The transcriptional activator MYC2 is targeted by PUB10 for ubiquitin-mediated proteolysis in jasmonic acid responses.
Abstract
MYC2 is an important regulator for jasmonic acid (JA) signaling, but little is known about its posttranslational regulation. Here, we show that the MYC2 C-terminal region interacted with the PLANT U-BOX PROTEIN10 (PUB10) armadillo repeats in vitro. MYC2 was efficiently polyubiquitinated by PUB10 with UBC8 as an E2 enzyme and the conserved C249 in PUB10 was required for activity. The inactive PUB10(C249A) mutant protein retained its ability to heterodimerize with PUB10, thus blocking PUB10 E3 activity as a dominant-negative mutant. Both MYC2 and PUB10 were nucleus localized and coimmunoprecipitation experiments confirmed their interaction in vivo. Although unstable in the wild type, MYC2 stability was enhanced in pub10, suggesting destabilization by PUB10. Moreover, MYC2 half-life was shortened or prolonged by induced expression of PUB10 or the dominant-negative PUB10(C249A) mutant, respectively. Root growth of pub10 seedlings phenocopied 35S:MYC2 seedlings and was hypersensitive to methyl jasmonate, whereas 35S:PUB10 and jin1-9 (myc2) seedlings were hyposensitive. In addition, the root phenotype conferred by MYC2 overexpression in double transgenic plants was reversed or enhanced by induced expression of PUB10 or PUB10(C249A), respectively. Similar results were obtained with three other JA-regulated genes, TAT, JR2, and PDF1.2. Collectively, our results show that MYC2 is targeted by PUB10 for degradation during JA responses.
INTRODUCTION
Regulated proteolysis plays important roles in plant signaling pathways in at least two important steps. In several pathways, signaling is restrained by repressors that are destroyed by ubiquitin-mediated proteolysis upon signal perception to initiate signaling (for a review, see Sadanandom et al., 2012). For example, the transcriptional repressors AUX/IAA are targeted by the F-box auxin receptor TRANSPORT INHIBITOR RESPONSE1 (Dharmasiri et al., 2005; Kepinski and Leyser, 2005). During jasmonic acid (JA) signaling, the JASMONATE ZIM DOMAIN PROTEIN family of transcriptional repressors is ubiquitinated by the F-box JA-Ile receptor CORONATINE INSENSITIVE1 for subsequent degradation by 26S proteasomes (Chini et al., 2007; Thines et al., 2007). In addition, DELLA transcriptional repressors in gibberellin signaling are destabilized by F-box SLEEPY1/GA INSENSITIVE DWARF2 (McGinnis et al., 2003; Dill et al., 2004). In other pathways, ubiquitin-mediated proteolysis is used to degrade positive regulatory components, e.g., transcription factors, to downregulate and attenuate signaling (for a review, see Sadanandom et al., 2012). For example, EIN3 BINDING F-BOX PROTEIN1/2 mediate degradation of the transcriptional activators ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3 LIKE1 in ethylene signaling (Gagne et al., 2004; An et al., 2010). In abscisic acid (ABA) signaling, the transcriptional activators ABA INSENSITIVE3 and ABA INSENSITIVE5 are destabilized by the RING-type E3 ligases ABI3 INTERACTING PROTEIN2 and KEEP ON GOING, respectively (Zhang et al., 2005; Liu and Stone, 2010). In addition, DEHYDRATION RESPONSIVE ELEMENT BINDING PROTEIN 2A (DREB2A) and INDUCER OF CBP EXPRESSION1, which function as transcriptional activators in abiotic stress signaling, are degraded by DREB2A INTERACTING PROTEIN1/2 and HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1, respectively (Dong et al., 2006; Qin et al., 2008).
Like other eukaryotes, ubiquitin-dependent protein degradation in plants requires the sequential action of three enzymes: E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme, and E3 ubiquitin ligase, with substrate specificity being conferred by the E3 ligase. More than 1300 genes encoding E3 ligases, including proteins with RING, HECT, F-box, and U-box domains, have been annotated in Arabidopsis thaliana (Vierstra, 2009). Among these various E3 categories, a large number of RING-motif proteins and SCF complexes have been investigated with respect to their molecular functions in plant growth and development (Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Lau and Deng, 2012).
Compared with RING proteins and SCF complexes, much less is known about the U-box proteins, which are referred to as PUB (PLANT U-BOX) proteins (Yee and Goring, 2009). The Arabidopsis genome encodes at least 64 PUBs and ∼40% of them have been shown to have E3 activities when associated with specific UBCs (Mudgil et al., 2004; Wiborg et al., 2008). Whereas the biochemical properties and NMR structure have been determined for a PUB protein (Andersen et al., 2004; Wiborg et al., 2008), the biological function of only a limited number of PUBs is known (Yee and Goring, 2009). For example, PUB9, 18, and 19 have been linked to ABA responses (Samuel et al., 2008; Bergler and Hoth, 2011; Seo et al., 2012), PUB12, 13, 17, 22, 23, and 24 play roles in various steps of the innate immunity pathway (Yang et al., 2006; Cho et al., 2008; Trujillo et al., 2008; Lu et al., 2011; Stegmann et al., 2012; Antignani et al., 2015), and PUB22 and 23 are also associated with drought response as plants overexpressing these proteins displayed drought hypersensitivity (Cho et al., 2008). Similarly, PUBs of other plant species have also been implicated in responses to biotic and abiotic stresses (for a review, see Yee and Goring, 2009).
Transcription factors are key regulators of signaling pathways, and their levels are generally low at steady state but may increase or decrease in response to signals. Accumulating evidence shows that transcription factor levels are tightly regulated by ubiquitin-mediated proteolysis, and the E3 ligases of many transcription factors have been identified. Although a large number of PUBs have been investigated, there is as yet no report on the identification of a transcription factor target of any PUB E3 ligase. An important transcription factor in plants is the basic helix-loop-helix (bHLH) protein MYC2, first identified as a positive regulator of ABA signaling. This factor has subsequently been implicated in JA signaling as well (Abe et al., 1997; Boter et al., 2004; Lorenzo et al., 2004).
In the past two decades, it has been shown that MYC2 acts as a master regulator to integrate signals from various pathways to coordinate plant defense and development (for a review, see Kazan and Manners, 2013). MYC2, MYC3, and MYC4 are very unstable proteins degraded by 26S proteasomes. MYC2 protein levels change in response to signals, e.g., circadian rhythm, methyl jasmonate (MeJA), and specific light conditions (Shin et al., 2012; Zhai et al., 2013; Chico et al., 2014). Dark, far-red light, shade, and CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) destabilize MYC2, MYC3, and MYC4, whereas MeJA, red light, and blue light stabilize them. Differential regulation of MYC2 stability by the change in the ratio of red to far-red light regulates JA-dependent defenses (Chico et al., 2014). TIME FOR COFFEE (TIC) represses MYC2 protein accumulation under the control of circadian clock. As a result, TIC regulates JA-mediated phenotypes, e.g., root growth inhibition, gene expression, and defense response, in a MYC2-dependent manner (Shin et al., 2012). Deletion of the destruction element in MYC2 renders it more stable, and MYC2 phosphorylation facilitates its own turnover (Zhai et al., 2013). Both destruction element-deleted and nonphosphorylatable mutants phenocopy myc2 mutant with respect to root growth inhibition, gene expression, and defense response (Zhai et al., 2013). However, the E3 ligase(s) involved in MYC2 degradation has not yet been identified. MYC2 levels are elevated in cop1 mutant, but there is no evidence that COP1 E3 ligase acts directly to destabilize MYC2 (Chico et al., 2014). The identification of an E3 ligase responsible for MYC2 ubiquitination would certainly advance our knowledge of how plant signaling is regulated.
In an attempt to expand our knowledge on regulated proteolysis in Arabidopsis in general and, the biological function of PUBs in particular, we have chosen to investigate PUB10, which is hitherto uncharacterized. Using yeast two-hybrid assays, we first identified MYC2 as a putative binding protein of PUB10. PUB10 was shown to have the capacity to autoubiquitinate and to efficiently polyubiquitinate MYC2 in vitro. MYC2 was very unstable in the wild type, but its half-life was prolonged in pub10 mutant plants and by inducible overexpression of the PUB10(C249A) mutant protein. By contrast, inducible overexpression of PUB10 accelerated the MYC2 decay rate. These results suggest MYC2 is a target of PUB10 in vivo. This notion was further supported by the observation that pub10 mutant phenocopied 35S:MYC2 plants, whereas 35S:PUB10 plants were similar to the jin1-9 (myc2) mutant in JA responses. Taken together, our results provide evidence that MYC2 levels are regulated by PUB10 in vivo.
RESULTS
PUB10 Interacts with MYC2 in Yeast Cells and in Vitro
We were interested to investigate the biological role of PUB10 in Arabidopsis. As a first step, we performed yeast two-hybrid assays using PUB10 as a bait to interrogate a small library of preys comprising ∼1500 transcription factors encoded by the Arabidopsis genome (Mitsuda et al., 2010). Preliminary experiments uncovered several candidate proteins that interacted with PUB10 in yeast cells. The strongest among these was MYC2, a bHLH protein (Abe et al., 1997; Boter et al., 2004; Lorenzo et al., 2004), which was further investigated.
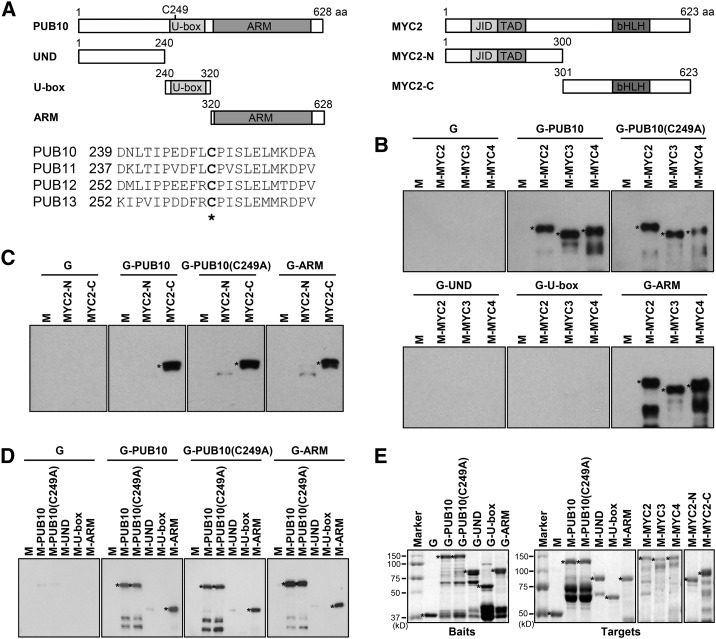
Supplemental Figure 1 shows that MYC2 interacted with PUB10 in yeast two-hybrid assays performed under stringent conditions. To confirm this result, we tested GST (G)-PUB10 fusion protein purified from Escherichia coli extracts for its capacity to bind to MBP (M)-MYC2 in vitro (Figure 1). Indeed, full-length wild-type G-PUB10 and its armadillo (ARM) repeats were able to bind M-MYC2 as well as two MYC2 homologs, MYC3 and MYC4, both tagged with MBP (Fernández-Calvo et al., 2011) (Figures 1A and 1B). By analyzing PUB10 and MYC2 deletion derivatives, we found that the PUB10 ARM repeats interacted with the C-terminal region of MYC2 (MYC2-C), although weak interaction was seen with MYC2-N as well (Figure 1C). These binding results were specific; negative control experiments showed that neither GST nor G-UND/U-box could bind M-MYC2 (Figures 1B and 1C).
Figure 1.
PUB10 Self-Associates and Interacts with MYC2, MYC3, and MYC4 through Its ARM Repeats in Vitro.
(A) Schematic diagrams of full-length PUB10, MYC2, and deletion derivatives used for in vitro pull-down assays. Amino acid alignment of a short sequence of the conserved U-box motif in Arabidopsis PUB10, PUB11, PUB12, and PUB13. Numbers refer to the positions of the first amino acid in the sequence. The asterisk indicates a conserved cysteine residue. In PUB10(C249A), Cys-249 of the PUB10 U-box was changed to Ala.
(B) PUB10 interacts with MYC2, MYC3, and MYC4. GST, GST-PUB10, GST-PUB10(C249A), and deletion derivatives were used as baits. MBP, MBP-MYC2, MBP-MYC3, and MBP-MYC4 were used as target proteins.
(C) PUB10 interacts with the C-terminal region of MYC2. GST, GST-PUB10, GST-PUB10(C249A), and GSTPUB10-ARM (G-ARM) were used as baits. MBP, MBP-MYC2-N, and MBP-MYC2-C were used as target proteins.
(D) Self-association of PUB10. GST, PUB10, GST-PUB10(C249A), and GST-PUB10-ARM (G-ARM) were used as baits. MBP, MBP-PUB10, MBP-PUB10(C249A), and deletion derivatives were used as target proteins.
(E) Input amounts of the bait and target proteins. SDS-PAGE gel was stained with Coomassie blue and used to monitor the load of bait and target proteins before pull-down assays. Asterisks indicate the bait and target proteins used in each experiment.
For (B) to (D), target proteins (2 μg) were pulled down with the indicated bait proteins (2 μg each) and detected by anti-MBP antibody. aa, amino acids; UND, large U-box N-terminal domain; JID, JAZ-interacting domain; TAD, transactivation domain; G, GST; M, MBP.
Because PUB10 is a putative E3 ligase, we mutated the conserved C249 of PUB10, which is known to compromise E3 activity in related PUBs (Lu et al., 2011) (Figure 1A). Figures 1B and 1C show that MYC2 binding was not affected by the C249A mutation. As E3 ubiquitin ligases are usually activated via dimerization (Xie et al., 2002; Seo et al., 2003; Zhang et al., 2005), we also investigated the capacity of PUB10 to self-associate. Figure 1D shows that PUB10 as well as the PUB10(C249A) mutant could homodimerize; moreover, wild-type PUB10 and PUB10(C249A) mutant could also form heterodimers in vitro. Deletion analysis identified the ARM repeat-containing region (320 to 628 amino acids) as the dimerization domain.
PUB10 Binds to Specific UBCs and Polyubiquitinates MYC2 in Vitro
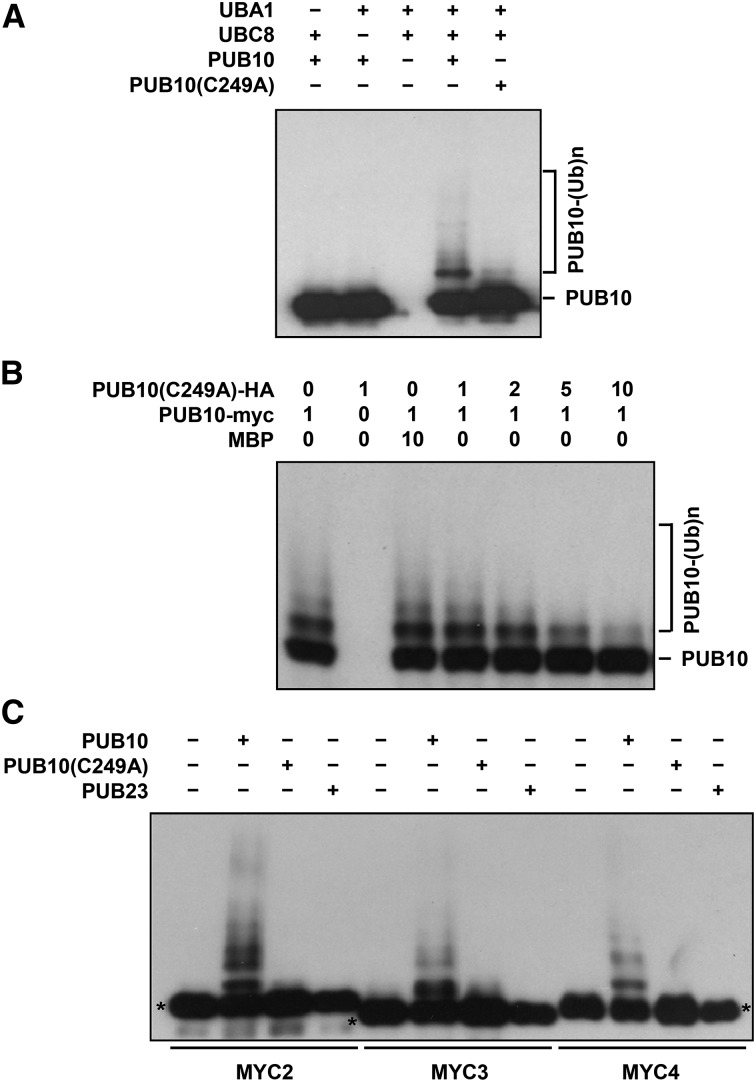
Because PUB10 is a plant U-box protein, it was reasonable to assume that it may have ubiquitin E3 ligase activity. PUB10 protein purified from E. coli extracts was used as a source of E3 enzyme for in vitro ubiquitination reactions (Figure 2). Figure 2A shows that PUB10 was able to perform autoubiquitination using the Arabidopsis UBC8 as an E2 and that the E3 activity was abrogated by the C249A mutation.
Figure 2.
PUB10 Is an E3 Ubiquitin Ligase and Polyubiquitinates MYC2, MYC3, and MYC4 in Vitro.
(A) PUB10 E3 activity depends on the integrity of its U-box motif. Epitope-tagged recombinant PUB10, PUB10(C249A), Arabidopsis UBA1, and Arabidopsis UBC8 proteins were purified from E. coli extracts. MBP-PUB10 and MBP-PUB10(C249A) were assayed for E3 activity in the presence or absence of Arabidopsis E1 (His-tagged UBA1) and Arabidopsis E2 (His-tagged UBC8). Polyubiquitinated PUB10 was detected by anti-MBP antibody.
(B) Excess PUB10(C249A) protein functions as a dominant negative mutant by blocking PUB10 E3 activity. Numbers indicate the relative amounts of proteins in the reaction mixture, where 1 represents 100 ng of MBP, MBP-PUB10-myc, and MBP-PUB10(C249A)-HA. Polyubiquitinated PUB10 was detected by anti-myc antibody.
(C) MYC2, MYC3, and MYC4 are substrates of PUB10 E3 ligase. Polyubiquitination of GST-MYC2, GST-MYC3, and GST-MYC4 was assayed in the presence of UBA1, UBC8, PUB10, and ubiquitin. MBP-PUB10(C249A) mutant protein and MBP-PUB23 were used as negative controls. Polyubiquitinated MYC2, MYC3, and MYC4 were detected by anti-GST antibody. Asterisks indicate the size of the unmodified substrates.
Since the PUB10(C249A) mutant had the capacity to heterodimerize with wild-type PUB10, we hypothesized that excess PUB10(C249A) mutant would titrate out wild-type PUB10 to form inactive wild-type/mutant dimers, thus blocking E3 activity. We performed in vitro ubiquitination assay using a fixed amount of PUB10 and varying amounts of the C249A mutant. The PUB10 E3 activity was progressively inhibited with an increasing ratio of PUB10(C249A)/PUB10 (Figure 2B). Near complete inhibition was seen when the mutant/wild type ratio was 10 to 1.
To identify other interacting Arabidopsis E2s, we performed yeast two-hybrid assays using PUB10 as a bait and 35 Arabidopsis UBCs as preys (Supplemental Figure 2) (Kraft et al., 2005). In addition to UBC8, PUB10 interacted with at least three other Arabidopsis UBCs (2, 31, and 36) in yeast cells (Supplemental Figure 2A). Each of these four UBCs was capable of interacting with PUB10 in vitro (Supplemental Figure 3) and of supporting autoubiquitination of PUB10 in vitro (Supplemental Figure 4), although with varying degrees of activity. The highest activity was obtained when UBC8 served as the ubiquitin conjugating enzyme. Therefore, UBC8 was used as a source of E2 in subsequent in vitro ubiquitination experiments.
The association of PUB10 with MYC2 suggested that the latter may be a substrate of the PUB10 E3 ligase. To examine this possibility, we used UBC8 and PUB10 as the E2 and E3 enzyme, respectively. PUB23, another U-box protein (Cho et al., 2008; Trujillo et al., 2008), was used as a negative control. Figure 2C shows that PUB10 efficiently polyubiquitinated MYC2 in vitro. The ubiquitination activity was greatly reduced in the PUB10(C249A) mutant, indicating the importance of this amino acid in maintaining the activity. No activity was detected with PUB23. We also tested MYC3 and MYC4 and found them to be substrates of PUB10 as well (Figure 2C).
PUB10 Associates with MYC2 in the Nucleus
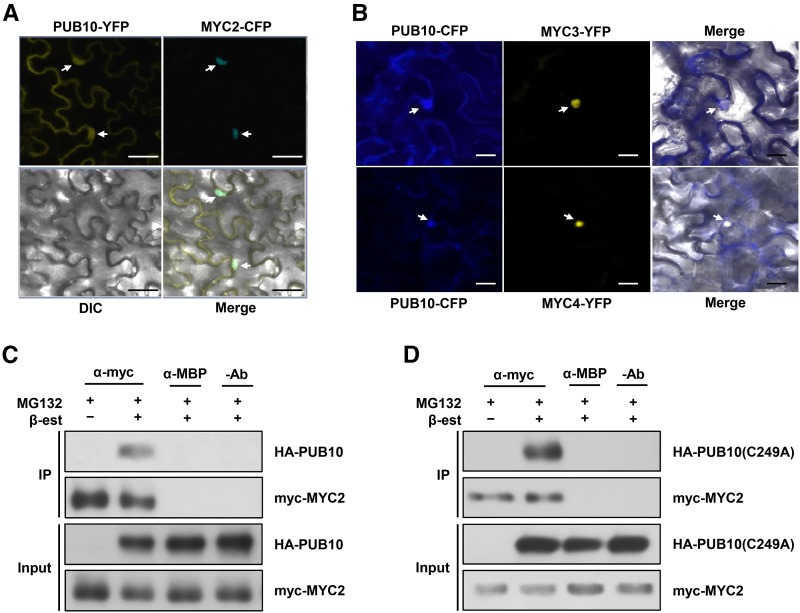
The PUB10/MYC2 interaction in vitro raised the question of whether they also interact in vivo. We examined the localization of PUB10-YFP and MYC2-CFP by transient expression in Nicotiana benthamiana leaf cells (Figure 3). PUB10-YFP was found largely localized to plasma membranes and nuclei, whereas MYC2-CFP was localized only to nuclei (Lorenzo et al., 2004) (Figure 3A). Similar nuclear localization was found with MYC3 and MYC4 (Figure 3B; Fernández-Calvo et al., 2011). The observation that PUB10 and MYC2 were both found in nuclei suggested possible interaction within this cellular compartment.
Figure 3.
PUB10 Colocalizes with MYC2, MYC3, and MYC4 and PUB10 Interacts with MYC2 in Vivo.
(A) Colocalization of PUB10-YFP and MYC2-CFP in the nucleus. Fluorescence fusion genes, 35:PUB10-YFP and 35S:MYC2-CFP, were transiently expressed in N. benthamiana leaves in the presence of 50 µM MG132 (n = 6, biological replicates).
(B) Colocalization of PUB10-CFP, MYC3-YFP, and MYC4-YFP in the nucleus. Fluorescence fusion genes, 35:PUB10-CFP, 35S:MYC3-YFP and 35S:MYC4-YFP, were transiently expressed in N. benthamiana leaves in the presence of 50 µM MG132.
For (A) and (B), confocal images were taken 2 d after infiltration. Colocalization is shown by merging YFP, CFP, and differential interference contrast (DIC) images (Merge). Arrows indicate nuclei. Bars = 20 µm.
(C) Coimmunoprecipitation of PUB10 with MYC2 in Arabidopsis. Two-week-old double transgenic Arabidopsis seedlings carrying XVE:HA-PUB10/35S:myc-MYC2 (line #8) were treated for 16 h with 50 μM MG132 in the absence or presence of 25 µM β-estradiol (β-est).
(D) Coimmunoprecipitation of PUB10(C249A) with MYC2 in Arabidopsis. Two-week-old double transgenic Arabidopsis seedlings carrying XVE:HA-PUB10(C249A)/35S:myc-MYC2 (line #5) were treated for 16 h with 50 µM MG132 in the absence or presence of 25 µM β-estradiol.
For (C) and (D), extracts were immunoprecipitated with anti-myc or anti-MBP antibodies. Input proteins and the immunoprecipitates were analyzed by immunoblots using anti-HA and anti-myc antibodies. Input refers to the starting protein amounts in extracts used for IP reactions.
To obtain evidence of PUB10/MYC2 interaction in vivo, we generated double-transgenic plants harboring XVE:HA-PUB10/35S:myc-MYC2 and XVE:HA-PUB10(C249A)/35S:myc-MYC2. Note that transgene expression in the XVE system is inducible and requires β-estradiol treatment (Zuo et al., 2000). Figure 3C shows that HA-PUB10, which was expressed only upon β-estradiol treatment, was pulled down by myc-MYC2. The interaction was clearly specific and dependent on myc-MYC2, as HA-PUB10 was not detected in the noninduced sample. Neither myc-MYC2 nor HA-PUB10 was detected in the absence of an antibody or when anti-MBP antibody was used as a negative control. Similar results were obtained with the HA-PUB10(C249A) mutant (Figure 3D).
Expression Profile of PUB10-GUS
We generated transgenic plants expressing PUB10pro and MYC2pro:GUS fusions to determine expression profile of its promoter (Supplemental Figure 5). Supplemental Figure 5A shows that in vegetative tissues PUB10 was strongly expressed in germinated seeds, primary and lateral roots, vascular tissues, mesophyll cells, and trichomes. Petals, stamen, and stigma also showed strong GUS expression and the same was true with embryos. Comparison of the expression profiles of PUB10 and MYC2 (Supplemental Figure 5B) showed considerable overlap in vascular bundles of roots and leaves. This coexpression result argued that the interaction of the two proteins in vivo has physiological relevance.
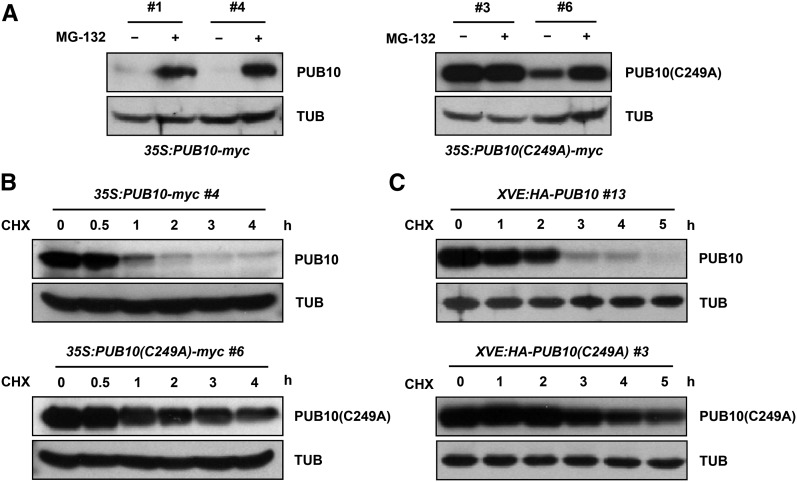
Self-Destruction of PUB10
Because PUB10 is able to autoubiquitinate, we examined whether this E3 ligase reduces its own half-life. To this end, we compared the stability of wild-type PUB10 with PUB10(C249A) mutant in transgenic plants expressing 35S:PUB10-myc or 35S:PUB10(C249A)-myc (Figure 4; Supplemental Figure 6). Immunoblot analysis showed that wild-type PUB10-myc protein was almost undetectable in the nontreated samples, but its level could be considerably elevated when protein degradation was blocked by MG132 (Figure 4A). By contrast, PUB10(C249A)-myc mutant protein was already expressed at high levels in nontreated sample. We conclude that the stability of PUB10 is regulated by 26S proteasomes and the ubiquitin E3 ligase activity of PUB10 is a major determinant of its own half-life.
Figure 4.
PUB10 Reduces Its Own Half-Life through Autoubiquitination.
(A) Steady state level of overexpressed PUB10 and PUB10(C249A). Ten-day-old seedlings of 35S:PUB10-myc (line #1 and #4) and 35S:PUB10(C249A)-myc (line #3 and #6) were incubated in liquid MS medium with or without 50 μM MG132 for 16 h. Protein levels were detected by anti-myc antibody.
(B) Degradation rate of overexpressed PUB10 and PUB10(C249A). Ten-day-old seedlings of 35S:PUB10-myc (line #4) and 35S:PUB10(C249A)-myc (line #6) were incubated in liquid MS medium with 50 μM MG132 for 16 h and washed five times before being transferred to liquid MS medium with 200 μM cycloheximide (CHX). Proteins were extracted at the indicated time points and detected by anti-myc antibody.
(C) Degradation rate of induced PUB10 and PUB10(C249A). Ten-day-old seedlings of XVE:HA-PUB10 (line #13) and XVE:HA-PUB10(C249A) (line #3) were incubated in liquid MS medium with 50 μM MG132 and 25 μM β-estradiol for 16 h and washed five times before being transferred to liquid MS medium with 200 μM cycloheximide. Proteins were extracted at the indicated time points and detected by anti-HA antibody.
For (A) to (C), α-tubulin levels as detected by anti-α-tubulin antibody were used as loading controls.
To further confirm the self-destruction of PUB10, we determined the time course of PUB10-myc and PUB10(C249A)-myc levels after de novo protein synthesis in transgenic seedlings was inhibited by cycloheximide. In the cycloheximide chase experiments, whereas wild-type PUB10-myc protein decayed with a half-life of ∼30 min, the half-life of the catalytically inactive PUB10(C249A)-myc mutant protein was prolonged to more than 3 to 4 h (Figure 4B). Similar results were obtained irrespective of whether the transgenes were expressed from a 35S (Figure 4B) or the XVE-inducible promoter (Figure 4C).
MYC2 Protein Levels in Plants of Different Genotypes
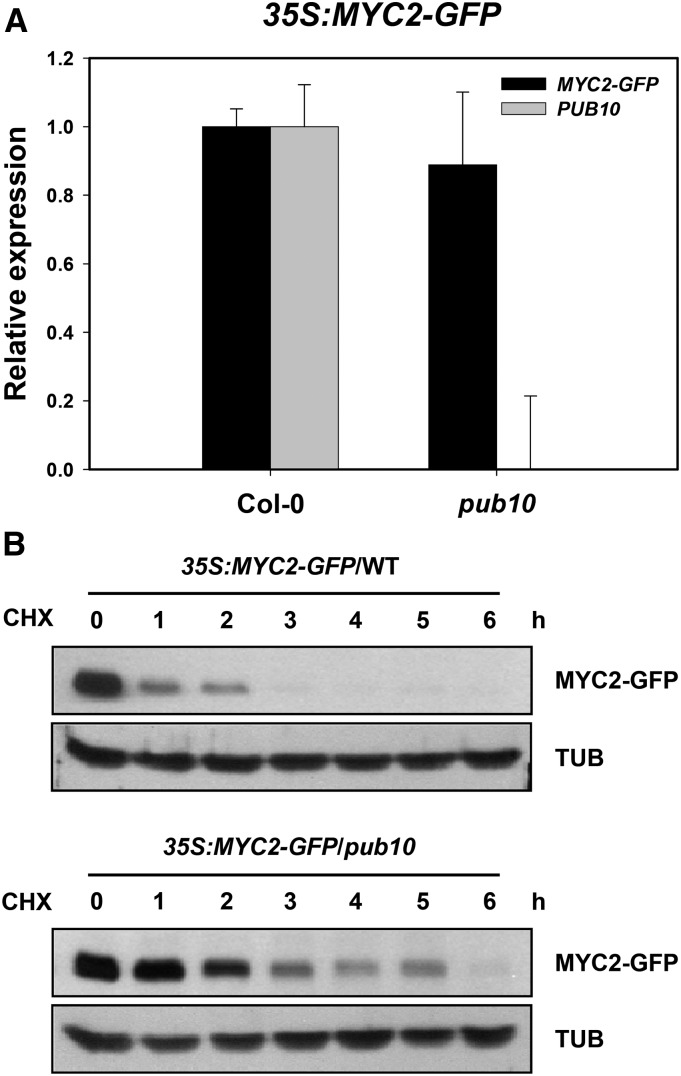
So far, we have shown that PUB10 and MYC2 proteins were expressed in the same cell types and tissues formed a complex in planta and MYC2 was ubiquitinated by PUB10 in vitro. Together, these results suggested that MYC2 may be targeted by PUB10 for ubiquitin-mediated degradation in vivo. If this hypothesis was correct, we would expect MYC2 to have a slower decay rate in the pub10 mutant compared with the wild type. We transformed wild-type and pub10 with 35S:MYC2-GFP (Figure 5; Supplemental Figure 6). Quantitative analysis showed that MYC2-GFP transcript was expressed to comparable levels in both genotypes (Figure 5A). We compared the half-life of MYC2-GFP in wild-type and pub10 plants in cycloheximide chase experiments. Figure 5B shows that in wild-type plants, the half-life of MYC2-GFP was <1 h, but this was prolonged to ∼2 h in the pub10 mutant, which is devoid of PUB10 activity.
Figure 5.
MYC2 Is Stabilized in pub10 Mutant.
(A) Real-time RT-PCR of MYC2-GFP and PUB10 transcript levels in 35S:MYC2-GFP/WT (line #2) and 35S:MYC2-GFP/pub10 (line #6) plants. Bars represent average values ± sd (n = 3 biological replicates).
(B) Degradation rate of overexpressed MYC2-GFP in the wild type and pub10 mutant. Ten-day-old seedlings of 35S:MYC2-GFP/WT (line #2) and 35S:MYC2-GFP/pub10 (line #6) were incubated in liquid MS medium with 50 µM MG132 for 16 h and washed five times before being transferred to liquid MS medium with 200 µM cycloheximide. Proteins were extracted at the indicated time points and detected by anti-GFP antibody. β-Tubulin levels as detected by anti-β-tubulin antibody were used as loading controls.
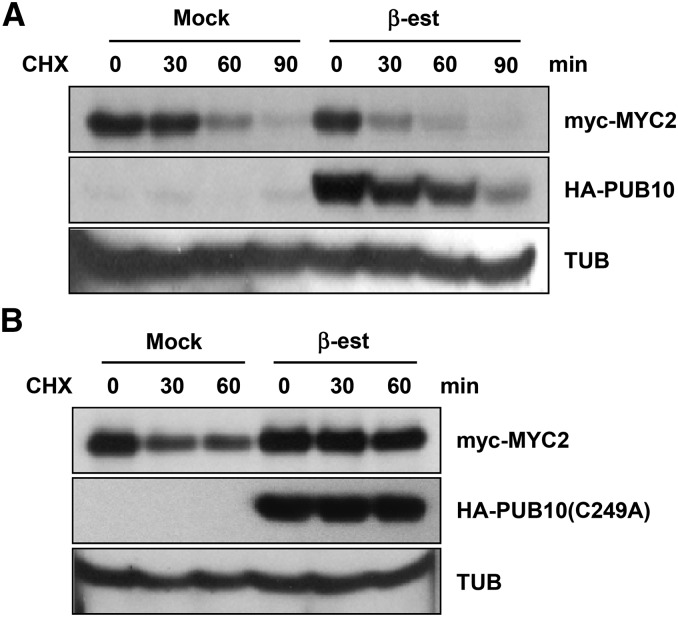
To further provide support for PUB10-mediated proteolysis of MYC2 in vivo, we investigated the half-life of MYC2 protein in transgenic plants in which PUB10 level or activity had been altered by transgene expression (Figure 6). Double transgenic plants expressing XVE:HA-PUB10/35S:myc-MYC2 were treated with MG132 to block 26S proteasome activity and accumulate myc-MYC2. One set of plants were treated with β-estradiol to induce HA-PUB10, whereas untreated plants were used as controls. After 16 h, treated and untreated seedlings were washed five times with liquid Murashige and Skoog (MS) medium and cycloheximide, a protein synthesis inhibitor, was added and the myc-MYC2 levels were monitored in time-course experiments. In this type of experiment using the β-estradiol inducer to activate PUB10 expression, the results obtained without the inducer served as an internal control. Figure 6A shows that induced overexpression of HA-PUB10 increased the decay rate of myc-MYC2 compared with the no-inducer control (mock treatment). Results from the same experiment also showed that HA-PUB10 was unstable, with a half-life of ∼30 to 60 min. In a separate but similar experiment, we overexpressed the dominant-negative HA-PUB10(C249A) mutant in transgenic plants to suppress the endogenous PUB10 activity. Without induction (mock), myc-MYC2 decayed with a half-life of <30 min (Figure 6B). Upon induced overexpression of the dominant-negative HA-PUB10(C249A) mutant, however, the decay rate of myc-MYC2 was clearly prolonged. In the same cycloheximide chase experiment, the half-life of the HA-PUB10(C249A) mutant was more than 60 min, in contrast to the 30-min half-life of the wild-type HA-PUB10. These results provide additional evidence that MYC2 is destabilized by PUB10 and that PUB10 itself is in part responsible for its own destruction in vivo.
Figure 6.
Wild-Type PUB10 Destabilizes MYC2, Whereas Dominant-Negative PUB10(C249A) Mutant Renders MYC2 More Stable in Vivo.
Ten-day-old double transgenic Arabidopsis seedlings carrying XVE:HA-PUB10/35S:myc-MYC2 (line #8) (A) or XVE:HA-PUB10(C249A)/35S:myc-MYC2 (line #5) (B) were treated with 50 μM MG132 alone or 25 µM β-estradiol (β-est) plus 50 µM MG132 for 16 h, washed five times, and then transferred to MS medium with 200 µM cycloheximide (CHX). Proteins were extracted at the indicated time points and detected by anti-HA and anti-myc antibodies. For (A) and (B), α-tubulin levels as detected by anti-α-tubulin antibody were used as loading controls.
Phenotypes of Mutants and Transgenic Plants
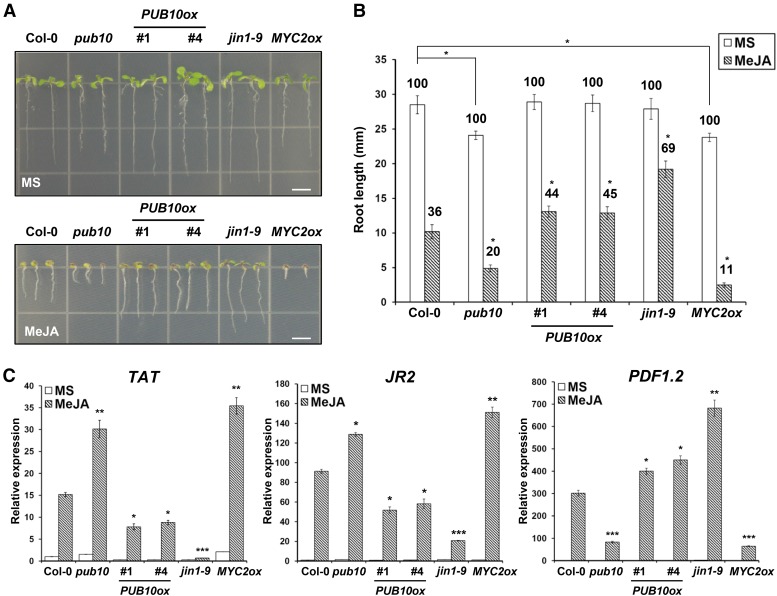
The reciprocal relationship between PUB10 activity and MYC2 protein levels suggested that the two proteins may play antagonistic roles in signaling pathways known to be regulated by MYC2. Although MYC2 was first characterized as a positive regulator of ABA signaling pathway (Abe et al., 1997, 2003), this transcription factor was subsequently found to be a key regulator of the JA response pathway (Berger et al., 1996; Boter et al., 2004; Lorenzo et al., 2004). To investigate JA phenotypes under PUB10 deficiency, we obtained T-DNA insertion mutant allele (pub10; SALK_017111) from the SALK collection and also produced transgenic plants expressing 35S:PUB10-myc. The jin1-9 mutant (MYC2 null) (Anderson et al., 2004) and a transgenic line overexpressing 35S:MYC2-GFP (MYC2ox) were analyzed in parallel as controls (Figure 7). Previous work has shown that primary root growth of Arabidopsis seedlings is inhibited by JA and that this inhibition is mediated by MYC2 (Dombrecht et al., 2007; Chen et al., 2011). Therefore, we compared the JA sensitivity of primary roots of wild-type, mutant, and transgenic lines. When grown on control medium, the MYC2ox line and pub10 (expected to have a higher MYC2 level) had shorter roots, compared with the wild type (Figure 7A). On MeJA medium, primary root growth of all genotypes was clearly inhibited, but the degree of inhibition varied. Compared with the wild type, the jin1-9 mutant and the two PUB10ox lines were clearly MeJA hyposensitive, whereas the reverse was true for the MYC2ox line and pub10 mutant (Figures 7A and 7B). These results confirmed previous findings that MYC2 is a positive regulator of JA signaling, which inhibits primary root growth of seedlings (Chen et al., 2011) and that PUB10 negatively regulates MYC2-mediated JA responses in vivo.
Figure 7.
The pub10 Mutant and PUB10 Overexpression Line Phenocopy MYC2 Overexpression Line and jin1 Mutant, Respectively, in MeJA Response.
(A) Primary root growth of the wild type (Col-0), pub10, two independent lines of 35S:PUB10-myc (PUB10ox, line #1 and #4), jin1-9, and 35S:MYC2-GFP (MYC2ox, line #2) on MS medium with or without 10 μM MeJA for 10 d. Bars = 5 mm.
(B) Quantitative effects of MeJA on root growth of various genotypes in (A). Average values of primary root length are given. The root length of each genotype in MS medium alone is designated as 100%. The relative root length with MeJA is shown as the percentage of that without MeJA. Bars represent average values ± sd of more than 50 seedlings of each genotype. Asterisks indicate significant change of root length in each genotype compared with that in Col-0. *P < 0.05; two-tailed t test.
(C) Real-time PCR analysis of JA-responsive gene expression of TAT, JR2, and PDF1.2 in the absence or presence of 50 μM MeJA for 12 h. Transcript levels were normalized to ACT2 expression. Bars represent average ± sd (n = 3 biological replicates). Asterisks indicate statistically significant differences compared with the wild type (Col-0). *P < 0.05, **P < 0.01, and ***P < 0.001; two-tailed t test.
In the same experiment, we also examined expression of three JA-responsive genes (TAT, JR2, and PDF1.2) in control and MeJA-treated seedlings. In wild-type seedlings, all three genes were upregulated by JA between 15- and 300-fold (Figure 7C). Of these, TAT and JR2 were induced by MeJA and positively regulated by MYC2, whereas PDF1.2 was also induced by MeJA but negatively regulated by MYC2. It has been shown that MYC2 differentially regulates two branches in JA signaling. Briefly, MYC2 activates JA/wound-responsive gene expression but represses JA/ET-responsive genes that are related to pathogen defense response (compared with the wild type, MYC2ox, and jin1-9 mutant) (Boter et al., 2004; Lorenzo et al., 2004). Further expression analysis of other genotypes showed that responses of TAT and JR2 displayed MeJA hypersensitivity in pub10 and MYC2ox but hyposensitivity in jin1-9 and PUB10ox lines. However, we found the opposite behavior in the case of PDF1.2 expression.
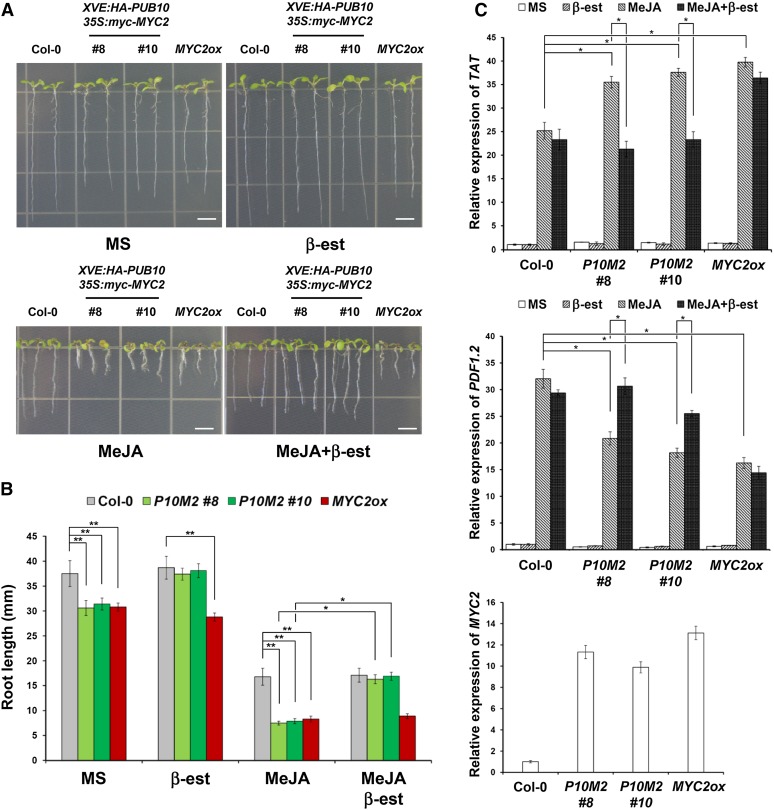
Our above results showed that transgenic plants overexpressing MYC2 (MYC2ox) were hypersensitive to MeJA with respect to primary root growth inhibition and gene expression. Inducible overexpression of PUB10 accelerated the MYC2 decay rate, whereas inducible overexpression of the dominant-negative PUB10(C249A) mutant had the opposite effect (Figure 6). If MYC2 levels are indeed regulated by PUB10 in vivo, inducible overexpression of PUB10 in transgenic plants overexpressing 35S-MYC2 should reverse the root growth inhibition phenotype, whereas inducible overexpression of the PUB10(C249A) mutant should enhance the inhibition. To see if these predicted responses could be observed at the phenotypic level, we examined root growth inhibition in double transgenic plants XVE:HA-PUB10/35S:myc-MYC2 and XVE:HA-PUB10(C249A)/35S:myc-MYC2 (Figure 8). In these plants, only the 35S:myc-MYC2 gene was expressed in the absence of inducer and this served as a convenient internal control. Figure 8C compares the MYC2 expression levels in wild-type, MYC2ox (35S:MYC2-GFP), and the two XVE:HA-PUB10/35S:myc-MYC2 double transgenic lines (#8 and #10). MYC2 transcript level in double transgenic plants was more than 10 times that of wild-type levels. On MS medium without any inducer, the MYC2ox line had slightly shorter root compared with the wild type and the same was true for the two inducible XVE:HA-PUB10/35S:myc-MYC2 double transgenic lines (Figure 8A, MS). In MeJA-supplemented medium, the MYC2ox line and the double transgenic lines displayed much shorter roots compared with the wild type (Figure 8A, MeJA), consistent with the elevated MYC2 transcript level. However, the short-root phenotype seen in the double transgenic lines on MeJA medium reverted to wild-type root length upon addition of β-estradiol, which activated the HA-PUB10 gene (Figure 8A, MeJA+β-est). Addition of the inducer without JA had a marginally significant effect on root length of the double transgenic lines. This reversal of the MYC2 overexpression phenotype by inducible expression of PUB10, which degrades MYC2, was most obvious on medium with MeJA plus inducer (Figures 8A and 8B). Analysis of PDF1.2 and TAT expression confirmed that the phenotypic reversal was paralleled at the gene expression level (Figure 8C).
Figure 8.
Wild-Type PUB10 Induction Reverses the Root and Gene Expression Phenotype of MYC2 Overexpression Line in the Presence of MeJA.
(A) Primary root growth of the wild type (Col-0), two independent lines of XVE:HA-PUB10/35S:myc-MYC2 (P10M2, line #8 and #10), and 35S:MYC2-GFP (MYC2ox line #2) on control medium (MS), 1 μM MeJA alone (MeJA), 10 μM β-estradiol alone (β-est), and 1 μM MeJA plus 10 μM β-estradiol (MeJA+β-est) for 15 d. Bars = 5 mm.
(B) Quantitative effect of MeJA on root growth of various genotypes in (A). Bars represent average ± sd of more than 50 seedlings of each genotype. *P < 0.05 and **P < 0.01; two-tailed t test.
(C) Real-time PCR analysis of JA-responsive gene expression of TAT and PDF1.2 in (A) and MYC2 expression in all genotypes grown on MS. Transcript levels were normalized to ACT2 expression. Bars represent average ± sd (n = 3 biological replicates). Asterisks indicate statistically significant differences compared with the wild type (Col-0). *P < 0.05 and **P < 0.01; two-tailed t test.
For (B) and (C), P10M2 is XVE:HA-PUB10/35S:myc-MYC2 and MYC2ox is 35S:MYC2-GFP.
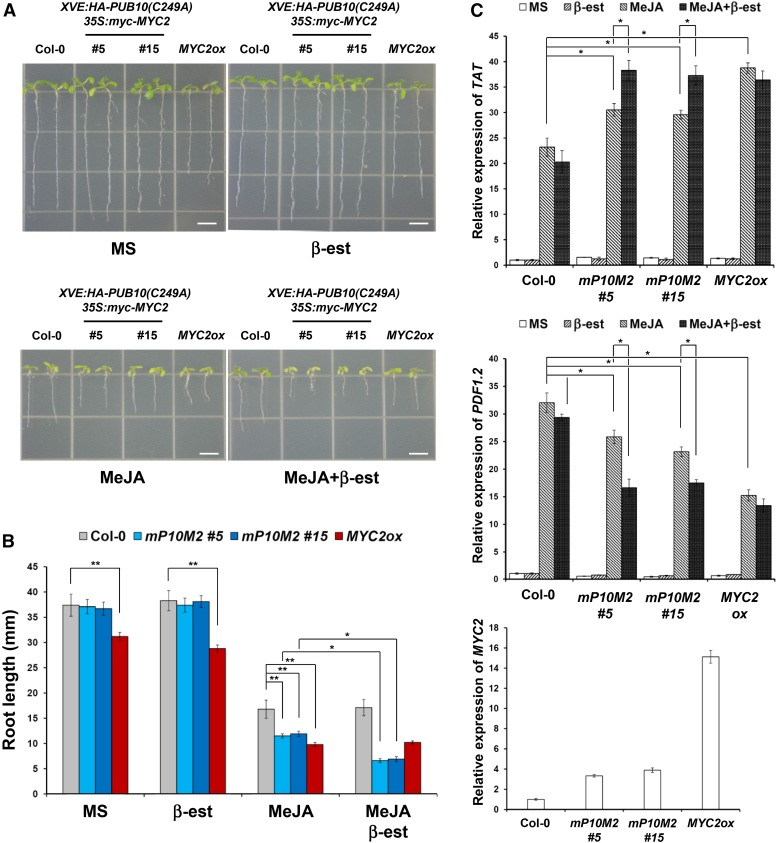
PUB10(C249A) functions as a dominant-negative mutant by delaying the degradation rate of MYC2 in vivo (Figure 7). Therefore, we analyzed root growth phenotype of XVE:HA-PUB10(C249A)/35S:myc-MYC2 double transgenic lines (#5 and #15) with or without MeJA and with or without inducer (Figure 9). Quantitative analysis showed that whereas MYC2-GFP transcript level in the 35S:MYC2-GFP line was more than 15-fold that of control, those in the double transgenic lines (#5 and #15) were 3- to 4-fold (Figure 9C). On MS medium, the MYC2ox line had a shorter primary root compared with the wild type, but root length of the two double transgenic lines was similar to that of the wild type. On MeJA-supplemented medium in the absence of inducer, root growth of all genotypes was inhibited but the MYC2ox line was hypersensitive to MeJA. The two double transgenic lines were less hypersensitive compared with the MYC2ox line, consistent with lower MYC2 transcript levels in these lines (Figures 9A and 9B). However, upon addition of inducer (Figure 9A, MeJA+β-est), the MeJA sensitivity of the double transgenic lines was further enhanced by the resultant induced expression of the HA-PUB10(C249A) dominant-negative mutant. The inducer had little or no significant effect on root length of the wild type and the 35S:MYC-GFP line. This enhanced JA hypersensitivity of induced double transgenic lines expressing the dominant-negative PUB10(C249A) mutant was also observed at the TAT and PDF1.2 gene expression level (Figure 9C).
Figure 9.
PUB10(C249A) Induction Enhances the Root and Gene Expression Phenotype of MYC2 Overexpression Line in MeJA.
(A) Primary root growth of the wild type (Col-0), two independent lines of XVE:HA-PUB10(C249A)/35S:myc-MYC2 (mP10M2, line #5 and #15), and 35S:MYC2-GFP (MYC2ox line #2) on control medium (MS), 1 μM MeJA alone (MeJA), 10 μM β-estradiol alone (β-est), and 1 μM MeJA plus 10 μM β-estradiol (MeJA+β-est) for 15 d. Bars = 5 mm.
(B) Quantitative effects of MeJA on root length of various genotypes in (A). Bars represent average ± sd of more than 50 seedlings of each genotype. *P < 0.05 and **P < 0.01; two-tailed t test.
(C) Real-time PCR analysis of JA-responsive gene expression of TAT and PDF1.2 in (A) and MYC2 expression in all genotypes grown on MS. Transcript levels were normalized to ACT2 expression. Bars represent average ± sd (n = 3 biological replicates). Asterisks indicate statistically significant differences compared with the wild type (Col-0). *P < 0.05 and **P < 0.01; two-tailed t test.
For (B) and (C), mP10M2 is XVE:HA-PUB10(C249A)/35S:myc-MYC2 and MYC2ox is 35S:MYC2-GFP.
The Role of PUB11 in JA Response
Among members of the PUB protein family, PUB11 is most closely related to PUB10, with 77% amino acid identity and 85% similarity. This close homology raised the issue of possible functional redundancy between PUB10 and PUB11 in JA responses. Similar to PUB10, we found that PUB11 was also able to bind to MYC2 in yeast cells (Supplemental Figure 1). Pull-down experiments verified that like PUB10, MYC2 was bound via the conserved ARM repeats of PUB11 (Supplemental Figure 7). In vitro experiments showed that PUB11 had the capacity to act as an E3 ligase to polyubiquitinate itself as well as MYC2, MYC3, and MYC4 and its catalytic activity required the conserved cysteine at Cys-247 (Supplemental Figure 8). Yeast two-hybrid assays, in vitro binding, and ubiquitination experiments showed that UBC2, 8, 10, and 36 could bind to PUB11 and mediate its E3 activity (Supplemental Figures 2 to 4). Analysis of transgenic plants carrying a PUB11 promoter-GUS fusion gene revealed that PUB11 displayed very similar expression profile as PUB10 (Supplemental Figure 9).
To study the function of PUB11, we obtained a loss-of-function mutant of PUB11 (SALK_029828) that did not express detectable PUB11 transcript (Supplemental Figure 10). We also generated pub10 pub11 double mutant. In contrast to pub10, pub 11 behaved similarly as the wild type with respect to root growth inhibition and gene expression in the presence of MeJA (Supplemental Figures 11 and 12). Moreover, in this assay, the phenotype of pub10 was not enhanced by PUB11 deficiency as a pub10 pub11 double mutant was similar to pub10 single mutant alone. Taken together, these results indicate that PUB11 does not play a major role in this MeJA-mediated response.
DISCUSSION
First identified as a positive regulator of ABA response (Abe et al., 1997, 2003), the bHLH transcription factor MYC2 has now been shown to function at a nodal point coordinating signal flux through JA response pathways (for a review, see Kazan and Manners, 2013). Recent reports showed that during JA signaling MYC2 becomes phosphorylated by an as yet unidentified kinase (Zhai et al., 2013). Like many phosphorylated transcription factors, the modified MYC2 is unstable and rapidly degraded. Here, we show that the plant U-box protein PUB10 serves as an E3 ligase for MYC2 in JA responses. Several lines of evidence support our claim: (1) PUB10 interacts with MYC2 in yeast and in vitro. Deletion analysis showed that the ARM repeats region of PUB10 binds to the C-terminal region of MYC2. (2) The PUB10 and MYC2 association can be recapitulated in vivo. (3) MYC2 and PUB10 are expressed in overlapping tissues and cell types. (4) In vitro, MYC2 can be polyubiquitinated by wild-type PUB10 but not by the dominant-negative mutant PUB10(C249A). However, both wild-type and mutant PUB10 are able to bind MYC2. (5) There is an inverse relationship between the MYC2 degradation rate and changes in PUB10 levels/activity.
PUB10 Level or Activity Determines MYC2 Stability in Vivo
If PUB10 is a cognate E3 ubiquitin ligase for MYC2 and targets the latter for destruction in vivo, we would expect a reciprocal relationship between the levels of the two proteins. MYC2 should be more stable and have a longer half-life in pub10 mutant compared with the wild type. Indeed, this was seen when the half-life of MYC2 was compared in cycloheximide chase experiments between wild-type and pub10 seedlings. The E3 ligase/substrate relationship between PUB10/MYC2 was further established in double transgenic plants expressing 35S:myc-MYC2 and inducible PUB10 or dominant-negative PUB10(C249A) mutant. In this system, the noninduced state serves as a convenient and powerful internal control, since plants of identical genotype were assayed. We found that induction of the dominant-negative PUB10(C249A) mutant, which blocks PUB10 activity, increases the half-life of myc-MYC2. This result is similar to that found in pub10 mutant. By contrast, induced overexpression of the wild-type PUB10 accelerates the degradation rate of MYC2 compared with the noninduced state.
In addition to MYC2, we also found that PUB10 can associate with two MYC2 homologs, MYC3 and MYC4. However, since MYC3/4 do not play a significant role in JA mediation inhibition of root growth (Fernández-Calvo et al., 2011), these proteins were not investigated in the same detail as MYC2. We also found that PUB11, the closest relative of PUB10 within the PUB protein family, has E3 ligase activity and can also bind to MYC2, 3, and 4. Analysis of the pub11 mutant and pub10 pub11 double mutant rules out a significant role of this E3 ligase in this JA response.
It should be emphasized that there is not a simple one-to-one relationship between a transcription factor and an E3 ligase. Although the half-life of MYC2 is prolonged in pub10 mutant, this factor continues to be unstable, suggesting that it is likely targeted by other E3 ligases. In addition, there have been several reports that a single transcription factor may be targeted by several E3 ligases (e.g., ABI5; Lee et al., 2010; Liu and Stone, 2010; Seo et al., 2014), whereas an E3 ligase may mediate degradation of several transcription factors (e.g., COP1; reviewed in Lau and Deng, 2012). Based on this scenario, we would expect MYC2 to be targeted by several E3 ligases and PUB10 to ubiquitinate other substrates.
JA Effects on Root Growth and Gene Expression
A major effect of MeJA on young seedlings grown on MS medium is inhibition of root growth, and this phenotype has been used as a readout for JA sensitivity (Berger et al., 1996). We used this assay to compare MeJA sensitivity of seedlings of various plant genotypes expressing different levels of PUB10 and MYC2. We found that pub10 mutant phenocopies MYC2ox in being hypersensitive to MeJA compared with the wild type. The degree of hypersensitivity of pub10 is not as severe as that of MYC2ox, and this is to be expected as the MYC2 transcript level in the MYC2ox transgenic line is elevated by ∼15-fold or more. By contrast, overexpression of PUB10, which is expected to suppress MYC2 levels, phenocopies the jin1-9 mutant, which is a MYC2-null mutant, and both display JA hyposensitivity. Here again, the phenotype of PUB10ox is not as strong as that of the jin1-9 mutant, and we expect these transgenic lines continue to have residual MYC2.
We also manipulated MYC2 levels in double transgenic plants by inducible overexpression of wild-type PUB10 or PUB10(C249A) dominant-negative mutant. In this series of experiments, the root growth phenotypes of noninduced plants served as internal controls. The MeJA-hypersensitive phenotype of these double transgenic plants is conferred by MYC2 overexpression. This hypersensitivity can be reversed by inducible overexpression of wild-type PUB10, which degrades MYC2, but enhanced by inducible overexpression of PUB10(C249A) dominant-negative mutant, which blocks MYC2 destruction.
We determined transcript levels of three JA-responsive genes (TAT, JR2, and PDF1.2) in plants treated with MeJA, and the results parallel those found with root growth inhibition experiments. MYC2ox and pub10 are hypersensitive, whereas PUP10ox and the jin1-9 mutant are hyposensitive. Similarly, the hypersensitive gene expression phenotype of MYC2ox in noninduced double transgenic plants can be reversed by inducible overexpression of wild-type PUB10 but enhanced by inducible overexpression of PUB10(C249A).
Dimerization Activates PUB10 E3 Ligase Activity
Using in vitro ubiquitination experiments, we showed here that PUB10 is an E3 ligase whose activity requires self-association as the E3 activity is attenuated by excess of the dominant-negative mutant PUB10(C249A), which presumably forms inactive heterodimers. The activation of E3 activity by ligase dimerization has been previously reported for RING motif E3 ligases (Xie et al., 2002; Seo et al., 2003; Zhang et al., 2005). We localized the dimerization domain to the PUB10 C-terminal region (320 to 628 amino acids; Figure 1), which contains six ARM repeats. This ARM repeat region also binds to MYC2. Among the 64 U-box proteins predicted to be encoded by the Arabidopsis genome (Azevedo et al., 2001; Wiborg et al., 2008), ARM repeats are found in 41 of them. However, there are variations in the ARM sequences and differences in copy number and the length of spacer sequences among family members. It is possible that the strength of substrate binding is determined by the ARM repeat sequence as well as by the copy number. We note that the ARM repeat sequences in PUB23 are different from those in PUB10. Moreover, the former contains five ARM repeats compared with six in the latter. These differences may account for the observation that PUB23 is unable to ubiquitinate MYC2.
PUB10 E3 activity depends on a cysteine residue (at position 249) conserved among several PUBs (Figure 1A; Lu et al., 2011) and a PUB10 mutant with the C249A mutation is catalytically inactive. Nevertheless, the inactive PUB10(C249A) mutant is still able to bind to MYC2 and to heterodimerize with wild-type PUB10, thereby interfering with the E3 activity of the latter. These abilities enable the PUB10(C249A) mutant to act as a dominant-negative mutant when it is expressed at a higher level (more than 5-fold) than the wild-type PUB10.
The ability to interfere with the in vivo E3 activity of PUB10 allows us to address the issue of how PUB10 itself is regulated. Our data show that autoubiquitination and self-destruction are major mechanisms in determining PUB10 in vivo levels. Among the 35 UBC2s tested, only four are able to associate with PUB10 and these four proteins are also able to function as E2 to support the E3 activity of PUB10 (Supplemental Figures 2 to 4). Although we have not investigated the expression profiles of these UBC2s, we hypothesize that they may function in a cell-type or tissue-specific manner.
In conclusion, we provided several lines of evidence to support the view that during JA signaling, MYC2 levels is regulated by PUB10 via ubiquitin-mediated proteolysis. PUB10 also regulates its own protein levels by self-ubiquitination. How changes in MYC2 levels are dynamically coordinated by changes in PUB10 levels are interesting issues to be addressed in future work.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia (Col-0), jin1-9 (SALK_017005) (Anderson et al., 2004), pub10 (SALK_017111), and pub11 (SALK_029828) mutant plants were used in this study. T-DNA insertion lines were obtained from the SALK collection. Homozygous plants for the T-DNA insertion were selected by genotyping progeny plants using PCR following standard procedures (Alonso et al., 2003). The absence of MYC2, PUB10, and PUB11 expression in the homozygous plants were further confirmed by RT-PCR. All genotypes were grown on 0.5% agar medium containing 0.5× MS salts (MP Biomedicals), 1% sucrose (Fisher), and 0.5 g/L MES hydrate (Sigma-Aldrich) in a growth room at 22°C under 16 h light/8 h dark with white fluorescent light (100 μmol·m−2·s−1).
Yeast Two-Hybrid Assays
The Matchmaker GAL4-bases two-hybrid system (Clontech) was used to perform yeast two-hybrid assays. Thirty-five entry clones of Arabidopsis UBCs were obtained from the ABRC (Kraft et al., 2005). Entry clones containing full-length MYC2 and 35 Arabidopsis UBCs were recombined into pGAD424-GW vector (Clontech) to generate activation domain (AD) constructs using Gateway LR Clonase II (Invitrogen). The entry clones containing full-length PUB10 and PUB11 were recombined into pGBT9-GW vector (Clontech) to generate binding domain (BD) constructs. All of the above constructs and empty vector controls were transformed into yeast stain AH109 by the modified lithium acetate method. Yeast transformants were screened on the selective medium SD/-Leu/-Trp/-His (BD) with 20 mM 3-amino-1,2,4-triazole to test for protein interactions.
Preparations of Recombinant Proteins
The cDNAs encoding full-length MYC2, MYC3, MYC4, PUB10, PUB11, and PUB23 were amplified by PCR and cloned into pENTR/D-TOPO vector (Invitrogen). Mutations (C249A and C247A) in the U-box motif of PUB10 and PUB11 were generated with QuickChange Lightning site-directed mutagenesis kit (Agilent Technologies) to disrupt E3 ubiquitin ligase activity (Xie et al., 2002; Seo et al., 2003; Zhang et al., 2005). All entry clones were recombined into pGEX-DC and pMAL-DC (Zhang et al., 2005). Deletion derivatives of MYC2, PUB10, and PUB11 were amplified from the corresponding entry clones by PCR, cloned into pENTR/D-TOPO, and then recombined into pGEX-DC and/or pMAL-DC to generate cDNAs for MBP-MYC2-N (1 to 300 amino acids), MBP-MYC2-C (301 to 623 amino acids), GST-/MBP-PUB10-UND (1 to 240 amino acids), GST-/MBP-PUB10-U-box (240 to 320 amino acids), GST-/MBP-PUB10-ARM (320 to 628 amino acids), and GST-PUB11-ARM (320 to 612 amino acids). cDNAs encoding full-length Arabidopsis UBC2, UBC8, UBC10, UBC16, UBC31, and UBC36 were amplified from the corresponding entry clones by PCR and ligated into pET-28a (Novagen) to generate His-tagged UBC constructs. In addition, the entry clones containing full-length UBC2, UBC8, UBC31, and UBC36 were recombined into pGEX-DC vector to obtain GST-UBC constructs. All constructs were transformed into Escherichia coli BL21 (DE3) pLysS strain and recombinant protein expression was induced by 0.1 mM isopropyl-β-d-thiogalactoside at 22°C overnight. Recombinant proteins were purified following manufacturer’s instructions.
In Vitro Pull-Down Assays
To determine the PUB10 domain responsible for interaction with MYC2, MYC3, and MYC4, 2 μg GST, GST-PUB10, GST-PUB10(C249A), GST-PUB10-UND, GST-PUB10-U-box, or GST-PUB10-ARM and 2 μg MBP, MBP-MYC2, MBP-MYC3, or MBP-MYC4 were mixed into 1 mL of pull-down buffer (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 0.5% Triton X-100, 0.5 mM β-mercaptoethanol, and proteinase inhibiter cocktail) and incubated at 25°C for 2 h. In the case of PUB11 and MYC2, MYC3, and MYC4, 2 μg GST, GST-PUB11, GST-PUB11(C247A), or GST-PUB11-ARM and 2 μg MBP, MBP-MYC2, MBP-MYC3, or MBP-MYC4 were mixed into 1 mL of pull-down buffer and incubated at 25°C for 2 h. To investigate which MYC2 domain was responsible for the interaction with PUB10, 2 μg GST, GST-PUB10, GST-PUB10(C249A), or GST-PUB10-ARM and 2 μg MBP, MBP-MYC2-N, or MBP-MYC2-C were mixed into 1 mL of pull-down buffer and incubated at 25°C for 2 h. To examine homodimerization of PUB10 and PUB10(C249A) mutant protein, 2 μg GST, GST-PUB10, GST-PUB10(C249A), or GST-PUB10-ARM and 2 μg MBP, MBP-PUB10, MBP-PUB10(C249A), MBP-PUB10-UND, MBP-PUB10-U-box, or MBP-PUB10-ARM were mixed into 1 mL of pull-down buffer and incubated at 25°C for 2 h. In addition, to confirm the interaction between PUB10 and PUB11 with UBC2, UBC8, UBC31, and UBC36, 2 μg GST, GST-UBC2, GST-UBC8, GST-UBC31, or GST-UBC36 and 2 μg MBP-PUB10 or MBP-PUB11 were mixed into 1 mL of pull-down buffer and incubated at 25°C for 2 h. After incubation, reaction mixtures were pulled down with glutathione-Sepharose 4 fast flow (GE Healthcare) for 2 h and then washed five times with the same pull-down buffer. After washing, pulled-down proteins were separated on 8% SDS-polyacrylamide gels and detected by immunoblotting using anti-MBP antibody (Santa Cruz).
In Vitro Ubiquitination Assays
In vitro ubiquitination assays were performed as previously described (Seo et al., 2003; Jang et al., 2010). The reaction mix (30 μL) contained 200 ng Arabidopsis UBA1, 200 ng Arabidopsis UBC2, UBC8, UBC10, UBC16, UBC31, or UBC36, 2 μg ubiquitin (Sigma-Aldrich), 500 ng MBP-PUB10, MBP-PUB10(C249), MBP-PUB11, MBP-PUB11(C247A), or MBP-PUB23, and with or without 500 ng GST-MYC2, GST-MYC3, or GST-MYC4. Reactions were incubated at 30°C for 2 h. The reaction mixtures were separated on 8% SDS-polyacrylamide gels. Polyubiquitinated MBP-PUB10, MBP-PUB11, GST-MYC2, GST-MYC3, and GST-MYC4 proteins were analyzed by immunoblots using anti-myc, anti-MBP, or anti-GST antibodies, respectively.
GUS Staining
For promoter-GUS fusion, the promoter fragments of PUB10 (2.2 kb), PUB11 (1.6 kb), and MYC2 (1.6 kb) were amplified by PCR, cloned into pENTR 3C vector (Invitrogen), and then recombined into pKGWFS7 vector (Karimi et al., 2002) to obtain the PUB10pro-, PUB11pro-, and MYC2pro-GFP-GUS fusion constructs, respectively. Arabidopsis (Col-0) plants were transformed by the floral dip method (Clough and Bent, 1998; Zhang et al., 2006). GUS staining was performed as previously described (Senecoff et al., 1996).
Subcellular Localization of PUB10, MYC2, MYC3, and MYC4
Full-length entry clones encoding PUB10, MYC2, MYC3, and MYC4 were recombined into pBCo-DC-YFP and/or pBCo-DC-CFP (Wu et al., 2010). All constructs were transformed into Agrobacterium tumefaciens strain GV3101 using freeze and thaw method. Cultured cells were harvested and resuspended in 10 mM MgCl2 plus 150 μM acetosyringone (Aldrich) and then kept at 25°C for at least 3 h without shaking. Agrobacterium suspensions containing 50 μM MG132 were infiltrated into leaves of Nicotiana benthamiana with a needleless syringe. Leaf cells were analyzed using LSM 780 confocal laser scanning microscope (Zeiss) 2 d after infiltration.
Generation of Overexpression Plants
The entry clone of MYC2 was recombined into pEarleyGate103 (Earley et al., 2006) and pBA-DC (Zhang et al., 2005). The entry clones of PUB10 and PUB10(C249A) were recombined into pBA-DC and pER8-DC (Zhang et al., 2005). All constructs were verified by sequencing and transformed into Agrobacterium strain GV3101 using freeze and thaw method and then infiltrated into Col-0 or the pub10 mutant using the floral dip transformation method (Clough and Bent, 1998; Zhang et al., 2006).
In Vivo Coimmunoprecipitation
Two-week-old seedlings of XVE:HA-PUB10/35S:myc-MYC2 or XVE:HA-PUB10(C249A)/35S:myc-MYC2 treated with 50 μM MG132 in the presence or absence of 25 μM β-estradiol (Sigma-Aldrich) for 16 h were ground in liquid nitrogen and homogenized in IP buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 0.1% Nonidet P-40, 1 mM PMSF, 10% glycerol, 1 mM DTT, 50 μM MG132, and protease inhibitor cocktail). After centrifugation, the supernatants were used for immunoprecipitation. Input controls were taken for immunoblot analysis. An aliquot of the supernatant containing 1 mg total protein was precleared by adding protein A-agarose (Millipore), and the mix was incubated for 1 h before being centrifuged at 15,000g for 1 min. The supernatant was incubated with anti-myc polyclonal antibody (Santa Cruz) or anti-MBP polyclonal antibody (Santa Cruz) at 4°C for 2 h. Antigen-antibody complexes were pulled down by protein A agarose at 4°C for 2 h and washed five times with IP buffer (without Nonidet P-40). Immunoprecipitated proteins were eluted by boiling at 95°C for 5 min in 2× NuPAGE LDS sample buffer (Life Technologies). Eluted proteins were separated on 8% SDS-polyacrylamide gels and then transferred to a polyvinylidene fluoride (PVDF) membranes (Millipore). Immunoblot analysis was performed with anti-HA and anti-myc monoclonal antibodies (Santa Cruz) and anti-mouse horseradish peroxidase-linked sheep antibody (GE Healthcare). Immunoblot signals were detected by Amersham ECL prime (GE Healthcare).
MG132 and Cycloheximide Treatments
Ten-day-old transgenic Arabidopsis seedlings of 35S:PUB10-myc, 35S:PUB10(C249A)-myc, 35S:MYC2-GFP, and 35S:MYC2-GFP/pub10 were transferred to liquid MS medium with or without 50 μM MG132 (EMD Millipore) for 16 h. Treated seedlings were collected for immunoblot analysis. To examine the half-life of PUB10-myc, PUB10(C249A)-myc, and MYC2-GFP, transgenic seedlings treated with MG132 as above were washed five times before being transferred to liquid MS medium containing 200 μM cycloheximide (Sigma-Aldrich) to block de novo protein synthesis. Treated seedlings were collected at the indicated time points for immunoblot analysis.
Effect of PUB10 and PUB10(C249A) Induction on MYC2 Protein Stability
Two-week-old seedlings of XVE:HA-PUB10/35S:myc-MYC2 or XVE:HA-PUB10(C249A)/35S:myc-MYC2 treated with 50 μM MG132 in the presence or absence of 25 μM β-estradiol (Sigma-Aldrich) for 16 h were washed five times before being transferred to liquid MS medium containing 200 μM cycloheximide to block de novo protein synthesis. Seedlings were collected at the indicated time points and analyzed by immunoblotting using anti-HA and anti-myc antibodies (Santa Cruz). The α-tubulin levels as detected by anti-α-tubulin antibody (Santa Cruz) were used as loading controls.
Root Growth Inhibition Assays
Seeds of the wild type (Col-0), pub10, pub11, pub10 pub11, 35S:PUB10-myc (PUB10ox), jin1-9, and 35S:MYC2-GFP (MYC2ox) were germinated after 4 d stratification and the seedlings grown vertically on MS medium with or without 10 µM MeJA. Root lengths were measured 10 d after germination. To test effects of inducible expression of PUB10 and PUB10(C249A) in the MYC2 overexpression background, seeds of the wild type (Col-0), XVE:HA-PUB10/35S:myc-MYC2, XVE:HA-PUB10(C249A)/35S:myc-MYC2, and 35S:MYC2-GFP (MYC2ox) were germinated and seedlings grown vertically on control medium (MS), 1 µM MeJA alone (MeJA), 10 µM β-estradiol alone (β-est), and 1 µM MeJA plus 10 µM β-estradiol (MeJA+β-est). Root length was measured 15 d after germination using ImageJ (NIH).
Real-Time RT-PCR Analysis
Total RNA was extracted from Arabidopsis seedlings using RNeasy plant mini kit (Qiagen) including DNase I treatment. Reverse transcription was performed using 2 μg each total RNA and oligo(dT)20 primers by SuperScript III reverse transcriptase (Invitrogen). Real-time RT-PCR was performed using SYBR premix Ex Taq (Tli RNaseH plus; TaKaRa) on the Bio-Rad CFX96 real-time system with gene-specific primers. Primer sequences used here are listed in Supplemental Table 1.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative data library under the following accession numbers: JR2 (At4g23600), MYC2 (At1g32640), MYC3 (At5g46760), MYC4 (At4g17880), PDF1.2 (At5g44420), PUB10 (At1g71020), PUB11 (At1g23030), PUB12 (At2g28830), PUB13 (At3g46510), PUB23 (At2g35930), TAT (At2g24850), UBA1 (At2g30110), UBC1 (At1g14400), UBC2 (At2g02760), UBC3 (At5g62540), UBC5 (At1g63800), UBC8 (At5g41700), UBC10 (At5g53300), UBC11 (At3g08690), UBC16 (At1g75440), UBC18 (At5g42990), UBC22 (At5g05080), UBC24 (At2g33770), UBC27 (At5g50870), UBC28 (At1g64230), UBC29 (At2g16740), UBC31 (At1g36340), UBC33 (At5g50430), UBC34 (At1g17280), and UBC36 (At1g16890).
Supplemental Data
Supplemental Figure 1. PUB10 and PUB11 Interact with MYC2 in Yeast.
Supplemental Figure 2. PUB10 and PUB11 Interact with Specific Arabidopsis UBCs in Yeast.
Supplemental Figure 3. PUB10 and PUB11 Interact with Specific Arabidopsis UBCs in Vitro.
Supplemental Figure 4. Specific UBCs Support Auto-ubiquitination of PUB10 and PUB11 in Vitro.
Supplemental Figure 5. Histochemical Localization of GUS Activity in Transgenic Plants Carrying PUB10pro-GUS and MYC2pro-GUS.
Supplemental Figure 6. Real-Time PCR Analysis of PUB10 and MYC2 Transcript Levels in Overexpression Plants.
Supplemental Figure 7. PUB11 Interacts with MYC2, MYC3, and MYC4 through Its Armadillo Repeats (ARM) in Vitro.
Supplemental Figure 8. PUB11 Is an E3 Ubiquitin Ligase and Polyubiquitinates MYC2, MYC3, and MYC4 in Vitro.
Supplemental Figure 9. Histochemical Localization of GUS Activity in Transgenic Plants Carrying PUB11pro-GUS.
Supplemental Figure 10. Molecular Validation of pub10 and pub11 Mutants.
Supplemental Figure 11. PUB11 Is Not Involved in Methyl Jasmonate Response in Roots.
Supplemental Figure 12. PUB11 Is Not Involved in Methyl Jasmonate-Responsive Gene Expression.
Supplemental Table 1. Oligonucleotide Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Bobby Williams, Cheng Lu, Enno Krebbers, Barbara Mazur, and Bongsoo Park for helpful suggestions; Maria Cruz de Carvalho for her critical reading of the article; and Brendan Kidd for the jin1-9 mutant. This work was supported in part by a grant from DuPont Company. N.M. was supported in part by a JSPS KAKENHI grant (25291055). P.Z. was a graduate student on leave from Nanjing University, China, and was supported in part by a scholarship from the Ministry of Education, China.
AUTHOR CONTRIBUTIONS
C.J., P.Z., S.D., and N.-H.C. designed the research plan. C.J., P.Z., J.S.S., and N.M. performed research. C.J., J.S.S., and N.-H.C. wrote the article.
Glossary
- JA
jasmonic acid
- ABA
abscisic acid
- bHLH
basic helix-loop-helix
- MeJA
methyl jasmonate
- MS
Murashige and Skoog
References
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosokawa D., Shinozaki K. (1997). Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- An F., et al. (2010). Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen P., Kragelund B.B., Olsen A.N., Larsen F.H., Chua N.H., Poulsen F.M., Skriver K. (2004). Structure and biochemical function of a prototypical Arabidopsis U-box domain. J. Biol. Chem. 279: 40053–40061. [DOI] [PubMed] [Google Scholar]
- Anderson J.P., Badruzsaufari E., Schenk P.M., Manners J.M., Desmond O.J., Ehlert C., Maclean D.J., Ebert P.R., Kazan K. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antignani V., Klocko A.L., Bak G., Chandrasekaran S.D., Dunivin T., Nielsen E. (2015). Recruitment of PLANT U-BOX13 and the PI4Kβ1/β2 phosphatidylinositol-4 kinases by the small GTPase RabA4B plays important roles during salicylic acid-mediated plant defense signaling in Arabidopsis. Plant Cell 27: 243–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C., Santos-Rosa M.J., Shirasu K. (2001). The U-box protein family in plants. Trends Plant Sci. 6: 354–358. [DOI] [PubMed] [Google Scholar]
- Berger S., Bell E., Mullet J.E. (1996). Two methyl jasmonate-insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 111: 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergler J., Hoth S. (2011). Plant U-box armadillo repeat proteins AtPUB18 and AtPUB19 are involved in salt inhibition of germination in Arabidopsis. Plant Biol (Stuttg) 13: 725–730. [DOI] [PubMed] [Google Scholar]
- Boter M., Ruíz-Rivero O., Abdeen A., Prat S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., et al. (2011). The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico J.M., Fernández-Barbero G., Chini A., Fernández-Calvo P., Díez-Díaz M., Solano R. (2014). Repression of jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. Plant Cell 26: 1967–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Cho S.K., Ryu M.Y., Song C., Kwak J.M., Kim W.T. (2008). Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 20: 1899–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. [DOI] [PubMed] [Google Scholar]
- Dill A., Thomas S.G., Hu J., Steber C.M., Sun T.P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.H., Agarwal M., Zhang Y., Xie Q., Zhu J.K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 103: 8281–8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J.M., Smalle J., Gingerich D.J., Walker J.M., Yoo S.D., Yanagisawa S., Vierstra R.D. (2004). Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. USA 101: 6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Henriques R., Seo H.S., Nagatani A., Chua N.H. (2010). Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2013). MYC2: the master in action. Mol. Plant 6: 686–703. [DOI] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451. [DOI] [PubMed] [Google Scholar]
- Kraft E., Stone S.L., Ma L., Su N., Gao Y., Lau O.S., Deng X.W., Callis J. (2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 139: 1597–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17: 584–593. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Yoon H.J., Terzaghi W., Martinez C., Dai M., Li J., Byun M.O., Deng X.W. (2010). DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 22: 1716–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Stone S.L. (2010). Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T.P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis K.M., Thomas S.G., Soule J.D., Strader L.C., Zale J.M., Sun T.P., Steber C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Ikeda M., Takada S., Takiguchi Y., Kondou Y., Yoshizumi T., Fujita M., Shinozaki K., Matsui M., Ohme-Takagi M. (2010). Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol. 51: 2145–2151. [DOI] [PubMed] [Google Scholar]
- Mudgil Y., Shiu S.H., Stone S.L., Salt J.N., Goring D.R. (2004). A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 134: 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanandom A., Bailey M., Ewan R., Lee J., Nelis S. (2012). The ubiquitin-proteasome system: central modifier of plant signalling. New Phytol. 196: 13–28. [DOI] [PubMed] [Google Scholar]
- Samuel M.A., Mudgil Y., Salt J.N., Delmas F., Ramachandran S., Chilelli A., Goring D.R. (2008). Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol. 147: 2084–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senecoff J.F., McKinney E.C., Meagher R.B. (1996). De novo purine synthesis in Arabidopsis thaliana. II. The PUR7 gene encoding 5′-phosphoribosyl-4-(N-succinocarboxamide)-5-aminoimidazole synthetase is expressed in rapidly dividing tissues. Plant Physiol. 112: 905–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo D.H., Ryu M.Y., Jammes F., Hwang J.H., Turek M., Kang B.G., Kwak J.M., Kim W.T. (2012). Roles of four Arabidopsis U-box E3 ubiquitin ligases in negative regulation of abscisic acid-mediated drought stress responses. Plant Physiol. 160: 556–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.S., Yang J.Y., Ishikawa M., Bolle C., Ballesteros M.L., Chua N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999. [DOI] [PubMed] [Google Scholar]
- Seo K.I., Lee J.H., Nezames C.D., Zhong S., Song E., Byun M.O., Deng X.W. (2014). ABD1 is an Arabidopsis DCAF substrate receptor for CUL4-DDB1-based E3 ligases that acts as a negative regulator of abscisic acid signaling. Plant Cell 26: 695–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Heidrich K., Sanchez-Villarreal A., Parker J.E., Davis S.J. (2012). TIME FOR COFFEE represses accumulation of the MYC2 transcription factor to provide time-of-day regulation of jasmonate signaling in Arabidopsis. Plant Cell 24: 2470–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M., Anderson R.G., Ichimura K., Pecenkova T., Reuter P., Žársky V., McDowell J.M., Shirasu K., Trujillo M. (2012). The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 24: 4703–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Trujillo M., Ichimura K., Casais C., Shirasu K. (2008). Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 18: 1396–1401. [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397. [DOI] [PubMed] [Google Scholar]
- Wiborg J., O’Shea C., Skriver K. (2008). Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem. J. 413: 447–457. [DOI] [PubMed] [Google Scholar]
- Wu H.W., Lin S.S., Chen K.C., Yeh S.D., Chua N.H. (2010). Discriminating mutations of HC-Pro of zucchini yellow mosaic virus with differential effects on small RNA pathways involved in viral pathogenicity and symptom development. Mol. Plant Microbe Interact. 23: 17–28. [DOI] [PubMed] [Google Scholar]
- Xie Q., Guo H.S., Dallman G., Fang S., Weissman A.M., Chua N.H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170. [DOI] [PubMed] [Google Scholar]
- Yang C.W., González-Lamothe R., Ewan R.A., Rowland O., Yoshioka H., Shenton M., Ye H., O’Donnell E., Jones J.D., Sadanandom A. (2006). The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18: 1084–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee D., Goring D.R. (2009). The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J. Exp. Bot. 60: 1109–1121. [DOI] [PubMed] [Google Scholar]
- Zhai Q., Yan L., Tan D., Chen R., Sun J., Gao L., Dong M.Q., Wang Y., Li C. (2013). Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet. 9: e1003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Garreton V., Chua N.H. (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19: 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.S., Niu Q.W., Chua N.H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646. [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.