division plane. Proc. Natl. Acad. Sci. USA 105: 18637–18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Brkljacic J., Meier I. (2008). Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell 20: 1639–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Meier I. (2014). Efficient plant male fertility depends on vegetative nuclear movement mediated by two families of plant outer nuclear membrane proteins. Proc. Natl. Acad. Sci. USA 111: 11900–11905. [DOI] [PMC free article] [PubMed] [Google Scholar]