Mutation of LONG AND BARBED AWN1, which encodes a cytokinin-activating enzyme, underlies the transition from the long, barbed awns in wild rice to the short, barbless awns in domesticated rice.
Abstract
Common wild rice (Oryza rufipogon), the wild relative of Asian cultivated rice (Oryza sativa), flaunts long, barbed awns, which are necessary for efficient propagation and dissemination of seeds. By contrast, O. sativa cultivars have been selected to be awnless or to harbor short, barbless awns, which facilitate seed processing and storage. The transition from long, barbed awns to short, barbless awns was a crucial event in rice domestication. Here, we show that the presence of long, barbed awns in wild rice is controlled by a major gene on chromosome 4, LONG AND BARBED AWN1 (LABA1), which encodes a cytokinin-activating enzyme. A frame-shift deletion in LABA1 of cultivated rice reduces the cytokinin concentration in awn primordia, disrupting barb formation and awn elongation. Sequencing analysis demonstrated low nucleotide diversity and a selective sweep encompassing an ∼800-kb region around the derived laba1 allele in cultivated rice. Haplotype analysis revealed that the laba1 allele originated in the japonica subspecies and moved into the indica gene pool via introgression, suggesting that humans selected for this locus in early rice domestication. Identification of LABA1 provides new insights into rice domestication and also sheds light on the molecular mechanism underlying awn development.
INTRODUCTION
Rice (Oryza sativa) is one of the earliest domesticated food crops and currently provides staple food for over one-third of the world’s population (Khush, 1997). Cultivated rice was domesticated from common wild rice (Oryza rufipogon) ∼8000 years ago (Zong et al., 2007; Fuller et al., 2010). The transition from wild to domesticated rice was accompanied by numerous morphological and physiological changes. To date, genetic factors underlying several rice domestication-related traits, such as seed shattering, prostrate growth habit, red pericarp, spreading panicle, dark hull color, and awn length have been cloned and characterized at the molecular level (Li et al., 2006; Konishi et al., 2006; Sweeney et al., 2006; Lin et al., 2007; Tan et al., 2008; Jin et al., 2008; Zhu et al., 2011; Ishii et al., 2013; Luo et al., 2013; Zhu et al., 2013).
Like other grasses, wild rice not only harbors a long awn at the tip of the seed, but the awn of wild rice is densely covered with upwardly angled barbs (Figures 1A and 1C). Long and barbed awns are evolutionarily beneficial in wild grass species because their ratcheting surface enables the seeds to capitalize on transient environmental stresses by transforming these fluctuations into directed movement, providing a selective advantage for seed dispersal and burial into soil (Kulić et al., 2009). In wild wheat, the hygroscopic motion of awns is capable of propelling seed into soil with the help of the awn barbs (Elbaum et al., 2007). Although awns of wild rice are not hygroscopic, they are still critical for efficient propagation and dissemination of wild rice seeds under natural conditions as they serve to protect seeds from bird and mammal predation, aid in seed dispersal by clinging to animal fur, and enabling self-planting (Supplemental Figures 1A to 1F). In contrast, cultivated rice is characterized by awnless seeds, or seeds with short, barbless awns (Figures 1B and 1D), that facilitate seed collection, storage, and processing by humans.
Figure 1.
The Awns of Wild Rice (O. rufipogon) and Cultivated Rice (O. sativa).
(A) Wild rice typically bears a long, barbed awn, which aids in seed dispersal and deters large predators, such as birds. A panicle of wild rice (Tk212) is illustrated in the lower right corner. Bar = 10 cm.
(B) Cultivated rice has either no awns or short and barbless awns and suffers from significant bird predation. A panicle of cultivated rice (93-11, indica) is illustrated in the lower left corner. Bar = 10 cm.
(C) The awn of wild rice (Tk212) is long and barbed. Scanning electron micrograph in the lower right corner shows the surface of the awn of wild rice in the boxed region. Bar = 200 μm.
(D) The awn of cultivated rice (93-11) is short and barbless. Scanning electron micrograph in the lower left corner shows the surface of the awn of cultivated rice in the boxed region. Bar = 200 μm.
Previous genetic studies identified many quantitative trait loci related to awn length in rice (Shigetoshi et al., 1996; Thomson et al., 2003; Xiong et al., 1999), and one major quantitative trait locus for awn length has been cloned and characterized at the molecular level (Luo et al., 2013). An-1 encodes a basic helix-loop-helix transcription factor that positively regulates the formation of awn primordia and negatively regulates grain number. The derived allele, an-1, which confers loss of awns and an increase in grain yield, was selected by humans during rice domestication (Luo et al., 2013). In addition, molecular mechanisms underlying awn traits are recently being addressed (Yuo et al., 2012; Toriba and Hirano, 2014). In this study, we focus on the transition from long, barbed awns to short, barbless awns during rice domestication and elucidate the molecular basis for this critical change.
RESULTS AND DISCUSSION
Fine-Mapping and Cloning of LABA1
To identify the genes controlling long, barbed awns, we developed a set of introgression lines using an accession of Yuanjiang common wild rice (YJCWR, O. rufipogon), which has long, barbed awns and was collected from Yuanjiang county of Yunnan province of China as a donor, and an elite indica rice variety, 93-11, which has short, barbless awns, as a recipient (Fu et al., 2010). Introgression line IL_9YIL304 carried four YJCWR introgressions on chromosomes 1, 3, 4, and 10 and exhibited long, barbed awns at the tip of the lemma and a higher proportion of awned seeds (Figures 2A to 2D; Supplemental Figure 2). Observation of the awn surface using scanning electron microscopy showed that the surface of IL_9YIL304’s awn was covered by numerous upwardly oriented barbs with a typical length of ∼200 μm, while the surface of 93-11’s awn was barbless (Figure 2E). Further examination at the longitudinal histological level showed that the barbs were derived from single epidermal cells on IL_9YIL304’s awn, and we observed no significant difference in cell size between the awns of 93-11 and IL_9YIL304, indicating that the long awn of IL_9YIL304 results from an increase in cell number (Figure 2F; Supplemental Figure 3).
Figure 2.
Phenotype of IL_9YIL304 and 93-11.
(A) Panicles of the introgression line IL_9YIL304 and 93-11. Bar = 10 cm.
(B) Comparison of awn length between IL_9YIL304 and 93-11. The awns of IL_9YIL304 (3.5 to 3.9 cm) were significantly longer than the awns of 93-11 (1.2 to 1.6 cm).
(C) Comparison of awn proportion between IL_9YIL304 and 93-11. Awn proportion of IL_9YIL304 was almost 100%, whereas only ∼30% grains in a panicle of 93-11 showed awns.
(D) Grains of IL_9YIL304 and 93-11. Grain of IL_9YIL304 exhibited a longer awn. Bar = 1 cm.
(E) Scanning electron micrographs showing the surface of the awn of IL_9YIL304 and 93-11 in boxed regions of (D). IL_9YIL304 had upwardly oriented barbs on the awn surface, whereas the awn of 93-11 was barbless. Bar = 200 μm.
(F) Longitudinal section in the boxed regions of (D) showed cell size in the awn of IL_9YIL304 and 93-11. Awn barb was derived from awn epidermal cell in IL_9YIL304. Arrows point to the layers of cells adjacent to vascular bundle whose size was measured. Bars = 200 μm.
In (B) and (C), sampled size was n = 20. The statistical significance was at P < 0.05 based on a two-tailed Student’s t test. Error bars represent the sd.
Genetic linkage analysis of 300 F2 individuals derived from a cross between IL_9YIL304 and 93-11 showed that the long awn was completely correlated with the presence of awn barbs and was controlled by a single dominant gene, LONG AND BARBED AWN1 (LABA1), located between simple sequence repeat markers RM1223 and RM17282 on the long arm of chromosome 4 (Figure 3A). Using 6655 F2 plants, we delimited LABA1 to a 34.6-kb region between markers M3 and RM17242, in which there were five predicted genes (Figures 3B and 3C). We then screened a genomic BAC library derived from YJCWR using markers M3 and RM17242, and a positive BAC clone (YJ0610808) was isolated and sequenced. When we compared the nucleotide sequences of wild and cultivated rice across the fine-mapped region, we discovered that a total of 21.8 kb was missing from the YJCWR genome, including three transposons that were present in 93-11. Reannotation of the YJCWR sequence using the Rice Genome Automated Annotation System identified two open reading frames (ORFs), including a possible lysine decarboxylase (LOC_Os04g43840) and an expressed protein (LOC_Os04g43880) (Figure 3D). Comparison of the coding sequences of these predicted genes in YJCWR and 93-11 revealed a 1-bp frame-shift deletion of cytosine at +69 bp in the first exon of LOC_Os04g43840, resulting in a premature stop codon in 93-11. No amino acid changes were found in LOC_Os04g43880. Therefore, we focused on LOC_Os04g43840 as a candidate for LABA1.
Figure 3.
Map-Based Cloning of LABA1.
(A) The target gene(s) for awn barbs and awn length mapped between RM1223 and RM17282 on the long arm of chromosome 4 based on linkage analysis using 300 F2 individuals.
(B) LABA1 was further delimited to a 34.6-kb region between the marker M3 and RM17242 based on analysis of recombinant plants identified from an F3 population of 6655 individuals.
(C) The Nipponbare genomic BAC clone (OSJNBb0061C13) shows five genes in this region.
(D) The Yuanjiang common wild rice (YJCWR) genomic BAC clone (YJ0610808) shows deletion of three segments containing transposon elements and retains two gene models in this region.
(E) An 8524-bp genomic fragment containing the entire ORF (4109 bp) of LOC_Os04g43840 was isolated by digesting the YJCWR genomic BAC (YJ0610808) DNA using EcoRI and BamHI and cloned into the binary vector pCAMBIA1300.
(F) to (H) Complementation phenotypes of Y179: transgenic line CTP-1 showed increased awn length (G) and awn barbs on awn surface (H) compared with that the transgenic control.
(I) to (K) RNAi phenotypes of IL_9YIL304: RNAi transgenic line RNAi-2 exhibited substantially reduced awn length (J) and shorter and smaller awn barbs (K) compared with IL_9YIL304.
(L) and (M) Comparison of awn length (L) and awn proportion (M) between Y179 and two independent transgenic lines CTP-1 and CTP-2.
(O) and (P) Comparison of awn length (O) and awn proportion (P) between IL_9YIL304 and two independent transgenic lines RNAi-1 and RNAi-2.
CTP, LABA1 complementation transgenic plants. RNAi, LABA1 RNA interference plants. In (L) to (O), sampled size was n = 10. The statistical significance was at P < 0.05 based on a two-tailed Student’s t test. Error bars represent the sd. Bars = 10 cm in (F) and (I), 1 cm in (G) and (J), and 200 μm in (H) and (K).
We next digested the positive BAC clone (YJ0610808) covering LOC_Os04g43840 using EcoRI and BamHI and isolated an 8524-bp fragment containing the entire ORF (4109 bp) of LOC_Os04g43840, a 2456-bp 5′-flanking region, and 1959 bp of the 3′-flanking region. This fragment was cloned into the binary vector pCAMBIA1300 to generate the pLABA1 construct (Figure 3E). Owing to the recalcitrance of the indica variety 93-11 to regeneration in tissue culture following transformation, we introduced the pLABA1 construct into a recipient line Y179 that had short, barbless awns (Supplemental Figure 4) and generated 11 complemented transgenic plants (CTPs). Of these, seven independent transformants showed longer awns and had obvious barbs on the surface of the awns, compared with control transgenic plants that harbored an empty vector (Figures 3F, 3G, 3L, and 3M). We also transformed an RNA interference (RNAi) construct that harbored a 286-bp specific region of the LOC_Os04g43840 mRNA into IL_9YIL304. Reverse transcription-quantitative PCR analysis showed that the expression of LOC_Os04g43840 was downregulated in RNAi transgenic plants compared with control plants (Supplemental Figure 5A). The length of the awn in independent RNAi transgenic lines was significantly less, and the awn barbs of RNAi transgenic lines were significantly shorter and smaller compared with control plants (Figures 3I to 3K, 3N, and 3O; Supplemental Figures 5B and 5C). From these results, we conclude that LOC_Os04g43840 is the LABA1 gene that controls both awn length and the presence of barbs on the awn.
Transcriptional Characterization of LABA1
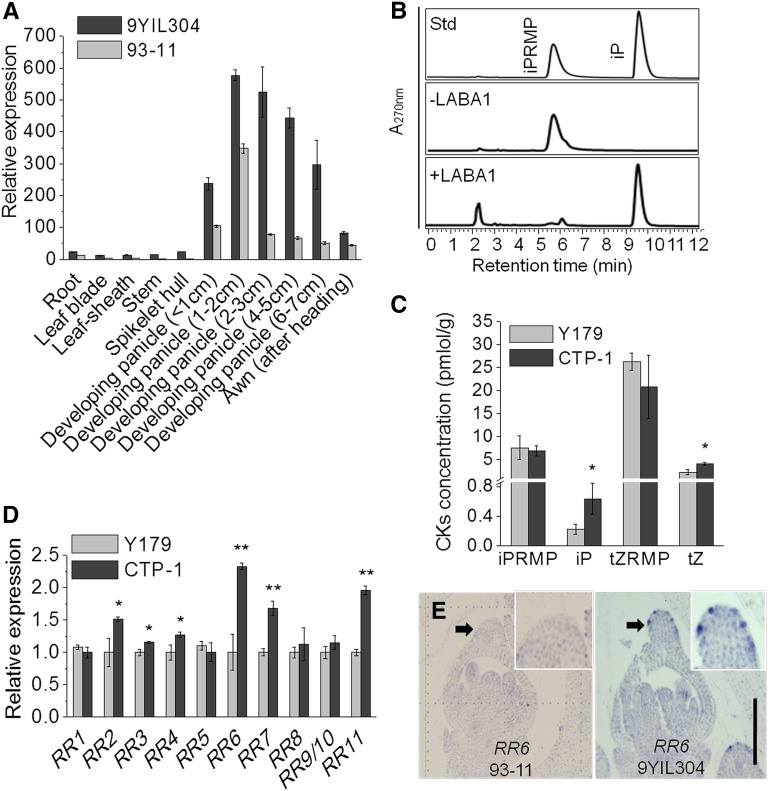
Sequence analysis of 5′- and 3′-rapid amplification of cDNA ends (RACE) cDNA products indicated that the LABA1 cDNA in O. rufipogon is 1062-bp long, with an ORF of 753 bp, a 119-bp 5′ untranslated region, and a 190-bp 3′ untranslated region, and it encodes a 250-amino-acid protein (Supplemental Figure 6). We constructed a LABA1-GFP fusion gene under the control of the CaMV35S promoter, and when it was introduced into onion inner epidermal cells, we observed that the LABA1-GFP fusion protein localized to the cytosol and nuclei (Supplemental Figure 7). The expression profiles of LABA1 were analyzed in different organs using reverse transcription-quantitative PCR, and the results showed that LABA1 was preferentially expressed in developing panicles, with the highest expression in panicles of 1 to ∼2 cm in length and less transcript accumulation in root, leaf blade, leaf sheath, culm, spikelet hull, and in the awn itself after heading (Figure 4A). The high levels of LABA1 expression in developing panicles is consistent with its role in controlling awn development, which occurs in the course of panicle development.
Figure 4.
Expression Pattern of LABA1 and Function of LABA1.
(A) The expression pattern of LABA1 in IL_9YIL304 and 93-11.
(B) Enzyme activity of LABA1 as a cytokinin nucleoside 5′-monophosphate phosphoribohydrolase was measured with iPRMP as substrate. Standards (Std) of iPRMP and iP and reaction products of iPRMP without LABA1 and with LABA1 were separated by HPLC.
(C) Comparison of cytokinin concentration in 1- to ∼2-cm young panicles of transgenic plants and control.
(D) Expression of rice RR1 to RR11 in 1- to ∼2-cm young panicles of Y179 and CTP-1. The expression of RR2, RR3, RR4, RR6, RR7, and RR11 was significantly increased in CTP-1.
(E) RNA in situ hybridization of RR6 in the awn of 93-11 and IL_9YIL304 at the Sp7 stage. The transcripts of RR6 were present in the epidermal cells of the awn primordia in IL_9YIL304, but not in 93-11. Bar = 100 μm.
LABA1 Encodes a Cytokinin-Activating Enzyme
A protein-BLAST at NCBI showed that LABA1 is a homolog of rice LONELY GUY (LOG), which encodes a cytokinin riboside 5′-monophosphate phosphoribohydrolase that directly converts inactive cytokinin nucleotide 5′-monophosphate (iPRMP and tZRMP) to the biologically active free-base form (iP and tZ) in the final step of bioactive cytokinin biosynthesis (Kurakawa et al., 2007). The deduced LABA1 protein is identical to rice LOG-like 6 with 76% amino acid identity to rice LOG (Kuroha et al., 2009). To determine whether LABA1 has cytokinin nucleotide phosphoribohydrolase activity, we assayed the enzyme activity of recombinant LABA1 with iPRMP and found that LABA1 was able to catalyze iPRMP to generate iP (Figure 4B). To confirm the function of LABA1 in vivo, we measured endogenous cytokinin concentrations in 1- to ∼2-cm young panicles of transgenic plants and observed that the concentration of bioactive cytokinins (iP and tZ) was significantly greater in the complementary transgenic plant CTP-1, compared with control plants (Figure 4C). We further examined the transcript levels of 11 cytokinin-responsive RESPONSE REGULATOR genes in 1- to ∼2-cm young panicles and found that six of them (RR2, RR3, RR4, RR6, RR7, and RR11) were significantly increased in complementary transgenic plant CTP-1 compared with the transgenic control (Figure 4D). RNA in situ hybridization of RR6 showed clearly that the induced transcripts were present in the epidermal cells of the awn primordia in IL_9YIL304, but not in 93-11 (Figure 4E). These results indicated that LABA1 encodes a cytokinin-activating enzyme that increased cytokinin content in epidermal cells of the awn primordia.
Specific Expression of LABA1 Promotes Awn Elongation and Awn Barb Formation
To understand the effect of LABA1 on awn development, we compared awn development between IL_9YIL304 and 93-11 (Figure 5). Based on the spikelet development (Sp) stages described previously (Itoh et al., 2005), we found no difference in spikelet development between IL_9YIL304 and 93-11 until the Sp8e stage (Figures 5A1, 5A2, 5B1, and 5B2). At the Sp7 stage, awn primordia of similar size initiated at the top of the lemma of IL_9YIL304 and 93-11 (Figures 5A2 and 5B2). By the Sp8e stage, all spikelets had formed awn primordia in the apex of lemma of 93-11, but only a few of them finally developed into macroscopic awns (>1 mm), indicating that most 93-11 awn primordial elongate very slowly during spikelet development. In contrast, the awn primordia of IL_9YIL304 had extended much farther and then formed more and longer awns than that of 93-11 (Supplemental Figure 8); in addition, a few epidermal cells on the top of IL_9YIL304’s awn swelled and bulged out into the hemisphere. However, no swollen epidermal cells were found on 93-11’s awn primordia (Figures 5A3 and 5B3). The swollen epidermal cells formed basipetally along the awn and they elongated and sharpened upwardly to form barbs at the Sp8 late stage in IL_9YIL304. However, no barb formed at any time during awn development in 93-11 (Figures 5A4 and 5B4).
Figure 5.
The Effect of LABA1 on Awn Development.
(A1) to (A4) Scanning electron microscopy images showed spikelet development of 93-11 from the Sp7 to Sp8l stage.
(B1) to (B4) Scanning electron microscopy images showed spikelet development of IL_9YIL304 from the Sp7 to Sp8l stage.
(C1) to (C4) RNA in situ hybridization of laba1 during spikelet development of 93-11 from the Sp7 to Sp8l stage.
(D1) to (D4) RNA in situ hybridization of LABA1 during spikelet development of IL_9YIL304 from the Sp7 to Sp8l stages.
(E1) to (E4) Expression of Histone H4 in developing spikelets of 93-11.
(F1) to (F4) Expression of Histone H4 in developing spikelets of IL_9YIL304.
(A1), (B1), (C1), (D1), (E1), and (F1) Sp6, formation of stamen primordia stage.
(A2), (B2), (C2), (D2), (E2), and (F2) Sp7, formation of carpel primordia stage.
(A3), (B3), (C3), (D3), (E3), and (F3) Sp8e (Sp8 early), differentiation of ovule and pollen stage.
(A4), (B4), (C4), (D4), (E4), and (F4) Sp8l (Sp8 late), differentiation of ovule and pollen stage.
Arrows point to awn primordia. Bars = 100 μm.
We further examined the expression of LABA1 during spikelet development using RNA in situ hybridization. At the Sp7 stage, the LABA1 signal starts to emerge in several cells at the top of the newly formed awn primordia. At the Sp8e and Sp8l stages, LABA1 transcripts were intensively localized in the young epidermal cells of awn primordia of IL_9YIL304, but not in the swollen epidermal cells at the Sp8e stage and developing barbs at the Sp8l stage (Supplemental Figure 9). In contrast, there was little expression of laba1 in the awn primordia of 93-11 (Figures 5C1 to 5C4 and 5D1 to 5D4). The specific expression of LABA1 in young (non-barbed) epidermal cells of the awn primordia was coincident with the increase in cytokinin content in epidermal cells and suggests that LABA1 expression is necessary for awn barbs to develop. However, LABA1 doesn’t express in developing barbs, implying that LABA1 regulates awn barb formation indirectly; thus, we conclude that LABA1 is necessary but not sufficient for the development of barbs on the awns.
To gain further insight into the role of LABA1 in awn elongation, we used RNA in situ hybridization to analyze the expression of Histone H4, a marker of active cell division (Marzluff and Duronio, 2002). We found that, at Sp8e and Sp8l stages, the transcripts of Histone H4 were more abundant in the awn primordia of IL_9YIL304 than in 93-11 (Figures 5E1 to 5E4 and 5F1 to 5F4), indicating more active cell division in the awn primordia of IL_9YIL304 than in 93-11. To further understand whether the activation of cell division was caused by cytokinin, we analyzed transcript levels of the cell cycle gene CYCD3, which plays an essential role in the activation of cell division induced by cytokinin (Dewitte et al., 2007). Our results showed that CYCD3 expression increased in complemented transgenic plants and decreased in RNAi transgenic plants (Supplemental Figure 11), suggesting that the long awn and high awn frequency phenotype in wild rice was caused by active cell division mediated by cytokinins in the awn primordia.
The laba1 Allele Is Unique to Cultivated Asian Rice
To determine whether the laba1 allele, which contains 1-bp deletion at +69-bp site (Chr. 4: 25959586) discovered in 93-11, existed in wild rice prior to domestication, we examined its frequency in wild and cultivated rice accessions using two panels of germplasm: (1) a Mini Diversity Panel 1 (MiniDP1) consisting of 98 accessions of O. sativa (23 temperate japonica, 16 tropical japonica, 7 aromatic, 22 indica, 23 aus, and 7 admix), 4 O. rufipogon, 7 Oryza glaberrima (cultivated African rice), and 7 Oryza barthii (wild African rice); and (2) Mini Diversity Panel 2 (MiniDP2), which consisted of 247 strains of cultivated rice (110 indica, 110 japonica, and 27 aus) and 39 accessions of wild rice (O. rufipogon) from 17 countries (Tan et al., 2008; Fan et al., 2009). We observed no occurrence of the laba1 allele in any of the Asian wild rice accessions (n = 43) nor in the 14 African rice accessions, but it did occur in 55% of O. sativa accessions in MiniDP1 and in 74% of O. sativa in MiniDP2. Because of the deep subpopulation structure in O. sativa, we were curious to know how laba1 was distributed across the five O. sativa subpopulations that had been characterized in MiniDP1. We observed the laba1 allele at the highest frequency in tropical japonica (100%) and indica (86%) and at lowest frequency in temperate japonica (13%) and aus (35%). To determine whether laba1 accounts for a barbless awn phenotype across diverse cultivated rice varieties, we phenotyped 62 awned accessions from the MiniDP1 (23 temperate japonica, 16 tropical japonica, 7 aromatic, 22 indica, and 23 aus) to quantify the presence or absence of barbs. The laba1 deletion was found in 32 awned varieties and was perfectly correlated with the absence of barbs. The LABA1 allele occurred consistently with the presence of barbs, but there were five exceptions, all from the aus subpopulation (cv Kasalath, Kalamkati, Kaukau, DD 62, and DZ 193). The five exceptional aus varieties carried the wild-type LABA1 allele but did not have barbs, indicating either that there is a second, aus-specific mutation in the LABA1 gene leading to a barbless phenotype or that variation in another gene mediates barb formation in the aus subpopulation.
Evidence of Positive Selection on laba1 in O. sativa
To determine whether laba1 was targeted by human selection during rice domestication, we sequenced an ∼5-kb genomic region containing the LABA1 gene (∼4.1 kb) and an ∼0.9-kb promoter region in the MiniDP2 and analyzed the data for patterns of variation. We found that overall, cultivated rice possesses only 8.4% of the nucleotide diversity observed in wild rice, far below the ∼80% genetic diversity retained in cultivated rice at the whole-genomic level (Huang et al., 2012). Tajima’s D was negative (P < 0.001), suggesting strong directional selection across this region of the genome in cultivated rice (Table 1). Using 19 single nucleotide polymorphisms (SNPs) with minor allele frequency >0.05 in either wild rice or cultivated rice, we constructed LABA1 haplotypes. We identified 19 haplotypes across the 39 wild accessions and only three haplotypes across all 247 cultivars (Supplemental Figure 12). Ninety-two of the accessions were awned (43 indica, 41 japonica, and 8 aus), so we were able to associate each cultivar gene haplotype with the barb phenotype.
Table 1. Nucleotide Diversity and Tajima’s D Test.
| Taxon | N | L | S | H | π | θw | Tajima’s D |
|---|---|---|---|---|---|---|---|
| O.sativa | 247 | 5054 | 46 | 28 | 0.00019 | 0.00153 | −2.54 P < 0.001 |
| O. rufipogon | 39 | 5036 | 71 | 35 | 0.00226 | 0.00342 | −1.23 P > 0.100 |
N, total number of sequences; L, average length (bp) of the sequences per taxon; S, number of polymorphic (segregating) sites; H, number of haplotypes; π, average number of pairwise nucleotide differences per site calculated based on the total number of polymorphic sites; θw, Watterson’s estimator of per base pair calculated based on the total number of polymorphic sites.
The most frequent haplotype in O. sativa was CH1, which contains the laba1 allele and is associated with barbless awns. CH1 was present in 74% of MiniDP2 varieties (105 indica, 73 japonica, and 5 aus). Haplotype CH2, which is associated with barbed awns (if an accession carries an awn), was detected in 17% of O. sativa varieties in the MiniDP2 (5 indica and 37 japonica). CH2 is identical to CH1 except that it contains a functional allele without 1-bp deletion at the +69-bp position (Chr. 4: 25959586). Haplotype CH3 differs from CH2 by only a single SNP at +154 bp (Chr. 4: 25959672) and was observed in 22 aus varieties, but in none of the other wild or cultivated groups. We conclude that both CH1 and CH3 were derived from CH2 via a single base-pair deletion (CH1) or substitution (CH3), given that CH2 is associated with the barbed awn phenotype and carries wild-type SNP alleles (i.e., identical to O. rufipogon) at both the +69-bp and +154-bp sites. The pattern and extent of haplotype variation at the LABA1 locus observed in O. sativa diverges from that observed in O. rufipogon, where 19 different haplotypes were detected in 39 O. rufipogon accessions (Supplemental Figure 12), all of which differed from those observed in cultivated rice.
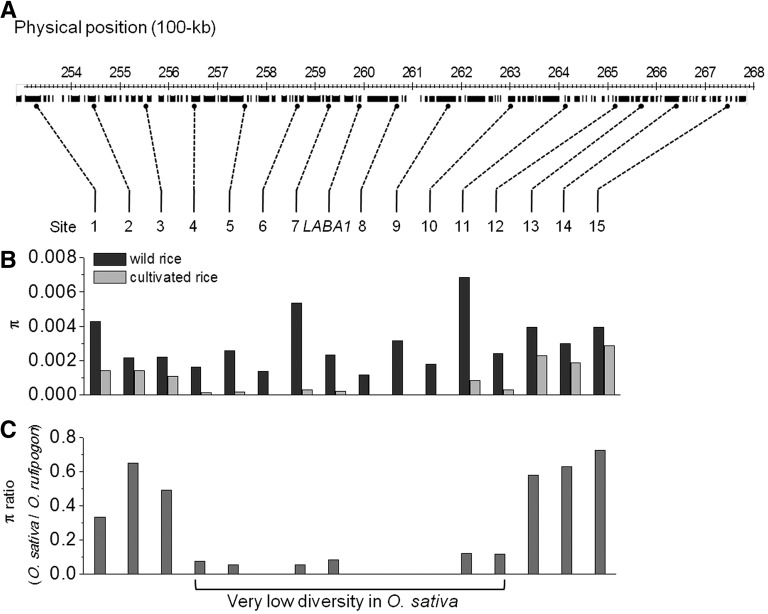
The contrasting distributions of haplotypes, CH1 through CH3, across indica, japonica, and aus, and their associations with the awn and barb phenotypes suggest that CH2 is a functional haplotype from which CH1 arose and was selected upon, resulting in its high frequency in cultivated rice today. To further investigate the possibility of positive selection on LABA1, we examined the nucleotide diversity of 15 loci that mapped within an ∼1.4-Mb interval surrounding the gene (Figure 6A) and identified an interval of ∼800-kb surrounding the LABA1 locus that exhibited significantly reduced nucleotide diversity in cultivated rice relative to wild rice (Figure 6B). This selective sweep is consistent with recent whole-genome investigations that identified regions associated with low nucleotide diversity and presumably, strong artificial selection in cultivated rice (He et al., 2011; Huang et al., 2012; Xu et al., 2012). Our results identify awn morphology as a target of selection and indicate that the laba1 allele conferring short, barbless awns in O. sativa was strongly selected by human beings during rice domestication.
Figure 6.
Nucleotide Diversity across LABA1 Genomic Region on Chromosome 4.
(A) The location of sampled loci in genomic region of ∼1.4 Mb around LABA1 on chromosome 4.
(B) Nucleotide diversity π of cultivated rice and wild rice at the sampled loci.
(C) The relative ratio of π in cultivated rice to wild rice shows a selective sweep of ∼800 kb surrounding laba1 in cultivated rice.
Extended Haplotype Analysis of LABA1
To explore the subpopulation origin of the laba1 allele (CH1) in O. sativa, we examined the genomic environment surrounding LABA1 by constructing extended haplotypes (EHs) across a 1.4-Mb region flanking LABA1. The region encompassed by the EH contained the 800-kb region defining the selective sweep. Using the 98 accessions of O. sativa from the MiniDP1 and a SNP data set generated using the high-density rice array (S.R. McCouch, unpublished data), we used 36 ancestrally informative polymorphisms (Kovach et al., 2009) to construct EHs around LABA1.
EHs were developed for all 98 accessions of O. sativa in the MiniDP1. These included 62 awned accessions that had been phenotyped for the barb trait as well as 36 accessions that lacked awns and therefore could not be phenotyped for the barb trait. Among the 62 awned accessions, we observed variable awn phenotypes that ranged from short awns found only on terminal spikelets on a panicle, to long and fully awned panicles.
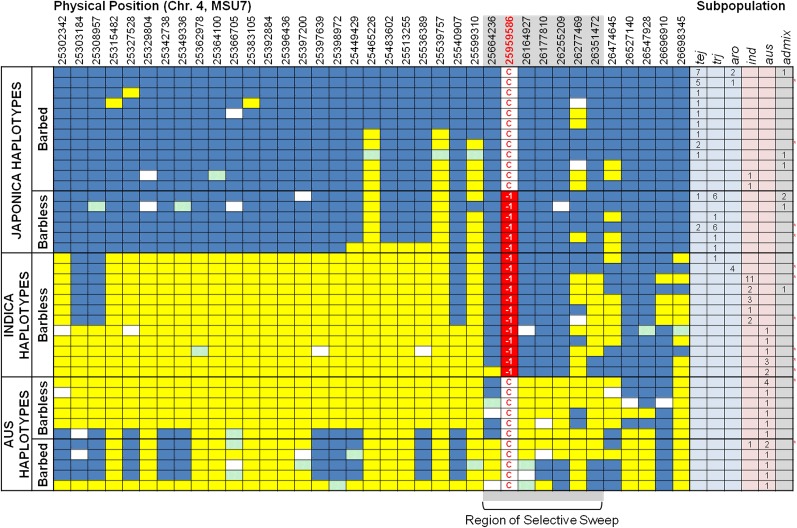
Similar to results discussed above that identified three gene haplotypes in O. sativa cultivars (CH1, CH2, and CH3), EH variation could also be clustered into three major haplotype groups based on shared polymorphisms (Figure 7). One EH group was strongly associated with temperate and tropical japonica varieties (hereafter referred to as the japonica-EH), one EH group was associated primarily with indica and some aus varieties (referred to as the indica-EH), and the third group was strongly associated with aus varieties (referred to as the aus-EH).
Figure 7.
EH Analysis of Awned and Awnless O. sativa Accessions across a 1.4-Mb Region Flanking LABA1.
Physical positions on chromosome 4 are indicated across the top of the figure in base pairs. Blue cells match the allele found in the reference genome, Nipponbare (temperate japonica), and yellow cells indicate a nonreference allele. All polymorphisms are SNP data, which were extracted from the high-density rice array data set. Haplotypes are clustered based on whether they were found more in the japonica varietal group (japonica haplotypes) or the indica varietal group (indica haplotypes). Haplotypes specific to the aus subpopulation are grouped separately (aus haplotypes). The allele found at the functional nucleotide polymorphism is indicated in white and red: the wild type C in white cells and derived laba1 in red cells. The numbers of accessions that harbored each haplotype are indicated in the right-hand table (tej = temperate japonica, trj = tropical japonica, aro = aromatic, ind = indica, aus = aus, admix = admixture, determined by HDRA genome-wide information). Awnless accessions are grouped with their predicted barb trait groups; red asterisks indicate the presence of at least one awnless accession with a specific haplotype.
In the japonica-EH group, we observed both functional and nonfunctional alleles at LABA1 (based on the presence or absence of the 1-bp deletion at +69 bp), and there was a clear frequency difference between temperate and tropical japonica varieties (Figure 7). Twenty out of 23 (87%) temperate japonica landrace varieties with this haplotype carried the functional allele (CH2) and had long, barbed awns, while 100% (15/15) of tropical japonica varieties had the nonfunctional allele (CH1) and lacked barbs or lacked awns altogether. We also identified two indica varieties that carried the japonica-EH and had the functional (CH2) LABA1 allele, along with the long, barbed awn phenotype.
Within the indica-EH group, 100% of varieties carried the nonfunctional (CH1) allele and none had barbed awns. Along with the 19 indica landrace varieties that fell into this category were one tropical japonica, four aromatic, and eight aus varieties. Thus, for both the japonica-EH and the indica-EH, which are found in diverse genetic backgrounds, the 1-bp deletion at +69 bp in the LABA1 gene was predictive of the barbed awn phenotype. Upon careful inspection, the indica-EH was characterized by an ∼690-kb region surrounding the LABA1 gene that matched the japonica-EH. There was also evidence of recombination that had restored indica genomic background outside the selective sweep region in different varieties within this group (Figure 7), as would be expected if the introgression were ancient. Thus, we hypothesize that the barbless-awn phenotype was introgressed into the indica gene pool via hybridization with a sympatric japonica ancestor, possibly a tropical japonica domesticate, in the early phases of O. sativa domestication. Consistent with this hypothesis, the introgression maps squarely within the 800-kb region of reduced O. sativa nucleotide diversity.
For the aus-EH group, which was represented in this study by 15 aus and 1 indica varieties, 100% of accessions carried the nondeletion LABA1 allele, but for this subpopulation, the presence of the C-SNP at position +69 bp in the LABA1 gene was not predictive of phenotype. Five aus varieties carrying the aus-EH had long, barbed awns and 11 had either no barbs or no awns at all (Figure 7). These observations indicate that there is either an additional mutation, within the gene itself or its promoter region, or variation at an entirely different locus responsible for the barbless phenotype in aus varieties.
A Second Gene Confers the Barbless Phenotype to aus Varieties
To test whether a different mutation in the LABA1 gene or promoter region might be associated with long and barbed awns, we examined resequencing data across LABA1 and a 2-kb region upstream of LABA1 in four awned aus varieties to look for potential explanatory polymorphisms. Of these four accessions, one individual had barbed awns (cv Sathi), while the other three were smooth (cultivars BJ1, Kalamkati, and Kasalath). Within the LABA1 genic region (Chr 4: 25959399-25963504) there were no additional indels and only one SNP at +154 bp, matching the CH3 gene haplotype. All four accessions harbored CH3 along with identical promoter sequences (Chr 4: 25957399-25959399), leading us to reject the hypothesis that aus has additional functional variation at LABA1. These results suggest that members of the aus subpopulation instead carry a unique mutation at a second locus conditioning the loss of barbs on awns.
We next examined individuals from a population of backcross introgression lines (BILs) derived from a cross between Nipponbare (temperate japonica) and Kasalath (aus), for which previously published genotype data are available (Ishimaru et al., 2001; Xu et al., 2009; http://rgp.dna.affrc.go.jp/E/publicdata/genotypedataBILs/genotypedata.html). Both parents of the BIL population harbor the wild-type LABA1 allele, but Nipponbare has short, terminal barbed awns while Kasalath has long, non-barbed awns. We speculated that barbless phenotypes found in progeny within this population must be due to a dysfunctional locus from Kasalath. Seeds from nine awned BC1F8 lines and six awned BC2F2 families were phenotyped for the presence of barbs. This gave us two perspectives on the inheritance of the barb phenotype: While the BC1F8 seeds reflect the genotypes of the original BILs, the BC2F2 seed phenotypes reflect their maternal BC2F1 genotype in which every Kasalath segment in the original BIL is heterozygous, allowing us to assess dominance of the barb trait. Out of the 15 total seed packets evaluated, nine completely lacked barbed awns. These nine barbless genotypes shared a region inherited from Kasalath on short arm of chromosome 4, located between markers RM6770-C946, while the remaining seven barbed genotypes harbored Nipponbare alleles in that region, as expected. Additionally, we noticed that while the barbless BC1F8 seeds were completely smooth, the BC2F2 seeds scored as barbless had very sparse, thin hairs, hinting at additive effects of the Nipponbare and Kasalath alleles at this novel barb locus. Because this candidate region on the short arm of chromosome 4 is located physically far from the LABA1 locus, we conclude that the aus subpopulation carries a second gene that is independent of LABA1 but also confers barbless awns.
Reduction of Awn Length and Loss of Barbs Is Associated with Increased Rice Yield
To test for additional traits affected by the presence of the derived laba1 allele, we measured husked rice yield in six YJCWR-derived introgression lines in the genetic background of 93-11 harboring awns of varying length (∼7, ∼4, and ∼2 cm), with and without barbs. We found that removal of the long awns could increase husked rice yield by up to ∼5% (Supplemental Figure 13A). Measurement of husked rice yield in the complemented transgenic plants (CTP-1, -2) and control plants (Y179) indicated that selection on laba1 significantly improved husked rice yield (Supplemental Figure 13B). Using a volumetric weight instrument, we discovered that awn length has a negative relationship with unit weight (kg/m3) and that for rice within the same awn length class, the loss of barbs increased unit weight (Supplemental Figure 13C). Comparison of unit weight between the complemented transgenic plants (CTP-1, -2) and control plant (Y179) suggested that selection on laba1 remarkably increased unit weight (Supplemental Figure 13D). From these experiments, we conclude that selection of laba1 increased the efficiency of rice processing and ease of rice storage in both indica and tropical japonica rice. It is interesting that laba1 is not fixed in temperate japonica, presumably because in the temperate region, the awnless phenotype had already been selected based on variation at a different locus (e.g., an-1). Another explanation is that long, barbed awns were found useful in these areas of the world to deter seed predation by animals; we see evidence in ancient Chinese texts that farmers continued to use rice landraces with long awns and, in fact, have given names to accessions based on these characteristics (e.g., the ability to “choke a boar”). Both possible reasons are consistent with the fraction of temperate japonica landraces that have retained long, barbed awns we see today.
Self-propagation and seed dispersal are vital to wild grasses, and long, barbed awns play an indispensible role in the self-propagation and seed dispersal of wild rice. However, as cultivators of grass species, human beings value the convenience of seed collection and the ease of seed storage. The selection on laba1 led to great reduction in awn length and to the elimination of awn barbs, facilitating seed collection, processing, and storage. This work has identified a domestication gene, LABA1, explored the evolutionary history and penetrance of the laba1 derived allele, and shed light on the molecular mechanism by which LABA1 conditions both awn length and the presence of barbs on the awns.
METHODS
Plant Materials
In positional cloning, spikelet development analysis, and expression pattern analysis, we used an indica cultivar 93-11 with short awn and an introgression line IL_9YIL304 with long and barbed awn. In LABA1 complementation and RNAi tests, Y179 and IL_9YIL304 were used as transgenic recipients, respectively. IL_9YIL304 was screened from the BC4F8 population derived from the cross between YJCWR (Yuanjiang common wild rice) and 93-11. Y179 harboring short and smooth awn was screened from the BC4F4 population derived from the cross between Tk212 (wild rice from India with long, barbed awns) and an awnless indica cultivar Teqing. The rice diversity panels MiniDP1 and MiniDP2 and the accessions used in selective sweep analysis are listed in Supplemental Data Set 1.
Primers
The primers used in this study are listed in Supplemental Table 1.
Generation of Constructs
The BAC clone YJ0610808, containing the LABA1 gene, was identified from the BAC library of Yuanjiang common wild rice. The 8524-bp fragment harboring the entire LABA1 gene was inserted into the binary vector pCAMBIA1300 to form the pLABA1 construct. The RNAi construct contained an inverted repeat harboring the 286-bp LABA1 cDNA fragment in vector RNAi-LABA1. These two plasmid constructs were introduced into Agrobacterium tumefaciens strain EHA105 and subsequently transformed into the introgression lines Y179 and IL_9YIL304, respectively.
Phenotypic Evaluation
Awn length was measured as the average awn length of apical spikelet of each primary branch on the main stem panicle. Awn proportion was measured as the percentage of awned spikelets on main stem panicle, and the awn of mature grains with >1 mm in length was recorded as awned phenotype. Twenty plants were used to measure the awn length and awn proportion of IL_9YIL304 and 93-11. Phenotypic measurements were conducted using two independent T2 homozygous lines (CTP-1, -2), two independent RNAi T2 homozygous lines (RNAi-1, -2), and 10 plants for each line. Ten Y179 and IL_9YIL304 plants harboring empty vector were used as the control.
Scanning Electron Microscopy
The young panicles were fixed in 2.5% glutaraldehyde-phosphate buffer saline fixative solution, dehydrated through an ethanol series, and dried using a carbon dioxide critical-point dryer. The awn of mature grain was cleaned with ethanol and dried at 45°C. The dry panicles and awn were gold plated and observed using a Hitachi S-2460 scanning electron microscope at 15 kV.
5′- and 3′-RACE and Quantitative RT-PCR
We extracted total RNAs using an RNeasy Plant Mini Kit (Qiagen). We conducted 5′- and 3′-RACE with the GeneRacer kit (Invitrogen Life Technologies) following the manufacturer’s instructions. First-strand cDNA synthesis was performed with the GoScript Reverse Transcription System (Promega). Quantitative RT-PCR was done on a CFX96 real-time system (Bio-Rad). Diluted cDNA was amplified using the SsoFast Evagreen Supermix (Bio-Rad). We normalized the levels of transcripts by endogenous Ubiquitin transcripts amplified with primers UbiF and UbiR. Each set of experiments was repeated three times, and the relative quantification method (2−△△CT (DDCT)) was used to evaluate quantitative variation (Livak and Schmittgen, 2001).
RNA in Situ Hybridization
Young panicles of 93-11 and IL_9YIL304 were fixed in 4% (w/v) paraformaldehyde, followed by a series of dehydration and infiltration, and were embedded in paraffin (Paraplast Plus; Sigma-Aldrich). The tissues were sliced into 8-mm sections with a microtome (Leica RM2145). The 286-bp 5′-region of LABA1 cDNA was amplified and used as the template to generate sense and antisense RNA probes. Digoxigenin-labeled RNA probes were prepared using a DIG Northern Starter Kit (catalog number 2039672; Roche) according to the manufacturer’s instruction. RNA in situ hybridization with sense and antisense probes was performed on sections of young panicles as described previously (Zhang et al., 2007). Slides were observed under bright field through a microscope (Leica DMR) and photographed with a micro color CCD camera (Apogee Instruments).
Subcellular Localization
Subcellular localization of LABA1 was performed using the coding sequence of a GFP fused in frame to the LABA1-coding sequence and transcribed from a CaMV35S promoter. The resulting plasmid was bombarded into onion epidermal cells using helium biolistic device (Bio-Rad PDS-1000). We examined the bombarded tissues with a confocal laser scanning microscope (Carl Zeiss LAM510).
Enzyme Assays
The coding region of LABA1 was inserted into pCOLD1 (Takara) to express His-tagged recombinant proteins. BL21 (DE3) harboring pG-Tf2 (Takara) was used as the Escherichia coli host. Enzyme activity of LABA1 as a cytokinin nucleoside 5′-monophosphate phosphoribohydrolase was measured by incubating 3 μg purified LABA1 protein in a reaction mixture (50 mM Tris-HCl, 1 mM MgCl2, and 1 mM DTT, pH 6.5) with 100 mM iPRMP at 30°C for 2 h. The reaction products were separated and monitored as described previously (Kurakawa et al., 2007).
Sequencing and Data Analysis
Multiple sequences were aligned with ClustalX (Thompson et al., 1997). Nucleotide diversity and Tajima’s D test was calculated and performed using DnaSP, version 5.0 (Rozas et al., 2003).
Adhesion Test and Self-Planting Test
IL_9YIL341, IL_9YIL90, IL_9YIL86, IL_9YIL205, IL_9YIL63, and IL_9YIL98 harboring ∼7-cm barbed awn, ∼7-cm barbless awn, ∼4-cm barbed awn, ∼4-cm barbless awn, ∼2-cm barbed awn, and ∼2-cm barbless awn, respectively, were screened from the BC4F8 population derived from the cross between YJCWR and 93-11. Grains of these lines and an awnless cultivar W9311 were used for both tests. In adhesion test, 50 grains with different type of awn were scattered randomly on a smooth table. We covered all grains with a piece of woolen cloth. After pressing woolen cloth by placing a 0.5 kg flat board on it for 1 min, we lifted and removed the sheep wool carefully; the number of grains left on the table was recorded. The percentage of seeds attached to the woolen cloth was calculated. Each test was repeated for 20 times. In the self-planting test, 50 rice grains with different types of awns were scattered from a height of 1 meter to 0.25% agar medium. The grains landing with >60° angle between awn and horizontal was recorded as self-planting. The percentage of self-planting was calculated. Each test was repeated 20 times.
Unit Weight and Husked Rice Yield Measurements
IL_9YIL341, IL_9YIL90, IL_9YIL86, IL_9YIL205, IL_9YIL63, and IL_9YIL98 harboring ∼7-cm barbed awn, ∼7-cm barbless awn, ∼4-cm barbed awn, ∼4-cm barbless awn, ∼2-cm barbed awn, and ∼2-cm barbless awn, respectively, were screened from the BC4F8 population derived from the cross between YJCWR and 93-11. Grains of these lines and the awnless cultivar W9311 were harvested and dried for 3 d at 45°C. Unit weight of each line was measured using a volumetric weight instrument (HGT-1000). In husked rice yield measurement, grains of 200 gram in each line were husked using a rice husking machine. We weighed husked rice and calculated the rate of output as husked rice yield. Both measurements were repeated for 10 times in each line.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: LABA1 cDNA from YJCWR (KR703212) and YJ0610808 (KR703211). The accession numbers of LABA1/laba1 genomic sequences in 247 cultivated and 39 wild rice varieties are listed in Supplemental Data Set 1b. The accession numbers of sequences used in selective sweep analysis in 60 cultivated and 35 wild rice varieties are listed in Supplemental Data Set 1c.
Supplemental Data
Supplemental Figure 1. Long and barbed awn aids seed dispersal.
Supplemental Figure 2. Graphic genotype of introgression line 9YIL304.
Supplemental Figure 3. Comparison of cell size of awn between IL_9YIL304 and 93-11.
Supplemental Figure 4. Graphic genotype of transgenic recipient Y179.
Supplemental Figure 5. Full-length sequence of LABA1 in Yuanjiang common wild rice (YJCWR) and sequence comparison between LABA1 and laba1 in 93-11.
Supplemental Figure 6. RNA interference of LABA1 in IL_9YIL304.
Supplemental Figure 7. The subcellular localization of LABA1 protein using onion inner epidermal cells.
Supplemental Figure 8. Awn primordia of IL_9YIL304 elongate faster than that of 93-11 at the Sp8 stage.
Supplemental Figure 9. The expression of LABA1 in developing barbs.
Supplemental Figure 10. In situ hybridization of LABA1 and RR6 sense probe at the sp7 stage.
Supplemental Figure 11. Cytokinin responsiveness of CYCD3 and relative expression of CYCD3 in transgenic plants.
Supplemental Figure 12. Reconstruction of LABA1 haplotypes in MiniDP2.
Supplemental Figure 13. Reduction of awn length and loss of barb is associated with increased rice yield.
Supplemental Table 1. Primers used in this study.
Supplemental Data Set 1a. Rice accessions of Mini Diversity Panel 1 (MiniDP1).
Supplemental Data Set 1b. Rice accessions of Mini Diversity Panel 2 (MiniDP2).
Supplemental Data Set 1c. Rice accessions used for selective sweep analysis.
Supplementary Material
Acknowledgments
We thank Xiaolan Zhang for the help with RNA in situ hybridization, Pengfei Liu for the help with enzyme assay, and Jing Zhao for providing seeds of Y179. We thank the International Rice Research Institute, Chinese Rice Research Institute, Institute of Crop Sciences of Chinese Academy of Agricultural Sciences, Guangxi Academy of Agricultural Sciences, and Guangdong Academy of Agricultural Sciences for providing the wild rice and cultivated rice samples. This research was supported by the National Natural Science Foundation of China (Grants 31071395 and 91335202), the China National High-tech Research and Development (‘863’) Program (Grant 2012AA10A301), and self-regulated projects of the State Key Laboratory of Plant Physiology and Biochemistry.
AUTHOR CONTRIBUTIONS
C.S. designed and supervised this study. L.H. conducted characterization of introgression line, map-based cloning, genetic transformation, gene expression analysis, protein functional analysis, and evolutionary analysis in MiniDP2. D.R.W. analyzed extended haplotype in MiniDP1 and mapping population. L.X. conducted cytokinin measurement. F.L. and Q.F. constructed the introgression lines. X.S. and Y.F. maintained plant materials and performed the field management. L.T., Z.Z., H.C., and P.G. conducted the collection of rice germplasm and phenotypic data. C.S., L.H., S.R.M., and D.R.W. analyzed the data and wrote the artilce.
Glossary
- ORF
open reading frame
- RNAi
RNA interference
- SNP
single nucleotide polymorphism
- EH
extended haplotype
- BIL
backcross introgression line
Footnotes
Articles can be viewed online without a subscription.
References
- Dewitte W., Scofield S., Alcasabas A.A., Maughan S.C., Menges M., Braun N., Collins C., Nieuwland J., Prinsen E., Sundaresan V., Murray J.A. (2007). Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. USA 104: 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum R., Zaltzman L., Burgert I., Fratzl P. (2007). The role of wheat awns in the seed dispersal unit. Science 316: 884–886. [DOI] [PubMed] [Google Scholar]
- Fan C., Yu S., Wang C., Xing Y. (2009). A causal C-A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor. Appl. Genet. 118: 465–472. [DOI] [PubMed] [Google Scholar]
- Fu Q., Zhang P., Tan L., Zhu Z., Ma D., Fu Y., Zhan X., Cai H., Sun C. (2010). Analysis of QTLs for yield-related traits in Yuanjiang common wild rice (Oryza rufipogon Griff.). J. Genet. Genomics 37: 147–157. [DOI] [PubMed] [Google Scholar]
- Fuller D., Sato Y., Castillo C. (2010). Consilience of genetics and archaeobotany in the entangled history of rice. Archaeol. Anthropol. Sci. 2: 115–131. [Google Scholar]
- He Z., Zhai W., Wen H., Tang T., Wang Y., Lu X., Greenberg A.J., Hudson R.R., Wu C.I., Shi S. (2011). Two evolutionary histories in the genome of rice: the roles of domestication genes. PLoS Genet. 7: e1002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T., Numaguchi K., Miura K., Yoshida K., Thanh P.T., Htun T.M., Yamasaki M., Komeda N., Matsumoto T., Terauchi R., Ishikawa R., Ashikari M. (2013). OsLG1 regulates a closed panicle trait in domesticated rice. Nat. Genet. 45: 462–465, e1–e2. [DOI] [PubMed] [Google Scholar]
- Ishimaru K., Yano M., Aoki N., Ono K., Hirose T., Lin S.Y., Monna L., Sasaki T., Ohsugi R. (2001). Toward the mapping of physiological and agronomic characters on a rice function map: QTL analysis and comparison between QTLs and expressed sequence tags. Theor. Appl. Genet. 102: 793–800. [Google Scholar]
- Itoh J., Nonomura K., Ikeda K., Yamaki S., Inukai Y., Yamagishi H., Kitano H., Nagato Y. (2005). Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46: 23–47. [DOI] [PubMed] [Google Scholar]
- Jin J., Huang W., Gao J.P., Yang J., Shi M., Zhu M.Z., Luo D., Lin H.X. (2008). Genetic control of rice plant architecture under domestication. Nat. Genet. 40: 1365–1369. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△CT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Khush G.S. (1997). Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 35: 25–34. [PubMed] [Google Scholar]
- Konishi S., Izawa T., Lin S.Y., Ebana K., Fukuta Y., Sasaki T., Yano M. (2006). An SNP caused loss of seed shattering during rice domestication. Science 312: 1392–1396. [DOI] [PubMed] [Google Scholar]
- Kovach M.J., Calingacion M.N., Fitzgerald M.A., McCouch S.R. (2009). The origin and evolution of fragrance in rice (Oryza sativa L.). Proc. Natl. Acad. Sci. USA 106: 14444–14449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulić I.M., Mani M., Mohrbach H., Thaokar R., Mahadevan L. (2009). Botanical ratchets. Proc. Biol. Sci. 276: 2243–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., Sakakibara H., Kyozuka J. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655. [DOI] [PubMed] [Google Scholar]
- Kuroha T., Tokunaga H., Kojima M., Ueda N., Ishida T., Nagawa S., Fukuda H., Sugimoto K., Sakakibara H. (2009). Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21: 3152–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Zhou A., Sang T. (2006). Rice domestication by reducing shattering. Science 311: 1936–1939. [DOI] [PubMed] [Google Scholar]
- Lin Z., Griffith M.E., Li X., Zhu Z., Tan L., Fu Y., Zhang W., Wang X., Xie D., Sun C. (2007). Origin of seed shattering in rice (Oryza sativa L.). Planta 226: 11–20. [DOI] [PubMed] [Google Scholar]
- Luo J., et al. (2013). An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 25: 3360–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W.F., Duronio R.J. (2002). Histone mRNA expression: Multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 14: 692–699. [DOI] [PubMed] [Google Scholar]
- Rozas, J., Sánchez-DelBarrio, J.C., Messeguer, X., and Rozas, R. (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Shigetoshi S., Setsuji I., Masaki S., Choyu S. (1996). Genetic studies on an awnness gene An-4 on ch8 in rice, Oryza sativa L. Breed. Sci. 46: 321–327. [Google Scholar]
- Sweeney M.T., Thomson M.J., Pfeil B.E., McCouch S. (2006). Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Li X., Liu F., Sun X., Li C., Zhu Z., Fu Y., Cai H., Wang X., Xie D., Sun C. (2008). Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40: 1360–1364. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson M.J., Tai T.H., McClung A.M., Lai X.H., Hinga M.E., Lobos K.B., Xu Y., Martinez C.P., McCouch S.R. (2003). Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor. Appl. Genet. 107: 479–493. [DOI] [PubMed] [Google Scholar]
- Toriba T., Hirano H.Y. (2014). The DROOPING LEAF and OsETTIN2 genes promote awn development in rice. Plant J. 77: 616–626. [DOI] [PubMed] [Google Scholar]
- Xiong L., Liu K., Dai X., Xu C., Zhang Q. (1999). Identification of genetic factors controlling domestication-related traits of rice using an F2 population of a cross between Oryza sativa and O. rufipogon. Theor. Appl. Genet. 98: 243–251. [Google Scholar]
- Xu X., et al. (2012). Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30: 105–111. [DOI] [PubMed] [Google Scholar]
- Xu Y., This D., Pausch R.C., Vonhof W.M., Coburn J.R., Comstock J.P., McCouch S.R. (2009). Leaf-level water use efficiency determined by carbon isotope discrimination in rice seedlings: genetic variation associated with population structure and QTL mapping. Theor. Appl. Genet. 118: 1065–1081. [DOI] [PubMed] [Google Scholar]
- Yuo T., Yamashita Y., Kanamori H., Matsumoto T., Lundqvist U., Sato K., Ichii M., Jobling S.A., Taketa S. (2012). A SHORT INTERNODES (SHI) family transcription factor gene regulates awn elongation and pistil morphology in barley. J. Exp. Bot. 63: 5223–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Madi S., Borsuk L., Nettleton D., Elshire R.J., Buckner B., Janick-Buckner D., Beck J., Timmermans M., Schnable P.S., Scanlon M.J. (2007). Laser microdissection of narrow sheath mutant maize uncovers novel gene expression in the shoot apical meristem. PLoS Genet. 3: e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B.F., et al. (2011). Genetic control of a transition from black to straw-white seed hull in rice domestication. Plant Physiol. 155: 1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Tan L., Fu Y., Liu F., Cai H., Xie D., Wu F., Wu J., Matsumoto T., Sun C. (2013). Genetic control of inflorescence architecture during rice domestication. Nat. Commun. 4: 2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y., Chen Z., Innes J.B., Chen C., Wang Z., Wang H. (2007). Fire and flood management of coastal swamp enabled first rice paddy cultivation in east China. Nature 449: 459–462. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.