Elucidation of the genetic determinants of maize primary metabolism and a metabolite-metabolite-agronomic trait network will promote efficient use of metabolites in maize improvement.
Abstract
Deciphering the influence of genetics on primary metabolism in plants will provide insights useful for genetic improvement and enhance our fundamental understanding of plant growth and development. Although maize (Zea mays) is a major crop for food and feed worldwide, the genetic architecture of its primary metabolism is largely unknown. Here, we use high-density linkage mapping to dissect large-scale metabolic traits measured in three different tissues (leaf at seedling stage, leaf at reproductive stage, and kernel at 15 d after pollination [DAP]) of a maize recombinant inbred line population. We identify 297 quantitative trait loci (QTLs) with moderate (86.2% of the mapped QTL, R2 = 2.4 to 15%) to major effects (13.8% of the mapped QTL, R2 >15%) for 79 primary metabolites across three tissues. Pairwise epistatic interactions between these identified loci are detected for more than 25.9% metabolites explaining 6.6% of the phenotypic variance on average (ranging between 1.7 and 16.6%), which implies that epistasis may play an important role for some metabolites. Key candidate genes are highlighted and mapped to carbohydrate metabolism, the tricarboxylic acid cycle, and several important amino acid biosynthetic and catabolic pathways, with two of them being further validated using candidate gene association and expression profiling analysis. Our results reveal a metabolite-metabolite-agronomic trait network that, together with the genetic determinants of maize primary metabolism identified herein, promotes efficient utilization of metabolites in maize improvement.
INTRODUCTION
Maize (Zea mays) is one of the world’s most important crops and has the highest total tonnage of any grain crop worldwide. Maize yield has dramatically increased in the last 100 years, in large part due to harnessing natural genetic variation to breed for improvements in desired traits (Haley, 2011). Maize has long been a focus of intensive breeding programs to improve quality traits due to its agricultural and economic value. Protein and oil contents in maize grain have been chief targets for improvement using breeding technologies (Moose et al., 2004). Breeding for not only high yield but also better quality maize is currently drawing more interest and is imperative to meet the increasing nutritional demand of the global population. Recently, progress has been made in the analysis of metabolites, which facilitates the study of plant metabolism on a broader scale and should promote the breeding of high yielding and nutritional crops (Martin et al., 2011; Riedelsheimer et al., 2012a).
Plant primary metabolism, also referred to as central metabolism, comprises the reactions that result in assimilation, respiration, transport, and differentiation processes that take place in most, if not all, cells (Fernie and Schauer, 2009). Primary metabolites include a wide range of intermediate compounds and end products of metabolic pathways that accumulate in sink organs (e.g., seeds, fruits, and tubers) and determine relevant crop quality traits related to nutritional content and composition, such as carbohydrates and amino and organic acids (Alonso-Blanco et al., 2009). Primary metabolism is moreover closely linked to plant growth and development. As such, uncovering the genetic determinants and the complex network(s) that underlie primary metabolism is fundamental to genetic improvement and metabolic engineering of either metabolic composition or plant primary production (Lisec et al., 2008). During the last few years, next-generation sequencing technologies have greatly helped efforts understand genomic diversity in maize (Huang and Han, 2012). Such advances in maize genomics have generated rich resources that markedly accelerate genetic analysis. For instance, high-resolution genome wide association studies have become practical owing to high-throughput genome and transcriptome sequencing (Fu et al., 2013; Hirsch et al., 2014), and the large number of molecular markers generated can also be applied in high-density genetic mapping and genomic selection (Ganal et al., 2012). Combining metabolite profiling and genetic mapping can be efficient for identifying the genomic loci responsible for metabolic variation (Schauer et al., 2006; Chan et al., 2011; Matsuda et al., 2012; Sauvage et al., 2014; Wen et al., 2014).
There have been several studies on maize metabolic traits using forward-genetic approaches, and a considerable number of quantitative trait loci (QTLs) have been identified (Zheng et al., 2008; Yan et al., 2010; Riedelsheimer et al., 2012b). However, since these studies were restricted to a single type of tissue or metabolite, a global view and systematic understanding of maize primary metabolite diversity as well as its association with phenotypic variation for morphology and growth-related traits is still lacking. In this study, we performed gas chromatography-mass spectrometry-based metabolite profiling across three tissues of a recombinant inbred line (RIL) population that has been much used for the dissection of maize kernel nutritional traits (Chander et al., 2008a, 2008b; Yang et al., 2010). One of the two parental lines of this population is the maize reference B73, while the other is a high-oil line By804.
Our aims were, first, to document the natural variation of a large number of primary metabolites in three tissues of a RIL population and, second, to dissect the genetic architecture of metabolic diversity and identify genes involved in the intricate metabolic network in various tissues. Also, third, we combined the genetic, metabolic, and agronomic data obtained from measuring the same material and examined the correlation between plant performance in the field and steady state metabolite levels. Finally, we assessed whether simultaneous analyses of primary metabolite levels and plant performance in the RIL population offers opportunities to identify the genetic determinants and biological mechanism of the latter.
Considerable genetic variation in maize primary metabolism across different tissues was evident from our analysis. Moreover, for a large proportion (76%) of metabolites, the changes in levels among different lines were highly correlated with each other. Linkage mapping using a high-density bin map led to the identification of a large number of metabolite QTL with moderate to major phenotypic effects. These QTL, accompanied by frequent epistatic interactions, contributed considerably to the metabolic variation present in the population. Furthermore, there was concordance between metabolite levels and agronomic traits. This study thus enhances our understanding of maize primary metabolism and provides genetic and biochemical insights that help uncover uncharacterized aspects of the metabolic network, dissect more complex phenotypic traits, and approach the ultimate goal of crop improvement.
RESULTS AND DISCUSSION
High-Density Linkage Map for Dissecting Large-Scale Metabolic Traits
A By804/B73 RIL population consisting of 197 recombinant lines was developed and used in this study. A high-density bin map was constructed based on the information of 15,285 single nucleotide polymorphism (SNP) markers. Briefly, 2496 recombinant bins were distributed throughout the genome, and 82% of which were <1 Mb in length. Considering each bin as a marker, a genetic linkage map on the basis of recombination frequency was constructed; the map was 1790.2 centimorgans (cM) in length, ∼0.72 cM per bin. Parental alleles were evenly distributed across the genome of the RIL population (Supplemental Figure 1). The RILs along with the map information were subsequently used for QTL mapping.
Natural Variation of Maize Primary Metabolites
Samples (i.e., leaves at seedling stage, leaves at reproductive stage, and kernels at 15 DAP) of the RIL population were harvested, extracted, and profiled using gas chromatography-time-of-flight-mass spectrometry (GC-TOF-MS). A total of 79 metabolites with known chemical structures were identified in the RILs, 33 of which were detected in all three tissue types. These metabolites can largely be classified as sugars, organic acids, and amino acids and cover multiple pathways including glycolysis, the tricarboxylic acid (TCA) cycle, and amino acid metabolism (Figure 1). Detailed information for each metabolite is shown in Supplemental Data Set 1A. The RILs manifested great diversity at the metabolite level (Table 1). Positive and negative transgressive segregation was observed for most metabolites within the RILs (Supplemental Figure 2). Significant changes were found in the levels of the majority of metabolites (94.5%) when comparing the metabolic profiles in different types of tissues (Supplemental Data Set 1B).
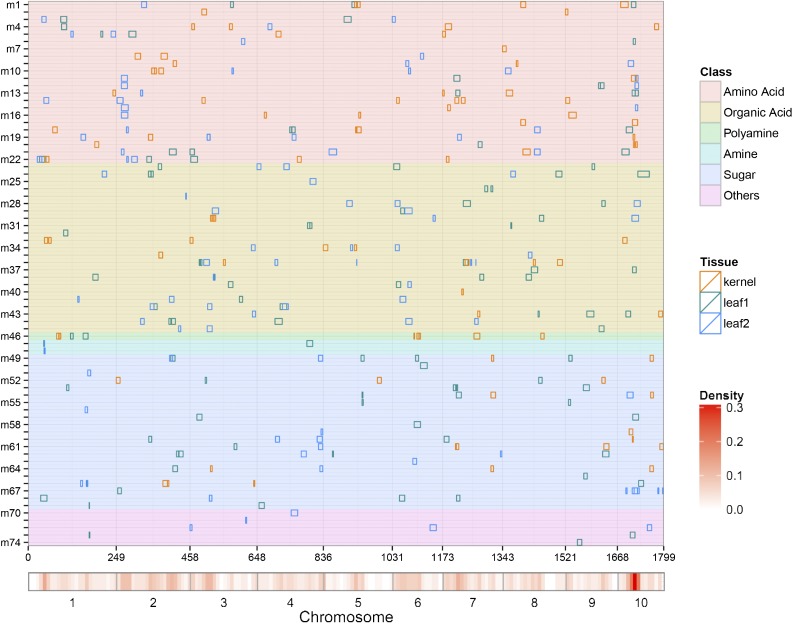
Figure 1.
Chromosomal Distribution of Metabolic QTLs Identified in This Study.
QTL regions (represented by the confidence interval) across the maize genome responsible for metabolite level from the three tissues are shown as green (leaf1, leaf at seedling stage), blue (leaf2, leaf at reproductive stage), and orange (kernel) boxes, respectively. The x axis indicates the genetic positions across the maize genome in cM. Heat map under the x axis illustrates the density of metabolic QTL across the genome. The window size is 10 cM. Detailed information of all detected QTLs is shown in Supplemental Data Set 1C. Metabolites from different chemical groups are marked by distinct colors as shown on the right. m1, alanine; m2, arginine; m3, asparagine; m4, aspartic acid; m5, GABA; m6, glutamic acid; m7, glutamine; m8, glycine; m9, histidine; m10, homoserine; m11, isoleucine; m12, lysine; m13, methionine; m14, ornithine; m15, phenylalanine; m16, proline; m17, serine; m18, threonine; m19, tryptophan; m20, tyrosine; m21, valine; m22, β-alanine; m23, 2-oxo-glutaric acid; m24, ascorbic acid; m25, cinnamic acid, 4-hydroxy-, trans; m26, cis-aconitic acid; m27, cis-caffeic acid; m28, citric acid; m29, dehydroascorbic acid; m30, fumaric acid; m31, galactonic acid; m32, galactonic acid-1,4-lactone; m33, glyceric acid; m34, isocitric acid; m35, lactic acid; m36, malic acid; m37, malic acid, 2-methyl; m38, nicotinic acid; m39, pyruvic acid; m40, quinic acid; m41, quinic acid-3-caffeoyl-, cis; m42, quinic acid-3-caffeoyl-, trans; m43, succinic acid; m44, threonic acid; m45, trans-caffeic acid; m46, putrescine; m47, dopamine; m48, tyramine; m49, fructose; m50, fructose-6-phosphate; m51, fucose; m52, galactinol; m53, glucoheptose; m54, glucose; m55, glucosone-3-deoxy; m56, isomaltose; m57, maltose; m58, mannose; m59, melezitose; m60, myo-inositol; m61, raffinose; m62, rhamnose; m63, squalene, all-trans; m64, sucrose; m65, tagatose; m66, threitol; m67, trehalose; m68, xylose; m69, xylulose; m70, glyceraldehyde-3-phosphate; m71, glycerol; m72, proline-4-hydroxy; m73, N-acetyl-serine; m74, urea.
Table 1. Range and Mean of Fold Changes of Metabolic Traits Measured in By804/B73 RIL Population.
QTL Identification and Genetic Epistasis for Maize Metabolic Variation
We mapped QTL associated with variation in the levels of primary metabolites in each aforementioned tissue of the RILs. In leaves of seedlings, leaves at the reproductive stage, and kernel, 107, 100, and 90 QTLs were identified for 52, 53, and 35 metabolites, respectively. The number of QTLs for each metabolic trait ranged from 1 to 7, with a mean of 1.9 to 2.6 across the three tissues (Table 2; Supplemental Data Set 1C). The confidence interval of each QTL varies from 0 to 32.5 cM with a median of 10.3 cM, when using 2-LOD (logarithm of odds) drop from the peak bin to delimit the region. If we only considered the peak bin, the QTL interval size ranged from 0.01 to 8.5 Mb with a mean of 0.8 Mb (Supplemental Data Set 1C). The percentage of phenotypic variation that each QTL could explain ranged from 2.4 to 49.0%, with a mean of 9.7, 9.6, and 14.7% in the three tissues, respectively (Table 2). A large number of metabolite QTLs were identified with moderate effects (86.2% of the mapped QTLs, R2 = 2.4 to 15%), which is in line with previous studies on primary metabolites (Lisec et al., 2008; Rowe et al., 2008). The magnitude of QTLs for some secondary metabolites is generally greater than those described for QTLs of primary traits (Alseekh et al., 2015). The varied explained phenotypic variance can partly reflect different genetic architecture between the natural variation of central metabolism and some secondary metabolism, which could also result from the different mapping populations. A total of 47 metabolites were found having QTLs that were detected in more than one tissue. A total of 254 QTL affecting variation in the levels of these 47 metabolites were identified, 10 of which overlapped, indicating that they likely partially share the same genetic control and/or coregulation across the different tissue types. However, the majority of QTLs were specific to each tissue, which may suggest distinct genetic and biochemical regulatory basis of the metabolic readout between tissues. Different tissues of an organism are well documented to use only a subset of the full capabilities that are encoded by the genome and to be partially dependent on other tissues for metabolic support in multicellular organisms (Gomes de Oliveira Dal’Molin et al., 2015). Moreover, the metabolic diversity among different tissues might arise from spatially or temporally different expression patterns of genes such as tissue-specific transcript expression and alternative splicing (Thatcher et al., 2014). In some cases, tissue-specific gene-metabolite association may represent a single metabolite but duplicate genes that display spatially or temporally distinct expression patterns. A similar observation was made in recent studies in tomato (Solanum lycopersicum) wherein QTLs for the primary metabolite content of seeds of an introgression line population were compared with those determined in the fruit pericarp (Toubiana et al., 2012). While an interesting observation, it is clear that in both instances further research will be required to fully elucidate the mechanistic reasons behind these differences.
Table 2. Summary of QTL for Metabolite Levels Identified in Three Tissues.
Five metabolite QTL hot spots were observed across the maize genome, which were determined using 1000 permutations at the level of 0.05 (Figure 1; Supplemental Data Set 1C). These QTLs were often shared by metabolites that are biochemically related, for instance, a hot spot at chromosome 10 was identified for about half of the amino acids detected in this study (Figure 1). Candidate genes underlying those QTLs could be central regulators of the pathway or influence the rate-limiting reaction. Principal component analysis results for the metabolic traits measured in leaves at seedling stage, leaves at reproductive stage, and kernels all showed that first five principal components cumulatively explained ∼50% of the phenotypic variation. Fifteen QTLs were identified for the five principal components across the three tissues, with explained phenotypic variance ranging between 6.4 and 19.0%. The QTL intervals for each principal component are summarized in Supplemental Data Set 1C. Seven of these 15 QTL overlapped with the QTL that were frequently identified for different metabolites, including the hot spot on chromosome 7 (Supplemental Data Set 1C).
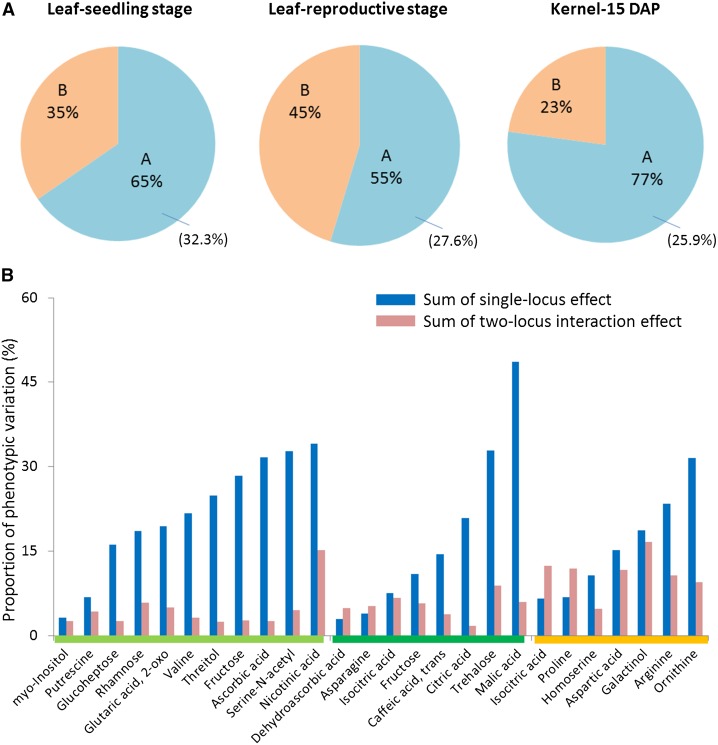
Pairwise epistatic interactions between QTLs of each metabolic trait were next investigated, as described in Methods. Significant epistatic interactions (P < 0.05) were detected for 11 out of 34 metabolites (32.4%) whose level in leaf at seedling stage was controlled by more than one QTL (Figure 2; Supplemental Table 1). Significant epistatic interactions were also found for a considerable percentage of metabolites that were identified in leaf at reproductive stage and in the kernel (i.e., 27.6 and 25.9%, respectively). The epistatic effect (i.e., sum of two-locus interaction effect) on metabolic variation ranged from 1.7 to 16.6% with an average of 6.6%, which implied epistasis may play an important role for some metabolites. In some cases, the epistatic effect was negligible compared with the main effects of the QTL (i.e., sum of single-locus effect), while in other cases, the former was comparable to or even greater than the latter (Figure 2; Supplemental Table 1 and Supplemental Data Set 1C). The stronger epistasis detected in kernels may be due to (1) the fact that more genes are expressed in the kernel, (2) the smaller sample size for analyzing the metabolites detected in the kernel, or (3) merely a false-positive observation.
Figure 2.
Schematic Summary of QTL Identification in This Study.
(A) Pie chart showing the proportions of metabolites that have different numbers of QTL. “A” represents the proportion of metabolites that have more than one QTL; “B” represents the proportion of metabolites that have only one QTL; number in the parenthesis represents the proportion of metabolites in “A” for which significant epistatic interactions were detected.
(B) Proportion of phenotypic variation explained by all the single QTLs and epistatic interactions. The bars above the metabolite names represent different tissue types: light green, leaf at seedling stage; dark green, leaf at reproductive stage; orange, kernel.
Metabolic variation can also be attributed to epistatic interactions according to a handful of previous studies on both specific metabolites and global metabolomes in plant species (Kliebenstein et al., 2002; Loudet et al., 2003; Calenge et al., 2006; Lisec et al., 2008; Rowe et al., 2008; Gong et al., 2013). In a QTL analysis of nitrogen use efficiency in Arabidopsis thaliana, Loudet et al. (2003) demonstrated that epistatic interactions between the identified QTLs play an important role in the control of the content of total free amino acids in the plant. As much as 15% of the total variance of total free amino acids is explained by epistatic interactions (Loudet et al., 2003). In this study, we identified effect of epistatic interaction for the content of several amino acids (asparagine, ornithine, proline, arginine, valine, and aspartate), which explained 3.19 to 11.97% of the phenotypic variation. Lisec et al. (2008) tested the epistatic interactions between metabolite QTLs (mQTLs) of 50 known metabolites and markers located elsewhere in the genome and found 38 interactions for 27 metabolites, with epistatic effects explaining 2.72% of the phenotypic variation on average. Out of these 27 metabolites, 15 were identified in our current study, and we also found epistatic effects for six of them, including ascorbic acid, putrescine, malate, galactinol, ornithine, and proline. These six metabolites are worth noting and the molecular mechanisms that underlie genetic epistasis warrant further investigation.
Although we found a large number of metabolite QTLs with moderate to main phenotypic effects, for ∼30% of metabolic traits, epistasis showed influences on the genetic architecture based on the pairwise analysis between these identified loci in this study (Figure 2). However, the contribution of epistasis to the metabolic variation may be still underestimated here, chiefly due to two reasons. First, analysis with the current sample size does not have enough power to detect minor and three- or higher-order epistatic interactions. Second, a biparental population is limited to cover the majority of the existing diversity. Nevertheless, the present result still suggests that epistasis is widely prevalent in the determination of metabolite levels within this RIL population. Possible mechanisms underlying genetic epistasis in natural populations could refer to physical or functional interactions between gene products or gene and gene products that are connected within a biochemical or regulatory pathway (Phillips, 2008; Kliebenstein, 2009; Bassel et al., 2012). Regulatory or enzymatic interactions can generate genetic epistasis through functional epistasis, for instance, transcription factors regulating enzymes, protein-protein interactions, and epigenetic modification. Metabolic traits might be different from those complex quantitative traits, such as flowering time, for which little epistasis has been detected and many different regulatory pathways may be involved (Buckler et al., 2009). Further analysis of specific epistatic interactions is expected to elucidate the connections between the observed genetic epistasis and molecular mechanisms.
A Maize Primary Metabolic Network Involving Key Genes and Metabolites
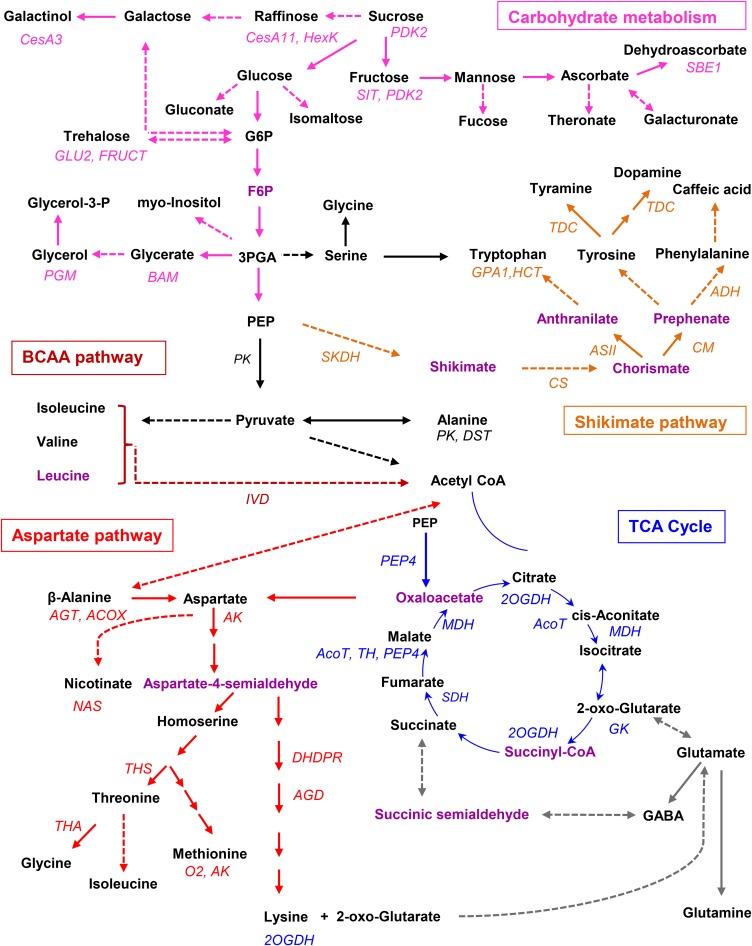
A maize primary metabolic network encompassing the key candidate genes and metabolites identified in this study is displayed in Figure 3 and covers carbohydrate metabolism, TCA cycle, and several important amino acids biosynthetic and catabolic pathways, including the aspartate, aromatic amino acid, and branched-chain amino acid (BCAA) metabolic pathways. Genes within the bin at the peak as well as within the confidence interval of each QTL were annotated and are listed in Supplemental Data Sets 1D and 1E, respectively. In the candidate list, genes encoding catalytic enzymes, transcription factors, and transporters were found. Notably, well characterized maize genes that are involved in primary metabolic processes were also identified, for instance, PEP4, CESA3, CESA11, O2, PDK2, glu2, and gpa1. PEP4 encodes phosphoenolpyruvate carboxylase (PEPc) and is located within the QTL responsible for malate level in this study. PEPc catalyzes the addition of bicarbonate (HCO3-) to phosphoenolpyruvate to form the four-carbon compound oxaloacetate and inorganic phosphate (Chollet et al., 1996; Paulus et al., 2013). PEPc plays major roles in plant and bacterial metabolism in the C4, TCA, and crassulacean acid metabolism cycles, with only the first two being relevant in maize. Phosphorylation by PEPc kinase activates the enzyme, whereas PEPc phosphatase inactivates it with both kinase and phosphatase being regulated at the level of transcription. It is further believed that malate acts as a feedback inhibitor of kinase expression levels and as an activator for phosphatase transcription (Nimmo, 2000). CESA genes encode cellulose synthases, which belong to the hexosyltransferase family of glycosyltransferases. In this study, CESA3 and CESA11 were found within the QTL interval for galactinol and raffinose, respectively. The functional alleles (O2) of the locus opaque endosperm 2 encode a basic leucine zipper protein transcription factor that regulates expression of several genes in the endosperm, notably those encoding the 22-kD α-zein storage proteins (Schmidt et al., 1992). Lysine catabolism in the maize endosperm is also affected in the o2 mutant (Kemper et al., 1999). In this study, O2 was found in the QTL region for methionine, which shares the same metabolic precursor as lysine.
Figure 3.
A Maize Primary Metabolic Network Involving Key Genes and Metabolites Identified in This Study.
Metabolites that were not identified in this study are shown in purple. Candidate genes identified in this study are shown in the respective pathway or under the corresponding associated metabolites. ACOX, acyl-CoA oxidase; AcoT, aconitase; ADH, arogenate dehydrogenase; AGD, diaminopimelate aminotransferase; AGT, alanine glyoxylate aminotransferase; AS, anthranilate synthase component II; AK, aspartate kinase; BAM, β-amylase; CesA, cellulose synthase; DHDPR, dihydrodipicolinate reductase; DST, dihydrolipoamide S-acetyltransferase; FRUCT, β-fructofuranosidase; gpa1, glyceraldehyde-3-phosphate dehydrogenase1; GK, glutamate kinase; HCT, hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyltransferase; HexK, hexokinase; IVD, isovaleryl-CoA-dehydrogenase; MDH, malate dehydrogenase; O2, Opaque2; PDK2, pyruvate dehydrogenase kinase isoform 2; PEP4, phosphoenolpyruvate carboxylase 4; PGM, phosphoglucomutase; PK, pyruvate kinase; SDH, succinate dehydrogenase; SIT, sugar/inositol transporter; SKDH, shikimate dehydrogenase; TDC, tyrosine/DOPA decarboxylase; TH, thioredoxin H-type; THA, threonine aldolase; THS, threonine synthase; 2OGDH, 2-oxoglutarate dehydrogenase.
In contrast to the above-mentioned genes with clearly defined function in maize, most candidate genes found here were merely putatively annotated by bioinformatic and comparative genomic approaches, while the function of the remaining candidate genes remains unknown. On the basis of biochemical knowledge (i.e., a complete understanding of genes involved in metabolism), it was apparent that our genetic mapping results coincided with predictions of gene function in many cases. For carbohydrate metabolism, we found genes that were predicted to be involved in, among other pathways, starch biosynthesis and degradation, sucrose degradation, and glycolysis within the QTL intervals. For instance, GRMZM2G175218 (annotated on the basis of sequence similarity as a β-amylase), a gene encoding a member of the glycoside hydrolase family that may display β-amylase activity, was found in the QTL interval of glycerate. Genes for β-fructofuranosidase (GRMZM2G018692 and GRMZM2G018716) were located in the QTL for trehalose. Genes encoding polygalacturonate 4-α-galacturonosyltransferase (GRMZM2G048008) and phosphoglycerate kinase (GRMZM2G003724) were within the QTL for threitol. All of these are promising candidate genes that require further experimental validation (Figure 3, Table 3; Supplemental Data Set 1D).
Table 3. Summary of Key Candidate Genes Involved in Maize Primary Metabolism Identified in This Study.
| Candidate Genea | Metabolite | Chromosome | R2 (%)b | Annotation |
|---|---|---|---|---|
| GRMZM2G056469 | Dopamine; tyramine | 1 | 32.37 | Tyrosine/DOPA decarboxylase |
| GRMZM2G093125 | Dopamine; tyramine | 1 | 32.37 | Tyrosine/DOPA decarboxylase |
| AC213521.3_FG005 | Tryptophan | 1 | 5.70 | Shikimate dehydrogenase |
| GRMZM2G024686 | Methionine | 1 | 8.59 | Aspartate kinase |
| GRMZM2G018716 | Trehalose | 2 | 7.68 | β-Fructofuranosidase |
| GRMZM2G018692 | Trehalose | 2 | 7.68 | β-Fructofuranosidase |
| GRMZM5G820287 | β-Alanine | 2 | 13.37 | Acyl-CoA oxidase |
| GRMZM2G120833 | Homoserine | 2 | 24.88 | Threonine synthase |
| GRMZM2G123652 | Quinicacid,3-caffeoyl-,trans | 2 | 13.09 | Shikimate dehydrogenase |
| GRMZM2G107469 | Homoserine | 2 | 17.81 | Threonine aldolase |
| GRMZM2G337113 | Tryptophan | 2 | 12.53 | gpa1; glyceraldehyde-3-phosphate dehydrogenase1 |
| GRMZM2G039454 | Galactinol | 3 | 27.58 | CesA-3;cellulose synthase (UDP-forming) |
| GRMZM2G159145 | Malate | 3 | 7.31 | Thioredoxin H-type |
| GRMZM2G156816 | Tryptophan | 3 | 8.86 | Anthranilate N-benzoyltransferase |
| GRMZM2G055795 | Raffinose | 3 | 7.09 | cesa11; cellulose synthase 11 |
| GRMZM5G856653 | Raffinose | 3 | 7.09 | Hexokinase |
| GRMZM2G025854 | Glycerol | 3 | 4.48 | Phosphoglucomutase |
| GRMZM2G323719 | Glutaric acid, 2-oxo | 4 | 11.06 | Glutamate 5-kinase |
| GRMZM2G364988 | Malate | 5 | 5.85 | Aconitase |
| GRMZM2G479325 | Malate | 5 | 5.85 | Acyl-CoA N-acyltransferase |
| GRMZM2G015132 | Alanine | 5 | 6.38 | Dihydrolipoamide acetyltransferase |
| GRMZM2G049372 | Fructose | 5 | 9.21 | Sugar/inositol transporter |
| GRMZM2G373717 | Dehydroascorbic acid | 6 | 11.84 | sbe1; 1,4-α-glucan branching enzyme |
| GRMZM2G365961 | Quinicacid,3-caffeoyl-,cis | 6 | 8.30 | Arogenate dehydrogenase |
| GRMZM2G473001 | Malate | 7 | 18.34 | PEP4; phosphoenolpyruvate carboxylase |
| GRMZM2G122780 | Citrate | 7 | 6.86 | Succinate dehydrogenase (ubiquinone) |
| GRMZM2G406672 | Succinate | 7 | 18.26 | Malic oxidoreductase |
| GRMZM2G141289 | Aconitic acid, cis | 7 | 16.52 | Malate dehydrogenase |
| AC217975.3_FG001 | Sucrose; fructose | 7 | 19.11 | Pyruvate dehydrogenase kinase isoform 2 |
| GRMZM2G059191 | Fumarate | 8 | 9.01 | Ferredoxin-NADP reductase |
| GRMZM2G104575 | Lysine | 9 | 5.63 | Dihydrodipicolinate reductase |
| GRMZM2G004534 | Alanine | 10 | 12.30 | Pyruvate kinase |
| GRMZM2G008247 | Trehalose | 10 | 6.11 | glu2; glycoside hydrolase, family 1 |
| GRMZM2G148508 | Phenylalanine; tyrosine | 10 | 8.92 | Anthranilate synthase |
| GRMZM2G119881 | Phenylalanine; tyrosine | 10 | 8.92 | Anthranilate synthase |
| GRMZM2G151041 | Lysine; citrate | 10 | 8.28 | 2-Oxoglutarate dehydrogenase, E1 component |
| GRMZM2G331861 | Lysine | 10 | 8.28 | ll-diaminopimelate aminotransferase |
The TCA cycle is a universal feature of the metabolism of aerobic organisms; it is a series of catabolic reactions that support the biosynthesis of ATP and reductant. At the same time, the TCA cycle is clearly embedded in a wider metabolic network that allows its activity to contribute to other aspects of metabolism (Sweetlove et al., 2010). In addition to genes encoding TCA cycle enzymes that have been well studied in Arabidopsis, such as aconitase, MDH (malate dehydrogenase), SDH (succinate dehydrogenase), and 2OGDH (2-oxoglutarate dehydrogenase; Huang et al., 2013; Araújo et al., 2014; Hooks et al., 2014), we also identified genes involved in the photosynthesis, oxidative-reductive reactions, and lipid biosynthesis that were within the QTL of metabolites in the TCA cycle. These included genes that were annotated as encoding ferredoxin-NADP reductase, thioredoxin, Acyl-CoA N-acyltransferase, malic oxidoreductase, and very-long-chain 3-ketoacyl-CoA synthase (Table 3; Supplemental Data Set 1D).
Amino acids serve as constituents of proteins, precursors for anabolism, and, in some cases, as signaling molecules in mammalians and plants (Häusler et al., 2014). l-tryptophan, l-phenylalanine, and l-tyrosine are aromatic amino acids that are derived from the shikimate pathway. They serve as precursors of numerous natural products, such as pigments, alkaloids, hormones, and cell wall components (Maeda and Dudareva, 2012). The aspartate-derived amino acid pathway in plants leads to the biosynthesis of lysine, methionine, threonine, and isoleucine (Jander and Joshi, 2009). Valine, leucine, and isoleucine form the small group of BCAAs classified by their small branched-hydrocarbon residues (Binder, 2010). These amino acids are essential components in the diets of humans and other animals. Unlike animals, plants are able to de novo synthesize these amino acids. On the basis of the above, we decided to focus on finding maize genes involved in the shikimate, aspartate, and BCAA pathways in this study, as well as some genes related to the level of other amino acids that were also found within the corresponding QTLs and indicated in Figure 3.
The aspartate-derived amino acid pathway has been relatively well characterized, which is beneficial for gene identification based on our genetic mapping. Indeed, we found six genes involved in the aspartate-derived amino acid biosynthetic pathway (Figure 3). Genes encoding two enzymes (DHDPR and AGD) that participate in two of five steps of lysine biosynthesis from asparatate-4-semialdehyde locate within the QTL interval of lysine (Figure 3, Table 3). Aspartate kinase (AK) catalyzes the first step of the aspartate-derived amino acid pathway, and the corresponding gene was found in the QTL of kernel methionine level (Figure 3, Table 3). In Arabidopsis, two AK genes encode bifunctional enzymes with both aspartate kinase and homoserine dehydrogenase activity (Paris et al., 2002; Rognes et al., 2003). They catalyze both the first and third steps in the biosynthesis of methionine, threonine, and isoleucine. Genes for threonine synthase, which catalyzes the final reaction of threonine biosynthesis, and threonine aldolase, which leads to glycine formation from threonine, were both within the QTLs of homoserine identified in the kernel (Figure 3, Table 3).
Duplicated candidate genes were found in the same metabolic QTL, for instance, two tandem genes (GRMZM2G056469 and GRMZM2G093125 with 98.9% identity in amino acid sequence) located at the short arm of chromosome 1 within the QTL for tyramine and dopamine level. Both genes were annotated to encode a tyrosine decarboxylase (TDC; EC 4.1.1.25) that catalyzes the substrate l-tyrosine into tyramine and CO2. TDC belongs to the family of lyases, specifically the carboxylyases, which participate in tyrosine metabolism and alkaloid biosynthesis. There are seven homologous genes of these two TDC genes (i.e., with identity of more than 70%; GRMZM5G806967, GRMZM2G009400, AC196412.3_FG001, GRMZM2G108514, GRMZM2G441632, GRMZM2G021388, and GRMZM2G021277) in the maize genome. These seven genes belong to the aromatic amino acid decarboxylase family; two genes (GRMZM2G021388 and GRMZM2G021277; annotated as tryptophan decarboxylase) of which have been found associated with tryptophan in maize kernel in our previous study (Wen et al., 2014). These duplicated genes may diverge from each other in the pattern of neofunctionalization, where one of the duplicates obtains a novel function, or subfunctionalization, in which case the duplicate copies obtain differential expression patterns in terms of tissue specificity or stress response (Kliebenstein, 2009). Despite the high identity of the two TDC genes in predicted amino acid sequence, large divergence was found in their 5′ untranslated region (UTR) and 3′ UTR (Supplemental Figure 3). Understanding the functional divergence of duplicated genes will necessitate further experimentation including cloning of the causal genes and marker development for metabolic improvement. Moreover, further study of metabolic QTLs may shed light on evolutionary questions regarding the fate of duplicated genes, as implicated by Kliebenstein (2009).
The majority of the above-mentioned candidate genes were contained within the peak bins of each QTL (Table 3). Matching the functional annotation of the gene with the biochemical properties of the corresponding metabolites facilitated the identification of potential causal genes. Using prior knowledge of the metabolic pathway and gene annotation, we could first delimit the search region to the peak bin of each QTL and then check if there were genes encoding enzymes that directly catalyze the reaction with the respective metabolites as either substrate or product. The search region could be enlarged to the confidence interval if there were no well annotated genes meeting this criterion in the peak bin, as indicated in previous studies (Salvi and Tuberosa, 2005; Price, 2006). For all the metabolites, we assigned the candidate genes within the confidence interval (2-LOD-drop region) and performed Gene Ontology term analysis of all resulting candidate genes (Supplemental Data Set 1F). This approach revealed significant enrichment in terms relating to metabolic process (P = 0.0006 after false discovery rate [FDR] correction), including the regulation of primary metabolic process (P = 0.0025 after FDR correction; Supplemental Data Set 1F).
Revealing the underlying gene(s) in each QTL will be greatly benefited from increased mapping power and resolution. In a highly polygenic setting with extensive epistasis and unknown error versus genetic variance structure, population size and recombination provide the majority of the mapping resolution and power, although high-density markers can depict recombination events more accurately. Due to relatively limited number of lines in the RIL population used here, the gene number within the confidence interval is large in many cases, which could make pinpointing the candidate genes challenging (Supplemental Data Set 1E). This difficulty can be addressed using QTL fine-mapping strategies and harnessing other tools such as expression profiling and candidate gene association analysis, especially for complex loci or metabolic traits of great importance and interest. In addition, genes within the QTL but with unannotated function might be easily ignored due to insufficient information, which complicates the identification and annotation of novel genes. Moreover, although the genome of one parental line (i.e., B73) is the reference for maize, fine-mapping approaches would omit information of a fraction of genes as a result of the numerous presence/absence genomic variations among maize lines (Springer et al., 2009). These situations should be taken into account for the candidate gene selection.
Validation of Candidate Genes and Identification of Potential Functional Variants Using Association Analysis
Linkage analysis is often used to complement genome-wide association studies. Here, we used association analysis to aid in validating the candidate genes identified using linkage mapping and to facilitate uncovering functional genetic variants. Taking AGT and TDC as examples, we found that the loci identified by linkage mapping also showed strong association with the same metabolic trait according to association analysis in a panel containing 513 maize inbred lines (Figures 4 and 5). Detailed information about this association panel is given in Methods. Potential functional genetic variants of AGT and TDC were identified by resequencing the two parental lines and tested in the association panel.
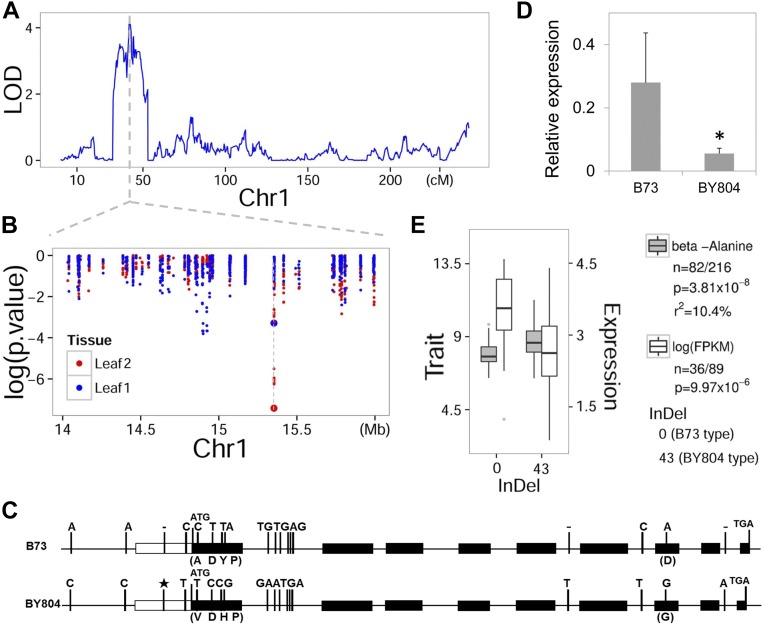
Figure 4.
Validation of Candidate Gene AGT within QTL Interval Using Association Analysis.
(A) LOD curves of QTL mapping for level of β-alanine in maize leaves at seedling stage on chromosome 1.
(B) Scatterplot of association results between polymorphic markers (i.e., SNPs and InDels) in the confidence interval and the level of β-alanine. The −log10-transformed P values from the association analysis are plotted against the genomic physical position. Physical position of candidate gene AGT is indicated by the vertical dashed line. The bigger dot represents the 43-bp InDel marker. Markers shown in blue were in the association analysis for level of β-alanine in maize leaves at seedling stage (leaf 1), while markers in red were in the association analysis for level of β-alanine in maize leaves at reproductive stage (leaf 2). Association analysis was performed using the mixed linear model controlling for the population structure (Q) and kinship (K).
(C) Gene structure of AGT and natural variation between alleles from By804 and B73. The star marks the 43-bp insertion in By804.
(D) Relative levels of AGT mRNA in B73 and By804. Expression levels were measured by reverse transcription-quantitative PCR and values for three biological replications were averaged (P = 0.04; t test).
(E) Box plot for level of β-alanine (gray) and expression of AGT (white) plotted as a function of genotypes at the site InDel_0/43. P value for the expression level was calculated based on ANOVA, and P value for the level of β-alanine was calculated using MLM controlling for population structure (Q) and kinship (K).
Figure 5.
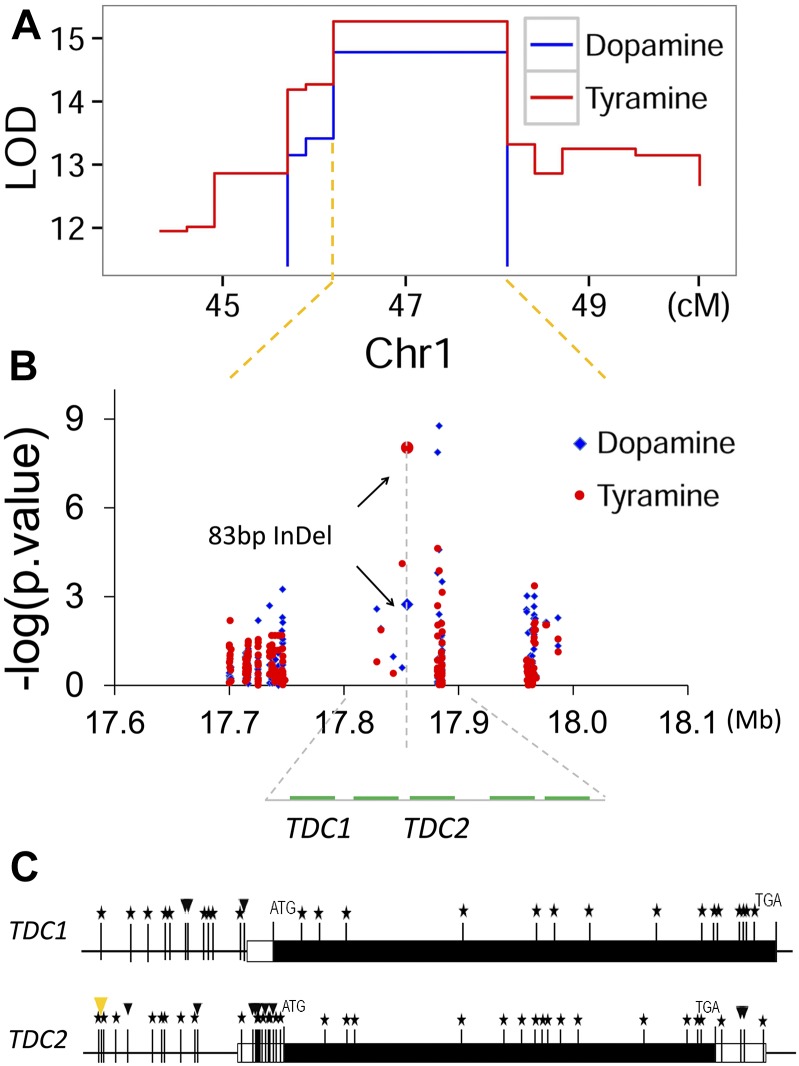
Validation of Candidate Genes TDC1 and TDC2 within QTL Interval Using Association Analysis.
(A) Diagram of linkage mapping result for the level of tyramine and dopamine in maize leaves at reproductive stage. LOD values of the bins at the peak of QTL interval are shown as a function of their genetic positions.
(B) Scatterplot of association results between polymorphic markers (i.e., SNPs and InDels) in the peak bin and the level of metabolites tyramine and dopamine in maize leaves at reproductive stage. The −log10-transformed P values from the association analysis are plotted against the genomic physical position. Genes located within the 17.8- to ∼17.9-Mb region on chromosome 1 are indicated as green bars. Association analysis was performed using the mixed linear model controlling for the population structure (Q) and kinship (K).
(C) Gene structure of TDC1 and TDC2 and sequence variation between alleles from By804 and B73. Stars mark SNPs, while arrowheads mark insertion-deletion polymorphisms (InDels) between the allelic sequence of By804 and B73. The bigger orange arrowhead in the promoter region of TDC2 represents the 83-bp InDel.
AGT (GRMZM2G124353) encodes an alanine glyoxylate aminotransferase and colocated with a QTL for β-alanine detected in leaf at seedling stage (Figure 4A). The association between its allelic variations and β-alanine is indicated in Figure 4B. After resequencing the AGT locus of By804 and B73, 39 polymorphic sites between the two parental lines were found, including 15 SNPs that were identified by RNA-sequencing in our previous study (Fu et al., 2013; Figure 4C; Supplemental Tables 2 and 3). Notably, a 43-bp InDel was detected in the 5′ UTR of AGT. We developed a PCR-based marker for detecting this InDel polymorphism and used it to genotype the association panel (Supplemental Table 2). A strong association signal was detected with this 43-bp InDel (P = 3.81 × 10−8, mixed linear model, n = 298; Figure 4B, Table 4). The relative mRNA level of AGT in B73 was significantly higher than that in By804 as indicated by reverse transcription-quantitative PCR (P = 0.04, t test; Figure 4D). Further expression profiling of AGT in seedling leaves of 125 maize inbred lines based on RNA-sequencing revealed that the 43-bp InDel in the 5′ UTR of AGT can significantly affect its own expression level (P = 9.97 × 10−6, n = 125, ANOVA; Figure 4E). In addition, the expression level of AGT was negatively correlated with the level of β-alanine in maize leaves at seedling stage (r = −0.15, P = 0.05, n = 116) and leaves at reproductive stage (r = −0.18, P = 0.03, n = 116). Taken together, we speculate that the 43-bp InDel can cause transcriptional variation of AGT in maize leaves, which results in changes of the level of β-alanine. Resequencing also revealed five SNPs between By804 and B73 in the coding region, three of which resulted in amino acid substitutions (i.e., Ala-Val, Tyr-His, and Asp-Gly). All three SNPs were significantly associated with the level of β-alanine (P = 3.8 × 10−8 ∼6.12 × 10−4, mixed linear model, n = 339; Table 4).
Table 4. Summary of Candidate Gene Association Results for AGT and TDC2 Based on Resequencing.
| Gene | Trait | Marker | Chromosome | Position (bp)a | P (leaf1)b | P (leaf2)c | Region | Codon change | Amino acid replacement |
|---|---|---|---|---|---|---|---|---|---|
| AGT | β-Alanine | InDel_15353417 | 1 | 15353417 | 5.19 × 10−4 (297) | 3.81 × 10−8 (298) | 5′ UTR | ||
| chr1.S_15353577 | 1 | 15353577 | 0.02 (339) | 1.11 × 10−6 (339) | Exon | C/T | Ala-Val | ||
| chr1.S_15353647 | 1 | 15353647 | 4.23 × 10−3 (339) | 10.05 × 10−7 (339) | Exon | T/C | |||
| chr1.S_15353690 | 1 | 15353690 | 4.33 × 10−4 (339) | 6.62 × 10−7 (339) | Exon | T/C | Tyr-His | ||
| chr1.S_15353704 | 1 | 15353704 | 4.33 × 10−4 (339) | 6.62 × 10−7 (339) | Exon | A/G | |||
| chr1.S_15355785 | 1 | 15355785 | 6.12 × 10−4 (339) | 0.01 (339) | Exon | A/G | Asp-Gly | ||
| TDC2 | Tyramine | InDel_17855186 | 1 | 17855186 | 2.6 × 10−8 (282) | 9.2 × 10−9 (272) | 5′ UTR | ||
| Dopamine | InDel_17855186 | 1 | 17855186 | 0.02 (205) | 0.002 (234) | 5′ UTR |
The physical position of each polymorphic site in the maize genome.
P value for the association between each marker and the corresponding metabolite level in leaves at seedling stage; association analysis was conducted using the MLM accounting for population structure (Q) and kinship (K); number of lines used in the association analysis is indicated in the parentheses.
P value for the association between each marker and the corresponding metabolite level in leaves at reproductive stage; association analysis was conducted using the MLM accounting for population structure (Q) and kinship (K); number of lines used in the association analysis is indicated in the parenthesis.
Locus TDC contains two paralogous genes (GRMZM2G056469 and GRMZM2G093125, referred to as TDC1 and TDC2 hereafter) that both encode a tyrosine decarboxylase, as mentioned above. This locus was located within the peak bin of the QTL for tyramine and dopamine detected in leaf at reproductive stage (Figure 5A). Associations between SNPs in the peak bin and the level of tyramine and dopamine in leaf at reproductive stage are shown in Figure 5B. Expression of both TDC1 and TDC2 in maize kernel is very low (Sekhon et al., 2011) and was not above the cutoff in our previous RNA-sequencing study (Fu et al., 2013). However, expression levels of both genes are much higher in anthers, leaves, and seedling shoots of B73 (Sekhon et al., 2011). We resequenced both genes in the two parental lines to find more sequence polymorphisms and potential functional variants. In TDC1, we identified 33 polymorphic sites between the two parental lines, including four small InDels in 5′ UTR and 29 SNPs in both the coding and untranslated regions, seven of which resulted in amino acid substitutions (i.e., Val-Ala, Asp-Ala, Lys-Asp, Ala-Thr, Val-Ala, Ala-Val, and Ile-Leu; Figure 5C; Supplemental Table 4). We identified 45 polymorphic sites, including 14 InDels and 31 SNPs, between the two parental lines by resequencing TDC2. Six of these polymorphisms resulted in amino acid substitutions (Supplemental Table 5). Dramatically, an 83-bp InDel was located in the 5′ UTR of TDC2 at site −478 that may affect the gene expression, thus affecting the phenotype (Table 4, Figure 5; Supplemental Table 5). A PCR-based marker for detecting this 83-bp InDel polymorphism was then designed and used to genotype the association panel (Supplemental Table 6). Subsequent association analysis revealed the 83-bp InDel was significantly associated with the level of tyramine and dopamine detected in leaf at reproductive stage (P = 9.19 × 10−9 and 0.02, n = 272 and 205, respectively; mixed linear model; Figure 5B, Table 4; Supplemental Table 5). To circumvent problems arising from the varied linkage disequilibrium decay levels across maize genome (varying from 1 to 100 kb with an average of 1 to 5 kb; Yan et al., 2011), prior knowledge of the metabolic pathway and gene annotation was additionally considered for calling candidate genes associated with level of tyramine or dopamine within this QTL. Among the genes within the QTL region, TDC1 and TDC2 did not show the strongest association with the level of dopamine (Figure 5B), but rather the strongest association signal was mapped to the gene (i.e., GRMZM2G093197) next to TDC2, which is annotated as uroporphyrinogen-III synthase involved in tetrapyrrole biosynthesis. However, a direct link between GRMZM2G093197 and dopamine is not evident. By contrast, tyramine is synthesized from tyrosine by the enzyme TDC (EC 4.1.1.25), as mentioned above and shown in Figure 3. Dopamine is also produced from tyrosine, which is converted into l-DOPA by the enzyme tyrosine hydroxylase, after which l-DOPA is converted into dopamine by the enzyme aromatic amino acid decarboxylase (Broadley, 2010). Gene annotation and pathway knowledge were combined to determine the most likely candidate genes in the target region in a manner that avoids the interferences of linkage disequilibrium between the strongest signal and the causal genomic variant(s).
Identification of potential functional genetic variants together with cross-validation can thus be achieved by combining linkage and association analysis. It may be challenging to find dramatic genetic diversity in the coding region of very crucial genes (e.g., genes that function as center hub) using naturally occurring variation due to the nature of primary metabolism. However, genetic variants that may result in regulatory variation frequently can be found when using natural populations, as implied from this study and others (Carroll, 2008; Albert and Kruglyak, 2015). For instance, a recent study indicated that a large portion of functional variation in maize probably stems from differences in copy number and gene regulation rather than in protein-coding sequence, implying that changes in gene regulation are a frequent driver of functional variation (Carroll, 2008; Wallace et al., 2014; Albert and Kruglyak, 2015).
Metabolite-Metabolite Network and Relationships between Metabolites and Agronomic Traits
Significant pairwise correlations (r = 0.5 and P = 0.05) between metabolites identified from each tissue are illustrated in Supplemental Figure 4 and Supplemental Data Set 1G. There were 122, 102, and 51 significant correlations between metabolites detected in leaf at seedling stage, leaf at the reproductive stage, and kernel, respectively. Metabolites of the same chemical class or involved in the same biochemical pathway tended to correlate with each other. Different tissue types exhibited distinct correlation networks in general; however, a part of significant metabolite-metabolite correlations and subnetworks were conserved across tissues. Notably, the link of a majority of amino acids was consistently strong in the three types of tissues. In leaf at seedling stage, threonine, serine, isoleucine, glycine, and valine all contained more than nine connectives with other metabolites, mainly amino acids. Valine, isoleucine, pyroglutamic acid, and threonine were more frequently correlated with other amino acids in the leaf at reproductive stage, whereas in the kernel, serine, threonine, and γ-aminobutyric acid correlated with more metabolites compared with the rest of the metabolites. Sucrose, raffinose, and myo-inositol were highly connected to each other (r >0.9) within the network in the leaf at reproductive stage. In the kernel, the same highly connected triangle (r > 0.9) was observed between sucrose, fructose, and glucose (Supplemental Figure 4 and Supplemental Data Set 1G). Strong connectivity between amino acids was also observed in previous studies in tomato and Arabidopsis (Schauer et al., 2006; Sulpice et al., 2010; Toubiana et al., 2015), and the amino acid metabolism network is subject to a high degree of metabolic regulation (Galili and Höfgen, 2002).
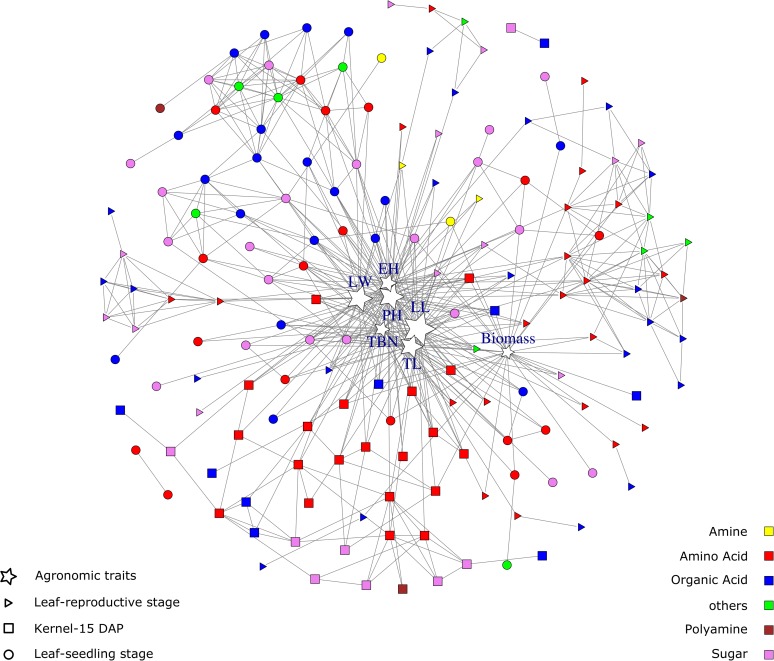
The relationships between the metabolite levels from each tissue and seven agronomic traits (i.e., plant height, ear height, length of ear leaf, width of ear leaf, tassel length, tassel branch number, and biomass) were built through elastic net regression. An overview of metabolite-metabolite-agronomic trait network across all three tissues was integrated in Figure 6. Metabolites involved in the regression model with each agronomic trait are shown in Supplemental Data Set 1H. For each agronomic trait, the number of metabolites that participated in the model ranged from 26 to 49, with an average of 37.4. The variance explained by the metabolites in the model for each agronomic trait ranged from 12 to 76% with an average of 40%. In some cases, the same metabolites detected in different tissues were in the same model for a single agronomic trait. For instance, plant height, length of ear leaf, and tassel length could be explained by succinate detected in all three tissues (leaf at seedling stage, leaf at reproductive stage, and kernel); tryptophan and malate detected from all the three tissues appeared in the model for tassel length; sucrose in leaf at seedling stage and kernel appeared in the model for plant height, length of ear leaf, tassel length, and tassel branch number. In addition, some metabolites appeared in the models for multiple agronomic traits, for instance, sucrose and cis-aconitate detected in leaf at seedling stage and rhamnose detected in the leaf at reproductive stage participated in the model for all seven agronomic traits.
Figure 6.
Metabolite-Metabolite-Agronomic Trait Association Network.
This illustration represents the union of metabolite-agronomic trait association network with the metabolic relevance networks obtained for each tissue (i.e., leaf at seedling stage, leaf at reproductive stage, and kernel). Nodes that correlate with each other are linked by gray edges. The color of the nodes represents the metabolite classes. The nodes with the star stand for agronomic traits, and the circles, triangles, and squares correspond to the metabolites in leaf at seedling stage, leaf at reproductive stage, and kernel, respectively. The r-square, which measures how well the data fitted to the regression model, was calculated for each model and represented in the network with the size of the corresponding node. The better the model fitted to the data, the bigger the size of the node.
For the length of ear leaf and plant height, the data were better fitted to the regression model compared with that for other agronomic traits, as indicated in Figure 6. This result may suggest differences in the complexity of the relationships between different agronomic traits and these primary metabolites identified in this study. In addition, the variance of plant morphology and growth-related traits were explained by combinational changes of a large number of metabolites in various tissues as inferred from the present result. The information from metabolites detected in multiple tissues is more comprehensive than that obtained from a single tissue. When considering metabolites for prediction of a complex trait and/or identification of potential biomarkers, it would be more efficient to take multiple tissues into account for sampling. However, choosing the type of tissue and metabolites may be largely dependent on the corresponding trait. For instance, for a couple of traits (i.e., biomass, plant height, ear height, and tassel length), metabolites detected in the kernel were not among the top 10% metabolites with the highest effect. This indicated that the role of the kernel in biomass, plant height, ear height, and tassel length may not be as important as that of the leaf.
To further reveal the relationship between the metabolism and agronomic traits and look for implications regarding the molecular mechanisms underlying the variation of complex traits, we analyzed the colocalization between the mQTLs and agronomic QTLs (pQTLs) identified in this study. We identified 23 QTLs for the seven agronomic traits, with one to six QTLs per trait (Supplemental Table 7). In total, 12.2% (35/287) of these mQTLs overlapped with pQTLs, significantly more than expected by chance (i.e., 8%; binomial test, P = 0.01). The effects of mQTLs are usually greater than those of the pQTL. Most intriguingly, some of these metabolites that shared QTLs with the agronomic traits were in the model for the corresponding agronomic traits according to our elastic net regression analysis. For instance, the level of alanine in the leaf at seedling stage was in the model of ear height, and they had a common QTL on chromosome 5; the same was observed between the level of myo-inositol in leaf at seedling stage and leaf width, as well as the level of galactonic acid from leaf at seedling stage and tassel branch number. Xylulose level in leaf at seedling stage, nicotinic acid and xylose level in the leaf at reproductive stage, and the level of sucrose and fumarate in the kernel were all in the model for length of ear leaf, and each of them had a QTL interval that overlapped with a QTL for the latter (Supplemental Data Sets 1C and 1H and Supplemental Table 7).
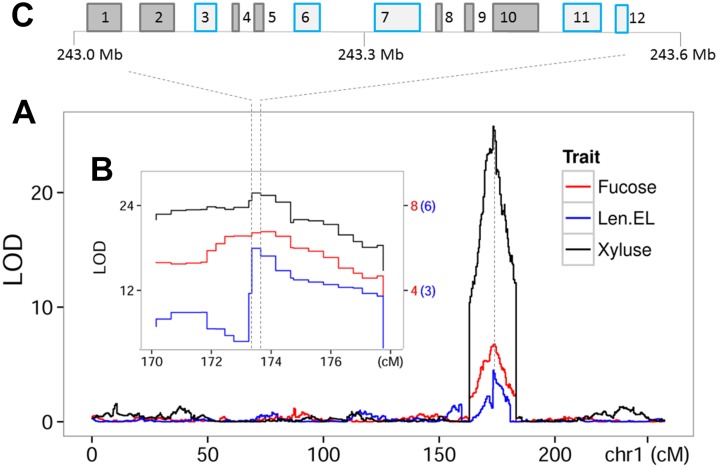
Although the metabolite-agronomic trait association based on the elastic net regression and the colocalization between mQTLs and pQTLs may explain each other, further dissection of the common QTLs between metabolic and agronomic traits is noteworthy. Importantly, metabolites might be regarded as intermediary traits for agronomic QTL cloning and biomarkers for molecular breeding. Here, we discuss an example of the relationship between metabolites and the length of ear leaf. A QTL on chromosome 1 for the length of ear leaf exactly colocalized with the QTL for level of both fucose measured in leaf at reproductive stage and xylulose measured in leaf at seedling stage (Figure 7). Xylulose level in leaf at seedling stage was associated with length of ear leaf according to the elastic net regression analysis (Supplemental Data Set 1H). Association between metabolite levels and the morphological traits is likely due to genetic coregulation. These sugars are both components of cell walls; hence, this colocalization could reflect an enhanced rate of cell wall biosynthesis. The overlapped QTL region provides a clue for finding the causal genetic components. Twelve genes were found within the ∼600-kb region of the peak bin. Five of these genes have putative function, encoding homeobox-leucine zipper protein (GRMZM2G097349), ribosomal L22e protein (AC199175.2_FG006), sucrose-proton symporter (GRMZM2G083248), galactose oxidase/kelch repeat superfamily protein (GRMZM2G019171), and NAD binding oxidoreductase family protein (GRMZM2G409133), whereas functions of the rest seven genes are unknown. It is evident that sucrose-proton symporter may influence the level of sugars identified here. Findings from previous studies about the proteins mentioned above provide helpful information. Sulpice et al. (2009) conducted metabolite profiling in 94 Arabidopsis accessions and revealed a significant correlation between biomass, starch, and protein content. Their work supported the possibility that regulators of starch metabolism or signals derived from starch act as integrators of plant metabolism and growth. They identified a kelch repeat F-box protein (At1g23390) whose transcript levels correlated with Arabidopsis rosette biomass. Association mapping further revealed that polymorphisms in this gene were significantly associated with the traits of fresh weight and levels of starch, protein, and sucrose. The conserved correlation between starch and protein might reflect the large energy costs associated with protein biosynthesis and maintenance (Gibon et al., 2009). According to this, the ribosomal L22e protein identified here might also be causal in addition to the kelch repeat protein. Further evaluation and validation of these genes will help to clone the QTLs affecting both metabolic and phenotypic traits as well as to dissect the molecular basis of complex traits and thus further enhance the crop breeding toolbox.
Figure 7.
Dissection of Metabolite and Candidate Genes Associated with Length of Ear Leaf.
(A) LOD curves of QTL mapping for length of ear leaf (Len.EL) and level of fucose and xylulose in maize leaves on chromosome 1. QTLs for these three traits overlap at ∼174 cM on chromosome 1.
(B) LOD values of the bins at the peak of QTL interval identified in (A) are shown as a function of their genetic positions.
(C) Graphical representation of genes within the genomic region spanning the single bin at the peak. Twelve genes represented by boxes were found within the ∼600-kb region. Five genes that have putative function are marked in light blue.
Conclusion
Great efforts have been devoted to increasing agricultural production with decreased inputs to meet the needs of the rising world population (Tester and Langridge, 2010). Increasing plant production and improving nutritional quality are of both economic and social interest (Martin et al., 2011; Fitzpatrick et al., 2012). Despite the success in generating, for instance, high-oil maize and quality protein maize lines using long-term selection, the efficiency of genetic improvement is relatively limited (Moose et al., 2004). A detailed understanding of genomic determinants of metabolic changes and the interaction between these pathways and regulatory networks that influence plant performance will enhance crop genetic improvement. There have been exhaustive studies on a single or several metabolic traits in maize; however, little effort has been dedicated to study the natural variation of primary metabolism and its role in the plant growth and morphology. Taking advantage of a valuable genetic resource, we revealed the genetic basis that underlies variation of primary metabolism in multiple tissues. This information could thus be of direct use in designing breeding strategies for the improvement of high value metabolites. Although a more complex genetic architecture was revealed for primary metabolism compared with secondary metabolism (Alseekh et al., 2015), the considerable number of metabolites with major QTLs (R2 > 15%), together with the relatively minor effects of epistasis identified in our study, suggests that we can still simplify breeding efforts for some metabolites by pyramiding favorable alleles of the major genes. Moreover, the hundreds of loci identified in this study will enable candidate gene identification and validation that will aid further dissection of molecular bases of metabolic variation.
The steady state level of primary metabolites in different tissues can be used as variables to correlate with and predict various plant traits. Reciprocal regulations between metabolite level and other cellular processes, such as changes in gene transcription, protein accumulation, and enzyme activity, complicate the elucidation of the metabolic network. Plant morphology and the source sink relationship can also affect the metabolite profile (Schauer et al., 2006; Do et al., 2010; Chitwood et al., 2013). Our ability to dissect the metabolic networks and explore their relationships with agronomic traits could greatly benefit from studying metabolic and agronomic traits in parallel. Metabolite profiling in various tissues or different developmental stages of the RIL population is helpful to gain insights into the regulation between metabolic network and physiological or developmental traits. Owing to the permanent nature of this RIL population, the large diversity of phenotypes investigated from different environments can be integrated. These phenotypic data together with other modern systems biology approaches, including transcriptomics and proteomics, as well as various molecular phenotypes can thus be further used to elucidate the complex system.
METHODS
Plant Materials and Growth Conditions
A maize (Zea mays) RIL population (By804/B73) derived from a cross between normal line B73 and high-oil line By804 was used for linkage analysis in this study (Chander et al., 2008a, 2008b). The two parental lines along with 196 recombinant lines were planted in one-row plots in an incompletely randomized block design at Huazhong Agricultural University field experiment station (Wuhan, E 109°51', N 18°25') in 2013. All lines were self-pollinated. Leaf samples of each line at both seedling stage (the fifth leaf, sampled at the same day, i.e., 50 d after sowing) and reproductive stage (leaf above the ear, sampled at the same day, i.e., 90 d after sowing) were collected and store at −80°C until use for extraction. Leaves from the same genotype were harvested from at least three plants and bulked. For each line, at least three ears from different plants were harvested 15 DAP, and the kernels were bulked and store at −80°C before extraction. We investigated agronomic traits of each line 98 d after sowing, including plant height, ear height, length of ear leaf, width of ear leaf, tassel length, and tassel branch number. The fresh shoot biomass (biomass) was recorded at 15 DAP for each line. An association panel containing 513 maize inbred lines (Yang et al., 2011) that was collected previously was grown under the same conditions as described above in 2013.
A total of 471 samples (192 samples from leaf at seedling stage, 184 samples from leaf at reproductive stage, and 95 samples from 15 DAP kernel) from the By804/B73 RILs and 490 samples from the association panel were obtained and extracted for metabolite profiling.
Genotyping and Linkage Map Construction
All recombinant lines together with two parental lines (n = 199) were genotyped using Illumina MaizeSNP50 BeadChip, which contains 56,110 SNP loci (Ganal et al., 2011). SNP genotyping was performed on the Illumina Infinium SNP genotyping platform at Cornell University Life Sciences Core Laboratories Center. After quality control, the SNPs with missing rate of <10% and heterozygosity of <10% were used to construct the genetic linkage map. A subset containing 368 lines of the association panel was genotyped using RNA-sequencing and Illumina MaizeSNP50 BeadChip previously (Fu et al., 2013). Briefly, 90-bp pair-end Illumina RNA-sequencing was subsequently performed on the immature seeds of 15 DAP for these 368 lines. In total, 1.06 million high-quality SNPs were identified and expression data of 28,679 genes were obtained in the whole panel (Fu et al., 2013).
To construct the genetic linkage map, we developed an economic go-wrong method integrating the Carthagene software (de Givry et al., 2005) in a Linux system with in-house Perl scripts (can be downloaded from https://github.com/panqingchun/linkage_map). Markers that completely colocalized were assigned into a chromosomal bin. Each bin was regarded as one marker. We obtained 15,285 polymorphic markers in total, which were incorporated into 2496 unique bins (markers) in this study.
Metabolite Profiling Based on GC-TOF-MS
Metabolites for GC-TOF-MS were extracted according to a protocol adapted from Lisec et al. (2006) and Roessner et al. (2001). Briefly, for each sample, 50 mg fresh powder was used and mixed with 700 μL 100% methanol and 30 μL ribitol (0.2 mg/mL stock in water) as an internal quantitative standard for the polar phase. After shaking the mix for 15 min at 70°C, the extract was centrifuged for 10 min at 20,817g. The supernatant from the centrifuged mixture was then mixed with 375 μL chloroform and 750 μL water. After centrifugation for 15 min at 1699g, a 150-μL aliquot from the upper polar phase was taken and dried in vacuum. The dried samples were shipped to Golm, Potsdam for metabolic profiling. After derivatization, 1 μL each sample was injected into a GC-TOF-MS system (Pegasus III; Leco). Gas chromatography was performed using a 30-m MDN-35 column. The injection temperature was 230°C, and the transfer line and ion source were set at 250°C. Chromatograms and mass spectra were evaluated and metabolite levels determined in a targeted fashion using a library derived from the Golm Metabolome Database (Kopka et al., 2005). Each metabolite is represented by the observed ion intensity of a selected unique ion that allows for a relative quantification between groups. The raw phenotypic and genotypic data for all lines as well as the genetic map are provided as supplemental data (Supplemental Data Sets 1I to 1M).
QTL Mapping and Candidate Gene Identification
We conducted QTL analysis using composite interval mapping implemented in Windows QTL Cartographer V2.5 for metabolite content measured in three tissues of the RIL population (Zeng et al., 1999; Wang et al., 2006). Zmap (model 6) with a 10-cM window and a walking speed of 0.5 cM were used. For each trait (metabolic trait as well as agronomic trait), a threshold for significant QTLs was determined by 1000 permutations (P = 0.05). The bins were clearly defined and a uniform LOD value was assigned for each bin. Confidence interval for each QTL was assigned as 2-LOD drop of the peak. The detailed information, including location, confidence interval, and explained phenotypic variance of each QTL for each trait, is shown in Supplemental Data Set 1C (for metabolic trait) and Supplemental Table 7 (for agronomic trait). The filtered working gene list of maize genome was downloaded from MaizeGDB (http://www.maizegdb.org) to identify possible candidate genes in each QTL. Candidate genes were annotated according to InterProScan (http://www.ebi.ac.uk/interpro/scan.html). We searched and selected the most likely candidate within the confidence interval by testing for either gene-metabolite association or association between the gene and the pathways in which the metabolite is involved. We also performed principal component analysis on the covariance matrix of metabolic traits measured in each tissue, which takes complex correlated data arranged in multidimensional space and reduces the dimensionality of the data into more simple, linearized axes while retaining as much original variation as possible (Wilson et al., 2004). QTL analysis for the principal components was performed as described above for the metabolic traits.
Epistasis Analysis
For each metabolic trait, we investigated the pairwise additive by additive epistatic interactions for all identified QTLs. Epistatic interactions were determined by two-way ANOVA (using P < 0.05 as significant threshold) using all QTLs in pairwise combinations (Yu et al., 1997). The proportion of variance explained by epistasis was tested by comparing the residual of the full model containing all single-locus effects and two-locus interaction effect with that of reduced model containing all single-locus effects but excluding two-locus interaction effect.
Network Construction and Visualization Based on Metabolite and Phenotypic Data
Metabolite levels in three different tissues from the RIL population as described above were measured and recorded. The obtained data sets were preprocessed as follows: (1) the lines with more than 90% missing for the metabolite levels were filtered out, (2) if the level of a specific metabolite was missing for more than half of the lines, (3) the metabolite was excluded from the analysis, and (4) the other missing values were imputed using random forest imputation (Liaw and Wiener, 2002). To robustly estimate the missing values, the imputation was repeated 10 times and the average of the imputed values was used. The metabolite relevance networks were obtained from each tissue-specific data set (i.e., leaf at seedling stage, leaf at reproductive stage, and kernel 15 DAP). To infer the networks, we applied pairwise Pearson correlation, and the coefficients above 0.5 at the significance level of 0.05 (FDR corrected) were selected to establish the edges (links) of the network. The same preprocessing procedure as for metabolite data was applied on the data for the agronomic traits (i.e., plant height, ear height, length of ear leaf, width of ear leaf, tassel length, tassel branch number, and biomass). The relationships between all measured metabolites across different tissues and the agronomic traits were obtained by employing elastic net regression (http://cran.r-project.org/package=elasticnet). The profiles of the metabolites from all tissues were considered as predictors (regressors). Regression models were then fitted for each agronomic trait separately. The regression coefficients were robustly estimated by 10-fold cross validation based on the optimum value for the penalty parameter from the set {0.01, 0.05, 0.1, 0.5, 1, 1.5, 2, 10, 100}. The r-square, which measures how well the data fitted to the regression model, was calculated for each model and represented in the network with the size of the corresponding node (the better the model fitted to the data, the bigger the size of the node).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: XP_008651482 (AGT), XP_008651749 (TDC1), and DAA43760 (TDC2). Scripts for constructing the genetic linkage map can be downloaded from https://github.com/panqingchun/linkage_map.
Supplemental Data
Supplemental Figure 1. Allele distribution within the B73/By804 RIL population.
Supplemental Figure 2. Distribution of metabolite level in the B73/By804 recombinant line population.
Supplemental Figure 3. Sequence differences between two TDC genes in B73.
Supplemental Figure 4. Metabolite relevance networks.
Supplemental Table 1. Epistatic interactions between QTL.
Supplemental Table 2. Primers used for resequencing AGT in B73 and By804 and PCR-based InDel detection.
Supplemental Table 3. Sequence polymorphisms between B73 and By804 in the candidate gene AGT identified by resequencing.
Supplemental Table 4. Sequence polymorphisms between B73 and By804 in the candidate gene TDC1 identified by resequencing.
Supplemental Table 5. Sequence polymorphisms between B73 and By804 in the candidate gene TDC2 identified by resequencing.
Supplemental Table 6. Primers used for resequencing TDC1 and TDC2 in B73 and By804 and PCR-based InDel detection.
Supplemental Table 7. Summary of QTL intervals for seven agronomic traits.
Supplemental Data Set 1A. Summary of metabolite information including tissue type of the RIL population in which metabolites were identified, metabolite name, metabolite class, retention time, and mass used for quantification.
Supplemental Data Set 1B. Significance of metabolite level changes between different types of tissues.
Supplemental Data Set 1C. Summary of QTL intervals for metabolites identified in three tissues.
Supplemental Data Set 1D. List of candidate genes and their detailed information in the peak bin for each QTL.
Supplemental Data Set 1E. List of candidate genes within the confidence interval for each QTL.
Supplemental Data Set 1F. GO term analysis for candidate genes within the QTL for all metabolites identified in three tissues.
Supplemental Data Set 1G. List of significant metabolite-metabolite correlations in different tissues.
Supplemental Data Set 1H. List of significant metabolite-agronomic trait associations based on elastic net regression.
Supplemental Data Set 1I. Data of metabolic traits detected in the seedling leaves of the By804/B73 RIL population.
Supplemental Data Set 1J. Data of metabolic traits detected in the leaves at reproductive stage of the By804/B73 RIL population.
Supplemental Data Set 1K. Data of metabolic traits detected in the kernels of the By804/B73 RIL population.
Supplemental Data Set 1L. Data of seven agronomic traits for the By804/B73 RIL population.
Supplemental Data Set 1M. Genotypic data and linkage map for the By804/B73 RIL population.
Supplementary Material
Acknowledgments
The research was financed by the National Natural Science Foundation of China (31201220, 31123009, and 31222041), the National Program on Key Basic Research Project of China (2014CB138202), the National Hi-Tech Research and Development Program of China (2012AA10A307), and within the framework of Deutsche Israeli Project FE 552/12-1, administrated by the Deutsche Forschungsgemeinschaft. W.W. was partly supported by a fellowship provided under the auspices of the EU-funded OPTICHINA project.
AUTHOR CONTRIBUTIONS
W.W., A.R.F., and J.Y. designed and supervised this study. W.W., K.L., S.A., Y.Z., L.Z., M.J., A.F., and W.L. performed the experiments. W.W., N.O., N.Y, Y.X., H.L., Q.P., and Z.N. performed the data analysis. W.W., A.R.F., and J.Y. prepared the article. All authors read, contributed, and approved the article.
Glossary
- QTL
quantitative trait loci
- RIL
recombinant inbred line
- SNP
single nucleotide polymorphism
- cM
centimorgan
- GC-TOF-MS
gas chromatography-time-of-flight-mass spectrometry
- TCA
tricarboxylic acid
- LOD
logarithm of odds
- mQTL
metabolite QTL
- BCAA
branched-chain amino acid
- PEPc
phosphoenolpyruvate carboxylase
- UTR
untranslated region
- FDR
false discovery rate
- pQTL
agronomic QTL
- DAP
days after pollination
Footnotes
Articles can be viewed online without a subscription.
References
- Albert F.W., Kruglyak L. (2015). The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 16: 197–212. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C., Aarts M.G., Bentsink L., Keurentjes J.J., Reymond M., Vreugdenhil D., Koornneef M. (2009). What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21: 1877–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alseekh S., et al. (2015). Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. Plant Cell 27: 485–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W.L., Martins A.O., Fernie A.R., Tohge T. (2014). 2-Oxoglutarate: linking TCA cycle function with amino acid, glucosinolate, flavonoid, alkaloid, and gibberellin biosynthesis. Front. Plant Sci. 5: 552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel G.W., Gaudinier A., Brady S.M., Hennig L., Rhee S.Y., De Smet I. (2012). Systems analysis of plant functional, transcriptional, physical interaction, and metabolic networks. Plant Cell 24: 3859–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S. (2010). Branched-chain amino acid metabolism in Arabidopsis thaliana. The Arabidopsis Book 8: e0137, doi/10.1199/tab.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadley K.J. (2010). The vascular effects of trace amines and amphetamines. Pharmacol. Ther. 125: 363–375. [DOI] [PubMed] [Google Scholar]
- Buckler E.S., et al. (2009). The genetic architecture of maize flowering time. Science 325: 714–718. [DOI] [PubMed] [Google Scholar]
- Calenge F., Saliba-Colombani V., Mahieu S., Loudet O., Daniel-Vedele F., Krapp A. (2006). Natural variation for carbohydrate content in Arabidopsis. Interaction with complex traits dissected by quantitative genetics. Plant Physiol. 141: 1630–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S.B. (2008). Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36. [DOI] [PubMed] [Google Scholar]
- Chan E.K., Rowe H.C., Corwin J.A., Joseph B., Kliebenstein D.J. (2011). Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol. 9: e1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chander S., Guo Y.Q., Yang X.H., Yan J.B., Zhang Y.R., Song T.M., Li J.S. (2008a). Genetic dissection of tocopherol content and composition in maize grain using quantitative trait loci analysis and candidate gene approach. Mol. Breed. 22: 353–365. [Google Scholar]
- Chander S., Guo Y.Q., Yang X.H., Zhang J., Lu X.Q., Yan J.B., Song T.M., Rocheford T.R., Li J.S. (2008b). Using molecular markers to identify two major loci controlling carotenoid contents in maize grain. Theor. Appl. Genet. 116: 223–233. [DOI] [PubMed] [Google Scholar]
- Chitwood D.H., Kumar R., Headland L.R., Ranjan A., Covington M.F., Ichihashi Y., Fulop D., Jiménez-Gómez J.M., Peng J., Maloof J.N., Sinha N.R. (2013). A quantitative genetic basis for leaf morphology in a set of precisely defined tomato introgression lines. Plant Cell 25: 2465–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet R., Vidal J., O’Leary M.H. (1996). Phosphoenolpyruvate carboxylase: A ubiquitous, highly regulated enzyme in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 273–298. [DOI] [PubMed] [Google Scholar]
- de Givry S., Bouchez M., Chabrier P., Milan D., Schiex T. (2005). CarthaGene: multipopulation integrated genetic and radiation hybrid mapping. Bioinformatics 21: 1703–1704. [DOI] [PubMed] [Google Scholar]
- Do P.T., Prudent M., Sulpice R., Causse M., Fernie A.R. (2010). The influence of fruit load on the tomato pericarp metabolome in a Solanum chmielewskii introgression line population. Plant Physiol. 154: 1128–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie A.R., Schauer N. (2009). Metabolomics-assisted breeding: a viable option for crop improvement? Trends Genet. 25: 39–48. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick T.B., Basset G.J., Borel P., Carrari F., DellaPenna D., Fraser P.D., Hellmann H., Osorio S., Rothan C., Valpuesta V., Caris-Veyrat C., Fernie A.R. (2012). Vitamin deficiencies in humans: can plant science help? Plant Cell 24: 395–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., et al. (2013). RNA sequencing reveals the complex regulatory network in the maize kernel. Nat. Commun. 4: 2832. [DOI] [PubMed] [Google Scholar]
- Gomes de Oliveira Dal’Molin C., Quek L.E., Saa P.A., Nielsen L.K. (2015). A multi-tissue genome-scale metabolic modeling framework for the analysis of whole plant systems. Front. Plant Sci. 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G., Höfgen R. (2002). Metabolic engineering of amino acids and storage proteins in plants. Metab. Eng. 4: 3–11. [DOI] [PubMed] [Google Scholar]
- Ganal M.W., Polley A., Graner E.M., Plieske J., Wieseke R., Luerssen H., Durstewitz G. (2012). Large SNP arrays for genotyping in crop plants. J. Biosci. 37: 821–828. [DOI] [PubMed] [Google Scholar]
- Ganal M.W., et al. (2011). A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE 6: e28334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y., Pyl E.T., Sulpice R., Lunn J.E., Höhne M., Günther M., Stitt M. (2009). Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant Cell Environ. 32: 859–874. [DOI] [PubMed] [Google Scholar]
- Gong L., Chen W., Gao Y., Liu X., Zhang H., Xu C., Yu S., Zhang Q., Luo J. (2013). Genetic analysis of the metabolome exemplified using a rice population. Proc. Natl. Acad. Sci. USA 110: 20320–20325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley C. (2011). A cornucopia of maize genes. Nat. Genet. 43: 87–88. [DOI] [PubMed] [Google Scholar]
- Hirsch C.N., et al. (2014). Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26: 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks M.A., Allwood J.W., Harrison J.K.D., Kopka J., Erban A., Goodacre R., Balk J. (2014). Selective induction and subcellular distribution of ACONITASE 3 reveal the importance of cytosolic citrate metabolism during lipid mobilization in Arabidopsis. Biochem. J. 463: 309–317. [DOI] [PubMed] [Google Scholar]
- Huang S., Taylor N.L., Ströher E., Fenske R., Millar A.H. (2013). Succinate dehydrogenase assembly factor 2 is needed for assembly and activity of mitochondrial complex II and for normal root elongation in Arabidopsis. Plant J. 73: 429–441. [DOI] [PubMed] [Google Scholar]
- Huang X., Han B. (2012). A crop of maize variants. Nat. Genet. 44: 734–735. [DOI] [PubMed] [Google Scholar]
- Häusler R.E., Ludewig F., Krueger S. (2014). Amino acids--a life between metabolism and signaling. Plant Sci. 229: 225–237. [DOI] [PubMed] [Google Scholar]
- Jander G., Joshi V. (2009). Aspartate-derived amino acid biosynthesis in Arabidopsis thaliana. The Arabidopsis Book 7: e0121, doi/10.1199/tab.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper E.L., Neto G.C., Papes F., Moraes K.C., Leite A., Arruda P. (1999). The role of opaque2 in the control of lysine-degrading activities in developing maize endosperm. Plant Cell 11: 1981–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D. (2009). Advancing genetic theory and application by metabolic quantitative trait loci analysis. Plant Cell 21: 1637–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D.J., Figuth A., Mitchell-Olds T. (2002). Genetic architecture of plastic methyl jasmonate responses in Arabidopsis thaliana. Genetics 161: 1685–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J., et al. (2005). GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics. 21: 1635–1638. [DOI] [PubMed] [Google Scholar]
- Liaw A., Wiener M. (2002). Classification and regression by random forest. R News 2: 18–22. [Google Scholar]
- Lisec J., Meyer R.C., Steinfath M., Redestig H., Becher M., Witucka-Wall H., Fiehn O., Törjék O., Selbig J., Altmann T., Willmitzer L. (2008). Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J. 53: 960–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J., Schauer N., Kopka J., Willmitzer L., Fernie A.R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 1: 387–396. [DOI] [PubMed] [Google Scholar]
- Loudet O., Chaillou S., Merigout P., Talbotec J., Daniel-Vedele F. (2003). Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis. Plant Physiol. 131: 345–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H., Dudareva N. (2012). The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 63: 73–105. [DOI] [PubMed] [Google Scholar]
- Martin C., Butelli E., Petroni K., Tonelli C. (2011). How can research on plants contribute to promoting human health? Plant Cell 23: 1685–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F., Okazaki Y., Oikawa A., Kusano M., Nakabayashi R., Kikuchi J., Yonemaru J., Ebana K., Yano M., Saito K. (2012). Dissection of genotype-phenotype associations in rice grains using metabolome quantitative trait loci analysis. Plant J. 70: 624–636. [DOI] [PubMed] [Google Scholar]
- Moose S.P., Dudley J.W., Rocheford T.R. (2004). Maize selection passes the century mark: a unique resource for 21st century genomics. Trends Plant Sci. 9: 358–364. [DOI] [PubMed] [Google Scholar]
- Nimmo H.G. (2000). The regulation of phosphoenolpyruvate carboxylase in CAM plants. Trends Plant Sci. 5: 75–80. [DOI] [PubMed] [Google Scholar]
- Paris S., Wessel P.M., Dumas R. (2002). Overproduction, purification, and characterization of recombinant bifunctional threonine-sensitive aspartate kinase-homoserine dehydrogenase from Arabidopsis thaliana. Protein Expr. Purif. 24: 105–110. [DOI] [PubMed] [Google Scholar]
- Paulus J.K. Schlieper, D., Groth, G. (2013). Greater efficiency of photosynthetic carbon fixation due to single amino-acid substitution. Nat. Commun. 4: 1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips P.C. (2008). Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 9: 855–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.H. (2006). Believe it or not, QTLs are accurate! Trends Plant Sci. 11: 213–216. [DOI] [PubMed] [Google Scholar]
- Riedelsheimer C., Czedik-Eysenberg A., Grieder C., Lisec J., Technow F., Sulpice R., Altmann T., Stitt M., Willmitzer L., Melchinger A.E. (2012a). Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nat. Genet. 44: 217–220. [DOI] [PubMed] [Google Scholar]
- Riedelsheimer C., Lisec J., Czedik-Eysenberg A., Sulpice R., Flis A., Grieder C., Altmann T., Stitt M., Willmitzer L., Melchinger A.E. (2012b). Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc. Natl. Acad. Sci. USA 109: 8872–8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U., Luedemann A., Brust D., Fiehn O., Linke T., Willmitzer L., Fernie A. (2001). Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes S.E., Dewaele E., Aas S.F., Jacobs M., Frankard V. (2003). Transcriptional and biochemical regulation of a novel Arabidopsis thaliana bifunctional aspartate kinase-homoserine dehydrogenase gene isolated by functional complementation of a yeast hom6 mutant. Plant Mol. Biol. 51: 281–294. [DOI] [PubMed] [Google Scholar]
- Rowe H.C., Hansen B.G., Halkier B.A., Kliebenstein D.J. (2008). Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell 20: 1199–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi S., Tuberosa R. (2005). To clone or not to clone plant QTLs: present and future challenges. Trends Plant Sci. 10: 297–304. [DOI] [PubMed] [Google Scholar]
- Sauvage C., Segura V., Bauchet G., Stevens R., Do P.T., Nikoloski Z., Fernie A.R., Causse M. (2014). Genome-wide association in tomato reveals 44 candidate loci for fruit metabolic traits. Plant Physiol. 165: 1120–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N., et al. (2006). Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24: 447–454. [DOI] [PubMed] [Google Scholar]
- Schmidt R.J., Ketudat M., Aukerman M.J., Hoschek G. (1992). Opaque-2 is a transcriptional activator that recognizes a specific target site in 22-kD zein genes. Plant Cell 4: 689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon R.S., Lin H., Childs K.L., Hansey C.N., Buell C.R., de Leon N., Kaeppler S.M. (2011). Genome-wide atlas of transcription during maize development. Plant J. 66: 553–563. [DOI] [PubMed] [Google Scholar]
- Springer N.M., et al. (2009). Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 5: e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R., et al. (2009). Starch as a major integrator in the regulation of plant growth. Proc. Natl. Acad. Sci. USA 106: 10348–10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R., et al. (2010). Network analysis of enzyme activities and metabolite levels and their relationship to biomass in a large panel of Arabidopsis accessions. Plant Cell 22: 2872–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove L.J., Beard K.F., Nunes-Nesi A., Fernie A.R., Ratcliffe R.G. (2010). Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci. 15: 462–470. [DOI] [PubMed] [Google Scholar]
- Tester M., Langridge P. (2010). Breeding technologies to increase crop production in a changing world. Science 327: 818–822. [DOI] [PubMed] [Google Scholar]
- Thatcher S.R., Zhou W., Leonard A., Wang B.B., Beatty M., Zastrow-Hayes G., Zhao X., Baumgarten A., Li B. (2014). Genome-wide analysis of alternative splicing in Zea mays: landscape and genetic regulation. Plant Cell 26: 3472–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubiana D., Batushansky A., Tzfadia O., Scossa F., Khan A., Barak S., Zamir D., Fernie A.R., Nikoloski Z., Fait A. (2015). Combined correlation-based network and mQTL analyses efficiently identified loci for branched-chain amino acid, serine to threonine, and proline metabolism in tomato seeds. Plant J. 81: 121–133. [DOI] [PubMed] [Google Scholar]
- Toubiana D., Semel Y., Tohge T., Beleggia R., Cattivelli L., Rosental L., Nikoloski Z., Zamir D., Fernie A.R., Fait A. (2012). Metabolic profiling of a mapping population exposes new insights in the regulation of seed metabolism and seed, fruit, and plant relations. PLoS Genet. 8: e1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace J.G., Bradbury P.J., Zhang N., Gibon Y., Stitt M., Buckler E.S. (2014). Association mapping across numerous traits reveals patterns of functional variation in maize. PLoS Genet. 10: e1004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Basten C.J., Zeng Z.B. (2006). Windows QTL Cartographer 2.5. (Raleigh, NC: Department of Statistics, North Carolina State University; ). [Google Scholar]
- Wen W., Li D., Li X., Gao Y., Li W., Li H., Liu J., Liu H., Chen W., Luo J., Yan J. (2014). Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat. Commun. 5: 3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L.M., Whitt S.R., Ibáñez A.M., Rocheford T.R., Goodman M.M., Buckler E.S. IV (2004). Dissection of maize kernel composition and starch production by candidate gene association. Plant Cell 16: 2719–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., et al. (2010). Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nat. Genet. 42: 322–327. [DOI] [PubMed] [Google Scholar]
- Yan J.B., Warburton M., Crouch J. (2011). Association mapping for enhancing maize (Zea mays L.) genetic improvement. Crop Sci. 51: 433–449. [Google Scholar]
- Yang X., Guo Y., Yan J., Zhang J., Song T., Rocheford T., Li J.S. (2010). Major and minor QTL and epistasis contribute to fatty acid compositions and oil concentration in high-oil maize. Theor. Appl. Genet. 120: 665–678. [DOI] [PubMed] [Google Scholar]
- Yang X.H., Gao S., Xu S., Zhang Z., Prasanna B.M., Li L., Li J., Yan J. (2011). Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol. Breed. 28: 511–526. [Google Scholar]
- Yu S.B., Li J.X., Xu C.G., Tan Y.F., Gao Y.J., Li X.H., Zhang Q., Saghai Maroof M.A. (1997). Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA 94: 9226–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z.B., Kao C.H., Basten C.J. (1999). Estimating the genetic architecture of quantitative traits. Genet. Res. 74: 279–289. [DOI] [PubMed] [Google Scholar]
- Zheng P., et al. (2008). A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nat. Genet. 40: 367–372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.