Nicotinate O-glucosylation, a Brassicaceae-specific metabolic trait, plays a key role in seed germination under stress conditions in Arabidopsis thaliana.
Abstract
The glycosylation of nicotinate (NA), a key intermediate of the NAD salvage pathway, occurs widely in land plants. However, the physiological function of NA glycosylation is not well understood in planta, and no gene encoding NA glycosyltransferase has been reported to date. NA glycosylation in Arabidopsis thaliana occurs at either the N- or the O-position of the NA molecule, and O-glucosylation appears to be unique to the Brassicaceae. Using gene-enzyme correlations focused on Family 1 glycosyltransferases (GTs; EC 2.4), we identified and characterized three Arabidopsis GTs, which are likely involved in NA glycosylation. These include one NAOGT (UGT74F2; previously identified as a salicylic acid glycosyltransferases) and two NANGTs (UGT76C4 and UGT76C5). Arabidopsis mutants of UGT74F2 accumulate higher levels of free NA, but not salicylic acid, than that of the wild type, and this inversely correlated with seed germination rates under various abiotic stresses. The germination defect of the ugt74f2-1 mutant could be fully complemented by overexpression of UGT74F2. These observations, together with comprehensive chemical analysis, suggest that NA glycosylation may function to protect plant cells from the toxicity of NA overaccumulation during seed germination. Combined with phylogenetic analysis, our results suggest that NAOGTs arose recently in the Brassicaceae family and may provide a fitness benefit. The multifunctionality of UGT74F2 in Arabidopsis is also investigated and discussed.
INTRODUCTION
NAD+ and its derivatives (e.g., NADH, NADP+, and NADPH) are essential cofactors for many of the more than 1800 redox reactions that widely occur across all kingdoms of life (Gossmann et al., 2012). Common redox reactions do not generally lead to net consumption of NAD [we use the term NAD to represent NAD(P)(H) hereafter]. On the contrary, a growing number of enzymes that degrade the core structure of NAD have recently been identified in various organisms (reviewed in Hashida et al., 2009; Imai, 2009). A common feature of these NAD-consuming reactions is the release of nicotinamide (NIM), which then feeds into the conserved NAD salvage pathway found in all organisms.
Among the NAD-consuming enzymes found in plants, poly(ADP-ribose) polymerase (PARP) is likely the primary contributor to NAD degradation in planta. This is due to its high turnover rate for NAD compared with other enzymes such as Sirtuins and NUDX (nucleoside diphosphate linked to a moiety-X) hydrolases (Briggs and Bent, 2011). PARP homologs are found in almost all eukaryotic organisms. The Arabidopsis genome contains three PARPs: PARP-1, PARP-2, and PARP-3, encoded by AT3G02390, AT2G31320, and AT5G22470, respectively. PARPs are known to be involved in abiotic stress tolerance in plants (e.g., high light, drought, and oxidative stress) (De Block et al., 2005); however, PARP-3 expresses specifically in seeds and was recently proposed to play an important role during seed germination (Hunt et al., 2007; Rissel et al., 2014).
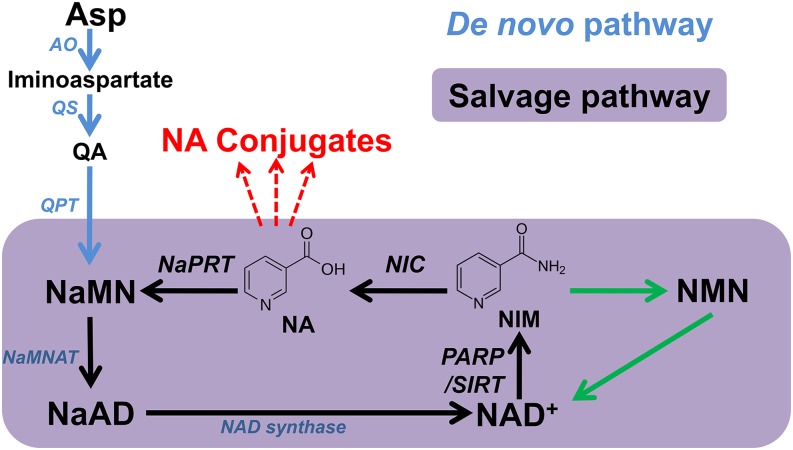
To maintain stable NAD pools, plants have evolved two different salvage pathways to recycle NIM back to NAD. One such pathway operates only in green algae, in which NIM is converted to nicotinamide mononucleotide (NMN) by nicotinamide phosphoribosyltransferase and further adenylated to form NAD (Lin et al., 2010). The second pathway is ubiquitous in all land plants in which NIM is initially converted to nicotinate (NA) by nicotinamidase (Hunt et al., 2007; Wang and Pichersky, 2007). The resulting NA is then used as the substrate in the three-step Preiss-Handler pathway for NAD production (Figure 1). The NAD salvage pathway is known to play a key role in plant abiotic stress tolerance. This conclusion was drawn from the evidence that nic1-1 mutant, in which the salvage pathway was blocked, is more sensitive to high salt and abscisic acid treatments compared with wild-type plants (Wang and Pichersky, 2007).
Figure 1.
Metabolism of NAD in Plants.
The steps involved in the de novo pathway are indicated by blue arrows, the four-step salvage pathway (start from NIM) is indicated by a black arrow, and two-step salvage pathway is indicated by green arrows. AO, aspartate oxidase; NaAD, nicotinate adenine dinucleotide; NAD, nicotinamide adenine dinucleotide; NaMN, nicotinate mononucleotide; NaMNAT, NaMN adenylyltransferase; NaPRT, nicotinate phosphoribosyltransferase; NIC, nicotinamidase; PARP, poly(ADP-ribose) polymerase; QPT, quinolinate phosphoribosyltransferase; QS, quinolinate synthase; SIRT, sirtuins (NAD dependent protein deacetylases).
Most of our knowledge about the NAD salvage pathway in plants has been obtained from feeding experiments, typically using carbonyl [14C]-nicotinamide ([14C]-NIM) as a tracer (Ashihara et al., 2005; Matsui et al., 2007; Wang and Pichersky, 2007). These experiments showed that, in addition to being a precursor for NAD replenishment in planta, NA can also be converted into a variety of pyridine-ring-containing metabolites. Two common NA conjugates are commonly detected at the N- position in most plant species: trigonelline (also known as N-methyl nicotinate) and nicotinate N-glucoside (NANG). The formation of trigonelline and NANG is catalyzed by S-adenosyl-l-methionine-dependent N-methyltransferases (EC 2.1.1.7) and nicotinate N-glucosyltransferase (NANGT; EC 2.4.1.196) respectively (Willeke et al., 1979; Ashihara et al., 2005, 2011, 2012; Matsui et al., 2007). Although NA conjugation has been proposed to function in detoxification, there is, to date, no genetic evidence to support this hypothesis. Genes encoding NA modification enzymes have not yet been identified from plants (Ashihara et al., 2005; Wang and Pichersky, 2007). Previously, we detected at least three different NA-derived metabolites in leaf tissue of Arabidopsis thaliana fed with exogenous [14C]-NIM, although no structural information was available for two of these three compounds at the time (Wang and Pichersky, 2007). One of these NA conjugates was not reported before, suggesting that it may not occur widely in plants other than Arabidopsis and relatives (Matsui et al., 2007; Wang and Pichersky, 2007).
To investigate the physiological functions and evolutionary trajectory of NA modifications in planta, we determined the structures of several NA glucosides in Arabidopsis. Moreover, we identified three putative NA glycosyltransferase genes from the Family 1 UGTs (which contains106 members) using gene-enzyme (G-E) correlation analysis. Whereas UGT74F2 catalyzes the O-glucosylation at the carboxyl position of NA, both UGT76C4 and UGT76C5 catalyze the N-glucosylation at the pyridyl nitrogen of NA in vitro. We further show that deficiency in UGT74F2 delayed seed germination rates under various abiotic stress conditions and demonstrate that this delay was caused by overaccumulation of free NA, but not of other substrates of UGT74F2 salicylic acid (SA), o-aminobenzoate (o-ABA), or benzoic acid (BA). Our comprehensive and combined phylogenetic and biochemical analysis of glycosyltransferase proteins demonstrates that the NAOGTs arose recently in the Brassicaceae and have acquired an important, physiological function during seed germination. The importance of multifunctionality of UGT74F2 in Arabidopsis is further discussed in the context of our results.
RESULTS
Structural Elucidation of Glycosylated NA in Arabidopsis
We previously reported that excised Arabidopsis leaf discs fed with [14C]-NIM accumulated [14C]-NA and two unknown labeled compounds (Wang and Pichersky, 2007). In this study, we used liquid chromatography/electrospray ionization-quadrupole time-of-flight mass spectrometry (LC/ESI-qTOF-MS) to characterize the structures of these two unknown compounds in Arabidopsis. We evaluated material from 7-d-old seedlings of wild type (Columbia-0 [Col-0]) and nic1-1 mutants that had been treated with 200 µM NIM for 24 h. We reasoned that the differentially accumulated metabolites detected in the wild-type plants versus the nic1-1 mutant are likely NA derivatives because nic1-1, which carries a T-DNA insertional mutation in the first committed enzyme in the NAD salvage pathway, is incapable of converting NIM to NA and its derivatives (Figure 2) (Wang and Pichersky, 2007). Two major compounds were identified in wild-type plants, which fulfill this criterion (peaks labeled as “unknown 1” and “unknown 2” in the LC chromatogram for Col-0; Figure 2). The [M+H]+ ions for unknown 1 and unknown 2 display monoisotopic mass-to-charge ratios (m/z) at 286.0932 and 286.0928, respectively. Both compounds also yielded similar daughter ions of m/z 124.0400, consistent with the fragmentation of an NA moiety (C6H6NO2+ with calculated m/z at 124.0398). The lost fragments for both unknown 1 and unknown 2 therefore have a predicted molecular weight of 162.0 (C6H10O5), consistent with a hexose moiety. These two compounds are likely hexosides of NA either at the O- or N- position corresponding to unknown 1 and 2, respectively, based on the polarity of their chemical structures and their behavior on thin-layer chromatography (TLC) and in our HPLC analytical system (Figure 2). Subsequently, we compared our liquid chromatography-mass spectrometry (LC-MS) results for the unknowns with results obtained during an analysis of enzymatically synthesized chemicals (see below) and thus concluded that the hexose group attached to NA in both of these compounds was glucose.
Figure 2.
Workflow for the Structural Characterization of the Two Major Unknown NA Derivatives in Arabidopsis Using LC/ESI-qTOF-MS.
The positions on the liquid chromatograph of Unknown 1 and Unknown 2 (top left) were determined by comparison of plant analytes extracted from Col-0 and the nic1-1 mutant after treatment with 200 μM NIM. The individual mass spectra of peaks of Unknown 1 and Unknown 2 (top right and middle, x axis units = m/z) were used for structural elucidation (bottom). NA and NIM authentic standards were analyzed and used for comparison with plant analytes.
NAOG Is Unique in the Brassicaceae Family
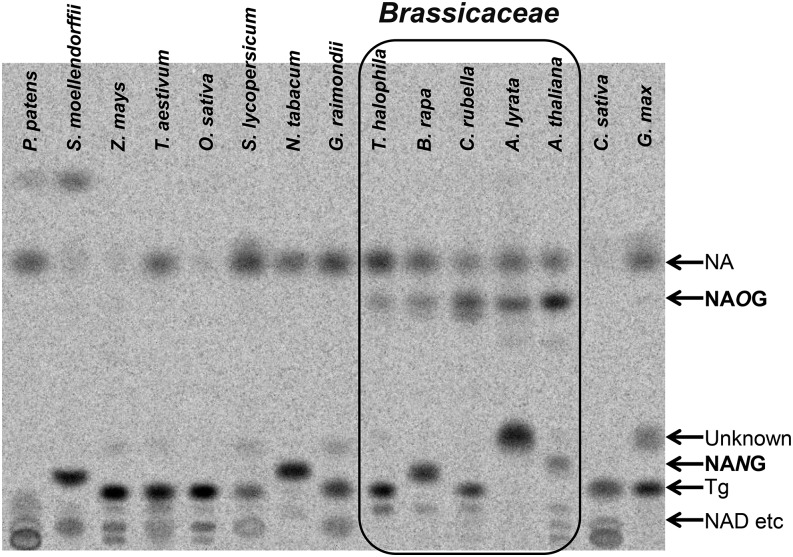
As NAOG has previously not been reported in other tested plant species (Willeke et al., 1979; Ashihara et al., 2005, 2011, 2012; Matsui et al., 2007), we explored the distribution of NAOG in various plants by feeding experiments using 10 µM [14C]-NIM. Our selected plants included several species from the Brassicaceae family: Arabidopsis lyrata, Capsella rubella, Brassica rapa, and Thellungiella halophila. The results showed that NAOG occurs exclusively in the Brassicaceae. The highest levels of NAOG accumulation were observed in A. thaliana and A. lyrata, based on the percentage distribution of labeled chemicals (Figure 3). We did not detect any NAOG in plant species outside the Brassicaceae family, including other rosids (Gossypium raimondii (Malvaceae), Glycine max (Fabaceae), and Cucumis sativa (Cucurbitaceae), asterids (Nicotiana tabacum and Solanum lycopersicum [Solanaceae]), monocots (Zea mays, Triticum aestivum, and Oryza sativa [Poaceae]), and in spikemosses and mosses (Selaginella moellendorffii or Physcomitrella patens) (Figure 3). It is worth noting that we observed an additional NA conjugate that predominated in A. lyrata; this finding suggested that A. lyrata contains an additional NA modification not present in the closely related A. thaliana.
Figure 3.
[14C]-Labeled Chemical Profiling of Leaf Extracts from Different Species Following Separation by TLC.
The leaf tissue was treated with 10 µM [14C]-NIM for 4 h. NAOG, nicotinate O-glucoside; Tg, trigonelline.
To compare the metabolic fate of the two glucosylated NAs in Arabidopsis, we placed 7-d-old Arabidopsis seedlings in liquid medium containing [14C]-NIM for 24 h. The [14C]-containing compounds in both the plant material and the liquid medium were separated and quantified. We observed that >95% of the labeled NAOG was retained in the plant cells. In contrast, more than 20% of labeled NANG was secreted into the medium (Supplemental Figures 1A and 1B). To further compare the biochemical properties of these two NA glucosides, we treated chemical extracts of [14C]-NIM-treated seedlings with β-glucosidase (0.05 units). The results clearly showed that NAOG was degraded to NA by β-glucosidase but NANG was not (Supplemental Figure 1C).
NA Glycosyltransferase Genes Discovered by G-E Correlation Analysis
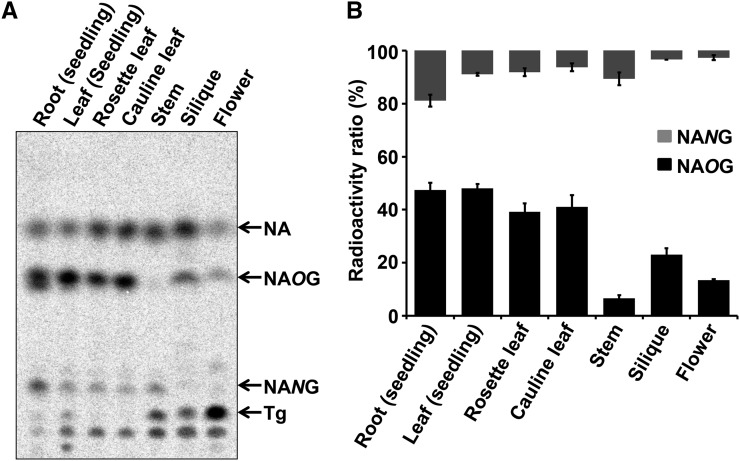
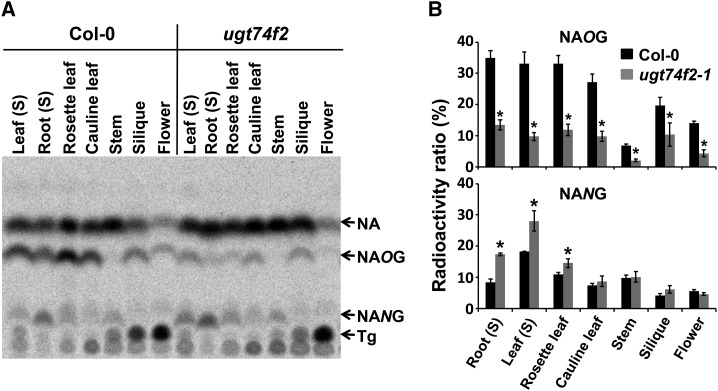
The tissue-specific accumulation pattern of a compound can help identify its biosynthetic genes, which may also exhibit similar expression patterns. To understand the spatial patterns of NA glycosylation in Arabidopsis, seven different Arabidopsis tissues were treated with [14C]-NIM; we measured NAOG and NANG accumulation in these tissues (for detailed information about the selected tissues; Supplemental Table 1). The accumulated levels of NAOG in seedlings, rosette leaves, and cauline leaves were all similar. NAOG levels were significantly lower in stems and flowers (Figure 4). NANG is more highly accumulated in roots (Figure 4). Notably, trigonelline, another common NA conjugate in plants, accumulated predominantly in Arabidopsis flowers.
Figure 4.
Quantitative Determination of [14C]-Labeled NAOG and NANG Content in Various Organs of Arabidopsis.
(A) Excised organs were immersed in 10 μM [14C]-NIM solution for 6 h. The radiolabeled chemicals were extracted and separated by TLC.
(B) Relative quantification of [14C]-NAOG and [14C]-NANG in different organs. Data are presented as means ± se (n = 3).
To identify enzymes involved in NA glycosylation in Arabidopsis, we first attempted to purify the NAOGT protein from crude protein extracts of Arabidopsis seedlings using various combinations of chromatographic columns, including phenyl-, Q-, Blue-Sepharose, and monoQ matrices. We detected NAOGT activity from desalted crude protein extracts of 7-d-old Arabidopsis seedlings using [14C]-labeled NA and UDP-Glc as substrates (the standard reaction included 20 μg of crude protein; extracts were incubated at 28°C for 6 h). Unfortunately, the crude protein completely lost NAOGT activity after two or three chromatographic steps due to its instability in our protein purification condition. Only the Km value of 0.36 ± 0.021 mM (n = 3) toward NA was determined with the partially purified crude protein, using UDP-Glc as a cosubstrate. We did not detect NANGT activity from the crude enzyme extracts from roots, even after incubating the reactions overnight using NA and UDP-Glc as substrates, probably due to its instability or inhibition by some unknown factors in the crude extract.
We postulated that members from the Family 1 GTs may catalyze the NA glycosylation reactions as the Family 1 GTs are known to be responsible for most, if not all, of the small-molecule glycosylations in plants (Bowles et al., 2006). In an attempt to identify NAOGT and NANGT candidate genes, we performed in silico screening of 106 Arabidopsis family 1 UGT genes. We integrated publicly available microarray data with [14C]-NAOG and [14C]-NANG accumulation patterns, which are indicators of corresponding enzyme activity, using two-tailed Pearson’s correlation analysis in selected tissues (see detailed information in Supplemental Table 1). To simplify our G-E correlation analysis, all transcript and metabolite signals were transformed to relative content (the highest data point was set as 1.0; there were seven data points for each variety, n = 7; Supplemental Data Set 1). G-E correlation analysis indicated that NAOG was correlated with four UGTs (we set the cutoff values for correlation as r > 0.75 and P < 0.05), of which, UGT74F2 was ranked highest.
We found that recombinant UGT74F2 (AT2G43820) catalyzed the conversion of NA to NAOG in vitro using UDP-Glc as cosubstrate (Figure 5). We further determined the kinetic properties of UGT74F2 for NA. UGT74F2 showed a strong preference for NA (Kcat/Km = 42.9 ± 1.32 mM−1 s−1, n = 3; Table 1). Given that many small molecule GTs are often able to accept identical substrates, we further evaluated the catalytic properties of seven UGT74F2 homologs from Arabidopsis (AT1G05530, UGT75B2; AT2G31750, UGT74D1; AT2G31790, UGT74C1; AT1G24100, UGT74B1; AT1G05560, UGT75B1; AT1G05680, UGT74E2; and AT2G43840, UGT74F1). Of these, only recombinant UGT75B1 exhibited weak NAOGT activity, which is <10% of the NAOGT observed for UGT74F2 (Figure 5). Previously, UGT74F2, and its close homolog and tandem duplicate UGT74F1, had been shown to exhibit activity toward various chemicals containing aromatic rings such as SA, BA, and o-ABA (also known as anthranilate) (Lim et al., 2002; Quiel and Bender, 2003; Dean and Delaney, 2008). To gain deeper insights into the biochemical properties of UGT74F2 and UGT74F1, we assessed their substrate specificities in the same assay conditions. UGT74F2 had 4-fold higher catalytic efficiency (Kcat/Km) toward o-ABA and BA than that of NA (Table 1); UGT74F2 only showed weak activity to SA and produced slightly more salicylic acid glucose ester (SGE) than salicylic acid 2-O-glucose (SAG) (Supplemental Figure 2). UGT74F1 showed similar activity to BA, o-ABA, and SA (only SAG was produced) (Supplemental Figure 2). Moreover, we did not detect NAOGT activity with the recently identified SAGT (AT3G11340, UGT76B1) from Arabidopsis, although the SAGT activity of UGT76B1 was confirmed (Noutoshi et al., 2012; Supplemental Figure 3).
Figure 5.
NA Glucosyltransferase Assays of 13 Recombinant Arabidopsis Proteins.
Radio-TLC chromatogram of NAOG and NANG generated by the recombinant Arabidopsis glucosyltransferases using [14C]-NA and UDP-Glc as substrates. Crude protein extracts from 7-d-old Arabidopsis seedlings were used as a positive control for the NAOGT assay. The proteins with obvious NAOGT or NANGT activity are highlighted in bold.
Table 1. Kinetic Parameters for NA Glucosyltransferases Cloned in This Study.
| Enzyme | Substrate | Km (mM) | Kcat/Km (mM−1 s−1) |
|---|---|---|---|
| Nicotinatea | 0.22 ± 0.02b | 42.9 ± 1.32 | |
| UDP-Gluc | 0.19 ± 0.04 | 32.8 ± 1.41 | |
| UGT74F2 | Benzoic acid | 0.50 ± 0.12 | 198.3 ± 25.4 |
| Salicylic acid | N.D. | N.D. | |
| o-ABA | 0.84 ± 0.42 | 80.4 ± 26.8 | |
| UGT76C4 | NA | 0.74 ± 0.30 | 5.09 ± 1.17 |
| UDP-Glu | 0.064 ± 0.022 | 15.88 ± 5.40 | |
| UGT76C5 | NA | 0.56 ± 0.078 | 7.99 ± 1.06 |
| UDP-Glu | 0.22 ± 0.065 | 6.41 ± 1.25 | |
| Aly483498 | NA | 0.17 ± 0.03 | 36.59 ± 3.00 |
| UDP-Glu | 0.19 ± 0.02 | 23.48 ± 3.96 | |
| Carubv10023202 | NA | 0.24 ± 0.060 | 59.36 ± 10.19 |
| UDP-Glu | 0.15 ± 0.011 | 57.78 ± 3.26 | |
| Thhalv10001440 | NA | 0.32 ± 0.045 | 29.90 ± 2.59 |
| UDP-Glu | 0.32 ± 0.014 | 13.74 ± 1.39 | |
| Bra004787 | NA | 0.24 ± 0.016 | 56.02 ± 0.10 |
| UDP-Glu | 0.47 ± 0.012 | 17.17 ± 0.68 |
N.D., not determined due to the low activity.
Kinetic parameters were determined with 2.0 mM UDP-Glu.
All data gained in this study are presented as mean ± sd from triplicate independent assays.
Kinetic parameters were determined with 1.0 mM NA.
In our G-E correlation analysis of NANG activity, we initially focused our attention on the top 10 candidate genes (r > 0.75 and P < 0.05; Supplemental Data Set 1) and UGT 76C subfamily (UGT76C1 to UGT76C5). We selected all UGT 76C members for two reasons: one, UGT76C4 and UGT76C5 ranked as the 10th and 8th positions, respectively, in the Pearson correlation analysis; and two, members of this subfamily (UGT76C1 and UGT76C2) were previously functionally characterized as cytokinin N-glucosyltransferases (Hou et al., 2004). We expressed a total of 13 candidate genes with tags in Escherichia coli cells and purified the recombinant enzymes by affinity chromatography (no purified recombinant UGT79B11 could be obtained). Among them, two recombinant proteins (UGT76C4 encoded by AT5G05880 and UGT76C5 encoded by AT5G05890) showed activity toward NA (Figure 5, right panel). Recombinant UGT76C4 had a Kcat of 3.57 ± 0.67 s−1 and a Km of 737 ± 301 μM (n = 3) toward NA (Table 1). UGT76C5 had a Kcat of 4.46 ± 0.027 s−1 and a Km of 564 ± 78 μM toward NA. No NANGT activity was detected for UGT76C1, UGT76C2, or UGT76C3 (Figure 5). No NANGT activity could be detected with other tested UGTs, including UGT73B4, UGT76E4, UGT83A1, UGT90A2, UGT79B10, UGT71C1, UGT79B8, and UGT76C1 to UGT76C3 (Figure 5; Supplemental Figure 4).
Further biochemical experiments demonstrated that UGT74F2, UGT76C4, and UGT76C5 did not show any detectable activity toward NA analogs such as [14C]-NIM or methyl nicotinate (MeNA). Furthermore, no NA conjugated products were detected with the three isolated NAGTs (UGT74F2, UGT76C4, and UGT76C5) when UDP-Gal, UDP-glucuronate, or UDP-N-acetylglucosamine was tested as sugar donors with the following exceptions: Low activity (<10% that of UDP-Glc) was observed for UGT74F2 using UDP-Gal and UGT76C4 had similarly low activity using UDP-N-acetylglucosamine (Supplemental Figure 5).
Spatial Expression Pattern and Subcellular Localization of UGT74F2, UGT76C4, and UGT76C5
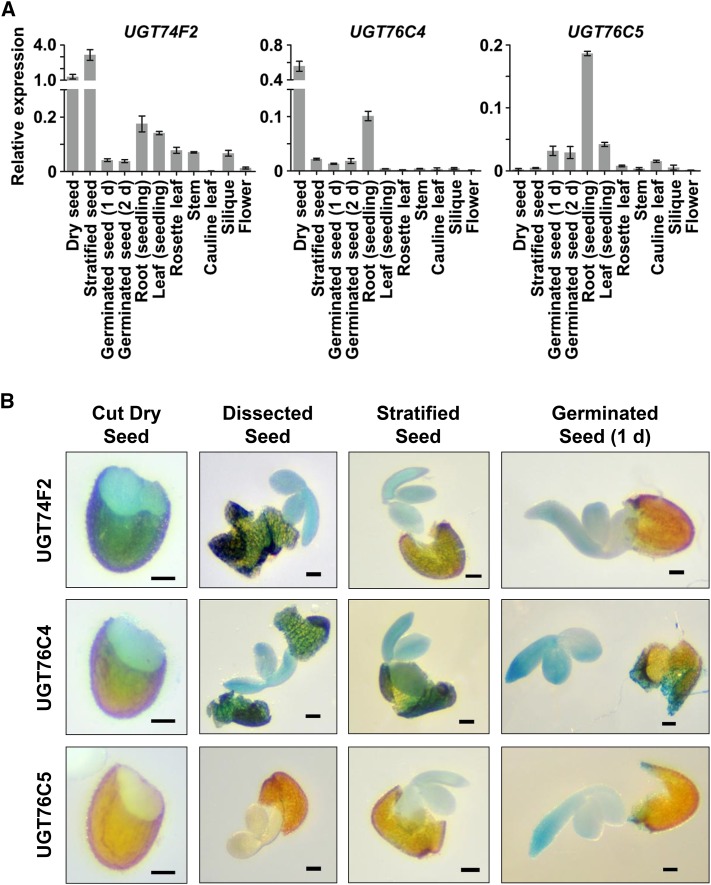
To gain more hints to the in planta physiological function(s) of the aforementioned NAGTs, real-time PCR was performed for each gene relative to actin (AT3G18780) as an internal control. As shown in Figure 6, both UGT74F2 and UGT76C4 were expressed predominately in dry seeds, and the highest level of UGT76C5 expression was detected in root tissue of 7-d-old seedlings. The expression profiles of these GTs were also examined in plants using β-glucuronidase (GUS) as a reporter gene, and the results largely confirmed the predominantly dry-seed patterns of expression for UGT74F2 and UGT76C4. The slight pattern differences between quantitative RT-PCR and GUS staining is probably caused by the UGT74F2 promoter elements not included in this experiment (Figure 6B). Altogether, these results suggest that UGT74F2 and UGT76C4 likely have a physiological function in seed development/germination.
Figure 6.
Expression Patterns of UGT74F2, UGT76C4, and UGT76C5 in Arabidopsis Tissues.
(A) qRT-PCR analysis of UGT74F2, UGT76C4, and UGT76C5 transcript levels in different tissues. Error bars represent the sd of three independent experiments.
(B) Histochemical GUS staining of Pro74F2:GUS, Pro76C4:GUS, and Pro76C5:GUS transgenic plants. All seed tissues had been stained for 4 h except the dissected seed (intact dry seeds had been stained in GUS solution for 48 h before dissection). Stratified seed means the seeds were stained after a 2-d stratification (4°C). Germinated seed means the seeds were stained after a 2-d stratification (4°C) and 1-d germination (22°C). Bar = 0.1 mm.
To investigate the subcellular localization of UGTs, we generated translational fusion constructs of UGT74F2-, UGT76C4-, and UGT76C5-GFP (green fluorescent protein) under the control of the CaMV35S promoter and introduced these into Arabidopsis leaf protoplasts. In all cases, GFP signal was detected exclusively in the cytosol, which is similar to the localization of GFP alone (Supplemental Figure 6).
UGT74F2, UGT76C4, and UGT76C5 Are Responsible for NA Glucosylations in Arabidopsis
To investigate the biochemical function of UGT74F2 in planta, we obtained a homozygous ethyl methanesulfonate (EMS)-mutagenized mutant in the Col-0 background for UGT74F2 that had been previously designated as ugt74f2i1a (one G-to-A mutation resulting in a splice junction mutant; Supplemental Figure 7A; Quiel and Bender, 2003; Dean and Delaney, 2008). We refer to this mutant as ugt74f2-1 hereafter for convenience. We treated different intact organs of ugt74f2-1 and wild-type plants with 10 μM [14C]-NIM and detected a significant decrease of [14C]-NAOG formation in the mutant compared with the wild type (Figure 7). However, the similar tissue-specific pattern of [14C]-NAOG observed in both the wild type and ugt74f2-1 suggested that residual NAOGT activity may remain in the ugt74f2-1 mutant (Figure 7). We resequenced the UGT74F2 transcripts, as ugt74f2-1 is a splice junction mutant, and found that the ugt74f2-1 plant generates three different UGT74F2 transcripts (Supplemental Figures 7B and 7C): The most abundant transcript was UGT74F2.3 (accounting for 72.5% of total UGT74F2 transcripts), which encodes a truncated UGT74F2 peptide of 219 amino acids without NAOGT activity; another transcript (UGT74F2.1, account for 17.5% of total UGT74F2 transcripts) encodes the wild-type 74F2 protein; and the third abundant UGT74F2 transcript (UGT74F2.2, accounting for 10.0% of total UGT74F2 transcripts) yields a protein with a one amino acid deletion (Glu at 209th position in wild-type UGT74F2). The purified recombinant UGT74F2.2 protein retains around 80% of the NAOGT activity of the wild-type protein (Supplemental Figure 7D).
Figure 7.
Metabolism of [14C]-NIM in Different Organs of Wild-Type and ugt74f2-1 Mutant Plants.
(A) Organs were excised from either 7-d-old seedlings or 8-week-old plants that had been immersed in 10 μM [14C]-NIM for 6 h. The radiolabeled chemicals were extracted and separated on a TLC plate (n-butanol:HOAc:H2O = 2:1:1). S, 7-d-old seedling. Arrows indicate the positions of NA, NAOG, NANG, and Tg (trigonelline).
(B) Relative quantification of [14C]-NAOG and [14C]-NANG in different organs compared with the wild type. Data are presented as means ± se (n = 3). Asterisks indicate significant difference from the wild type at P < 0.05.
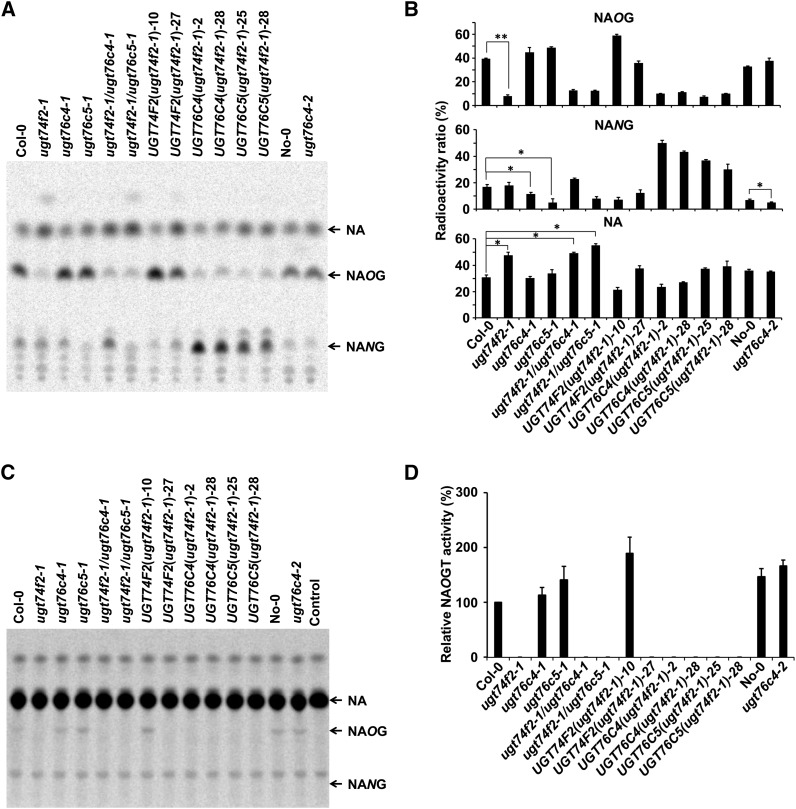
Similarly, two homozygous T-DNA/transposon insertion lines of UGT76C4, CS25037 and PSH11927 (in the No-0 background), designated as ugt76c4-1 and ugt76c4-2, respectively, were obtained and characterized (Supplemental Figure 8). A null allele mutant of UGT76C5 (SALK_140437; designated as ugt76c5-1) was obtained from ABRC and also characterized (Supplemental Figure 9). No transcripts of UGT76C4 or UGT76C5 were detected by RT-PCR in any of the corresponding homozygous mutants. All these mutants also do not display any obvious visible phenotypes. The accumulation patterns of NANG in different mutant lines at the 7-d-old growth stage were evaluated by radio-TLC. ugt76c5-1 accumulated much less NANG, accounting for 40% of the wild-type level (Figure 8). The analysis of complemented lines [UGT76C4(ugt76c4-1) and UGT76C5(ugt76c5-1); both are in Col-0 background] further confirmed that UGT76C4 and UGT76C5 were responsible for NA N-glucosylation in Arabidopsis (Supplemental Figure 10). We also examined the profiling of [14C]-labeled NA glucosylations in knockout lines of UGT74F1 (ugt74f1-1 [FLAG_454G04], Wassilewskija background [Dean and Delaney, 2008]; Supplemental Figure 11), UGT75B1 (ugt75b1-1, Landsberg erecta background [Eudes et al., 2008]; and ugt75b1-2 [PSH21231], No-0 background; Supplemental Figure 12), and UGT76C3 (ugt76c3-1 [SALK_137521]; Supplemental Figure 13). No mutant showed significant differences in accumulation of glucosylated NA, suggesting that none of these GTs was main contributors to NA glucoside accumulation in planta (Supplemental Figure 14).
Figure 8.
Characterization of the Transgenic Lines Used in This Study.
(A) Seven-day-old seedlings of each line were immersed in 10 μM [14C]-NIM solution for 6 h. The radiolabeled chemicals were then extracted and separated by TLC. Arrows indicate the positions of NA, NAOG, and NANG.
(B) Relative quantification of [14C]-NAOG, [14C]-NANG, and [14C]-NA for different lines. Data are presented as means ± se (n = 3). Asterisks indicate significant difference from the wild-type line; *P < 0.05, **P < 0.01.
(C) Radio-TLC chromatogram of NAOG generated by the crude proteins from 1-d germinated seeds of different transgenic lines using [14C]-NA and UDP-Glc as substrates. Five micrograms of crude proteins were used for each reaction, and the reactions were incubated at 25°C for 16 h. No protein was added in the control reaction. The positions of NA and NANG are indicated with arrows.
(D) Relative quantification of NAOGT activity for different transgenic lines. The NAOGT activity of 1-d germinated seeds of Col-0 was set as 100%. Data are presented as means ± se (n = 3).
We also generated transgenic lines with different combinations of NAOGT and NANGT genes, including UGT74F2 overexpression lines (in ugt74f2-1 background), ugt74f2 ugt76c4 and ugt74f2 ugt76c5 double mutants, as well as overexpression lines of NANGT in the ugt74f2-1 background. Feeding experiments clearly showed that there was a significantly higher concentration of free NA in lines with the ugt74f2-1 background than in lines of other backgrounds (Figure 8). It is also noteworthy that, although the expression level of UGT74F2 was upregulated 10- to 90-fold in dry seeds, 1-d-old and 7-d-old seedlings of UGT74F2(ugt74f2-1) plants (Supplemental Figures 15A to 15C), the NAOGT activity in UGT74F2(ugt74f2-1)-10, as indicated by feeding experiments, was only comparable to that of the wild type. The UGT74F2(ugt74f2-1)-27 line had even lower NAOGT activity than that of the wild type (Figures 8A and 8B). The difficulty of [14C]-NIM uptake by seeds prevents us from investigating NAGTs in the seed using the tracer technique. We thus used the crude protein from 1-d germinated seeds of these transgenic plants for NAGT comparison. In this experiment, only UGT74F2(ugt74f2-1)-10 had slightly higher (around 2-fold) NAOGT activity than the wild type. No clear [14C]-NAOG signal could be found in other ugt74f2 complemented lines, and no NANGT activity could be detected in this experiment (Figures 8C and 8D).
Deficiency of UGT74F2 in Arabidopsis Delayed the Germination Rate under Stress Conditions
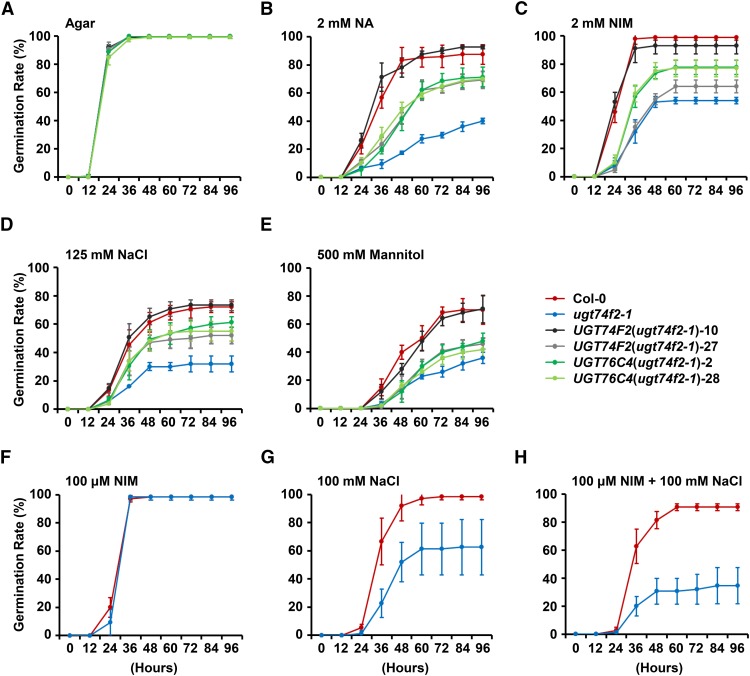
The predominant expression of UGT74F2 and UGT76C4 in dry seeds suggests a possible physiological function for these genes in seed development/germination. We performed a seed germination experiment using exogenous NA or NIM (all seeds used for the germination assays were harvested at the same time and stored for 3 months). Wild-type and mutant plants did not show significant differences in seed germination rates when grown on agar medium. However, the germination rate of ugt74f2-1 seeds was greatly inhibited compared with that of the wild-type seeds when grown on agar plates supplemented with NA (2.0 mM) or NIM (2.0 mM) (Figures 9A to 9C). ugt76c4-1 and ugt76c5-1 did not have phenotypes similar to that of ugt74f2-1, which suggested that NAOG is a more efficient detoxification strategy than N-glucosylation for responding to exogenous NA treatment at the seed germination stage (Supplemental Figure 16). Although knockout of NANGT did not cause any visible phenotypic change, overexpression of NANGT (UGT76C4) in the ugt74f2-1 background partially rescued the lowered seed germination rate of ugt74f2-1 under NA or NIM treatments (Figure 9). We did not include double mutants (ugt74f2 ugt76c4 and ugt74f2 ugt76c5) for further physiological experiments, as no phenotypic changes were observed between double mutant and the ugt74f2-1 single mutant when treated with NA (2.0 mM) or NIM (2.0 mM) (Supplemental Figure 16). This result again substantiates the conclusion that NAOG is the main detoxification strategy for exogenous NA treatment during seed germination.
Figure 9.
Germination Assay of Seeds from Col-0, the ugt74f2-1 Mutant, and Complemented Lines.
Germination percentages were scored every 12 h, and the data were calculated from at least triplicate tests. Emergence of visible radicles was the indicator for germination. Values of each data point represent means ± sd (n = 3; 75 seeds per genotype were used for each assay).
We performed additional germination assays using UGT74F2 transgenic plants (including mutant and complemented lines) and Col-0 plants under different stress conditions. In the presence of NaCl (125 mM) and mannitol (500 mM), the germination rate of ugt74f2-1 seeds was significantly lowered when compared with Col-0. This phenotypic change could be completely recovered by ectopically overexpressing UGT74F2 in the ugt74f2-1 mutant and partially recovered by overexpressing UGT76C4 (Figures 9D and 9E). The NAOGT assays among ugt74f2-1 and its complemented lines under stress conditions revealed that there is a correlation between NAOGT activity and seed germination rate (Supplemental Figure 17).
We also examined the combined effect of NIM and salinity on seed germination. NIM (100 μM) alone did not cause any difference in the germination rate of Col-0 and ugt74f2-1. However, the application of 100 μM NIM and 100 mM NaCl together caused a much larger difference in germination rate compared with NaCl alone, from 36 to 56% at 96 h after germination (Figures 9F to 9H). However, 100 μM SA did not cause the same additive effect on seeds germination (Supplemental Figure 18).
Perturbation of NA Homeostasis, Not SA, Affects Seed Germination
Because tracer experiments and enzymatic activity comparison often do not reflect the endogenous content of the chemicals of interest, we developed a new analytical method using liquid chromatography coupled with triple quadrupole mass spectrometry for monitoring and comparing the endogenous content of NA, its conjugates, and NAD(P)(H) during seed germination under control and abiotic stress conditions (see Methods for details). The nic1-1 mutant was also included in this experiment because nic1-1 was the only identified NAD salvage pathway mutant thus far. As expected, the nic1-1 seeds contained 10-fold higher NIM than wild-type seeds (Wang and Pichersky, 2007). Unexpectedly, the nic1-1 seeds had a comparable level of free NA to that of the wild type, which suggested there was an alternative pathway to generate free NA (e.g., NaMN to NaR to NA; NaMN could be generated from both a de novo pathway from aspartate and a salvage pathway) in Arabidopsis seeds besides the step from NIM to NA catalyzed by nicotinamidase (Figure 10). We also observed that the NIM content nic1-1 mutant was abruptly decreased 1 d after germination (Figure 10).
Figure 10.
Measurement of Endogenous NIM, NA (and Its Glucosides), NaMN, and NAD under Different Growth Conditions (Dry Seeds, Treatment with Water, 125 mM NaCl, or 500 mM Mannitol for 24 h after a 2-d Stratification).
All lines used in the experiment are in the Col-0 background. GH, glycoside hydrolase. nmol/g dry weight is the unit for all chemical content. Values of each data point represent means ± sd (n = 5). Asterisks indicate significant difference from the wild-type line under the same germination condition; *P < 0.01 (two-tailed Student’s t test).
In dry seeds, ugt74f2-1 had significantly less NAOG content (14.20 ± 2.51 nmol/g dry weight [DW]) and higher NANG content (302.70 ± 23.59 nmol/g DW) than the wild type (35.3 ± 2.48 nmol/g DW for NAOG and 23.15 ± 2.05 for NANG). UGT74F2(ugt74f2-1)-10 showed a reverse pattern of NAOG and NANG accumulation: slightly higher NAOG content (39.72 ± 4.02 nmol/g DW), although not significantly, and lower NANG content (10.86 ± 1.11 nmol/g DW) when compared with the wild type. UGT76C4(ugt74f2-1)-2 had a much higher NANG content (3851.63 ± 118.90 nmol/g DW) and lower NAOG content (11.52 ± 1.27 nmol/g DW). The ugt76c4-1 mutant had much less NANG (10.43 ± 0.62 nmol/g DW) and higher NAOG (44.46 ± 2.35 nmol/g DW). Altogether, these results suggested that UGT74F2 and UGT76C4 competed for NA in seeds tissues and the NA content was not significant changed among these transgenic lines except UGT74F2(ugt74f2-1)-10 had lower NA (52.51 ± 4.14 nmol/g DW versus 105.56 ± 18.4 nmol/g DW in the wild type).
In 1-d germinated seeds, LC-MS analysis showed that NAD content was increased, accompanying the decrease of NIM, NA, NAOG, and NaMN in all tested lines under water condition when compared with those in dry seeds (Figure 10). ugt74f2-1 seeds contained around 3- to 4-fold more NA than did wild-type seeds under various conditions (26.31 ± 4.40 nmol/g DW versus 7.82 ± 1.14 nmol/g DW under water condition; 33.80 ± 6.78 nmol/g DW versus 9.72 ± 0.92 nmol/g DW under NaCl treatment; 157.60 ± 35.98 nmol/g DW versus 50.52 ± 21.06 nmol/g DW under mannitol treatment; n = 4). Overexpressing UGT74F2 reduced the NA content back to the level of the wild type [17.59 ± 7.05 nmol/g DW and 32.43 ± 17.12 nmol/g DW in UGT74F2(ugt74f2-1)-10]. There was no significant difference in NAOG between the wild type, ugt74f2-1, or UGT74F2 overexpressors under any germination conditions (Figure 10). It is noteworthy that while the NA content in UGT76C4(ugt74f2-1)-2 was higher than that of the wild type (23.85 ± 3.02 nmol/g DW and 101.11 ± 8.09 nmol/g DW in 76C4OE), the NANG content in the 76C4OE line was ∼200-fold higher than in the wild type (Figure 10). Meanwhile, no significant differences in NaMN and NAD between different genotypes under the same treatment were observed in this assay. Measurements of endogenous SA, BA, o-ABA and their glucosides (SAG, SGE, BAGE, and oABAGE) are also included in this analysis due to the fact that UGT74F2 recognized all these aromatic acids as substrate (Table 1; Supplemental Figure 2). The results showed that there is no significant difference in the content of SA or its glucosides (SAG and SGE) between Col-0 and the transgenic plants, including the nic1-1 mutant, under the same stress condition. No clear signal for BA, o-ABA, and their glucosides could be detected in all tested seeds (Supplemental Figure 19). It should be noted that the endogenous NA content (105.6 ± 18.4 nmol/g DW) is around 120-fold higher than SA content (0.85 ± 0.16 nmol/g DW) in the dry seeds of wild-type plants (Figure 10; Supplemental Figure 19).
UGT74F2 Exhibited Other Biological Functions in Arabidopsis under Specific Conditions
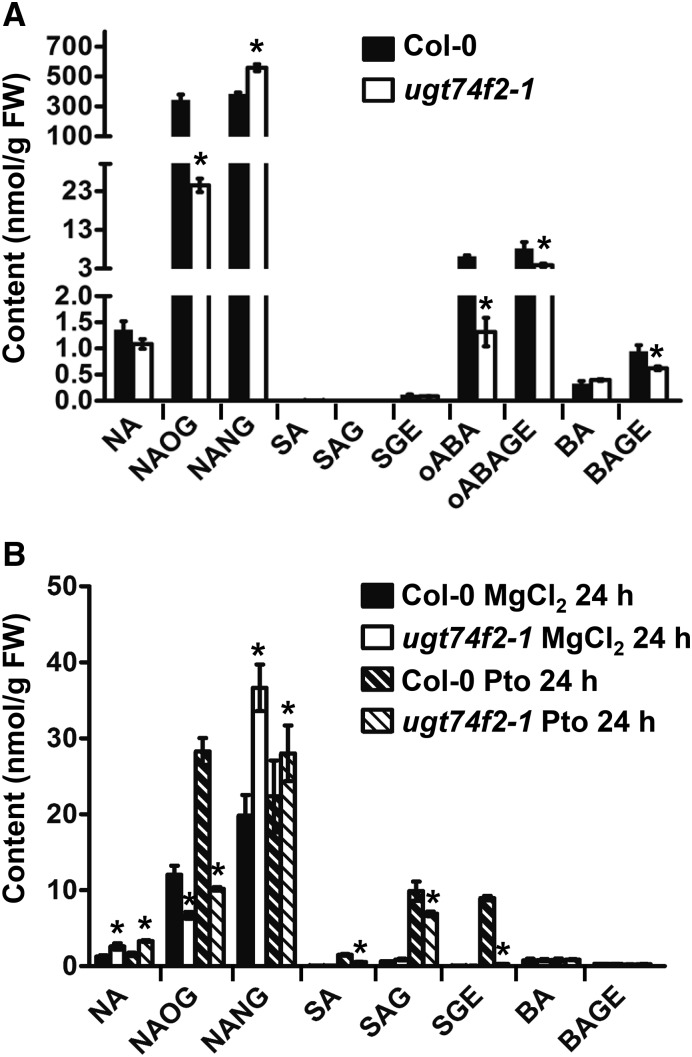
Previous studies showed that UGT74F2 exhibited activity toward SA and o-ABA in planta (Quiel and Bender, 2003; Dean and Delaney, 2008; Eudes et al., 2008; Song et al., 2009). To determine the biological functions of UGT74F2 in other tissues, we also compared the chemical profiling (NA, SA, BA, o-ABA, and their glucosides) in 7-d-old seedlings and pathogen-infected rosette leaves in Col-0 and the ugt74f2-1 mutant. NAOG content was decreased, accompanying the increase of NANG, in all tested tissues of the ugt74f2-1 mutant when compared with the wild type (Figure 11). In 7-d-old seedlings, the content of both oABAGE and BAGE was lower in the ugt74f2-1 mutant than in the wild type, which suggested that UGT74F2 also catalyzed the glucosylation of o-ABA and BA in young seedlings. Meanwhile, SGE content was significantly lower in pathogen infected leaves of ugt74f2-1 mutant plants than in the wild type, which suggested that UGT74F2 also catalyzed the glucosylation of SA in response to pathogen infection (Figure 11).
Figure 11.
Analysis of Metabolites NA, SA, BA, o-ABA, and Their Glucosides in Arabidopsis.
Measurement of endogenous NA, SA, BA, o-ABA, and their glucosides in 7-d-old seedlings (A) and Arabidopsis rosette leaves infected by PtoDC3000 for 24 h (B). FW, fresh weight. Values of each data point represent means ± sd (n = 4). Asterisks indicate significant difference from the wild-type line under the same germination condition; *P < 0.01 (two-tailed Student’s t test).
Phylogenetic Analysis and Biochemical Identification of NAOGT Homologs
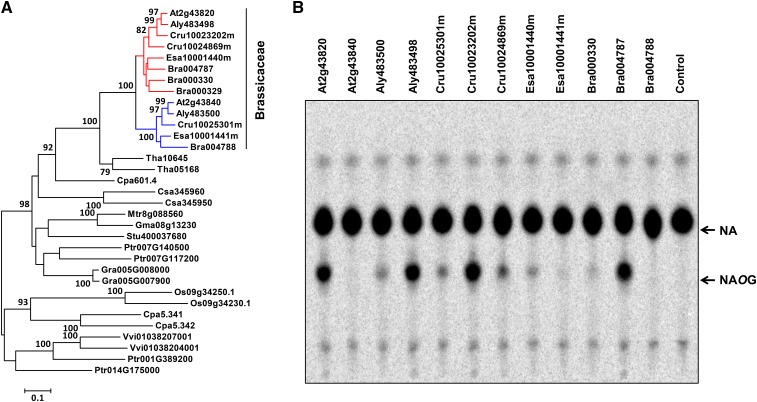
As shown in Figure 3, NAOG appears to be restricted to the Brassicaceae. To better understand the evolutionary history of NAOGT, we performed phylogenetic analysis including NAOGT and its homologs (>40% identity at the protein sequence level; for sequences and alignment, see Supplemental Data Set 2). Interestingly, UGT74F2 and its close homologs (>80% identity) form a clade, which contain protein sequences only from the Brassicaceae family. The Brassicaceae-specific distribution of 74F2 homologs is consistent with the NAOG distribution in the plant kingdom. To test whether the UGT74F2-like proteins are responsible for NAOG formation in Brassicaceae plants, we cloned all of the genes belonging to the UGT74F clade (Figure 12A). In vitro biochemical assays showed that the UGT74F2-like proteins from A. lyrata, C. rubella, B. rapa, and T. halophila all showed NAOGT activity similar to that of UGT74F2 (Figure 12B, Table 1). In contrast, only weak NAOGT activity was detected for some of the UGT74F1 homologs (Figure 12B). We did not detect positive selection for any residues in the UGT74F2 clade when we performed codon based z-testing of selection (Yang, 2007; Tamura et al., 2013).
Figure 12.
Phylogenetic Analysis and Biochemical Assays of UGT74 Proteins.
(A) Maximum likelihood tree of UGT74F2 and homolog proteins from other plant species (>40% identity). Bootstrap values (based on 1000 replications) >75% are shown for corresponding nodes. The clade of UGT74F1-like proteins and that of UGT74F2-like proteins are highlighted in blue and red, respectively. The scale measures evolutionary distance in substitutions per site. Aly, Arabidopsis lyrata; Bra, Brassica rapa; Cpa, Carica papaya; Cru, Capsella rubella; Csa, Cucumis sativus; Esa, Eutrema salsugineum; Gma, Glycine max; Gra, Gossypium raimondii; Mtr, Medicago truncatula; Os, Oryza sativa; Ptr, Populus trichocarpa; Stu, Solanum tuberosum; Tha, Tarenaya hassleriana; Vvi, Vitis vinifera.
(B) In vitro biochemical assays of proteins belonging to the UGT74F subfamily.
DISCUSSION
NAD, a common electron carrier in redox reactions, can be degraded by several kinds of NAD-consuming enzymes in all plant species (Briggs and Bent, 2011). To maintain NAD homeostasis, land plants (from mosses to flowering plants) evolved the Preiss-Handler pathway for reutilization of NIM (Noctor et al., 2006). Accompanying the occurrence of NA in the Preiss-Handler pathway, various NA modification systems (either glycosylation or methylation) were established (Figure 3). However, the physiological functions of NA modification have long been overlooked. In this study, we functionally identified three genes encoding NAGTs from Arabidopsis. Our results clearly show that NAOG, which specifically occurs in the Brassicaceae, plays an important role in seed germination under abiotic stress conditions in Arabidopsis.
G-E Correlation Analysis Implicated Candidate Genes Involved in NA Modifications in Arabidopsis
Family 1 GTs transfer sugar moieties from UDP activated sugars to a wide range of small molecules, including plant hormones, plant specialized metabolites, and xenobiotics, etc. Recent bioinformatic methods, including gene-gene and gene-metabolite correlation analysis, together with reverse genetics have been applied to identify genes involved in phytochemical biosynthesis pathways (Matsuda et al., 2010; Saito and Matsuda, 2010). Several GTs involved in the biosynthesis of flavonol glycosides and procyanidins have been identified using these strategies. However, the proportion of total UGTs in the Arabidopsis genome that have been functionally characterized remains at <40% (41/112) (Supplemental Data Set 1). There are two explanations for this: (1) The lineage-specific expansion of UGTs and independent acquisition of functions have occasionally led to imprecise prediction only based on the sequence and function of UGTs (Yonekura-Sakakibara and Hanada, 2011). In this study, only UGT74F2 (there are three identified SAGTs, including UGT74F1, UGT74F2, and UGT76B1, in Arabidopsis thus far) had the NAOGT activity. (2) Even more importantly, there is very limited information about the glycosylated compounds that occur in model plant species. For example, in a recent metabolomics study (Matsuda et al., 2010), only a few glycosides have been annotated with accurate structural information. It is well known that high-resolution mass spectrometry is susceptible to ion suppression or enhancement due to the unpredictable matrix effects (Lei et al., 2011). Traditional techniques, such as the radio-TLC method used in this study (the sensitivity of radio-TLC was tested up to 1 pmol of [14C]-labeled chemical), can provide reliable enzyme activity information for screening candidate genes. In this study, the correlation for NANGTs was not ranked in the top 5 (ranked at 8th, 10th, and 12th positions in Pearson correlation analysis), likely due to the fact that there are two NANGT genes in the Arabidopsis genome; however, the G-E correlation gave a reliable prediction for discovering candidate NAOGTs. Inclusion of more growth stages and/or organs in the feeding experiments used for the correlation analysis would be one plausible option to improve the utility of correlation analysis. In this study, we used the separated tissues for feeding experiments to gain the information of enzymatic activity, which excludes the effect of metabolite transportation between tissues. Such transport is common in plant specialized metabolism (PSM; Yazaki, 2005) and is known to frequently weaken gene-metabolite correlations. This method is applicable for dissecting other biosynthetic pathways if an appropriate labeled precursor is available.
NA Is an in Vivo Substrate for UGT74F2 and UGT76C4/5 in Arabidopsis
In vitro biochemical assays previously showed that UGT74F2 recognized SA, BA, and o-ABA as substrates (Lim et al., 2002; Quiel and Bender, 2003; Dean and Delaney, 2008; Eudes et al., 2008; Song et al., 2009). Although NA is not the best substrate for UGT74F2 in vitro, the high catalytic efficiency of UGT74F2 toward NA still strongly suggests that NA is an important substrate for UGT74F2 in Arabidopsis (Table 1; Supplemental Figure 2). Other supporting evidence for this hypothesis includes (1) the positive correlation between NAOGT activity and the expression profile of UGT74F2 (r > 0.75 and P < 0.05); (2) the ugt74f2-1 mutant has much lower NAOGT activity, indicated by our feeding experiment (Figures 7 and 8) and ugt74f2-1 is more sensitive to exogenous NA and NIM treatment than the wild type (Figure 9); and (3) endogenous chemical analysis showed that the content of free NA and NANG was increased and/or the NAOG content (except the NAOG content in seed tissue; Figure 10) was decreased in ugt74f2-1 mutant at various developmental stages and growth conditions (Figures 10 and 11). It would not be surprising if UGT74F2 could catalyze the glycosylation of other aromatic chemicals containing a carboxyl group (e.g., SA, BA, and o-ABA) in Arabidopsis (Table 1; Supplemental Figure 2), as multifunctionality is a common property of enzymes, especially those involved in PSM. This catalytic multifunctionality serves as the raw materials for the evolution of new enzyme function (Aharoni et al., 2005; Weng et al., 2012). Unlike UGT74F2, UGT76C4 and UGT76C5 both showed a narrow substrate spectrum and neither GT recognized NIM or MeNA as substrate. This result is consistent with our previous observation that no NIM conjugates could be detected in the nic1-1 mutant when treated with [14C]-NIM (Wang and Pichersky, 2007). NANG was still detectable in all tested tissues of the ugt76c4 or ugt76c5 single mutant plants (Figure 8), confirming that UGT76C4 and UGT76C5 have overlapping roles in NANG production. The tissue-specific expression profiles of UGT76C4 and UGT76C5 suggest that UGT76C4 is mainly responsible for NANG formation in seed stage, whereas UGT76C5 is mainly responsible for NANG formation in young seedlings. As expected, much lower NANG level (10.43 ± 0.62 nmol/g DW) was detected in the dry seeds of the ugt76c4-1 mutant when compared with the NANG content in the dry seeds of wild-type plants (23.15 ± 2.05 nmol/g DW), and very high level of NANG (3851.63 ± 118.90 nmol/g DW) was detected in the seeds of UGT76C4 overexpressor line (Figure 10).
NA O-Glycosylation Plays an Important Role in Seed Germination under Abiotic Stress Conditions
A major finding from this study is that the partial disruption of UGT74F2 led to a lower germination rate compared with the wild type under salt/mannitol stress conditions. It is well known that rehydration during seed germination is associated with high levels of oxidative stress, resulting in DNA damage (Waterworth et al., 2011). Damaged DNA is repaired through a process involving PARP, which is likely a prerequisite for proper embryonic and seedling morphogenesis. In Arabidopsis, inactivation of seed-specific PARP3 results in a lowered germination rate compared with that of Col-0, which suggests that PARP3 is an important component for seed viability and germination (Rissel et al., 2014). NIM, a catalytic product of PARP, is released and accumulated during seed germination. To date, NIC is the only known enzyme in plants that uses NIM as a substrate. The high NIM content in dry seeds, along with the observed abrupt decrease of NIM content in imbibed seeds (soaked in water for 24 h) of the nic1-1 mutant, suggest that hydrolysis of NIM is probably required for normal seed germination, as NIM is an endogenous inhibitor of PARP enzymes in plants (De Block et al., 2005). These findings also suggested that both the constitutively expressed NIC1 and the seed-specific NIC2 are together responsible for the hydrolysis of NIM during seed germination (Hunt et al., 2007; Wang and Pichersky, 2007). Hydrolysis of NIM subsequently leads to free NA production, which is toxic to plant cells at high concentrations (Wang and Pichersky, 2007). The toxic effect of free NA was also observed when the NAD salvage pathway was investigated in embryos of germinating mung bean (Phaseolus aureus) and lettuce (Lactuca sativa) cells (Zheng et al., 2005; Sasamoto and Ashihara, 2014).
Based on our experimental data, we propose that NAOG functions as an accessible storage form of NA because NAOG can be easily hydrolyzed by β-glucosidase to release NA for NAD production (Supplemental Figure 1). There are 379 glycoside hydrolases in the Arabidopsis genome, though the identities of the glycoside hydrolases involved in PSM remain largely unknown (Minic, 2008). The inability to overaccumulate NAOG in UGT74F2-overexpressing plants also suggests a balance between NAOGT and the corresponding glycoside hydrolases. This phenomenon is similar to what is known of SA glucosides in Arabidopsis: SGEs can be detected transiently at the early stages of pathogen infection. SAG is then induced to higher levels upon pathogen infection (Dempsey et al., 2011; Noutoshi et al., 2012). In the case of NA modifications in Arabidopsis, the content of NANG can increase up to around 200-fold higher than that of the wild type, suggesting that NANG is a stable end product in Arabidopsis. Surprisingly, NA content in the UGT76C4-overexpressing line (ugt74f2-1 background) was higher than that of the wild type under various abiotic growth conditions. It is likely that NANG is an inhibitor to other NA-utilizing enzymes (NAOGT or NaPRT; there are two genes encoding NaPRT in the Arabidopsis genome; namely, NaPRT1 and NaPRT2), and the blockage of these enzymes may subsequently result in the accumulation of free NA. Additional experiments will be needed to validate this hypothesis. Another possibility that whether NAOG is vacuolar storage form of NA in Arabidopsis also merits further investigation.
The catalytic promiscuity of UGT74F2 makes it more difficult to assign the phenotype to one of the substrates. However, our comprehensive chemical comparison (including NA, SA, BA, o-ABA, and their glucosides) between Col-0 and ugt74f2-1 clearly showed that the delay of seed germination of ugt74f2-1 was mainly caused by overaccumulation of NA, not SA, in the Arabidopsis seeds under stress conditions. Our results also demonstrate that the substrate availability (e.g., NA in all tissues, o-ABA and BA in young seedlings, and SA in pathogen infected leaves) plays a determining role for the biological function of UGT74F2 in Arabidopsis. The novel NAOGT activity of UGT74F2 discovered in this study (and/or NAD salvage pathway) may provide an alternative explanation for the different pathogen response between ugt74f1 and ugt74f2 mutants (Boachon et al., 2014).
Evolutionary Origin of NAOGT in Plants
To estimate the initial occurrence of NAOG in the plant kingdom, we constructed phylogenetic trees for PARPs, NICs, NaPRTs, and NAOGTs (Supplemental Figure 20). The results of this analysis showed that PARP, NaPRT, and NIC are all ancient genes that are preserved in all land plants. No close homologs of PARP, NaPRT, or NIC could be detected in microalgae (e.g., Chlamydomonas reinhardtii and Volvox carteri). This result is consistent with a previous report (Lin et al., 2010); algae has a two-step NAD salvage pathway (from NIM to NMN, then NMN is catalyzed by NMNAT back to NAD). When land plants started to use the four-step NAD salvage pathway, free NA was added to chemical context of plant metabolism. Several studies in yeast and bacteria have shown that the rate-limiting enzymatic step of the NAD salvage pathway is catalyzed by NaPRT (Wubbolts et al., 1990; Anderson et al., 2002), which provides an explanation for why NA can be detected in almost all land plants (Matsui et al., 2007). Because an overdose of free NA is known to be toxic to plant cells (Zheng et al., 2005; Wang and Pichersky, 2007; Sasamoto and Ashihara, 2014), an enzyme that can further metabolize NA might have become necessary to release endogenous NA stress (e.g., glucosyltransferase or methyltransferase).
The recruitment of a seed-specific promoter for UGT74F2, which is similar to that of Arabidopsis PARP3 and NIC2, further implies the selective advantage of NAOG to the plant cell (Supplemental Figure 21). It is believed that the salvage pathway of NAD is a way of conserving energy that would otherwise be needed for the de novo synthesis of a pyridine ring from aspartate (Noctor et al., 2006). In view of energy conservation, the reuse of the NA moiety of NAOG for NAD biosynthesis clearly has a selective advantage over NANG, which is secreted out of plant cells (Supplemental Figure 1). Moreover, the carboxyl group is often essential for the bioactivity of many compounds, such as phytohormones (abscisic acid, indole-3-acetic acid, jasmonic acid, gibberellic acid, and salicylic acid, etc.). Only blockage of the carboxyl group leads to completely inactive chemicals, which gives NAOG another advantage over NANG for releasing “endogenous NA stress” in plant cells.
The fact that the UGT74F1/UGT74F2 pair was present in all of the sequenced plants of the Brassicaceae suggests that an ancestral tandem duplication event occurred before these plants diverged from each other. Gene duplication is a major driving force for the acquisition of new functions in PSM (Pichersky and Gang, 2000). We assumed that the evolutionarily new NAOGT function (UGT74F2 homologs) was derived from the duplication of one UGT74F gene in an ancestral Brassicaceae plant. The occurrence of SAGTs (glucosylation at the hydroxyl group of SA), which are encoded by UGT74 genes, in rice and tobacco suggests that SAGT is probably an ancestral activity of UGT74F proteins (Lee and Raskin, 1999; Umemura et al., 2009). Biochemical assays revealed that Arabidopsis UGT74F2 had much weaker the SAGT activity of UGT74F1 (Supplemental Figure 2; Lim et al., 2002) and some of 74F1 homologs had weak NAOGT activity (Figure 12B). This observation is consistent with the theory that catalytic promiscuity serves as the starting point for the acquisition of new enzymatic functions: The evolutionally new enzyme here adopted one of the minor activities of its ancestor (Aharoni et al., 2005; Weng et al., 2012). The important role of UGT74F1 in the plant innate immune system was recently demonstrated (Noutoshi et al., 2012). This result, together with the important role of UGT74F2 in seed germination proved in this study, provides an explanation for the maintenance of the UGT74F1/UGT74F2 pairs in the genomes of current Brassicaceae plants. However, we cannot exclude the possibility NAOGT may evolved independently in other plant clades that are not included in our study.
To date, little is known about the physiological function of NA conjugates in the context of natural ecosystems. Nicotine is the most well known NA derivative in tobacco plants. In addition to nicotine, tobacco plants also produce NANG. In view of NA hemostasis, both NANG and nicotine could be functioned as detoxification to release the endogenous NA stress in tobacco. Although a role of nicotine in plant defense has been demonstrated (Mithöfer and Boland, 2012), many experiments are needed to elucidate whether other NA conjugates, such as NAOG and NANG, also function as defensive chemicals against biotic stresses. The transgenic plants generated in this study will enable evaluation of plant performance in such environments.
In summary, we conclude that UGT74F-type NAOGT is evolutionarily recent (restricted to the Brassicaceae) and has an important molecular function in fine-tuning NA homeostasis during seed germination. The identification and functional characterization of the Arabidopsis genes encoding NAOGT and NANGT in this study pave the way for further understanding of the chemical diversity of NA modifications in the Brassicaceae.
METHODS
Plant Materials and Chemicals
The Arabidopsis thaliana lines (Col-0, Wassilewskija, No-0, and transgenic plants) and other plant species mentioned in this study were grown on soil at 22°C under a 16-h-light/8-h-dark cycle. The rosette leaves of 4-week-old Arabidopsis were used for PtoDC3000 (Pseudomonas syringae pv tomato strain DC3000) infection by following the protocol described previously (Mengiste et al., 2003).
All chemicals used in this study were purchased from Sigma-Aldrich, except the radiolabeled [14C]-NIM, -NA, -SA, and -MeNA (55 mCi/mmol), which were purchased from American Radiolabeled Chemicals.
Analysis of [14C]-NIM or -NA Metabolism in Planta
The feeding experiments with 10 µM [14C]-NIM or -NA solution and the subsequent extraction and radio-TLC analysis were performed according to previously described protocols (Wang and Pichersky, 2007). Plant extracts containing 3000 cpm of [14C]-labeled metabolites were loaded on TLC plates for analysis.
G-E Correlation Analysis
Pearson’s correlation analysis (two tailed) was performed using SPSS software (version 19.0; IBM Software). Transcript expression data for 106 Family 1 GTs was downloaded from the Gene Expression Omnibus database (Barrett et al., 2011). The metabolite accumulation signals (the indicator of corresponding enzyme activity) were determined directly from radio-TLC analysis using ImageJ 1.38e software (downloaded from National Institutes of Health; http://rsb.info.nih.gov/ij/). To simplify the analysis procedure, all transcript and metabolite accumulation data were transformed into relative format, meaning the value of the largest data point was set as 1.0.
Generation, Expression, and Purification of Recombinant Glycosyltransferases
The protein expression constructs of the Arabidopsis GT genes were generated using a previously described strategy (Xu et al., 2013). Twelve NANGT candidate genes (AT1G10400, AT1G64910, AT2G15490, AT2G22930, AT2G29750, AT3G02100, AT3G46690, AT5G05860, AT5G05870, AT5G05880, AT5G05890, and AT5G05900) and eight UGT74F2 homolog genes (AT1G05530, AT1G05560, AT1G05680, AT1G24100, AT2G31790, AT2G31750, AT2G43820, and AT2G43840) were subcloned into the pEXP5-CT TOPO vector (Invitrogen) or pMAL-c2x vector (New England Biolabs) (see Supplemental Data Set 3 for primer information). All constructs were transformed into Escherichia coli BL21(+) cells for prokaryotic expression, after being verified by DNA sequencing. The resulting tagged fusion proteins were purified using Ni-NTA (for His-tagged proteins) or Dextrin Sepharose (for MBP-tagged proteins) affinity chromatography. Quantification and evaluation of the relative purity of the recombinant proteins was performed using SDS-PAGE with BSA as a standard.
Glycosyltransferase Assays and Enzyme Characterization
For enzymatic assays with [14C]-NA as the substrate, 100 µM [14C]-NA was used for each reaction and incubated for 120 min at 30°C (50 μL in a reaction system including 30 mM HEPES, pH 7.0, 0.2 mM UDP-Glc, 1 mM DTT, and 5 mM MnSO4). Five microliters of the enzyme reaction was spotted on a silica TLC plate, developed with an n-butanol/HOAc/H2O system (2:1:1), and compared with both radiolabeled and nonlabeled standards. The pH optimum for GTs in the study was determined using 50 mM Bis-Tris or Tris-only buffer with pH ranging from 5.0 to 9.0. Glycosyltransferase assays were also performed with one of following cations at 5 mM: Na+, K+, NH4+, Zn2+, Mn2+, Mg2+, Ca2+, Fe2+, and Fe3+. A NA concentration series of 0.02, 0.05, 0.1, 0.2, 0.5, and 1.0 mM, together with 2 mM UDP-Glc in each assay, was used in NA kinetics experiments. A UDP-Glc concentration series of 0.02, 0.05, 0.1, 0.2, 0.5, and 1.0 mM, together with 1 mM NA in each assay, was used in the UDP-Glc kinetics experiments. Kinetic parameters were calculated using a Hanes plot (Hyper32, version 1.0.0).
Quantitative RT-PCR Analysis and Promoter:GUS Staining
RNA extraction, reverse transcription, and quantitative RT-PCR were performed as described previously (Wang et al., 2008). The annealing temperature, specificity, and amplification efficiency of all primers used in the study were tested and are listed in Supplemental Data Set 3. Three traditional reference genes (Tubulin 2 [AT5G62690], Actin 2 [AT3G18780], and GAPDH [AT3G26650]) were tested, and Actin 2 was chosen due to its relative stable expression across all tested samples.
The promoter fragments for three NAGT genes (970 bp for UGT74F2 [from 983 to 14 bp upstream of the ATG codon], 384 bp for UGT76C4 [from 384 to 1 bp upstream of the ATG codon], and 1051 bp [from 1051 to 1 bp upstream of the ATG codon] for UGT76C5) were subcloned into pMDC162 Gateway binary vector using a PCR-mediated technique (see Supplemental Data Set 3 for primer information). Arabidopsis transformation, screening for homozygous plants, and GUS staining were performed as described before (Wang and Pichersky, 2007).
Subcellular Localization of UGT74F2, UGT76C4, and UGT76C5
Constructs of GFP fusion protein (pJIT163-hGFP vector), Arabidopsis leaf protoplast preparations, transformation, and image collection using a confocal laser scanning microscope were all performed as described previously (Xu et al., 2013).
Seed Germination Assays
All of the seeds that were used in the germination assays were harvested at the same time and stored for 3 months. Approximately 50 seeds for each line were sown on agar plates (pH was adjusted to 5.8 using MES-KOH) supplemented with various concentrations of different compounds (NA, NIM, NaCl, or mannitol) after sterilization in bleach (1% NaClO with 0.05% Tween 20) for 10 min and washing five times with sterilized water. Germination assays were performed in a growth room with a 16-h-light /8-h-dark cycle after stratification at 4°C for 2 d in the dark. Germination rates were scored every 12 h, and the results presented here were calculated from at least three separate assays. Emergence of visible radicles was used as the indicator for germination.
LC-MS Analysis
To elucidate structures of the two unknown NA derivatives in Arabidopsis, 100 mg fresh weight of 7-d-old seedlings (Col-0 and nic1-1), after treatment with 200 µM NIM for 24 h, was ground into fine a powder in liquid nitrogen. The NA derivatives were extracted with 1 mL of 50% aqueous methanol at 4°C. Following 10 min of maximum speed centrifugation with a microcentrifuge, the supernatant was filtered through a 0.22-µm syringe filter. Analyses were conducted using an Agilent 1260 LC pump coupled to an Agilent 6510 Q-TOF LC-MS system equipped with a dual electrospray ion source operated in positive mode. Two microliters of the extract was separated on an Agilent Zorbax RX-SIL column (150 × 2.1 mm, 5 µm). The column was at room temperature, and the flow rate was 0.3 mL/min. The chromatographic separation was achieved with a linear gradient running 95 to 1% A (solvent A, 0.1% formic acid in water; solvent B, acetonitrile) for 20 min, maintained for 2 min, and then returned to the initial conditions for 10 min for reequilibration. The mass spectrometry conditions were as follows: scan range 100 to 1000 m/z, drying gas temperature 350°C (nitrogen), drying gas flow 8 L/min, nebulizer pressure 38 p.s.i., capillary voltage 4 kV, fragmentor voltage 175 V, skimmer 1 voltage 65 V, and octopole RF peak voltage 750 V.
To determine the endogenous levels of NA, SA, BA, o-ABA, their glucosides, and NAD-related chemicals, 30 mg of dry seeds or 50 mg of fresh tissue (seedlings or leaves) was collected, ground into a fine powder in liquid nitrogen, and extracted with 1 mL of 50% aqueous acetonitrile (v/v) solution and 0.5 mL precooled CHCl3. After shaking for 60 min at 4°C, the supernatant was spun in a microcentrifuge at top speed for 10 min, and the resulting solution was filtered with a 0.22-µm syringe filter. After lyophilization, the residue was redissolved in 90% acetonitrile and analyzed immediately with a UPLC-MS/MS systems consisting of an Agilent 1290 Infinity LC pump and a 6460 triple quadrupole mass spectrometer. The positive mode analysis was performed in multiple reaction monitoring mode. The mass spectrometry parameters were optimized with standards of the metabolites; the optimized tandem mass spectrometry ion transitions monitored and mass spectrometry parameters are listed in Supplemental Table 2. An Agilent ZORBAX RX-SIL column (100 × 2.1 mm, 1.8 µm) was used for separation in hydrophilic interaction chromatography mode, and the column temperature was maintained at 40°C. Solvent A was 50 mM aqueous ammonium acetate and 0.1% formic acid (v/v) in water, and solvent B was acetonitrile. The chromatograph program was as follows at a flow of 0.30 mL/min: 0 to 1.5 min, 98% solvent B; 1.5 to 7.0 min, a linear gradient to 70% of B; 7.0 to 9.0 min, a linear gradient to 20% of B; 9.0 to 11.0 min, 20% of B; 11.0 to 11.1 min, a linear gradient to 98% of B and keep for 5 min. The ion source parameters were set as follows: drying gas temperature 325°C (nitrogen), drying gas flow 7 liters/min, nebulizer 40 p.s.i., sheath gas heater 350°C, sheath gas flow 12 liter/min, capillary voltage 3 kV (ESI+) and 3.5 kV (ESI−). The concentrations of glycosylated chemicals (NAOG, NANG, SAG, SGE, BAGE, and oABAGE) were calculated using the standard curve of corresponding free acid. Other chemical concentrations were quantified based on standard curves prepared with authentic reference standards.
Phylogenetic Analysis
The sequences of UGT74F1/2 and GT proteins with high identity (>40%) to UGT74F1/2 from other plant species were extracted from the TAIR (http://www.Arabidopsis.org) and Phytozome 9.0 databases (http://www.phytozome.net). Two Tarenaya glucosyltransferase sequences (Th10645 and Th05168) were extracted from http://genomevolution.org/CoGe/CoGeBlast.pl. A maximum likelihood tree was constructed using MEGA6 software with all default parameters (Tamura et al., 2013).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: AT2G43820 (UGT74F2), AT2G43840 (UGT74F1), AT1G05530 (UGT75B2), AT2G31750 (UGT74D1), AT2G31790 (UGT74C1), AT1G24100 (UGT74B1), AT1G05560 (UGT75B1), AT1G05680 (UGT74E2), AT3G11340 (UGT76B1), AT5G05870 (UGT76C1), AT5G05860 (UGT76C2), AT5G05900 (UGT76C3), AT5G05880 (UGT76C4), and AT5G05890 (UGT76C5).
Supplemental Data
Supplemental Figure 1. Comparison of the metabolic fates of two NA glucosides in Arabidopsis.
Supplemental Figure 2. Substrate specificity of UGT74F2 and UGT74F1.
Supplemental Figure 3. Enzymatic assays of UGT76B1 in vitro.
Supplemental Figure 4. NA N-glucosyltransferase assays of seven purified recombinant GT proteins.
Supplemental Figure 5. Sugar donor specificity of NA glucosyltransferase from Arabidopsis.
Supplemental Figure 6. Subcellular localization of UGT74F2, UGT76C4, and UGT76C5.
Supplemental Figure 7. Characterization of the ugt74f2-1 mutant.
Supplemental Figure 8. Characterization of the ugt76c4 mutants.
Supplemental Figure 9. Characterization of the ugt76c5-1 mutant.
Supplemental Figure 10. Characterization of NANGT T-DNA insertion mutants and their complemented lines.
Supplemental Figure 11. Characterization of the ugt74f1-1 mutant.
Supplemental Figure 12. Characterization of the ugt75b1 mutants.
Supplemental Figure 13. Characterization of the ugt76c3-1 mutant.
Supplemental Figure 14. Relative quantification of [14C]-NAOG and [14C]-NANG in different mutants.
Supplemental Figure 15. qRT-PCR analyses of UGT74F2 abundance in dry seeds, 1-d germinated seeds, and 7-d-old seedlings of Col-0, ugt74f2-1, and two complemented lines [UGT74F2(ugt74f2-1)-10 and UGT74F2(ugt74f2-1)-27].
Supplemental Figure 16. Seed germination assay of Col-0, ugt76c4-1, ugt76c5-1, ugt74f2-1, ugt74f2-1/ugt76c4-1, and ugt74f2-1/ugt76c5-1.
Supplemental Figure 17. NAOGT activity comparison in 1-d germinated seeds of different transgenic lines under three germination conditions.
Supplemental Figure 18. Germination assay of seeds from Col-0 and the ugt74f2-1 mutant on agar supplemented with SA, NaCl, or combination of SA and NaCl.
Supplemental Figure 19. Measurement of endogenous SA, SAG, and SGE in seeds.
Supplemental Figure 20. Phylogenetic analysis of PARPs, NICs, and NaPRTs of plant origin.
Supplemental Figure 21. Transcript levels of PARP3 and NIC2 in different tissues of Arabidopsis.
Supplemental Table 1. List of selected tissues for [14C]-NIM feeding experiments and publicly available microarray data set IDs.
Supplemental Table 2. The MRM settings for metabolites analyzed in this study.
Supplemental Data Set 1. Pearson correlation analysis for candidate NAGT gene selection and general information for Arabidopsis Family 1 glycosyltransferases.
Supplemental Data Set 2. The sequences and alignment used for the phylogenetic analysis shown in Figure 12.
Supplemental Data Set 3. Primers used in this study.
Supplementary Material
Acknowledgments
We thank John V. Dean (DePaul University, Chicago, IL) for providing the ugt74f1-1 and ugt74f2-1 seeds, Andrew D. Hanson (University of Florida) for providing the ugt75b1-1 seeds, and Dingzhong Tang (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing the PtoDC3000 strain. We also thank Eran Pichersky (University of Michigan) for his comments on the article. This work was financially supported by the National Program on Key Basic Research Projects (The 973 Program: 2012CB113900 and 2013CB127000), the National Natural Sciences Foundation of China (Grant 31270336), and the State Key Laboratory of Plant Genomics of China (Grant2014B0227-05) to G.W.
AUTHOR CONTRIBUTIONS
G.W. and W.L. designed the research. W.L., F.Z., Y.C., and T.Z. performed research. W.L., F.Z., Y.C., and G.W. analyzed data. G.W. wrote the article. M.E.S. edited the article.
Glossary
- NIM
nicotinamide
- NMN
nicotinamide mononucleotide
- NANG
nicotinate N-glucoside
- NANGT
nicotinate N-glucosyltransferase
- G-E
gene-enzyme
- SA
salicylic acid
- o-ABA
o-aminobenzoate
- BA
benzoic acid
- NA
nicotinate
- Col-0
Columbia-0
- LC-MS
liquid chromatography-mass spectrometry
- SGE
salicylic acid glucose ester
- SAG
salicylic acid 2-O-glucose
- TLC
thin-layer chromatography
- DW
dry weight
- PSM
plant specialized metabolism
- MeNA
methyl nicotinate
Footnotes
Articles can be viewed online without a subscription.
References
- Aharoni A., Gaidukov L., Khersonsky O., McQ Gould S., Roodveldt C., Tawfik D.S. (2005). The ‘evolvability’ of promiscuous protein functions. Nat. Genet. 37: 73–76. [DOI] [PubMed] [Google Scholar]
- Anderson R.M., Bitterman K.J., Wood J.G., Medvedik O., Cohen H., Lin S.S., Manchester J.K., Gordon J.I., Sinclair D.A. (2002). Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 277: 18881–18890. [DOI] [PubMed] [Google Scholar]
- Ashihara H., Stasolla C., Yin Y.L., Loukanina N., Thorpe T.A. (2005). De novo and salvage biosynthetic pathways of pyridine nucleotides and nicotinic acid conjugates in cultured plant cells. Plant Sci. 169: 107–114. [Google Scholar]
- Ashihara H., Yin Y., Katahira R., Watanabe S., Mimura T., Sasamoto H. (2012). Comparison of the formation of nicotinic acid conjugates in leaves of different plant species. Plant Physiol. Biochem. 60: 190–195. [DOI] [PubMed] [Google Scholar]
- Ashihara H., Yin Y., Watanabe S. (2011). Nicotinamide metabolism in ferns: formation of nicotinic acid glucoside. Plant Physiol. Biochem. 49: 275–279. [DOI] [PubMed] [Google Scholar]
- Barrett T., et al. (2011). NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res. 39: D1005–D1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boachon B., Gamir J., Pastor V., Erb M., Dean J.V., Flors V., Mauch-Mani B. (2014). Role of two UDP-glycosyltransferases from the L group of Arabidopsis in resistance against Pseudomonas syringae. Eur. J. Plant Pathol. 139: 707–720. [Google Scholar]
- Bowles D., Lim E.K., Poppenberger B., Vaistij F.E. (2006). Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 57: 567–597. [DOI] [PubMed] [Google Scholar]
- Briggs A.G., Bent A.F. (2011). Poly(ADP-ribosyl)ation in plants. Trends Plant Sci. 16: 372–380. [DOI] [PubMed] [Google Scholar]
- Dean J.V., Delaney S.P. (2008). Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol. Plant. 132: 417–425. [DOI] [PubMed] [Google Scholar]
- De Block M., Verduyn C., De Brouwer D., Cornelissen M. (2005). Poly(ADP-ribose) polymerase in plants affects energy homeostasis, cell death and stress tolerance. Plant J. 41: 95–106. [DOI] [PubMed] [Google Scholar]
- Dempsey, D.A., Vlot, A.C., Wildermuth, M.C., and Klessig, D.F. (2011). Salicylic acid biosynthesis and metabolism. The Arabidopsis Book 9: e0158, doi/10.1199/tab.0158. [DOI] [PMC free article] [PubMed]
- Eudes A., Bozzo G.G., Waller J.C., Naponelli V., Lim E.K., Bowles D.J., Gregory J.F. III, Hanson A.D. (2008). Metabolism of the folate precursor p-aminobenzoate in plants: glucose ester formation and vacuolar storage. J. Biol. Chem. 283: 15451–15459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossmann T.I., Ziegler M., Puntervoll P., de Figueiredo L.F., Schuster S., Heiland I. (2012). NAD(+) biosynthesis and salvage—a phylogenetic perspective. FEBS J. 279: 3355–3363. [DOI] [PubMed] [Google Scholar]
- Hashida S.N., Takahashi H., Uchimiya H. (2009). The role of NAD biosynthesis in plant development and stress responses. Ann. Bot. (Lond.) 103: 819–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou B., Lim E.K., Higgins G.S., Bowles D.J. (2004). N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J. Biol. Chem. 279: 47822–47832. [DOI] [PubMed] [Google Scholar]
- Hunt L., Holdsworth M.J., Gray J.E. (2007). Nicotinamidase activity is important for germination. Plant J. 51: 341–351. [DOI] [PubMed] [Google Scholar]
- Imai S. (2009). The NAD World: a new systemic regulatory network for metabolism and aging—Sirt1, systemic NAD biosynthesis, and their importance. Cell Biochem. Biophys. 53: 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.I., Raskin I. (1999). Purification, cloning, and expression of a pathogen inducible UDP-glucose:salicylic acid glucosyltransferase from tobacco. J. Biol. Chem. 274: 36637–36642. [DOI] [PubMed] [Google Scholar]
- Lei Z., Huhman D.V., Sumner L.W. (2011). Mass spectrometry strategies in metabolomics. J. Biol. Chem. 286: 25435–25442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E.K., Doucet C.J., Li Y., Elias L., Worrall D., Spencer S.P., Ross J., Bowles D.J. (2002). The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J. Biol. Chem. 277: 586–592. [DOI] [PubMed] [Google Scholar]
- Lin H., Kwan A.L., Dutcher S.K. (2010). Synthesizing and salvaging NAD: lessons learned from Chlamydomonas reinhardtii. PLoS Genet. 6: e1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T., Chen X., Salmeron J., Dietrich R. (2003). The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15: 2551–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F., Hirai M.Y., Sasaki E., Akiyama K., Yonekura-Sakakibara K., Provart N.J., Sakurai T., Shimada Y., Saito K. (2010). AtMetExpress development: a phytochemical atlas of Arabidopsis development. Plant Physiol. 152: 566–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A., Yin Y., Yamanaka K., Iwasaki M., Ashihara H. (2007). Metabolic fate of nicotinamide in higher plants. Physiol. Plant. 131: 191–200. [DOI] [PubMed] [Google Scholar]
- Minic Z. (2008). Physiological roles of plant glycoside hydrolases. Planta 227: 723–740. [DOI] [PubMed] [Google Scholar]
- Mithöfer A., Boland W. (2012). Plant defense against herbivores: chemical aspects. Annu. Rev. Plant Biol. 63: 431–450. [DOI] [PubMed] [Google Scholar]
- Noctor G., Queval G., Gakière B. (2006). NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J. Exp. Bot. 57: 1603–1620. [DOI] [PubMed] [Google Scholar]
- Noutoshi Y., Okazaki M., Kida T., Nishina Y., Morishita Y., Ogawa T., Suzuki H., Shibata D., Jikumaru Y., Hanada A., Kamiya Y., Shirasu K. (2012). Novel plant immune-priming compounds identified via high-throughput chemical screening target salicylic acid glucosyltransferases in Arabidopsis. Plant Cell 24: 3795–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E., Gang D.R. (2000). Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci. 5: 439–445. [DOI] [PubMed] [Google Scholar]
- Quiel J.A., Bender J. (2003). Glucose conjugation of anthranilate by the Arabidopsis UGT74F2 glucosyltransferase is required for tryptophan mutant blue fluorescence. J. Biol. Chem. 278: 6275–6281. [DOI] [PubMed] [Google Scholar]
- Rissel D., Losch J., Peiter E. (2014). The nuclear protein Poly(ADP-ribose) polymerase 3 (AtPARP3) is required for seed storability in Arabidopsis thaliana. Plant Biol (Stuttg) 16: 1058–1064. [DOI] [PubMed] [Google Scholar]
- Saito K., Matsuda F. (2010). Metabolomics for functional genomics, systems biology, and biotechnology. Annu. Rev. Plant Biol. 61: 463–489. [DOI] [PubMed] [Google Scholar]
- Sasamoto H., Ashihara H. (2014). Effect of nicotinic acid, nicotinamide and trigonelline on the proliferation of lettuce cells derived from protoplasts. Phytochem. Lett. 7: 38–41. [Google Scholar]
- Song J.T., Koo Y.J., Park J.B., Seo Y.J., Cho Y.J., Seo H.S., Choi Y.D. (2009). The expression patterns of AtBSMT1 and AtSAGT1 encoding a salicylic acid (SA) methyltransferase and a SA glucosyltransferase, respectively, in Arabidopsis plants with altered defense responses. Mol. Cells 28: 105–109. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura K., Satou J., Iwata M., Uozumi N., Koga J., Kawano T., Koshiba T., Anzai H., Mitomi M. (2009). Contribution of salicylic acid glucosyltransferase, OsSGT1, to chemically induced disease resistance in rice plants. Plant J. 57: 463–472. [DOI] [PubMed] [Google Scholar]
- Wang G., Pichersky E. (2007). Nicotinamidase participates in the salvage pathway of NAD biosynthesis in Arabidopsis. Plant J. 49: 1020–1029. [DOI] [PubMed] [Google Scholar]
- Wang G., Tian L., Aziz N., Broun P., Dai X., He J., King A., Zhao P.X., Dixon R.A. (2008). Terpene biosynthesis in glandular trichomes of hop. Plant Physiol. 148: 1254–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth W.M., Drury G.E., Bray C.M., West C.E. (2011). Repairing breaks in the plant genome: the importance of keeping it together. New Phytol. 192: 805–822. [DOI] [PubMed] [Google Scholar]
- Weng J.-K., Philippe R.N., Noel J.P. (2012). The rise of chemodiversity in plants. Science 336: 1667–1670. [DOI] [PubMed] [Google Scholar]
- Willeke U., Heeger V., Meise M., Neuhann H., Schindelmeiser I., Vordemfelde K., Barz W. (1979). Metabolism of nicotinic acid in plant cell suspension cultures. 6. Mutually exclusive occurrence and metabolism of trigonelline and nicotinic acid arabinoside in plant cell cultures. Phytochemistry 18: 105–110. [Google Scholar]
- Wubbolts M.G., Terpstra P., van Beilen J.B., Kingma J., Meesters H.A., Witholt B. (1990). Variation of cofactor levels in Escherichia coli. Sequence analysis and expression of the pncB gene encoding nicotinic acid phosphoribosyltransferase. J. Biol. Chem. 265: 17665–17672. [PubMed] [Google Scholar]
- Xu H., Zhang F., Liu B., Huhman D.V., Sumner L.W., Dixon R.A., Wang G. (2013). Characterization of the formation of branched short-chain fatty acid:CoAs for bitter acid biosynthesis in hop glandular trichomes. Mol. Plant 6: 1301–1317. [DOI] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yazaki K. (2005). Transporters of secondary metabolites. Curr. Opin. Plant Biol. 8: 301–307. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Hanada K. (2011). An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 66: 182–193. [DOI] [PubMed] [Google Scholar]
- Zheng X.Q., Hayashibe E., Ashihara H. (2005). Changes in trigonelline (N-methylnicotinic acid) content and nicotinic acid metabolism during germination of mungbean (Phaseolus aureus) seeds. J. Exp. Bot. 56: 1615–1623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.