The activity of the mitochondrial dihydrolipoyl dehydrogenase improves photorespiration and in turn stimulates photosynthetic carbon assimilation and plant growth.
Abstract
Mitochondrial dihydrolipoyl dehydrogenase (mtLPD; L-protein) is an integral component of several multienzyme systems involved in the tricarboxylic acid (TCA) cycle, photorespiration, and the degradation of branched-chain α-ketoacids. The majority of the mtLPD present in photosynthesizing tissue is used for glycine decarboxylase (GDC), necessary for the high-flux photorespiratory glycine-into-serine conversion. We previously suggested that GDC activity could be a signal in a regulatory network that adjusts carbon flux through the Calvin-Benson cycle in response to photorespiration. Here, we show that elevated GDC L-protein activity significantly alters several diagnostic parameters of cellular metabolism and leaf gas exchange in Arabidopsis thaliana. Overexpressor lines displayed markedly decreased steady state contents of TCA cycle and photorespiratory intermediates as well as elevated NAD(P)+-to-NAD(P)H ratios. Additionally, increased rates of CO2 assimilation, photorespiration, and plant growth were observed. Intriguingly, however, day respiration rates remained unaffected. By contrast, respiration was enhanced in the first half of the dark phase but depressed in the second. We also observed enhanced sucrose biosynthesis in the light in combination with a lower diel magnitude of starch accumulation and breakdown. These data thus substantiate our prior hypothesis that facilitating flux through the photorespiratory pathway stimulates photosynthetic CO2 assimilation in the Calvin-Benson cycle. They furthermore suggest that this regulation is, at least in part, dependent on increased light-capture/use efficiency.
INTRODUCTION
Mitochondrial dihydrolipoyl dehydrogenase (mtLPD; L-protein, EC 1.8.1.4) is a structurally conserved homodimeric flavoenzyme of the pyridine nucleotide-disulfide oxidoreductase enzyme family that is ubiquitous in aerobic organisms. In plants, mtLPD is a crucial component of two mitochondrial multienzyme complexes of or closely associated to the tricarboxylic acid (TCA) cycle, namely, the pyruvate dehydrogenase complex (mtPDHC; Millar et al., 1998), which additionally has a plastidial isoform, ptPDHC, and 2-oxoglutarate dehydrogenase (ODHC; Millar et al., 1999), as well as the branched-chain 2-oxoacid dehydrogenase complex (BCDHC) and the glycine decarboxylase complex (GDC; Oliver et al., 1990).
In contrast with GDC, which forms relatively labile complexes, the 2-oxoacid dehydrogenase complexes are stably built from multiple copies of the so-called E1 subunit (2-oxoacid dehydrogenase; EC 1.2.4.1-2-4), E2 subunit (dihydrolipoyl acyltransferase; EC 2.3.1.12), and E3 subunit (LPD). The large structures resulting from the close association of these polypeptides allow active site coupling that, in the case of mtPDHC, involves the transfer of acetyl groups from one E2 lipoyl residue to another such that the whole core can be acetylated from pyruvate entering via one E1 enzyme (Zhou et al., 2001; Marrott et al., 2014). Active site coupling was not demonstrated for GDC; however, formation of the GDC complex also requires multiple copies of each of the three enzyme components, the P-protein (the actual glycine decarboxylase; EC 1.4.4.2), T-protein (aminomethyltransferase; EC 2.1.2.10), and L-protein (mtLPD; EC 1.8.1.4). A fourth, nonenzymatic component, the H-protein, interacts with all three enzymes to convey lipoyl-bound intermediates of the GDC reaction cycle from the P- via the T- to the L-protein. In each of these multienzyme systems, mtLPD is needed for the NAD+-dependent reoxidation of a protein-bound (E2 or H-protein) lipoyl group that becomes reduced in course of the respective reaction cycles (Douce et al., 2001; reviewed in Reed, 2001; Mooney et al., 2002).
The mtPDHC and ODHC are crucial components of the TCA cycle and metabolic regulatory networks (Bunik and Fernie, 2009), while BCDHC is an important enzyme of branched-chain amino acid catabolism (Araújo et al., 2010). By contrast, GDC serves two functions: (1) It is essential for providing one-carbon units as biosynthetic precursors for a multitude of pathways, and (2) it is crucial for the photorespiratory glycine-to-serine conversion (Engel et al., 2007). Due to the very high photorespiratory flux in green tissues, GDC comprises ∼50% of the total mitochondrial matrix protein of photosynthesizing cells (Douce et al., 2001). It is thus present very much in excess of the other three enzyme complexes. In earlier works studying either P-protein antisense or H-protein overexpressor plants (Heineke et al., 2001; Timm et al., 2012a), we demonstrated that the activity of GDC can be an important determinant of the photosynthetic performance of plants. However, the precise mechanism by which GDC activity exerts its effect on photosynthetic processes is as yet unknown. We previously suggested that at least some of the crosstalk between mitochondria and chloroplasts underlying this response may be mediated via feedback inhibition from intermediates of the photorespiratory pathway (Timm et al., 2012a).
One of the four GDC proteins, mtLPD, is particularly interesting since it operates in three different branches of plant carbon metabolism, the high-flux bearing photorespiratory, the medium-flux bearing TCA cycle, and the low-flux bearing branched-chain amino acid degradation pathway. In Arabidopsis thaliana, mtLPD is encoded by two genes, mtLPD1 and mtLPD2, which are, to at least some degree, redundant (Lutziger and Oliver, 2001). In addition, two further LPD genes encode plastidial LPD, which serves as a ptPDHC component protein to provide acetyl-CoA for fatty acid biosynthesis (Lutziger and Oliver, 2000).
In light of the multifaceted role of LPD in cellular carbon metabolism, we examined the effects invoked by mtLPD overexpression in Arabidopsis mitochondria. In particular, we wanted to ascertain whether increased mtLPD levels would facilitate photosynthetic-photorespiratory metabolism and improve plant growth in a similar manner to that occurring following an increase in the amount of GDC H-protein.
RESULTS
Generation of Arabidopsis mtLPD Overexpression Lines
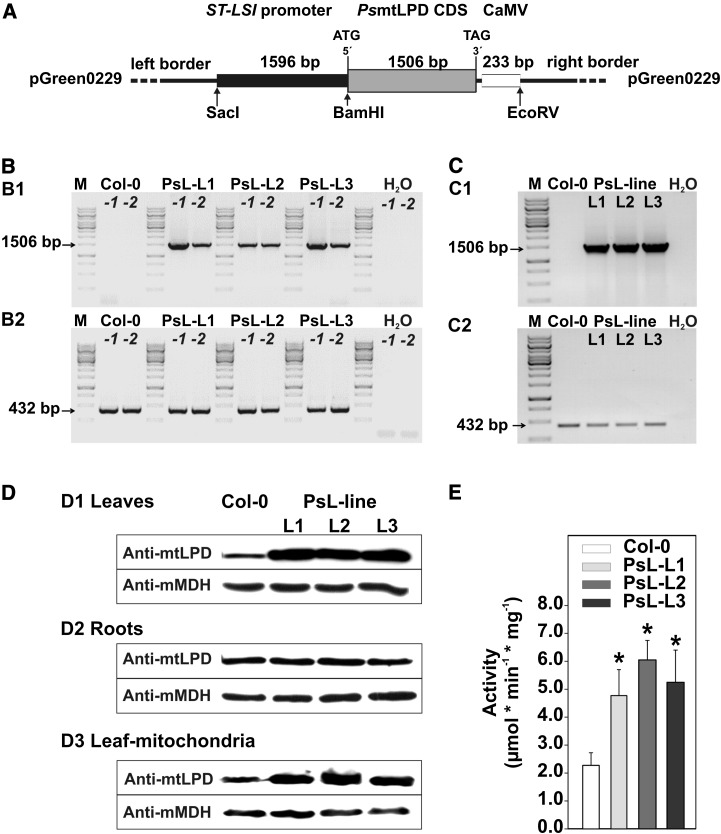
To overexpress mtLPD, a cDNA encoding pea (Pisum sativum) mtLPD (PsL; 1506 bp; Bourguignon et al., 1992) was ligated between the ST-LSI mesophyll-specific promoter (Stockhaus et al., 1989) and the CaMV terminator in a variant of the transformation vector pGREEN0229 (http://www.pgreen.ac.uk/) (Figure 1A). We used the pea sequence to avoid suppression of the endogenous mtLPD genes and the ST-LSI promoter as it drives expression in a light-dependent manner and only in chloroplast-containing tissue (Stockhaus et al., 1989), to avoid bias from metabolic alterations in nonphotosynthesizing organs. Resulting transformants of Arabidopsis (Col-0) were validated at the levels of DNA (Figure 1B), mRNA (Figure 1C), mtLPD protein (Figure 1D), and mitochondrial mtLPD activity (Figure 1E). These analyses confirmed proper integration and expression of the transgene and demonstrated 2- to 3-fold elevated LPD activity in leaf mitochondria. As anticipated, pea mtLPD expression was restricted to leaves and not observed in root protein extracts (Figure 1D). Three independent lines (PsL-L1, PsL-L2, and PsL-L3) were propagated to obtain stable T4 generation lines for subsequent analyses.
Figure 1.
Generation and Verification of mtLPD Overexpressors.
(A) Schematic overview of the mtLPD overexpression construct.
(B) PCR verification of the transformed construct into the genome of transgenic lines (B1) and the corresponding loading control (B2).
(C) RT-PCR verification of the full-length mtLPD transcript (C1) using signals of the constitutively expressed 40S ribosomal protein S16 gene as calibration control (C2).
(D) Immunoblot of leaves, roots, and isolated mitochondria using a specific antibody against mtLPD and mitochondrial malate dehydrogenase (mMDH) as loading control.
(E) Enzymatic activity of mtLPD in overexpressor and wild-type mitochondria. Values are means ± sd from four technical replicates. Asterisks indicate values that were significantly different from the wild-type control based on Student’s t test (*P < 0.05).
Visual Phenotype and Biomass Accumulation of the PsL Lines
We next examined whether enhanced mtLPD activity modifies plant growth under different photoperiods and CO2 concentrations. Plant growth was distinctly improved under short-day conditions (8/16- to 10/14-h day/night cycle), and this effect was most visible after 10 weeks of cultivation (Supplemental Figure 1A). Leaf biomass accumulation was significantly higher than in the wild type on both fresh weight (PsL-L1 ∼10%, PsL-L2 ∼18%, and PsL-L3 ∼14%) and dry weight (PsL-L1 ∼19%, PsL-L2 ∼47%, and PsL-L3 ∼40%) bases (Supplemental Figure 1B). During growth under elevated CO2 (2000 μL L−1), which inhibits photorespiration, the overexpressor lines consistently started flowering 2 weeks earlier than the wild type, although they did not display the accelerated growth detected in normal air (Supplemental Figure 1C). Moreover, no significant growth enhancement was observed under photoperiods of 12 h and longer (data not shown). This observation is consistent with other reports (Kebeish et al., 2007; Maier et al., 2012; Timm et al., 2012a), but it is currently unknown why longer photoperiods suspend the growth improvement observed in short days.
mtLPD Overexpression Alters Metabolite Levels in the TCA Cycle and the Photorespiratory Pathway
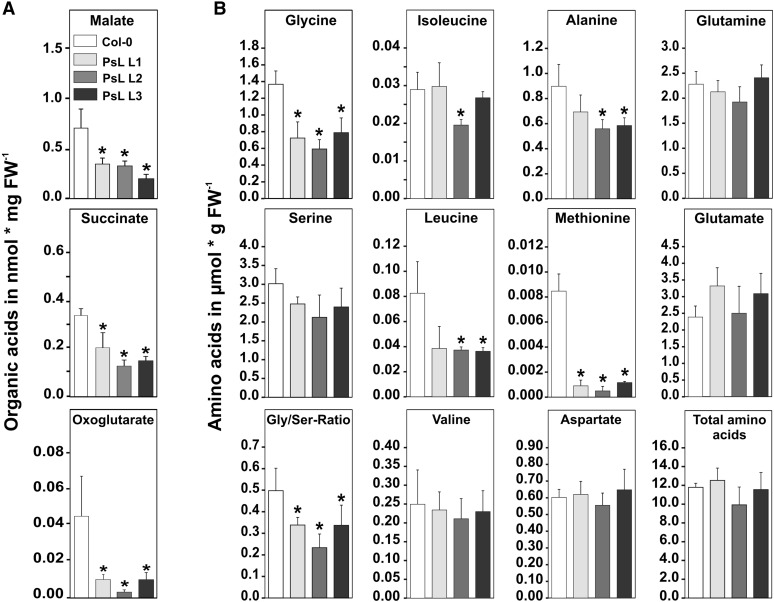
mtLPD is required for three metabolic branches of mitochondrial carbon metabolism: photorespiration, the TCA cycle, and the degradation of branched-chain amino acids. To find out how much mtLPD overexpression affects operation of these different pathways, we examined the levels of several diagnostic metabolites in leaves by gas chromatography (2-oxoglutarate, malate, and succinate) and by HPLC (amino acids). The steady state contents of 2-oxoglutarate, malate, and succinate were significantly decreased in all overexpressor lines (Figure 2A), indicating effects of elevated mtLPD activity on the carbon flux through the TCA cycle. Concerning the photorespiratory carbon flux, leaf glycine but not serine contents were significantly reduced resulting in significantly lower glycine/serine ratios in all of the transgenic lines (Figure 2B). Steady state contents of glutamate and glutamine remained rather unaffected (Figure 2B). Levels of methionine, an important acceptor and donor of one-carbon units, were distinctly lower than in the wild type. Among the branched-chain amino acids, we detected reduced levels for leucine in all three overexpressors, whereas isoleucine and valine remained unaffected. Total amino acid contents also did not show significant variations (Figure 2B; Supplemental Table 1). In summary, the observed changes suggest that elevated mtLPD activity facilitates carbon flow through photorespiration and the TCA cycle, without substantial pleiotropic effects on other pathways. Apart from that, we did not observe strong alterations on the intermediates of the degradation of branched-chain amino acids by BCDHC, which is upregulated under stress conditions but is less active less active under standard growth conditions (Taylor et al., 2004).
Figure 2.
Changes in Mitochondrial Metabolites in mtLPD Overexpressors.
Leaf material was harvested at the end of the light period from plants at growth stage 5.1 according to Boyes et al. (2001). Absolute steady state contents of three representatives of the TCA cycle (A) and selected amino acids (B) (complete list are given in Supplemental Table 1). Values are means ± sd of five independent biological replicates. Asterisks indicate values that were significantly different from the wild-type control based on Student’s t test (*P < 0.05). FW, fresh weight.
Enhanced Turnover of TCA Cycle and Photorespiratory Intermediates Results in the Accumulation of Soluble Sugars
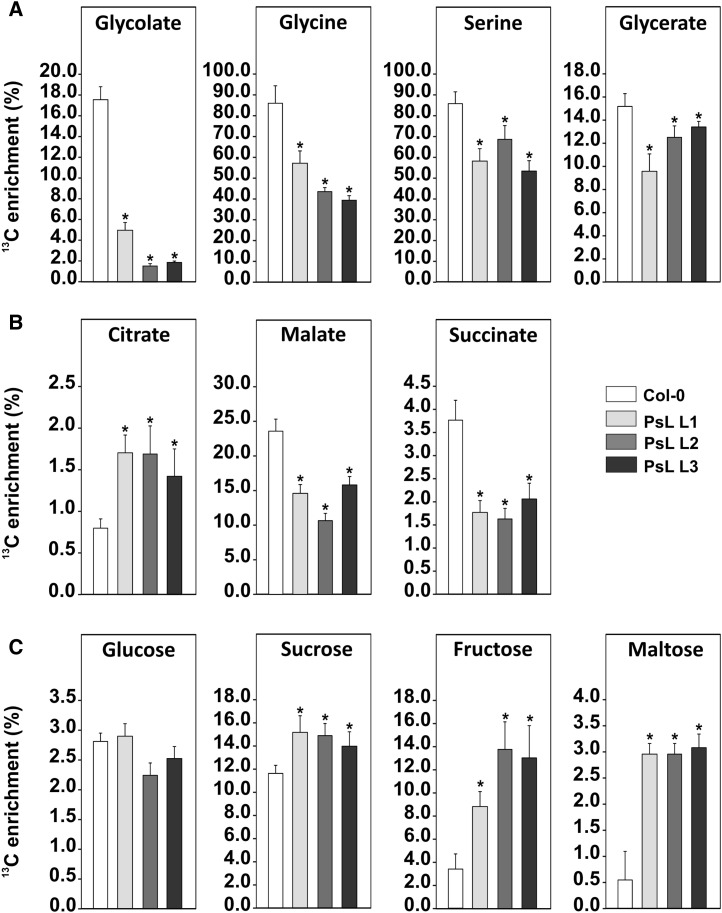
To further explore the metabolic changes that eventually result in the improved growth of the mtLPD overexpressor lines, we used a 13C-labeling approach (Timm et al., 2008; Araújo et al., 2010). Briefly, leaf discs were incubated with 13C-labeled glycine followed by gas chromatography-mass spectrometry (GC-MS) analysis of the isotope enrichment in intermediates of the photorespiratory pathway, the TCA cycle, and soluble sugars. In comparison with the wild type, all three overexpressor lines showed significantly lower 13C enrichment in several photorespiratory intermediates (glycolate ∼0.1- to 0.3-fold, glycine ∼0.4- to 0.6-fold, serine ∼0.6- to 0.8-fold, glycerate ∼0.6- to 0.8-fold), indicating faster turnover thereof, leading to a decrease in their overall pool size (Figure 3A; Supplemental Table 2). Lower 13C enrichment was also observed in two TCA cycle intermediates (malate ∼0.5- to 0.6-fold) and succinate (∼0.4- to 0.5-fold). Corresponding with the steady state metabolite data above, the 13C-labeling enrichment study also suggests a facilitated carbon flow through the photorespiratory pathway and the TCA cycle.
Figure 3.
13C Isotope Enrichment in Selected Intermediates of Photorespiration, the TCA Cycle, and Soluble Sugars.
Leaf discs were harvested from plants at growth stage 5.1 according to Boyes et al. (2001) after 6 h of illumination. 13C glycine labeling (3 h) was performed under growth conditions, leaf discs were harvested, and 13C enrichment of selected intermediates analyzed by GC-MS. Shown are representatives of photorespiration (A), the TCA cycle (B), and soluble sugars (C). Values are given as means ± sd from at least five biological and two technical replicates. Asterisks indicate values that were statistically significant based on Student’s t test (P < 0.05). For a comprehensive analysis of 13C enrichment see Supplemental Figure 2 and Supplemental Table 2.
By contrast, higher 13C enrichment was found for citrate (∼1.8- to 2.1-fold). Concerning carbohydrate end products, all three overexpressors displayed a significantly increased accumulation of 13C in sucrose (∼20 to 30%) and fructose (∼160 to 300%) compared with the control, whereas there were no distinct changes in glucose labeling, which is likely due to a large inactive pool in the vacuole (Tohge et al., 2011) that prevents complete labeling during the short time period. Interestingly, the transgenic plants displayed a significantly higher labeling of maltose (∼450%), indicating that starch degradation initiates already in the light. From these data, we conclude that higher mtLPD activity in leaves leads to a considerable metabolic shift of carbon from amino and organic acids toward soluble sugars, which could indicate an enhanced source capacity.
It would appear that the rapid operation of the photorespiration results in very high transfer of 13C to sucrose in spite of the fact that the pools of the photorespiratory intermediates are small. These data likely imply a coordinate upregulation of photorespiratory flux since label accumulation is lower in each of the intermediates. The sum effect of this is an enhanced sucrose production; moreover, given that sucrose export is effectively inhibited in the leaf disc system, this appears to be coupled to enhanced sucrose degradation (note the higher labeling in fructose) in the transgenic lines. To further substantiate this hypothesis without limiting of sucrose export, we next quantified absolute contents of selected sugars (sucrose, glucose, and fructose) of adult plants by gas chromatography. To allow for direct comparison between both approaches, leaf material was harvested at the middle of the day, which in principle correlates with the harvesting time point of the labeling approach. All three transgenic lines did accumulate those sugars in comparison to the wild type (Supplemental Figure 2).
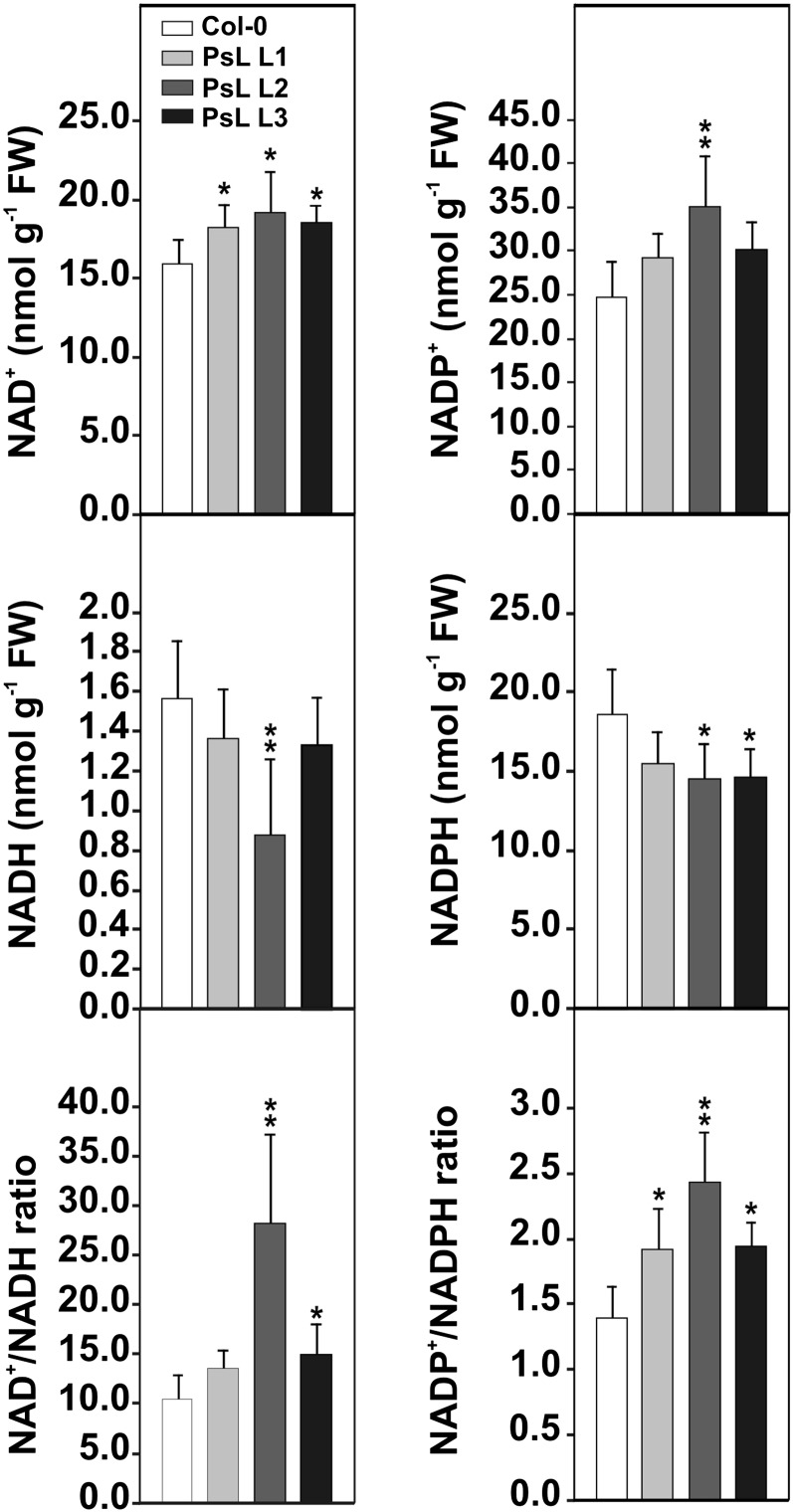
Overexpression of mtLPD Induces Higher Cellular NAD(P)+-to-NAD(P)H Ratios
Particularly in the photorespiratory pathway, mtLPD generates massive amounts of NADH in green tissues in the light. We therefore examined whether higher mtLPD activity in addition to the altered metabolic fluxes shown above would also have an effect on the cellular redox balance. To this end, we analyzed the levels of pyridine nucleotides in leaves at the end of the light period (9 h of illumination). The NAD+ and NADP+ contents were significantly elevated in all overexpressors, whereas the NADH and NADPH contents displayed a slight downward tendency compared with the wild type (Figure 4). These changes summed to an elevation in the deduced NAD+/NADH and NADP+/NADPH ratios with the largest change in line PsL-L2, which also displayed the highest mtLPD activity and increase in growth.
Figure 4.
Pyridine Nucleotide Levels in Leaves of mtLPD Overexpressors.
Leaf material was harvested at the end of the light period (9 h light) from plants at growth stage 5.1 according to Boyes et al. (2001). Values are mean ± sd of five independent biological replicates. Asterisks indicate values significantly different from the wild-type control based on Student´s t test (*P < 0.05; **P < 0.01). FW, fresh weight.
We next examined leaf contents of O-acetyl serine, Cys, GSH, ADP, and ATP (Table 1). These analyses did not reveal consistent alterations, with the possible exception of an increase in Cys (significant in lines PsL-L2 and PsL-L3) and GSH (significant in Line PsL-L3). Similarly, ADP contents were not changed, although we measured significantly elevated ATP contents in all lines; however, this did not translate to a significant increase in the deduced ATP/ADP ratios.
Table 1. Products of Sulfide Metabolism and Energy Equivalents in Wild-Type (Col-0) and mtLPD Overexpression Plants.
| Compound | Col-0 | PsL-L1 | PsL-L2 | PsL-L3 |
|---|---|---|---|---|
| O-acetylserine | 10.45 ± 1.46 | 10.28 ± 3.21 | 13.07 ± 3.36 | 12.66 ± 3.95 |
| Cysteine | 6.18 ± 1.62 | 7.13 ± 1.27 | 10.04 ± 1.35 | 11.57 ± 1.40 |
| Glutathione | 278.44 ± 51.29 | 237.53 ± 22.33 | 305.77 ± 19.77 | 342.89 ± 19.70 |
| ADP | 10.68 ± 2.38 | 10.20 ± 1.91 | 11.35 ± 1.90 | 12.68 ± 3.27 |
| ATP | 34.60 ± 5.88 | 45.35 ± 4.14 | 44.82 ± 1.86 | 57.06 ± 9.99 |
| ATP/ADP ratio | 3.40 ± 0.93 | 4.54 ± 0.58 | 4.05 ± 0.65 | 4.67 ± 1.30 |
Leaf tissue samples from plants at growth stage 5.1 (Boyes et al., 2001) were taken at the end of the light period (9 h illumination) and metabolite contents analyzed by HPLC. Values presented are means ± sd (pmol*mg−1 fresh weight) of measurements from at least five biological replicates per genotype. Numbers in bold indicate values statistically significant from the wild type based on Student’s t test (P < 0.05).
Elevated mtLPD Activity Influences Respiratory Rates
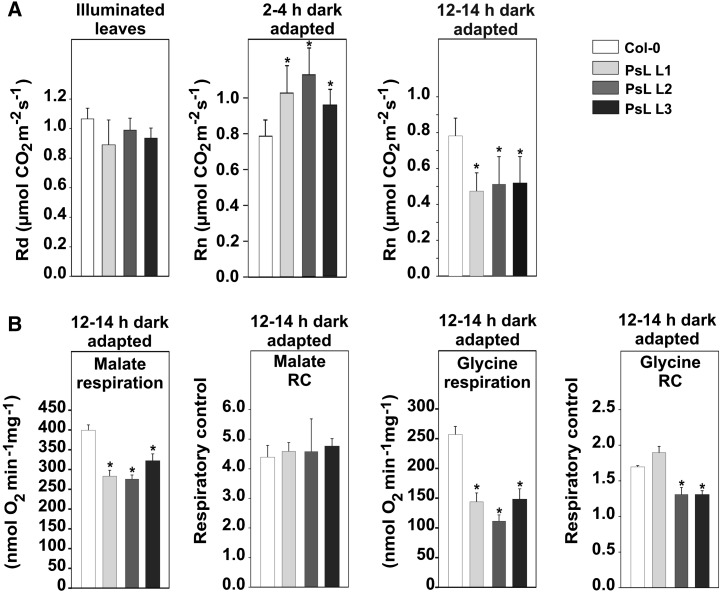
Several studies have shown that genetic manipulation of the TCA cycle or in the photorespiratory pathway results in altered rates of mitochondrial respiration (Raghavendra and Padmasree, 2003; Nunes-Nesi et al., 2005, 2007). We therefore next examined whether such changes also occur in mtLPD overexpressor plants. First, we measured the respiratory CO2 release using leaves from plants adapted to light and from plants in the first (2 to 4 h darkness) and second (12 to 14 h darkness) half of the night . We detected no significant alterations in the rate of day respiration (Rd) in any of the transgenic lines (Figure 5A). After 2 to 4 h of darkness, however, all three transgenic lines displayed significantly higher night respiration rates (RN, ∼20 to 40%) compared with wild-type plants. By contrast, after 12 to 14 h darkness, we observed significantly lower respiration (∼50 to 60% decrease) in leaves of the mtLPD overexpressors. This biphasic alteration in the night respiration rate of the PsL lines suggests that these plants rapidly respire a greater proportion of carbohydrates than the wild type in the first half of the night; thereafter, metabolism adapts to lower respiration rates, possibly to avoid carbon starvation as indicated by the reduced amounts of sucrose in transgenic lines (Supplemental Figure 2). Such an adaptation was confirmed by measuring respiration rates of well-coupled mitochondria isolated from the overexpressor lines after 12 to 14 h of darkness (Figure 5B). Here, we also observed significantly lower respiration rates in the transgenic lines relative to the control, both when malate and even more dramatically when glycine were supplied as respiratory substrate. In good correlation with these findings, PDHC and ODHC in isolated mitochondria were less active but displayed unaltered subunit abundances as tested with the respective E2 subunits (Supplemental Figure 3). Collectively, these results suggest that both complexes become inhibited, likely by posttranslational modification, at the end of the night. GDC protein amounts were also not changed but, in distinct contrast to PDHC and ODHC, GDC activity was higher in the transgenic lines (Supplemental Figure 3).
Figure 5.
Respiratory Gas Exchange and Mitochondrial Respiration of mtLPD Lines.
(A) Fully expanded leaves from plants at growth stage 5.1 (Boyes et al., 2001) were used for determination of the respiratory CO2 release during illumination and after dark adaptation (2 to 4 and 12 to 14 h, respectively; n > 4) by gas-exchange measurements.
(B) Malate and glycine respiration of intact mitochondria from mtLPD overexpressors and the wild type (n = 4). Respiratory coupling of mitochondria was verified by determination of respiratory control values (RC) from the transition from state 3 to state 4. RC values were in the range of previous reports (Keech et al., 2005) confirming that isolated organelles were intact and of good quality. Asterisks indicate values significantly different from the wild-type control based on Student’s t test (*P < 0.05).
Increased mtLPD Activity Stimulates Photosynthetic and Photorespiratory Gas Exchange
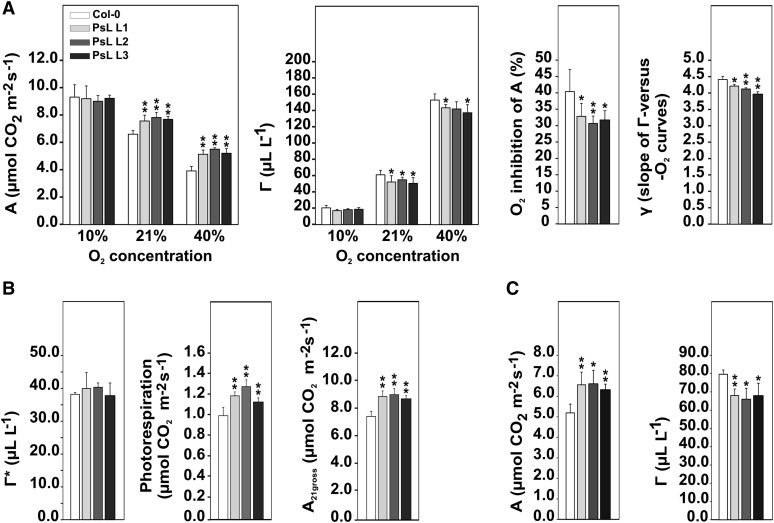
Artificial constraints on respiratory and photorespiratory metabolism can have distinct effects on photosynthetic parameters (Nunes-Nesi et al., 2005, 2007; Cousins et al., 2011; Timm et al., 2011, 2012a; Eisenhut et al., 2013). To find out whether such effects were also apparent in the mtLPD overexpressor plants, we examined the photosynthetic-(photo)respiratory gas exchange in more detail. In comparison with the wild type, we found significantly increased rates (∼15 to 18%) of net photosynthetic CO2 uptake in normal air (A21), which were accompanied with significantly lower (∼10 to 17%) CO2 compensation points (Γ21) at 21% oxygen (Figure 6A). Corresponding data obtained at 40% oxygen displayed the same tendencies, whereas differences from the wild type were not significant at 10% oxygen. In good agreement to that, photosynthesis in mtLPD overexpressors was significantly less inhibited by oxygen (Col-0, 40.44% ± 6.77%; PsL-L1, 32.88% ± 3.98%; PsL-L2, 30.73% ± 2.25%; PsL-L3, 31.81% ± 2.81%). Similarly, the oxygen effect on Γ, calculated as the slope (γ) of the Γ-versus-oxygen response curves, was significantly lower in all three overexpressor lines (Figure 6A).
Figure 6.
Photosynthetic Parameters of mtLPD Overexpression Lines.
For gas-exchange measurements plants were grown to growth stage 5.1 (Boyes et al., 2001) under normal air conditions (21% O2, 390 ppm CO2) with a photoperiod of 10/14-h day/night.
(A) Shown are net CO2 uptake (A), CO2 compensation points (Γ), oxygen inhibition, and slopes of the oxygen response curves (γ) of mtLPD lines compared with the wild type (n > 5; *P < 0.05, **P < 0.01).
(B) CO2 compensation points in the absence of dark respiration (Γ*), rates of photorespiration, and gross photosynthesis (n = 5; *P < 0.05, **P < 0.01). All measurements were performed using a light intensity of 1000 µmol m−2 s−1.
(C) A and Γ determination at a light intensity of 250 µmol m−2 s−1.
Furthermore, we examined the rates of photorespiration (Sharkey, 1988). While the CO2 compensation points in the absence of dark respiration (Γ*) were not significantly different between the wild type and mtLPD overexpressors (Figure 6B), all of the transgenic lines displayed significant increases in their deduced rates of photorespiration (Figure 6B). This clearly shows that the higher photosynthetic rates in transgenic lines are accompanied with elevated rates of photorespiration, which is fully consistent with the results obtained from 13C labeling. Taken together, the observed effects indicate that most of the metabolic changes induced by higher activity of mtLPD are related to photosynthetic-photorespiratory metabolism.
Overexpression of mtLPD Improves Photosynthetic Light-Use Efficiency
We next wanted to check whether and how these metabolic changes alter the efficiency of photosynthetic electron transport. This was done by measuring chlorophyll fluorescence (DUAL-PAM-100; Walz). We found that mtLPD overexpression had no effect on the total chlorophyll a and b contents (Table 2). In close agreement with this observation, neither the maximum quantum yields (Y) nor the maximum relative electron transport rates (rETRmax) of photosystem I (PSI) and photosystem II (PSII) varied between genotypes. Similarly, we did not detect any significant alterations in either the PSI acceptor (Y[NA]) or donor (Y[ND]) side limitation (Supplemental Data Set 1). Nevertheless, all three transgenic lines displayed better light-use efficiency than the wild type. For example, mtLPD overexpressors show optimal rates of rETR over both photosystems at considerably lower light intensities (Table 2). Accordingly, the light compensation points, which in general are in the range of previous reports (Takemiya et al., 2005), are reduced and the efficiency of photosynthetic light conversion (α) at lower light intensities (around growth conditions) is higher in the mtLPD overexpressors (Table 2). To further substantiate these findings, we additionally determined net photosynthetic CO2 uptake rates and CO2 compensation points at similar light intensities (250 µmol m−2 s−2) by gas exchange and detected the same improvement of photosynthesis as observed with measurements under saturating light intensities (Figure 6C).
Table 2. Chlorophyll Contents and Chlorophyll Fluorescence Parameters of Wild-Type (Col-0) and mtLPD Overexpression Plants.
| Parameter | Col-0 | PsL-L1 | PsL-L2 | PsL-L3 |
|---|---|---|---|---|
| Chlorophyll a | 1.46 ± 0.15 | 1.46 ± 0.18 | 1.69 ± 0.21 | 1.59 ± 0.24 |
| Chlorophyll b | 0.51 ± 0.03 | 0.51 ± 0.07 | 0.52 ± 0.07 | 0.50 ± 0.09 |
| Chlorophyll a and Chlorophyll b | 1.97 ± 0.15 | 1.97 ± 0.22 | 2.21 ± 0.23 | 2.09 ± 0.32 |
| Chlorophyll a/chlorophyll b | 2.91 ± 0.33 | 2.91 ± 0.49 | 3.27 ± 0.52 | 3.18 ± 0.35 |
| PSI | ||||
| rETRmax | 55.86 ± 5.75 | 52.11 ± 4.11 | 49.88 ± 3.92 | 50.91 ± 6.08 |
| Alpha | 0.38 ± 0.03 | 0.44 ± 0.03** | 0.43 ± 0.02** | 0.45 ± 0.03** |
| Lopt | 341.14 ± 22.94 | 293.66 ± 20.12** | 285.57 ± 17.76** | 299.17 ± 28.47* |
| PSII | ||||
| Fv/Fm | 0.77 ± 0.01 | 0.78 ± 0.01 | 0.76 ± 0.02 | 0.77 ± 0.02 |
| rETRmax | 41.23 ± 2.63 | 37.63 ± 2.78 | 36.71 ± 2.12 | 38.33 ± 4.77 |
| Alpha | 0.40 ± 0.02 | 0.42 ± 0.01 | 0.43 ± 0.01* | 0.44 ± 0.02* |
| Lopt | 246.12 ± 15.08 | 220.88 ± 14.87* | 218.42 ± 13.12** | 215.24 ± 19.64* |
| Lcp# | 11.96 ± 0.81 | 11.01 ± 1.56 | 9.48 ± 1.29* | 9.66 ± 0.91* |
Plants were grown under environmental controlled conditions (10/14-h day/night, 390 ppm CO2, 20/18°C day/night) to growth stage 5.1 (Boyes et al., 2001) and subsequently used for chlorophyll determination and chlorophyll fluorescence measurements. Values presented are means ± sd of measurements from at least five biological replicates per genotype (µmol*mg−1 fresh weight). Values in bold indicate values statistically significant from the wild type based on Student’s t test (*P < 0.05, **P < 0.01). Further chlorophyll fluorescence parameters (Y[I], Y[II], Y[NA], and Y[ND]) are given in Supplemental Data Set 1. Fv/Fm, maximum quantum yield of PSII (dark adapted); Lopt, optimal light intensity; Lcp, light compensation point (#determined by gas-exchange measurements); rETRmax, relative maximal electron transport rate.
Overexpression of mtLPD Has Only Minor Effects on Rubisco Activity
Enzymes involved in photosynthetic carbon assimilation (triosephosphate isomerase and phosphofructokinase) as well as Rubisco activity are inhibited by some photorespiratory intermediates at least in vitro (Anderson, 1971; Kelly and Latzko, 1976; Campbell and Ogren, 1990). Since our metabolite analyses point to an improved functioning of the photorespiratory pathway and bearing in mind the higher rates of photosynthesis observed, we next measured Rubisco protein amounts, activities, and activation states. Notably, we did not detect any significant alterations in the total amounts of Rubisco (Table 3) but found that a higher fraction of Rubisco (significant in PsL-L1 and PsL-L2) is activated in the mtLPD overexpressors.
Table 3. Rubisco Activity in mtLPD Overexpressors Compared to the Wild Type.
| Parameter | Col-0 | PsL-L1 | PsL-L2 | PsL-L3 |
|---|---|---|---|---|
| Total protein | 9.88 ± 0.47 | 10.64 ± 1.23 | 10.74 ± 0.60 | 10.01 ± 1.12 |
| Total Rubisco | 20.49 ± 0.64 | 20.79 ± 0.82 | 21.18 ± 0.77 | 20.63 ± 0.56 |
| Rubisco/protein | 47.40 ± 1.53 | 47.82 ± 1.88 | 48.84 ± 1.61 | 47.52 ± 1.24 |
| Rubisco (initial) | 143.63 ± 12.36 | 137.92 ± 19.87 | 150.11 ± 12.81 | 135.43 ± 13.13 |
| Rubisco (total) | 202.91 ± 23.39 | 175.37 ± 17.31 | 186.14 ± 23.61 | 176.96 ± 8.53 |
| Rubisco activation | 70.09 ± 3.26 | 78.40 ± 4.24 | 81.14 ± 6.61 | 76.61 ± 7.64 |
Leaves from plants at growth stage 5.1 (Boyes et al., 2001) were harvested in the middle of the light period (5 h illumination) as used for all other physiological experiments (10/14-h day/night, 390 ppm CO2, 20/18°C day/night, 120 µmol m−2 s−1 light intensity, and ∼70% relative humidity). Values presented are means ± sd of measurements from at least five biological and two technical replicates per genotype. Numbers in bold indicate values statistically significant from the wild type based on Student’s t test (*P < 0.05). Units are as follows: total protein (µg mg−1 fresh weight), total Rubisco (µg cm−2 leaf area), Rubisco/protein and Rubisco activation (%), and Rubisco activity (nmol min−1 cm−2 leaf area).
Elevated mtLPD Activity Strongly Reprograms Diurnal Starch Metabolism
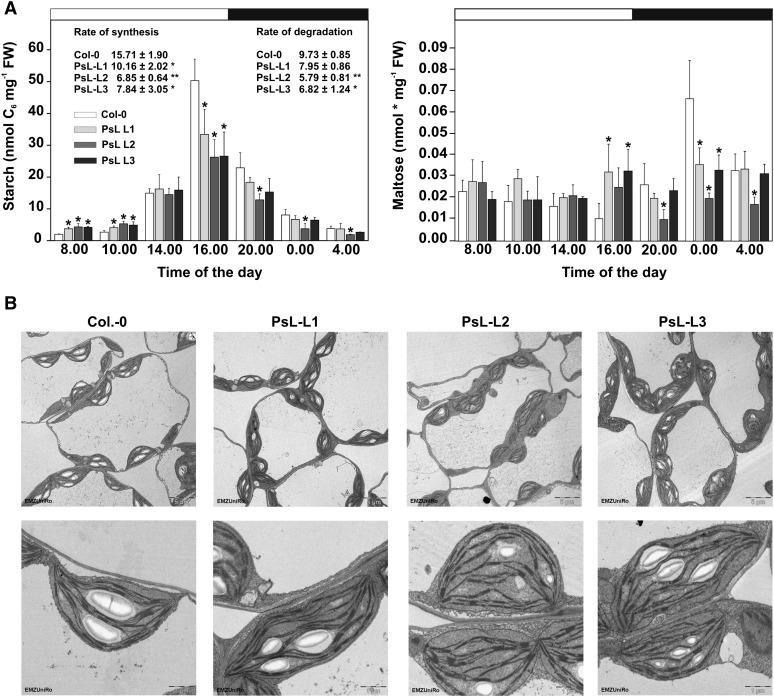
It could be anticipated that the increase in photosynthetic CO2 assimilation in mtLPD overexpressors leads to higher stocks of transitory starch accumulated at the end of the day. For that reason, we next quantified the levels of starch and its major breakdown product, maltose (Lu et al., 2005). Surprisingly, all three transgenic lines do not accumulate more starch than the wild type but rather contain reduced amounts (∼40 to 50%) at the end of the day (Figure 7A). This corresponds to higher amounts of maltose (∼3-fold) at this time point, indicating premature starch degradation in the light as also indicated by our 13C-enrichment studies. To further substantiate these findings, we visualized starch grana in chloroplasts of mtLPD overexpressor and wild-type plants by transmission electron microscopy. The starch grana, in close agreement with the quantitative data, were much smaller in all three transgenic lines compared with the wild type (Figure 7B). Taken together, it appears that the higher maltose contents observed in transgenic lines at the end of the light period are more likely due to the early degradation of transitory starch than to enhanced de novo biosynthesis of maltose in the light since this latter process is assumed to be of low flux capacity (Szecowka et al., 2013).
Figure 7.
Starch Metabolism and Visualization in mtLPD Lines.
(A) Leaf material was harvested from plants at growth stage 5.1 (Boyes et al., 2001) in a diurnal rhythm (4-h interval) and analyzed enzymatically for starch contents and by gas chromatography for maltose. Rates of starch biosynthesis or degradation are slopes of a linear regression of all daytime or nighttime values over the duration of the day and night, respectively. Given are means ± sd (n = 5), and asterisks indicate values significantly different from the wild type based on Student’s t test (*P < 0.05, **P < 0.01).
(B) Transmission electron micrographs of sections of leaves of 10-week-old wild-type and mtLPD1 overexpression plants. Representative sections are shown at magnifications of 1400× (upper panel) and 7100× (lower panel).
At the diurnal level, during the following dark phase, mtLPD overexpressors were able to break down stored starch like the wild type with the exception that the transgenic plants adapted their rates of starch breakdown to the reduced availability (Figure 7A). All three lines showed reduced rather than elevated contents of maltose, corresponding to the decreased breakdown rates. During the light period, all three overexpressors show higher levels of starch at the beginning of the day (2 to 4 h light) but wild-type-like amounts around the middle of the day, with no significant changes in their maltose contents. Accordingly, the rate of starch biosynthesis was also found to be reduced in mtLPD overexpressors compared with the wild type (Figure 7A). In conclusion, the metabolic reprogramming in mtLPD overexpression lines shifts the primary fixed carbon from amino acids and organic acids toward soluble sugar components rather than into transitory starch, as supported by the 13C labeling experiment and the absolute quantification of soluble sugars (Figure 3; Supplemental Figure 2).
DISCUSSION
The mitochondrial dihydrolipoyl dehydrogenase is an essential multitasking enzyme of mitochondrial metabolism. It is necessary for the functioning of the mtPDHC, ODHC, GDC, and BCDHC multienzyme complexes and thus is required for maintaining the metabolic fluxes through the TCA cycle, photorespiration, and branched-chain amino acid degradation. Here, we focused on the question whether and how increased mtLPD activity would affect the performance of these pathways. Particularly, we wanted to know whether such changes facilitate the photosynthetic-photorespiratory carbon flow and improve plant growth in a similar manner to that reported for plants overexpressing the GDC-H protein (Timm et al., 2012a).
Overexpression of mtLPD Mainly Alters the Pathway Fluxes through Photorespiration and the TCA Cycle with Minor Effects on the Degradation of Branched-Chain Amino Acids
The steady state metabolite measurements and 13C-enrichment studies indicate that overexpression of mtLPD in Arabidopsis dominantly enhances the metabolic capacity of photorespiration and alters the carbon flow through the TCA cycle, while branched-chain amino acid degradation appears to be much less affected (Figures 2 and 3; Supplemental Tables 1 and 2). Notably, the effects on the photorespiratory pathway do not require more of the other GDC proteins (Supplemental Figure 3), which is very similar to the observations made with plants that overexpress H-protein (Timm et al., 2012a). In other words, GDC activity in vivo can be elevated by a surplus of H-protein, alternatively, mtLPD. It is likely, but remains to be shown, that overexpression of P-protein or T-protein will produce corresponding results.
In contrast to the fragile GDC, mtPDHC and ODHC are much more stably built, and it is not very likely that overexpression of mtLPD alone would result in a higher enzymatic capacity of these two multienzyme complexes. We tested the possibility that overexpression of mtLPD could trigger enhanced biosynthesis of other subunits but did not find clear changes, at least for the E2 subunits (Supplemental Figure 3). Hence, the observed effects of mtLPD overexpression on the TCA cycle are likely more indirect as discussed further below in the respiration section.
Overexpression of mtLPD Strongly Shifts Pyridine Nucleotide Ratios but Has Minor Effects on the Overall Cellular Redox Homeostasis
Several studies of mutants impaired in the TCA cycle and photorespiration provided experimental evidence that the contents of pyridine nucleotides and their respective ratios are rather stable and do not undergo large changes in response to mutations within both pathways (Queval et al., 2007; van der Merwe et al., 2009; Araújo et al., 2012). However, despite reports on direct mutations of the TCA cycle and photorespiration, it was shown that impairments of the mitochondrial electron transport chain or associated components such as alternative mitochondrial NAD(P)H dehydrogenases result in considerable alterations in pyridine nucleotide contents (Liu et al., 2008; Wallström et al., 2014). It should be noted that these overall measurements do not necessarily reflect the subcellular levels of these metabolites; however, such measurements can be achieved only by highly challenging approaches (Kasimova et al., 2012). To circumvent this problem, the relative levels of metabolite pairs for compartment specific reactions are often analyzed (Queval et al., 2007; Vivancos et al., 2011; Araújo et al., 2012). Taking this indirect approach and in light of the above-mentioned reports, it seems likely that increases in the NAD+/NADH ratios, as found in mtLPD overexpressors, indicate that the NADH amounts mainly generated through the increased GDC and, to a lesser extent, TCA cycle activity are directly used by the mitochondrial electron transport chain to support ATP production, as reflected in the increased amounts of ATP found in mtLPD overexpression lines (Table 1). However, given the absence of accurate subcellular information at the mitochondrial level, we nevertheless strongly favor this hypothesis due to the physiological impact of the mtLPD manipulation, although the data presented here are obtained from total leaf extracts. Notwithstanding, increases observed in the absolute ATP contents could also be a consequence from photosynthetic light reactions since mtLPD overexpressors exhibit an improved light capture and light-use efficiency (Table 2) that eventually leads to higher production rates of energy equivalents. Aside from this, explanations for shifts of NADP+/NADPH ratios and its contents observed could at least be 2-fold. On one hand, reductions in the NADPH amounts could be a consequence of an increased turnover of the Calvin-Benson cycle due to the elevated rates of photosynthesis in mtLPD overexpressors as previously reported for GDC H-protein overexpressors (Timm et al., 2012a). On the other hand, increased activities of GDC and, in turn, higher rates of photorespiration (Figure 5) would also be anticipated to cause an increased release of ammonia in mitochondria. As a consequence, the higher demand of ammonia refixation for reducing equivalents in the chloroplast would eventually result in an increased drain of reduced ferredoxin into this pathway, competing with ferredoxin-NADP oxidoreductase (Linka and Weber, 2005). Nevertheless, shifts in the contents and ratios of both cofactors are in a physiologically tolerable range since we did not find major consequences on the levels of intermediates involved in balancing the cellular redox homeostasis (Table 1).
Differential Regulation of Mitochondrial Respiration during the Day-Night Cycle
Considering the fact that the basic pathways of respiration cover the functioning of glycolysis, the TCA cycle, and the oxidative phosphorylation, possibilities for regulation of respiration are multifaceted (Sweetlove and Fernie, 2008). Apart from the metabolic effects on the TCA cycle, we observed that the respiratory CO2 release differs during the day-night cycle in mtLPD overexpressors with no change in day respiration. Intuitively, these results might appear contradictory; however, there is not a prerequisite for a link between the rates of the TCA cycle and the rate of respiration, particularly given the fact that multiple flux modes are possible for the former. More precisely, there is strong evidence that the TCA cycle in leaves is used to produce carbon skeletons for biosynthesis of intermediates such as 2-oxoglutarate for amino acid biosynthesis rather than to completely oxidize pyruvate (Sweetlove et al., 2010). We also observed higher rates of respiration in the first half of the night and a decrease at the end of the night (Figure 5). These alterations indicate a higher respiratory consumption of primary stored photosynthates (Figures 3 and 7; Supplemental Figure 2) that accumulate during the day, whereas the decrease at the end of the night indicates decreased availability of these metabolites (Supplemental Figure 2).
In addition to substrate limitation, regulatory substrate sensing by a yet unknown mechanism could also result in lower mitochondrial respiratory activity. This hypothesis is supported by our data on glycine and malate respiration by isolated mitochondria that showed that both rates are decreased at the end of the night. Apparently, this effect cannot be explained simply by the reduced availability of carbohydrate resources for respiration but suggests that key enzymes of the respiratory process may be downregulated in the second half of the night to avoid carbon starvation. In good agreement with this hypothesis, we found that PDHC and ODHC activities in isolated mitochondria from dark adapted leaves are significantly reduced (Supplemental Figure 3) and therefore are likely targets for inactivation. Since both complexes do not show any significant alterations in their protein abundance (Supplemental Figure 3), we assume that the downregulation of the respective activities occurs on the posttranslational level or due to the action of a specific regulatory factor. The very recent demonstration that thioredoxin is a master regulator of the TCA cycle (Daloso et al., 2015) provides one possible mechanism by which this is achieved; however, it is worth noting that several other potential regulatory mechanisms have been proposed (Nunes-Nesi et al., 2013). Further experiments will be required to delineate the exact mechanism by which the mtLPD overexpressors regulate their night respiration.
Overexpression of mtLPD Enhances CO2 Fixation Due to Increased Light-Use Efficiency and Results in More Photorespiration, Increasing Net Photosynthesis
Photorespiration has often been classified as a process that reduces CO2 fixation and crop yields (Foyer et al., 2009; Maier et al., 2012; Peterhänsel et al., 2013). This view is changing and the repair function of photorespiration becomes more appreciated, for example, in light of new data on the evolution of photorespiration (Eisenhut et al., 2008; Hagemann et al., 2013) and field studies that demonstrated a positive correlation between photorespiration and yield for a number of highly productive wheat cultivars (Aliyev, 2012). Building upon our initial H-protein study (Timm et al., 2012a) and the data presented in this article, we envisage the following cause/effect relationship. Overexpression of L-protein in the mitochondria raises GDC activity and in turn improves the efficiency of photorespiratory carbon cycling (Figures 2 and 3). This allows increasing the performance of the Calvin-Benson cycle to enhance the regeneration of ribulose-1,5-bisphosphate (RuBP) by facilitation of the possibly existing negative feedback from photorespiratory metabolites and, in turn, photosynthetic carbon assimilation. Relaxation of this negative regulation likely involves increasing the efficiency of light capture and use rather than changes in Rubisco activity (Tables 2 and 3). Currently we assume that the higher light-use efficiency is due to a higher consumption on NADPH in the Calvin-Benson cycle and, therefore, an optimized acceptor supply of the photosynthetic electron transport chain. Subsequently this translates into faster photosynthesis and faster photorespiration, enhancing net CO2 fixation, growth, and biomass production (Figure 6; Supplemental Figure 1). This regulatory circuit seems to be rather specific and has no apparent negative side effects on metabolism as suggested by the unaltered day respiration and the minor effects on the relative chloroplast electron transport rates (Figure 5, Table 2). The fact that the highest effects on photosynthesis (Figure 6), the NAD(P)+-to-NAD(P)H ratio (Figure 4), and other diagnostic parameters were observed with overexpressor line PsL L2, which shows highest mtLPD activity (Figure 1), further substantiates this interpretation. The suggested scenario is compatible with other studies in which improved plant growth was achieved by the genetic engineering of glycolate metabolism (Kebeish et al., 2007; Maier et al., 2012) and with more recent modeling approaches (Xin et al., 2015).
Enhanced Growth Is Mediated by the Adjustment of Carbon Metabolism in mtLPD Overexpression Lines
From the onset of illumination, plants carry out photosynthesis that supports biosynthesis of sucrose and other photosynthates, which are thereafter exported to the remainder of the plant, thus facilitating metabolism, carbon storage, and plant growth. However, a considerable proportion of the generated photosynthates are alternatively stored as starch and remobilized during the following dark period to maintain plant metabolism during the night (Gibon et al., 2004; Bläsing et al., 2005). The rate of starch biosynthesis therefore needs to be highly regulated, a task that is coordinated by the balance of photosynthetic carbon assimilation and sucrose biosynthesis. It has been recently demonstrated that the availability, the turnover, and the proportion of export of triose phosphates (TPs) from the chloroplast exerts control over the amount of starch biosynthesized (Mugford et al., 2014). Since mtLPD overexpressors show enhanced rates of photosynthesis, we expected higher stocks of transitory starch to be accumulated at the end of the light period. Instead, we found starch decreased at this time point and moreover detected that both its rates of biosynthesis and degradation are reduced in the transgenic lines (Figure 7). This corresponds to our observation that mtLPD lines do show a higher redistribution of 13C to soluble sugars (Figure 3) and an increase in the total contents (Supplemental Figure 2), which indicates a higher demand for carbon to biosynthesize and export sucrose. Given that the supply of carbohydrates is crucial for high rates of growth (Graf and Smith, 2011; Ruts et al., 2012), it seems reasonable to assume that this reallocation of carbon toward sucrose biosynthesis results in the higher growth rates (Supplemental Figure 1). Collectively, we currently favor the explanation that the adjustment of carbon metabolism is due to (1) an enhanced TP utilization within the Calvin-Benson cycle to maintain the higher needs of RuBP and (2) a higher export of TP to the cytosol for sucrose biosynthesis. In combination, this would lead to a drop of TP and therefore reduce the amount of carbon used for starch biosynthesis. It is equally conceivable that the feed-forward model for sucrose biosynthesis (Stitt et al., 1984) explains, at least in part, the reprogramming of the rates of starch biosynthesis and degradation.
Conclusion
Overexpression of mtLPD improves photosynthesis and, in turn, biomass accumulation in Arabidopsis. Our present model to explain this effect envisages that higher GDC activity alleviates carbon flow through the photorespiratory pathway, lowering the accumulation of photorespiratory metabolites including those which impair Rubisco activation and possibly the activity of other enzymes of the Calvin-Benson cycle. Enhanced CO2 fixation goes along with increased efficiency of light capture and elevated photorespiration where the ratio of both processes is determined by the Rubisco’s specificity factor and the CO2-to-O2 ratio as is generally accepted. The detailed effects of higher GDC activity and particularly higher mtLPD activity on leaf carbohydrate metabolism and the operation of the TCA cycle remain to be studied.
METHODS
Materials and Plant Growth
Radiolabeled substrates were purchased from Isotec (13C-glycine) and Hartmann Analytic (NaH14CO3). We used Arabidopsis thaliana ecotype Col-0 as wild-type reference. Seeds were surface sterilized with hypochloric acid, sown on soil, and incubated at 4°C for at least 2 d to break dormancy. Except where mentioned, plants were grown side-by-side in controlled environment chambers (Percival; 10/14-h day/night cycle, 20/18°C, ∼120 µmol m−2 s−1 irradiance, 390 μL L−1 CO2, and 70% relative humidity) on a 4:1 mixture of soil (Type Mini Tray; Einheitserdewerk) and vermiculite and regularly watered with 0.2% Wuxal liquid fertilizer (Aglukon). If not specifically stated otherwise, we used plants at growth stage 5.1 according to Boyes et al. (2001) for the experiments.
cDNA Cloning and Expression
The entire coding sequence (1506 bp) of mtLPD was PCR amplified from pea (Pisum sativum) cDNA using oligonucleotides PsGDCL-SacI-S (5′-GAGCTCATGGCTATGGCGAACTTGGCT-3′) and PsGDCL-EcoRI-AS (5′-GAATTCTCAAATGTGAATGGGCTTGTC-3′). The resulting fragment (PsL) was ligated into vector pGEMT (Invitrogen) and sequenced for verification. The SacI-EcoRI fragment was excised and ligated in front of the CaMV poly(A) site of the pGreen 35S-CaMV cassette (http://www.pgreen.ac.uk/) to generate PsL:CaMV, which then was excised via BamHI and EcoRV and further introduced into the binary plant transformation vector pGREEN0229 containing the light-inducible ST-LSI promoter used previously (Timm et al., 2012b). The resulting construct (Figure 1 A) was introduced into Agrobacterium tumefaciens strain GV3101 and used for transformation (Clough and Bent, 1998) of Arabidopsis thaliana ecotype Col-0 (Arabidopsis). The resulting phosphinotricine (Basta)-resistant plants were preselected according their leaf mtLPD content by immunoblotting, and stable T4 lines from three independent transformation events (designated as PsL line 1, 2 and 3, respectively) were used for all physiological and biochemical analysis.
Validation of Transgenic Lines, RT-PCR, and Immunological Studies
To verify the genomic integration of the mtLPD overexpression construct, leaf DNA was PCR amplified (1 min at 94°C, 1 min at 58°C, and 1.5 min at 72°C; 35 cycles) with primers specific for the exogenous mtLPD fragment and the S16 gene (1 min at 94°C, 1 min at 58°C, 30 s at 72°C; 35 cycles) serving as a loading control. The functionality of the integrated overexpression construct was first verified by RT-PCR using 2.5 µg leaf RNA for cDNA synthesis (Nucleospin RNA plant kit [Macherey-Nagel] and RevertAid cDNA synthesis kit [MBI Fermentas]) and the oligonucleotide combination PsGDCL-SacI-S (sense) and PsGDCL-EcoRI-AS (antisense) mentioned above, yielding a 1506-bp PCR product for the full-length PsL transcript. Prior to PCR analysis, cDNA amounts were calibrated according to signals from 432-bp fragments of the constitutively expressed 40S ribosomal protein S16 gene, with oligonucleotides 40S-fw-RT (5′-GGCGACACAACCAGCTACTGA-3′) and 40S-rev-RT (5′-CGGTAACTCTTCTGGTAACGA-3′). Second, overexpression of the transgene was further verified by immunoblotting. Briefly, whole leaf, root, and mitochondrial protein extracts were separated by SDS-PAGE and gel blotting experiments performed according to standard protocols. Overexpression was detected using specific antibodies for mtLPD (Timm et al., 2013) and mMDH (Gietl et al., 1996) serving as a calibration control.
Isolation of Mitochondria, Respiratory Rates, and mtLPD Activity
Mitochondria were isolated from leaves of wild-type and transgenic plants according to Keech et al. (2005). Respiratory rates of intact mitochondria with glycine and malate as substrates were determined exactly as described by Ewald et al. (2007). To detect total mtLPD activity in mitochondrial extracts, we followed the protocol described by Yan et al. (2008) using the spectrophotometric enzyme test. Briefly, the reverse reaction of mtLPD was measured spectrophotometrically as the reduction of lipoamide at the expense of NADH. The final reaction volume was 1 mL containing 100 mM potassium phosphate (pH 6.3), 1.5 mM EDTA, 0.6 mg/mL BSA, 0.6 mM lipoamide, and 0.2 mM NADH and the reaction initiated by adding 10 to 20 µg mL−1 mitochondrial protein. The mtLPD activity was assayed at 22°C as change in absorbance at 340 nm.
Determination of Metabolite Levels
For GC-MS and HPLC analysis, samples were taken at the end of the day (9 h light). Leaf material was harvested from fully expanded rosette leaves using at least five biological replicates (growth stage 5.1 according to Boyes et al. [2001]), immediately frozen in liquid nitrogen, and stored at −80°C until analysis. For gas chromatography analysis of organic acids and soluble sugars (∼100 mg leaf tissue), we followed the method described by Sievers et al. (2013). Absolute amino acid contents were determined from ∼100 mg leaf material by HPCL as described previously (Hagemann et al., 2005). NAD(H) and NADP(H) contents were determined from 25 mg leaf tissue (end of the day, 9 h light) as described by Schippers et al. (2008). Derivatization, separation, and quantification of thiols, O-acetyl serine, and adenosine triphosphates were performed from 100 mg leaf tissue as described by Heeg et al. (2008).
13C-Glycine Feeding and Determination of Isotope Accumulation
For this purpose, we used plants that were grown under standard conditions mentioned above to growth stage 5.1 (Boyes et al., 2001). Leaf discs were generated from fully expanded rosette leaves after 3 h of illumination during a normal day/night cycle and incubated in a 10 mM MES-KOH solution (pH 6.5), containing either 10 mM [U-12C]-glycine or [U-13C]-glycine for 3 h (in principle 6 h after onset of illumination) under plant growth conditions (20°C, ∼120 µmol m−2 s−1, 390 ppm CO2 and 21% O2, and 70% relative humidity). Thereafter leaf discs were harvested, washed thrice with 10 mM MES-KOH solution (pH 6.5), and subsequently frozen in liquid nitrogen until analysis. Metabolite extraction and analysis were performed as described by Lisec et al. (2006) and the molecular accumulation of isotope determined following the method specified by Sienkiewicz-Porzucek et al. (2008).
Gas-Exchange Measurements
Standard leaf gas exchange was performed on fully expanded rosette leaves of plants at growth stage 5.1 (Boyes et al., 2001) as described previously (Timm et al., 2011) with the following conditions: photon flux density = 1000 µmol m−2 s−1, chamber temperature = 25°C, flow rate = 300 µmol s−1, and relative humidity = 60 to 70%. All photosynthetic parameters were determined in a 6-h time period between 2 h after onset and 2 h prior offset of illumination. Inhibition of CO2 assimilation rates (A) was calculated from measurements at 21 and 40% oxygen concentration as follows: O2 inhibition = (A21 − A40)/A21*100. Calculation of γ was performed by linear regression of the Γ-versus-oxygen concentration curves and given as slopes of the respective functions. Rates of night respiration (Rn) were determined with plants adapted to darkness for 2 to 4 h and 12 to 14 h during a normal day/night cycle. CO2 compensation points in the absence of dark respiration (Γ*) and day respiration (Rd) were determined as described by Häusler et al. (1999). Rates of photorespiration were calculated according to the model proposed by Sharkey (1988): photorespiration = 0.5 {(A+Rn)/[(Ci/2Γ*) − 0.5]), in which Ci stands for the intercellular CO2 concentration. Prior to each measurement plants were adapted to the respective condition for at least 15 min.
Chlorophyll and Chlorophyll Fluorescence Measurements
Leaf chlorophyll contents from 100 mg of leaf tissue were determined following the protocol from Porra (2002) and Porra et al., (1989). For the determination of in vivo chlorophyll fluorescence, we used a PAM fluorometer (DUAL-PAM-100; Walz) and dark-adapted plants (30 min) at growth stage 5.1 (Boyes et al., 2001) as for all other experiments. Maximum quantum yields (Y) and relative electron transport rates of PSI and PSII were measured at varying PPFDs and calculated using the WinControl software package (Walz). The respective data curves obtained were further used to calculate the light use efficiency (α), the optimal light intensity (Lopt), and the light compensation point (Lcp) following the model proposed by Eilers and Peeters (1988).
Rubisco Quantification and Carboxylase Activity Measurements
Total soluble leaf protein was extracted (50 mM Bicine/KOH, pH 8.0, 20 mM MgCl2, 50 mM β-mercaptoethanol, and 2 mM phenylmethylsulfonyl fluoride) from fully expanded rosette leaves of plants at growth stage 5.1 (Boyes et al., 2001) and quantified according to Bradford (1976). The total amount of Rubisco was determined by immunoblotting experiments using purified Rubisco (Andrews et al., 1973; Harris and Stern, 1977) from pea as standard. Carboxylase activity was analyzed according to the method described by Parry et al. (1997). Briefly, enzyme activity was determined in buffer (100 mM Bicine/KOH, pH 8.2, 20 mM MgCl2, 33 mM NaH14CO3, and 0.66 mM RuBP) at 25°C, using 20 µg total leaf protein. Initial activity was measured immediately after adding protein extract to the assay buffer. Total activity was determined after in vitro activation (5 min at 25°C) in the absence of RuBP and subsequently initiated by addition of RuBP. The reaction was quenched after 20, 40, and 60 s by adding 100 μL 10 M formic acid. Samples than were oven dried, the respective residues rehydrated (400 μL water), and the acid stable 14C determined by liquid scintillation counting after addition of 5 mL liquid scintillation cocktail (Ultima Gold; Perkin-Elmer). The Rubisco activation state was further calculated as relative ratio of initial to total activity (Perchorowicz et al., 1981).
Starch Content
Starch contents were determined from 20 mg leaf tissue using plants at growth stage 5.1 (Boyes et al., 2001) and spectrophotometrically analyzed using enzymatic assays in ethanolic extracts described by Cross et al. (2006). Rates of starch biosynthesis and degradation were determined as slopes from linear regression of all daytime and nighttime values, respectively, over the duration of one day as described by Lu et al. (2005).
Transmission Electron Microscopy
Leaf tissues were fixed and stored at 4°C in a mixture of 2% glutaraldehyde and 1% paraformaldehyde in 0.1 M sodium phosphate buffer, pH 7.3. After washes in 0.1 M sodium phosphate buffer (pH 7.3), processing for transmission electron microscopy continued with postfixation in an aqueous solution of 1% osmium tetroxide for 1 h, followed by washes in distilled water. Leaf tissues were dehydrated through an increasing acetone series to 100% acetone, followed by infiltration with a 1:1 mixture of acetone and epoxy resin (Epon 812; Serva) overnight. Specimens were infiltrated with pure resin for 4 h, transferred to rubber molds, and cured at 60°C in an oven for 2 d.
Areas of interest were exposed from the embedded leaves with a mill (Leica EM Trim 2; Leica Microsystems) prior to microtome cutting (Ultracut S; Reichert) with a diamond knife (Diatome). Semithin sections (∼0.5 µm) were stained with Toluidine blue to select appropriate areas for thin sectioning and ultrastructural examination. Thin sections (∼50 to 70 nm) were transferred to copper grids, stained with uranyl acetate and lead citrate, and examined on a Zeiss EM902 electron microscope operated at 80 kV (Carl Zeiss). Digital images of leaf areas and chloroplasts were acquired with a side-mounted 1x2k FT-CCD camera (Proscan) using iTEM camera control and imaging software (Olympus Soft Imaging Solutions).
Statistical Analysis
Statistical tests were performed using the two-tailed Student’s t test (Microsoft Excel 10.0) and by ANOVA for multiple genotypes using the Holm and Sidak test for comparisons (Sigma Plot 11; Systat Software). The term significant is used here only if the change in question has been confirmed to be significant at the level of *P < 0.05 or **P < 0.01.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: X63464 (PsL), At4g33010 (GDCP1), At2g26080 (GDCP2), At2g35370 (GDCH1), At2g35120 (GDCH2), At1g32470 (GDCH3), At1g11860 (GDCT), At3g17240 (GDCL1), At1g48030 (GDCL2), At4g37930 (SHMT1), At3g52200 (PDHCE2-1), At5g55070 (ODHCE2) At3g06850 (BCDHCE2), At1g5320 (mMDH1), and At2g09990 (40S ribosomal protein S16).
Supplemental Data
Supplemental Figure 1. Visual phenotype and biomass accumulation of mtLPD overexpressors.
Supplemental Figure 2. Absolute sugar contents in mtLPD overexpressors.
Supplemental Figure 3. Enzyme abundances and activities in mtLPD overexpressors at different times of the day/night cycle.
Supplemental Table 1. Absolute amino acid and organic acid contents in mtLPD overexpressors.
Supplemental Table 2. 13C enrichment in selected metabolites of primary metabolism.
Supplemental Data Set 1. Chlorophyll fluorescence parameters of wild-type and mtLPD overexpression plants.
Supplementary Material
Acknowledgments
We gratefully acknowledge the excellent technical assistance from Kathrin Jahnke, Klaudia Michl, Manja Henneberg, Ute Schulz (Rostock), and Ina Krahnert (Potsdam-Golm). We thank the Metabolomics Core Technology Platform of the Excellence cluster “CellNetworks” (University of Heidelberg) for support with liquid chromatography-based metabolite quantification. We also thank Martin Hagemann (Rostock) for critical discussions and Julia Walter (Rostock) for initial contributions to this project. We thank Hendrik Schubert and Christian Porsche (Rostock) for introduction into the use of the Dual-PAM-100 and Alfred J. Keys (Harpenden) for providing protocols for Rubisco assays. This work was funded by the Deutsche Forschungsgemeinschaft through the Forschergruppe FOR 1186 (Promics).
AUTHOR CONTRIBUTIONS
S.T., H.B., and A.R.F. designed the research. S.T., M. Wittmiß, S.G., R.E., A.F., M. Wirtz, and M.F. performed the research. S.T., M. Wittmiß, S.G., M. Wirtz, A.F., H.B., and M.F. analyzed the data. S.T., H.B., A.R.F., and R.H. wrote the article.
Glossary
- mtLPD
mitochondrial dihydrolipoyl dehydrogenase
- mtPDHC
mitochondrial pyruvate dehydrogenase complex
- TCA
tricarboxylic acid
- ODHC
2-oxoglutarate dehydrogenase
- BCDHC
branched-chain 2-oxoacid dehydrogenase complex
- GC-MS
gas chromatography-mass spectrometry
- GDC
glycine decarboxylase complex
- PSII
photosystem II
- PSI
photosystem I
- RuBP
ribulose-1,5-bisphosphate
- TP
triose phosphate
References
- Aliyev J.A. (2012). Photosynthesis, photorespiration and productivity of wheat and soybean genotypes. Physiol. Plant. 145: 369–383. [DOI] [PubMed] [Google Scholar]
- Anderson L.E. (1971). Chloroplast and cytoplasmic enzymes. II. Pea leaf triose phosphate isomerases. Biochim. Biophys. Acta 235: 237–244. [DOI] [PubMed] [Google Scholar]
- Andrews T.J., Lorimer G.H., Tolbert N.E. (1973). Ribulose diphosphate oxygenase. I. Synthesis of phosphoglycolate by fraction-1 protein of leaves. Biochemistry 12: 11–18. [DOI] [PubMed] [Google Scholar]
- Araújo W.L., Ishizaki K., Nunes-Nesi A., Larson T.R., Tohge T., Krahnert I., Witt S., Obata T., Schauer N., Graham I.A., Leaver C.J., Fernie A.R. (2010). Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W.L., Tohge T., Osorio S., Lohse M., Balbo I., Krahnert I., Sienkiewicz-Porzucek A., Usadel B., Nunes-Nesi A., Fernie A.R. (2012). Antisense inhibition of the 2-oxoglutarate dehydrogenase complex in tomato demonstrates its importance for plant respiration and during leaf senescence and fruit maturation. Plant Cell 24: 2328–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bläsing O.E., Gibon Y., Günther M., Höhne M., Morcuende R., Osuna D., Thimm O., Usadel B., Scheible W.R., Stitt M. (2005). Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon J., Macherel D., Neuburger M., Douce R. (1992). Isolation, characterization, and sequence analysis of a cDNA clone encoding L-protein, the dihydrolipoamide dehydrogenase component of the glycine cleavage system from pea-leaf mitochondria. Eur. J. Biochem. 204: 865–873. [DOI] [PubMed] [Google Scholar]
- Boyes D.C., Zayed A.M., Ascenzi R., McCaskill A.J., Hoffman N.E., Davis K.R., Görlach J. (2001). Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Bunik V.I., Fernie A.R. (2009). Metabolic control exerted by the 2-oxoglutarate dehydrogenase reaction: a cross-kingdom comparison of the crossroad between energy production and nitrogen assimilation. Biochem. J. 422: 405–421. [DOI] [PubMed] [Google Scholar]
- Campbell W.J., Ogren W.L. (1990). Glyoxylate inhibition of ribulosebisphosphate carboxylase/oxygenase activation in intact, lysed, and reconstituted chloroplasts. Photosynth. Res. 23: 257–268. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cousins A.B., Walker B.J., Pracharoenwattana I., Smith S.M., Badger M.R. (2011). Peroxisomal hydroxypyruvate reductase is not essential for photorespiration in Arabidopsis but its absence causes an increase in the stoichiometry of photorespiratory CO2 release. Photosynth. Res. 108: 91–100. [DOI] [PubMed] [Google Scholar]
- Cross J.M., von Korff M., Altmann T., Bartzetko L., Sulpice R., Gibon Y., Palacios N., Stitt M. (2006). Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions. Plant Physiol. 142: 1574–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daloso D.M., et al. (2015). Thioredoxin, a master regulator of the tricarboxylic acid cycle in plant mitochondria. Proc. Natl. Acad. Sci. USA 112: E1392–E1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce R., Bourguignon J., Neuburger M., Rébeillé F. (2001). The glycine decarboxylase system: a fascinating complex. Trends Plant Sci. 6: 167–176. [DOI] [PubMed] [Google Scholar]
- Eilers P., Peeters J. (1988). A model for the relationship between light intensity and the rate of photosynthesis in phytoplankton. Ecol. Modell. 42: 199–215. [Google Scholar]
- Eisenhut M., Ruth W., Haimovich M., Bauwe H., Kaplan A., Hagemann M. (2008). The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc. Natl. Acad. Sci. USA 105: 17199–17204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M., et al. (2013). Arabidopsis A BOUT DE SOUFFLE is a putative mitochondrial transporter involved in photorespiratory metabolism and is required for meristem growth at ambient CO2 levels. Plant J. 73: 836–849. [DOI] [PubMed] [Google Scholar]
- Engel N., van den Daele K., Kolukisaoglu U., Morgenthal K., Weckwerth W., Pärnik T., Keerberg O., Bauwe H. (2007). Deletion of glycine decarboxylase in Arabidopsis is lethal under nonphotorespiratory conditions. Plant Physiol. 144: 1328–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald R., Kolukisaoglu U., Bauwe U., Mikkat S., Bauwe H. (2007). Mitochondrial protein lipoylation does not exclusively depend on the mtKAS pathway of de novo fatty acid synthesis in Arabidopsis. Plant Physiol. 145: 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C.H., Bloom A.J., Queval G., Noctor G. (2009). Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annu. Rev. Plant Biol. 60: 455–484. [DOI] [PubMed] [Google Scholar]
- Gibon Y., Bläsing O.E., Palacios-Rojas N., Pankovic D., Hendriks J.H.M., Fisahn J., Höhne M., Günther M., Stitt M. (2004). Adjustment of diurnal starch turnover to short days: depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J. 39: 847–862. [DOI] [PubMed] [Google Scholar]
- Gietl C., Seidel C., Svendsen I. (1996). Plant glyoxysomal but not mitochondrial malate dehydrogenase can fold without chaperone assistance. Biochim. Biophys. Acta 1274: 48–58. [DOI] [PubMed] [Google Scholar]
- Graf A., Smith A.M. (2011). Starch and the clock: the dark side of plant productivity. Trends Plant Sci. 16: 169–175. [DOI] [PubMed] [Google Scholar]
- Hagemann M., Fernie A.R., Espie G.S., Kern R., Eisenhut M., Reumann S., Bauwe H., Weber A.P.M. (2013). Evolution of the biochemistry of the photorespiratory C2 cycle. Plant Biol (Stuttg) 15: 639–647. [DOI] [PubMed] [Google Scholar]
- Hagemann M., Vinnemeier J., Oberpichler I., Boldt R., Bauwe H. (2005). The glycine decarboxylase complex is not essential for the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Biol (Stuttg) 7: 15–22. [DOI] [PubMed] [Google Scholar]
- Harris G.C., Stern A.I. (1977). Isolation and some properties of ribulose-1,5-bisphosphate carboxylase-oxygenase from red kidney bean primary leaves. Plant Physiol. 60: 697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häusler R.E., Kleines M., Uhrig H., Hirsch H.J., Smets H. (1999). Overexpression of phosphoenolpyruvate carboxylase from Corynebacterium glutamicum lowers the CO2 compensation point (Γ*) and enhances dark and light respiration in transgenic potato. J. Exp. Bot. 336: 1231–1242. [Google Scholar]
- Heeg C., Kruse C., Jost R., Gutensohn M., Ruppert T., Wirtz M., Hell R. (2008). Analysis of the Arabidopsis O-acetylserine(thiol)lyase gene family demonstrates compartment-specific differences in the regulation of cysteine synthesis. Plant Cell 20: 168–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineke D., Bykova N., Gardeström P., Bauwe H. (2001). Metabolic response of potato plants to an antisense reduction of the P-protein of glycine decarboxylase. Planta 212: 880–887. [DOI] [PubMed] [Google Scholar]
- Kasimova M.R., Grigiene J., Krab K., Hagedorn P.H., Flyvbjerg H., Andersen P.E., Møller I.M. (2006). The free NADH concentration is kept constant in plant mitochondria under different metabolic conditions. Plant Cell 18: 688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebeish R., Niessen M., Thiruveedhi K., Bari R., Hirsch H.J., Rosenkranz R., Stäbler N., Schönfeld B., Kreuzaler F., Peterhänsel C. (2007). Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nat. Biotechnol. 25: 593–599. [DOI] [PubMed] [Google Scholar]
- Keech O., Dizengremel P., Gardeström P. (2005). Preparation of leaf mitochondria from Arabidopsis thaliana. Physiol. Plant. 124: 403–409. [Google Scholar]
- Kelly G.J., Latzko E. (1976). Inhibition of spinach-leaf phosphofructokinase by 2-phosphoglycollate. FEBS Lett. 68: 55–58. [DOI] [PubMed] [Google Scholar]
- Linka M., Weber A.P. (2005). Shuffling ammonia between mitochondria and plastids during photorespiration. Trends Plant Sci. 10: 461–465. [DOI] [PubMed] [Google Scholar]
- Lisec J., Schauer N., Kopka J., Willmitzer L., Fernie A.R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 1: 387–396. [DOI] [PubMed] [Google Scholar]
- Liu Y.J., Norberg F.E.B., Szilágyi A., De Paepe R., Åkerlund H.E., Rasmusson A.G. (2008). The mitochondrial external NADPH dehydrogenase modulates the leaf NADPH/NADP+ ratio in transgenic Nicotiana sylvestris. Plant Cell Physiol. 49: 251–263. [DOI] [PubMed] [Google Scholar]
- Lu Y., Gehan J.P., Sharkey T.D. (2005). Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol. 138: 2280–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutziger I., Oliver D.J. (2000). Molecular evidence of a unique lipoamide dehydrogenase in plastids: analysis of plastidic lipoamide dehydrogenase from Arabidopsis thaliana. FEBS Lett. 484: 12–16. [DOI] [PubMed] [Google Scholar]
- Lutziger I., Oliver D.J. (2001). Characterization of two cDNAs encoding mitochondrial lipoamide dehydrogenase from Arabidopsis. Plant Physiol. 127: 615–623. [PMC free article] [PubMed] [Google Scholar]
- Maier A., Fahnenstich H., von Caemmerer S., Engqvist M.K., Weber A.P., Flügge U.I., Maurino V.G. (2012). Transgenic introduction of a glycolate oxidative cycle into A. thaliana chloroplasts leads to growth improvement. Front. Plant Sci. 3: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrott N.L., Marshall J.J., Svergun D.I., Crennell S.J., Hough D.W., van den Elsen J.M., Danson M.J. (2014). Why are the 2-oxoacid dehydrogenase complexes so large? Generation of an active trimeric complex. Biochem. J. 463: 405–412. [DOI] [PubMed] [Google Scholar]
- Millar A.H., Hill S.A., Leaver C.J. (1999). Plant mitochondrial 2-oxoglutarate dehydrogenase complex: purification and characterization in potato. Biochem. J. 343: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.H., Knorpp C., Leaver C.J., Hill S.A. (1998). Plant mitochondrial pyruvate dehydrogenase complex: purification and identification of catalytic components in potato. Biochem. J. 334: 571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney B.P., Miernyk J.A., Randall D.D. (2002). The complex fate of alpha-ketoacids. Annu. Rev. Plant Biol. 53: 357–375. [DOI] [PubMed] [Google Scholar]
- Mugford S.T., Fernandez O., Brinton J., Flis A., Krohn N., Encke B., Feil R., Sulpice R., Lunn J.E., Stitt M., Smith A.M. (2014). Regulatory properties of ADP glucose pyrophosphorylase are required for adjustment of leaf starch synthesis in different photoperiods. Plant Physiol. 166: 1733–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A., Araújo W.L., Obata T., Fernie A.R. (2013). Regulation of the mitochondrial tricarboxylic acid cycle. Curr. Opin. Plant Biol. 16: 335–343. [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A., Carrari F., Lytovchenko A., Smith A.M., Loureiro M.E., Ratcliffe R.G., Sweetlove L.J., Fernie A.R. (2005). Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol. 137: 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A., Sweetlove L.J., Fernie A.R. (2007). Operation and function of the tricarboxylic acid cycle in the illuminated leaf. Physiol. Plant. 129: 45–56. [Google Scholar]
- Oliver D.J., Neuburger M., Bourguignon J., Douce R. (1990). Interaction between the component enzymes of the glycine decarboxylase multienzyme complex. Plant Physiol. 94: 833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry M.A.J., Andralojc P.J., Parmar S., Keys A.J., Habash D., Paul M.J., Alred R., Quick W.P., Servaites J.C. (1997). Regulation of Rubisco by inhibitors in the light. Plant Cell Environ. 20: 528–534. [Google Scholar]
- Perchorowicz J.T., Raynes D.A., Jensen R.G. (1981). Light limitation of photosynthesis and activation of ribulose bisphosphate carboxylase in wheat seedlings. Proc. Natl. Acad. Sci. USA 78: 2985–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterhänsel C., Krause K., Braun H.P., Espie G.S., Fernie A.R., Hanson D.T., Keech O., Maurino V.G., Mielewczik M., Sage R.F. (2013). Engineering photorespiration: current state and future possibilities. Plant Biol (Stuttg) 15: 754–758. [DOI] [PubMed] [Google Scholar]
- Porra R.J. (2002). The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 73: 149–156. [DOI] [PubMed] [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophyll a and chlorophyll b extracted with 4 different solvents - Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394. [Google Scholar]
- Queval G., Issakidis-Bourguet E., Hoeberichts F.A., Vandorpe M., Gakière B., Vanacker H., Miginiac-Maslow M., Van Breusegem F., Noctor G. (2007). Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 52: 640–657. [DOI] [PubMed] [Google Scholar]
- Raghavendra A.S., Padmasree K. (2003). Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci. 8: 546–553. [DOI] [PubMed] [Google Scholar]
- Reed L.J. (2001). A trail of research from lipoic acid to alpha-keto acid dehydrogenase complexes. J. Biol. Chem. 276: 38329–38336. [DOI] [PubMed] [Google Scholar]
- Ruts T., Matsubara S., Wiese-Klinkenberg A., Walter A. (2012). Diel patterns of leaf and root growth: endogenous rhythmicity or environmental response? J. Exp. Bot. 63: 3339–3351. [DOI] [PubMed] [Google Scholar]
- Schippers J.H.M., Nunes-Nesi A., Apetrei R., Hille J., Fernie A.R., Dijkwel P.P. (2008). The Arabidopsis onset of leaf death5 mutation of quinolinate synthase affects nicotinamide adenine dinucleotide biosynthesis and causes early ageing. Plant Cell 20: 2909–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey T.D. (1988). Estimating the rate of photorespiration in leaves. Physiol. Plant. 73: 147–152. [Google Scholar]
- Sienkiewicz-Porzucek A., Nunes-Nesi A., Sulpice R., Lisec J., Centeno D.C., Carillo P., Leisse A., Urbanczyk-Wochniak E., Fernie A.R. (2008). Mild reductions in mitochondrial citrate synthase activity result in a compromised nitrate assimilation and reduced leaf pigmentation but have no effect on photosynthetic performance or growth. Plant Physiol. 147: 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers N., Muders K., Henneberg M., Klähn S., Effmert M., Junghans H., Hagemann M. (2013). Establishing glucosylglycerol synthesis in potato (Solanum tuberosum L. cv. albatros) by expression of the ggpPS gene from Azotobacter vinelandii. J. Plant Sci. Mol. Bree. 2: 1. [Google Scholar]
- Stitt M., Kürzel B., Heldt H.W. (1984). Control of photosynthetic sucrose synthesis by fructose 2,6-bisphosphate. I. Coordination of CO2 fixation and sucrose synthesis. Plant Physiol. 75: 554–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockhaus J., Schell J., Willmitzer L. (1989). Correlation of the expression of the nuclear photosynthetic gene ST-LS1 with the presence of chloroplasts. EMBO J. 8: 2445–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove L.J.,. and Fernie A.R. (2008). Plant Respiration. In Encyclopedia of Life Science. (Chichester, UK: John Wiley & Sons; ), doi/10.1002/9780470015902.a0001301.pub2. [Google Scholar]
- Sweetlove L.J., Beard K.F.M., Nunes-Nesi A., Fernie A.R., Ratcliffe R.G. (2010). Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci. 15: 462–470. [DOI] [PubMed] [Google Scholar]
- Szecowka M., Heise R., Tohge T., Nunes-Nesi A., Vosloh D., Huege J., Feil R., Lunn J., Nikoloski Z., Stitt M., Fernie A.R., Arrivault S. (2013). Metabolic fluxes in an illuminated Arabidopsis rosette. Plant Cell 25: 694–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemiya A., Inoue S., Doi M., Kinoshita T., Shimazaki K. (2005). Phototropins promote plant growth in response to blue light in low light environments. Plant Cell 17: 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N.L., Heazlewood J.L., Day D.A., Millar A.H. (2004). Lipoic acid-dependent oxidative catabolism of α-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiol. 134: 838–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T., Ramos M.S., Nunes-Nesi A., Mutwil M., Giavalisco P., Steinhauser D., Schellenberg M., Willmitzer L., Persson S., Martinoia E., Fernie A.R. (2011). Toward the storage metabolome: profiling the barley vacuole. Plant Physiol. 157: 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm S., Nunes-Nesi A., Pärnik T., Morgenthal K., Wienkoop S., Keerberg O., Weckwerth W., Kleczkowski L.A., Fernie A.R., Bauwe H. (2008). A cytosolic pathway for the conversion of hydroxypyruvate to glycerate during photorespiration in Arabidopsis. Plant Cell 20: 2848–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm S., Florian A., Jahnke K., Nunes-Nesi A., Fernie A.R., Bauwe H. (2011). The hydroxypyruvate-reducing system in Arabidopsis: multiple enzymes for the same end. Plant Physiol. 155: 694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm S., Florian A., Arrivault S., Stitt M., Fernie A.R., Bauwe H. (2012a). Glycine decarboxylase controls photosynthesis and plant growth. FEBS Lett. 586: 3692–3697. [DOI] [PubMed] [Google Scholar]
- Timm S., Florian A., Wittmiß M., Jahnke K., Hagemann M., Fernie A.R., Bauwe H. (2013). Serine acts as a metabolic signal for the transcriptional control of photorespiration-related genes in Arabidopsis. Plant Phys. 162: 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm S., Mielewczik M., Florian A., Frankenbach S., Dreissen A., Hocken N., Fernie A.R., Walter A., Bauwe H. (2012b). High-to-low CO2 acclimation reveals plasticity of the photorespiratory pathway and indicates regulatory links to cellular metabolism of Arabidopsis. PLoS ONE 7: e42809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe M.J., Osorio S., Moritz T., Nunes-Nesi A., Fernie A.R. (2009). Decreased mitochondrial activities of malate dehydrogenase and fumarase in tomato lead to altered root growth and architecture via diverse mechanisms. Plant Physiol. 149: 653–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivancos P.D., Driscoll S.P., Bulman C.A., Ying L., Emami K., Treumann A., Mauve C., Noctor G., Foyer C.H. (2011). Perturbations of amino acid metabolism associated with glyphosate-dependent inhibition of shikimic acid metabolism affect cellular redox homeostasis and alter the abundance of proteins involved in photosynthesis and photorespiration. Plant Physiol. 157: 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallström S.V., Florez-Sarasa I., Araújo W.L., Escobar M.A., Geisler D.A., Aidemark M., Lager I., Fernie A.R., Ribas-Carbó M., Rasmusson A.G. (2014). Suppression of NDA-type alternative mitochondrial NAD(P)H dehydrogenases in arabidopsis thaliana modifies growth and metabolism, but not high light stimulation of mitochondrial electron transport. Plant Cell Physiol. 55: 881–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin C.P., Tholen D., Devloo V., Zhu X.G. (2015). The benefits of photorespiratory bypasses: how can they work? Plant Physiol. 167: 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L.J., Thangthaeng N., Forster M.J. (2008). Changes in dihydrolipoamide dehydrogenase expression and activity during postnatal development and aging in the rat brain. Mech. Ageing Dev. 129: 282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.H., McCarthy D.B., O’Connor C.M., Reed L.J., Stoops J.K. (2001). The remarkable structural and functional organization of the eukaryotic pyruvate dehydrogenase complexes. Proc. Natl. Acad. Sci. USA 98: 14802–14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.