Phytophthora sojae produces the xyloglucan-degrading enzyme XEG1 as a virulence factor; this apoplastic effector is also recognized via the plant’s PAMP recognition machinery.
Abstract
We identified a glycoside hydrolase family 12 (GH12) protein, XEG1, produced by the soybean pathogen Phytophthora sojae that exhibits xyloglucanase and β-glucanase activity. It acts as an important virulence factor during P. sojae infection but also acts as a pathogen-associated molecular pattern (PAMP) in soybean (Glycine max) and solanaceous species, where it can trigger defense responses including cell death. GH12 proteins occur widely across microbial taxa, and many of these GH12 proteins induce cell death in Nicotiana benthamiana. The PAMP activity of XEG1 is independent of its xyloglucanase activity. XEG1 can induce plant defense responses in a BAK1-dependent manner. The perception of XEG1 occurs independently of the perception of ethylene-inducing xylanase. XEG1 is strongly induced in P. sojae within 30 min of infection of soybean and then slowly declines. Both silencing and overexpression of XEG1 in P. sojae severely reduced virulence. Many P. sojae RXLR effectors could suppress defense responses induced by XEG1, including several that are expressed within 30 min of infection. Therefore, our data suggest that PsXEG1 contributes to P. sojae virulence, but soybean recognizes PsXEG1 to induce immune responses, which in turn can be suppressed by RXLR effectors. XEG1 thus represents an apoplastic effector that is recognized via the plant’s PAMP recognition machinery.
INTRODUCTION
Oomycetes are a major class of destructive plant pathogens, related to diatoms. As an important part of plants’ defense against pathogens such as oomycetes, host receptors can recognize conserved pathogen-associated molecular patterns (PAMPs) and microbe-associated molecular patterns (Kline, 2009; Ronald and Beutler, 2010). PAMPs were originally defined as highly conserved molecules that have an essential function in microbial fitness or survival within a class of microbes (Medzhitov and Janeway, 1997; Nürnberger and Brunner, 2002; Thomma et al., 2011). Plants can recognize many different PAMPs from bacteria and fungi (Newman et al., 2013). PAMPs identified so far from oomycetes include β-glucans, heptaglucoside, transglutaminase (Pep13), cellulose binding elicitor lectins, and elicitins (Sharp et al., 1984; Yu, 1995; Enkerli et al., 1999; Klarzynski et al., 2000; Brunner et al., 2002; Gaulin et al., 2006; Zhang et al., 2014). Nevertheless, the mechanisms that allow plants to perceive oomycetes as non-self and to defend against infection remain only partly known.
The hydrolytic enzymes of pathogens are important contributors to plant pathogenesis through degradation of host macromolecules and in some cases through detoxification of secondary metabolites (Morrissey and Osbourn, 1999; Kikot et al., 2009). Carbohydrate active enzymes can degrade plant cell wall polymers to permit the pathogen to invade plant tissue and to provide the pathogen with nutrients (Walton, 1994; Hématy et al., 2009). These enzymes include pectinases, polygalacturonases, glucanases, cellulases, and xyloglucanases. The effects of targeted disruption of some individual genes encoding carbohydrate active enzymes support their direct involvement in infection and disease. For example, deletion of the xyn11A gene had a marked effect on the ability of Botrytis cinerea to infect tomato (Solanum lycopersicum) leaves and grape (Vitis vinifera) berries (Brito et al., 2006), and a mutation in the xynB endoxylanase gene affected the virulence of Xanthomonas oryzae pv oryzae (Rajeshwari et al., 2005). In Colletotrichum coccodes, deletion of the pectate lyase gene CcpelA resulted in a substantial loss of virulence on green tomato fruit (Ben-Daniel et al., 2012), while in Colletotrichum gloeosporoides, deletion of the pectate lyase gene PelB reduced virulence on avocado (Persea americana) fruit (Yakoby et al., 2001). Nevertheless, the specific roles of the majority of cell wall-degrading enzymes (CWDEs) in virulence remain largely unknown, especially in oomycetes.
The innate immune systems of plants have evolved to detect CWDEs as inducers of immune responses (Misas-Villamil and van der Hoorn, 2008). Some plant cell wall fragments produced by CWDEs can elicit defense responses in plants (Hématy et al., 2009). For example, degradation of pectins by polygalacturonase generates oligogalacturonic acid, which can trigger plant defenses (Prade et al., 1999; D’Ovidio et al., 2004). Some CWDEs also are recognized directly as PAMPs (Nürnberger et al., 2004). For instance, fungal endopolygalacturonases (PGs) can be recognized as PAMPs via the Arabidopsis thaliana RLP42 protein (Zhang et al., 2014). Another hydrolytic enzyme, ethylene-inducing xylanase (EIX), is recognized as a PAMP via the tomato LRR-RLPs, Eix1 and Eix2, of which only Eix2 mediates a cell death response (Nürnberger et al., 2004; Ron and Avni, 2004). However, the full range of hydrolytic enzymes of microbes that can be detected by the innate immune systems of plants remains mostly undiscovered.
In this study, we identified a PAMP, XEG1, from the soybean pathogen Phytophthora sojae, that triggers cell death in several plant species. XEG1 is a glycoside hydrolase family 12 (GH12) member with xyloglucanase activity and is required for virulence.
RESULTS
XEG1 from P. sojae Is an Apoplastic Elicitor of Cell Death
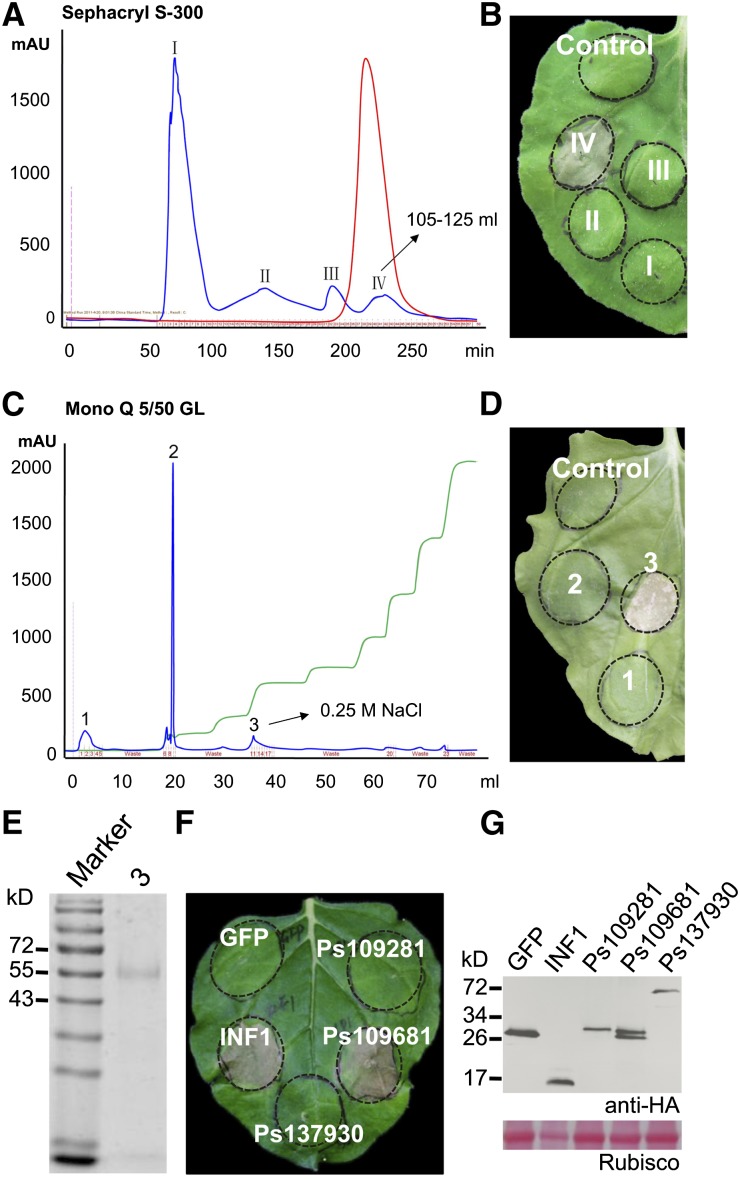
To identify defense response elicitor proteins from P. sojae, we fractionated proteins from culture filtrates and assayed them for induction of cell death in Nicotiana benthamiana leaves. Fractionation of a 70% ammonium sulfate precipitate by column chromatography on Sephacryl S-300 yielded four main peaks (Figure 1A). Proteins corresponding to peak IV could induce cell death (Figure 1B). SDS-PAGE analysis of the proteins of peak IV showed more than one band; therefore, peak IV was fractionated on a mono-Q anion exchange column using a NaCl gradient. Of three peaks of UV-absorbing material (Figure 1C), fractions corresponding to peak 3 could induce cell death (Figure 1D). Peak 3 showed a single band of 55 kD by SDS-PAGE (Figure 1E).
Figure 1.
XEG1 Purified from P. sojae Culture Supernatants Induces Cell Death in N. benthamiana.
(A) Gel permeation chromatography of supernatant proteins, resulting in four main peaks (I, II, III, and IV). Blue curve, protein concentration; red curve, salt concentration.
(B) N. benthamiana leaves from 2-month-old plants 48 h after infiltration with fractions from the Sephacryl peaks (10 mg/L protein solution) or buffer control (buffer “purified” in the same way as the secreted proteins).
(C) Anion-exchange chromatography of peak IV proteins, yielding three main peaks (1, 2, and 3). Blue curve, protein concentration; green curve, salt concentration.
(D) Representative N. benthamiana leaves from 2-month-old plants 48 h after infiltration with fractions from the Mono Q 5/50 peaks (10 mg/mL) or buffer control.
(E) SDS-PAGE of peak 3 proteins, stained with Coomassie blue. Left lane, molecular mass markers; right lane, peak 3 proteins.
(F) Representative N. benthamiana leaves 5 d after inoculation with Agrobacterium strains carrying the indicated genes in vector pGR107.
(G) Immunoblot analysis of proteins from N. benthamiana leaves transiently expressing GFP, INF1, Ps109681, Ps109281, or Ps137930 fused with 3*HA tags.
To identify the protein(s) in peak 3, tryptic digestion was performed, and the peptides generated were analyzed by mass spectrometry. Peptides were identified matching proteins encoded by the genes Ps109281 (four peptides), Ps109681 (four peptides), Ps158318 (two peptides), and Ps137930 (one peptide) (Supplemental Table 1). These genes encode proteins of 24, 26, 110, and 62 kD, respectively. To identify which protein induced cell death in N. benthamiana, we used Agrobacterium tumefaciens-mediated transformation (agroinfiltration) to transiently express these genes in N. benthamiana leaves. The coding sequences of Ps109281, Ps109681, and Ps137930, including their secretion signals, were cloned separately into the binary PVX vector pGR107, which adds a C-terminal 3*HA tag to the proteins (we failed to clone the cDNA of Ps158318). pGR107:GFP and pGR107:INF1 plasmid constructs served as negative and positive controls, respectively. Transient expression of the genes in N. benthamiana demonstrated that Ps109681 triggered cell death in N. benthamiana 5 d after infiltration (Figure 1F). Immunoblot analysis using α-HA antibody detected Ps109681 protein in the leaves as a pair of polypeptides close to the predicted size of 27 kD (Figure 1G). Since the protein encoded by Ps109681 is approximately half the size of the 55-kD band purified from the culture supernatant, we speculate that dimeric Ps109681 protein was present in the 55-kD band observed in fraction 3. Alternatively, the amount of Ps109681 protein present in fraction 3 might have been sufficient to trigger cell death but insufficient to be observed following PAGE. Based on its sequence, Ps109681 (designated XEG1) was identified as a protein elicitor secreted by P. sojae that is distinct from other cell death elicitors previously identified from Phytophthora species.
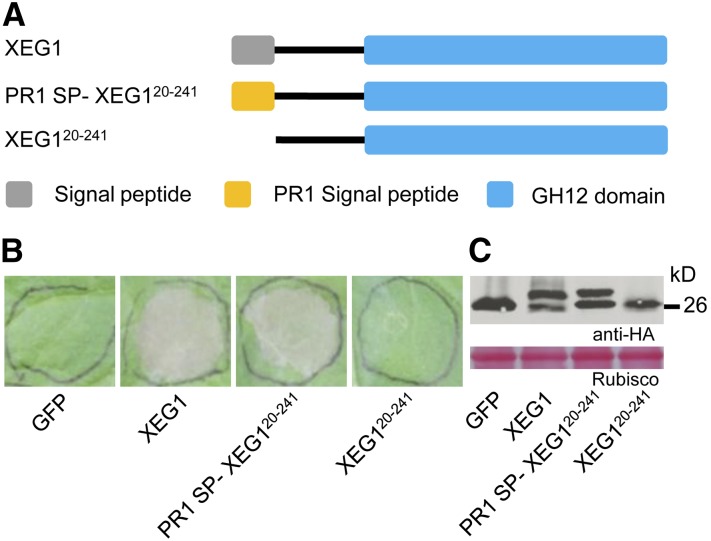
To determine whether XEG1 must be targeted to the apoplast in order to trigger cell death, we deleted the N-terminal signal peptide to produce PsXEG120–241 and then added the signal peptide from pathogenesis-related protein 1 (PR1) protein to produce PR11-30-XEG120–241 (Figure 2A). Both these proteins were expressed in N. benthamiana, and immunoblot analysis confirmed that the proteins produced were of the expected size (Figure 2C). Expression of XEG120–241 did not trigger cell death in N. benthamiana (Figure 2B), whereas expression of PR11–30-XEG120–241 did trigger cell death. These data indicated that XEG1 must be targeted to the extracellular space of N. benthamiana tissue to induce cell death.
Figure 2.
A Signal Peptide Is Required for XEG1-Induced Cell Death.
(A) Regions of XEG1 examined for cell death activity.
(B) Representative N. benthamiana leaves 5 d after agroinfiltration using constructs encoding the indicated proteins.
(C) Immunoblot of proteins from N. benthamiana leaves transiently expressing the indicated proteins from a pGR107-3*HA vector. The doublets evident in the secreted forms of the proteins may represent unprocessed proteins retaining their signal peptide or else may represent posttranslational modification.
XEG1 Protein Induces Cell Death in Tobacco, Tomato, Pepper, and Soybean
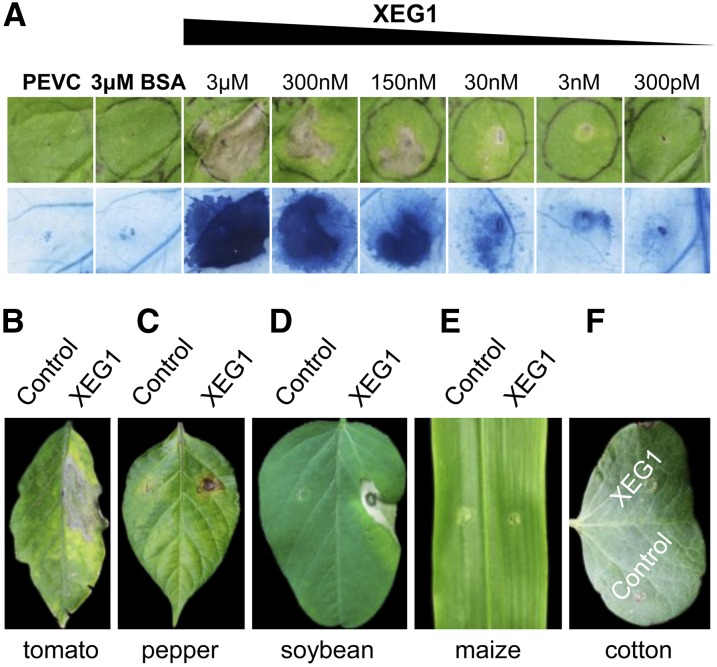
To further confirm that XEG1 protein could induce cell death in N. benthamiana, XEG1 was produced in the yeast Pichia pastoris using the pPICZaA vector (pPICZaA:PsXEG1), which targeted the protein for secretion into the culture medium. After recovery from the P. pastoris culture supernatant using Ni-NTA resin, the recombinant protein, XEG1rec, with a size of 27 kD (Supplemental Figure 1), was tested for cell death activity by infiltrating 300 pM to 3 μM protein solution into the mesophyll of N. benthamiana leaves. XEG1rec induced cell death in N. benthamiana (Figure 3A) 5 d after protein infiltration. Moreover, cell death was increased with increasing concentrations of XEG1 from 300 pM to 3 μM. To examine the host specificity of XEG1, we infiltrated XEG1rec (300 nM) into expanded leaves of various plant species. XEG1rec induced localized cell death in tomato, pepper (Capsicum annuum), and soybean (Glycine max) (Figures 3B to 3D), but not in maize (Zea mays) or cotton (Gossypium hirsutum) (Figures 3E and 3F). Thus, XEG1 can induce cell death in diverse plant families in addition to the cognate host, soybean.
Figure 3.
XEG1 Induces Cell Death in Various Plant Species.
(A) Representative N. benthamiana leaves infiltrated with purified XEG1 protein (300 pM to 3 μM). PEVC (P. pastoris culture supernatant from an empty vector control strain, purified in the same way as PsXEG1) is the culture supernatant control. Upper pictures, directly photographed 2 d postinfiltration. Lower pictures, photographed 2 d postinfiltration after staining with trypan blue.
(B) to (F) Cell death response in four other species of plants triggered by 300 nM XEG1rec or culture supernatant control. Representative leaves are shown of tomato (B), pepper (C), soybean (D), maize (E), and cotton (F).
XEG1 Belongs to the Glycoside Hydrolase GH12 Family, Which Is Widely Distributed across Microbial Taxa
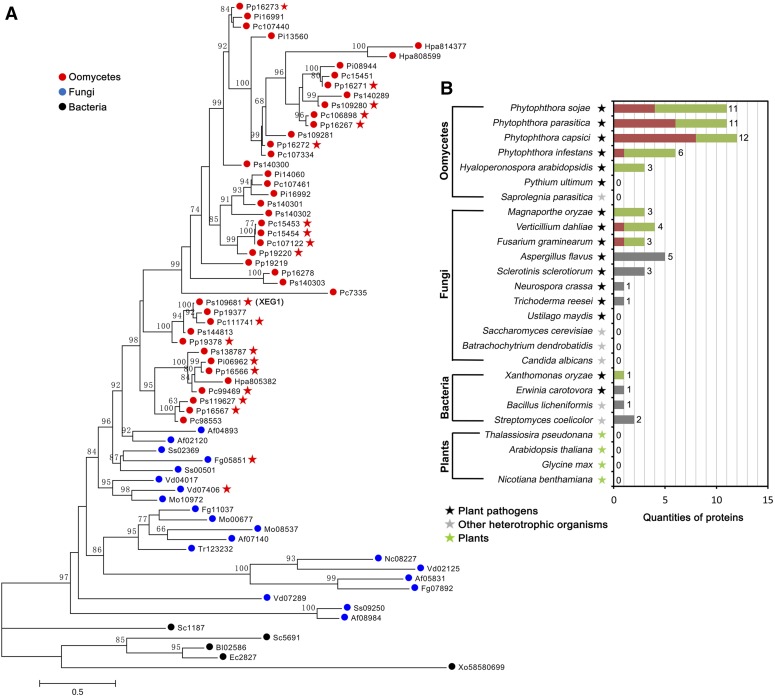
To study the phylogenetic distribution of XEG1 homologs in various organisms, we mined the genomes of 7 oomycete, 11 fungal, 4 bacterial, and 4 plant species. A total of 68 genes encoding homologous protein sequences were identified (Figure 4A). The protein sequences of XEG1 and homologs contained a GH12 domain predicted by the GenBank Conserved Domain Database as well as a signal peptide predicted by SignalP 4.1 (Supplemental Data Sets 1 and 2). These GH12 protein sequences are present in prokaryotic and eukaryotic microorganisms, but not in higher organisms such as plants (Figure 4B), demonstrating that GH12 proteins are common across microbial taxa, especially in plant-associated microbes (Figure 4B). Among oomycete plant pathogens, we identified a large number of GH12 proteins (6 to 12 per species) within the Phytophthora genus (Figures 4A and 4B), as well as three in the obligate biotroph Hyaloperonospora arabidopsidis. Surprisingly, none were found in the necrotrophic oomycete Pythium ultimum. No GH12 genes were found in the animal pathogens Saprolegnia parasitica, Batrachochytrium dendrobatidis, or Candida albicans.
Figure 4.
XEG1 Belongs to a GH12 Family That Is Distributed Widely across Microbial Taxa.
(A) Phylogeny of XEG1 and 68 related sequences from selected species. Bootstrap percentage support for each branch is indicated. The scale bar represents 50% weighted sequence divergence. The proteins that induce cell death in N. benthamiana are indicated by a red star.
(B) Distribution of GH12 proteins that do or do not induce cell death, as measured by agroinfiltration in N. benthamiana. The quantities of genes that induce cell death in N. benthamiana are indicated by crimson bars and those that do not by green bars. Numerals indicate total number of genes.
To determine whether these diverse oomycete and fungal GH12 proteins could trigger cell death in N. benthamiana, 54 genes (Figure 4B; Supplemental Data Set 3) were tested by agroinfiltration. Of the 54 genes, 21 genes from both oomycete and fungal species triggered visible cell death (Figure 4B; Supplemental Data Set 3). Thus, a wide diversity of GH12 proteins from different species could be recognized by N. benthamiana.
Tobacco Serk3/Bak1 Is Required for XEG1-Triggered Cell Death in N. benthamiana
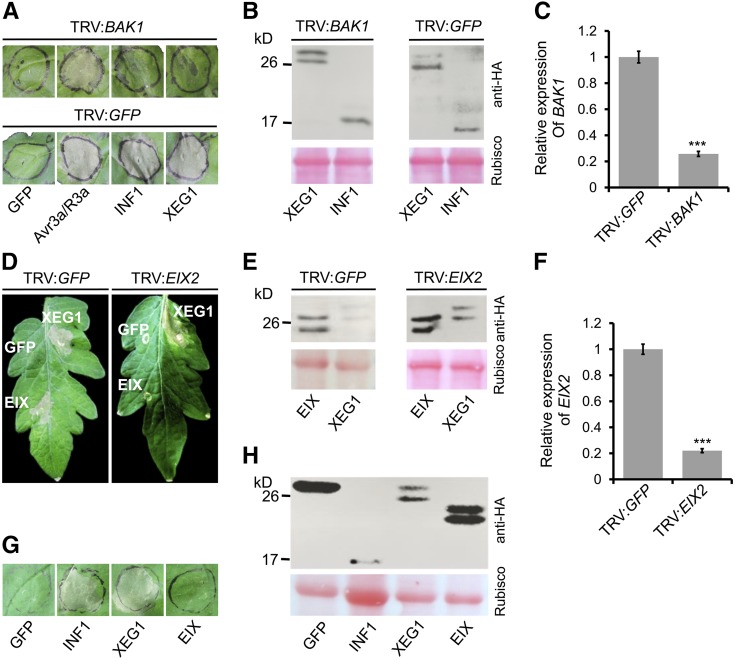
The RLP kinase protein Serk3/Bak1 participates in multiple pattern recognition receptor pathways, including cell death induction by the oomycete PAMP INF1 (Heese et al., 2007). To determine whether Serk3 participated in induction of cell death by XEG1, we generated virus-induced gene silencing (VIGS) constructs based on recombinant tobacco rattle virus (TRV) to target Serk3 expression in tobacco. Three weeks after viral inoculation, plants were agroinfiltrated with XEG1 or INF1 expression constructs. Our results showed that, as expected, INF1 did not trigger cell death in Serk3-silenced plants (Figure 5A). XEG1 also failed to trigger cell death in these plants, similar to INF1 (Figure 5A). However, Serk3-silenced plants were still capable of responding with cell death to transient coexpression of the avirulence gene Avr3a and the corresponding resistance gene R3a (Figure 5A). Immunoblotting confirmed that INF1 and XEG1 were successfully expressed at the expected size in N. benthamiana inoculated with TRV:Serk3 or TRV:GFP (Figure 5B). Reverse transcription-quantitative PCR (RT-qPCR) analysis confirmed that Serk3 expression was markedly reduced upon inoculation with TRV:Serk3 compared with inoculation with TRV:GFP (Figure 5C). From these results, we inferred that Serk3 was required for XEG1-triggered cell death in N. benthamiana.
Figure 5.
Cell Death Triggered by XEG1 Requires BAK1 but Not Eix2.
(A) BAK1 is required for XEG1-triggered cell death in N. benthamiana. N. benthamiana plants were subjected to VIGS by inoculation with TRV constructs (TRV:GFP or TRV:NbBAK1). Three weeks after inoculation, GFP, Avr3a/R3a, INF1, and XEG1 were transiently expressed in the gene-silenced leaves and then leaves were photographed 5 d later. The experiment was performed five times with five plants for each TRV construct. Representative photographs are shown.
(B) Immunoblot analysis of INF1 or XEG1 protein fused with 3*HA tags transiently expressed in VIGS-silenced N. benthamiana leaves.
(C) Tobacco BAK1 expression levels after VIGS treatment determined by qRT-PCR analysis. EF1α was used as an endogenous control. Means and standard errors from three biological replicates are shown. Asterisks indicate significant differences (P ≤ 0.001).
(D) Eix2 is not required for XEG1-triggered cell death in tomato. Tomato plants were subjected to VIGS by inoculation with TRV constructs (TRV:GFP or TRV:LeEIX2). Four weeks after inoculation, GFP, INF1, and PsXEG1 were transiently expressed in the gene-silenced leaves and then photographed 10 d later. The experiment was performed five times with five plants for each TRV construct. Representative photographs are shown.
(E) Immunoblot analysis of XEG1 or EIX protein fused with 3*HA tags transiently expressed in VIGS-silenced tomato leaves.
(F) Eix2 expression levels after VIGS treatment determined by RT-qPCR analysis. Actin gene was used as the endogenous control. Means and standard errors from three biological replicates are shown. Asterisks indicate significant differences (P ≤ 0.001).
(G) Representative N. benthamiana leaves 5 d after inoculation with Agrobacterium strains carrying the indicated genes in vector pGR107.
(H) Immunoblot analysis of proteins from N. benthamiana leaves transiently expressing GFP, INF1, XEG1, or EIX fused with 3*HA tags.
XEG1-Triggered Cell Death in Tomato Does Not Depend on Eix2
Tomato Eix2 was previously shown to transmit the cell death induction signal produced in tomato by the fungal Glycoside Hydrolase 11 protein EIX (Ron and Avni, 2004). To determine whether Eix2 was required for perception of XEG1 in tomato, we generated VIGS constructs based on TRV to suppress Eix2 expression in tomato. Four weeks after viral inoculation, plants were agroinfiltrated with XEG1 or EIX expression constructs. As expected, EIX did not trigger cell death in Eix2-silenced plants (Figure 5D). However, XEG1 still could trigger cell death in those plants (Figure 5D). Immunoblots confirmed that XEG1 and EIX were successfully expressed in tomato inoculated with TRV:Eix2 or TRV:GFP (Figure 5E). Quantitative RT-PCR (qRT-PCR) analysis confirmed that Eix2 expression was markedly reduced upon inoculation with TRV:Eix2 compared with inoculation with TRV:GFP (Figure 5F). Moreover, as Figure 4G shows, EIX could not induce HR in N. benthamiana leaves (as previously shown; Ron and Avni, 2004), whereas XEG1 could induce cell death in N. benthamiana leaves. Immunoblotting confirmed that XEG1 and EIX were successfully expressed in N. benthamiana (Figure 5H). The transient expression of XEG1 in N. benthamiana and Eix2-silenced tomato indicates that XEG1 is perceived independently of EIX and thus constitutes a novel PAMP.
The Enzymatic Activity of XEG1 Is Not Required for Elicitor Activity
GH12 proteins from fungi were previously shown to degrade xyloglucan (Master et al., 2008) or β-glucan (Karlsson et al., 2002). Purified XEG1rec was assayed for hydrolase activity using xyloglucan and β-glucan separately as substrates. XEG1rec could produce reducing sugars from both substrates, with somewhat higher activity toward xyloglucan (Table 1).
Table 1. Hydrolysis Activity of Wild-Type and Mutant Forms of XEG1 toward Xyloglucan and β-Glucan.
| Enzyme | Xyloglucana | β-Glucana | |
|---|---|---|---|
| Units/mg | (%) | Units/mg | |
| Wild type | 10.5 ± 0.6 | 100 ± 6 | 3.6 ± 0.4 |
| E136Drec | 0.10 ± 0.03 | 0.98 ± 0.26 | – |
| E222Drec | 0.14 ± 0.05 | 1.35 ± 0.46 | – |
Values represent the averages of three independent measurements with three replicates each. Standard errors are shown.
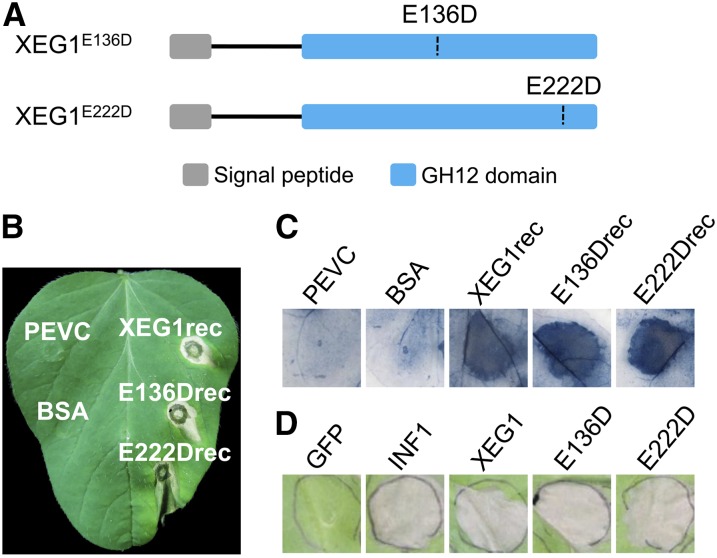
The catalytic site of GH12 from Trichoderma reesei includes the catalytic residues Glu-116 and Glu-200 (Sandgren et al., 2001), which are present in diverse members of the GH12 family (Sandgren et al., 2005). The corresponding catalytic residues in P. sojae XEG1 are Glu-136 and Glu-222 (Supplemental Figure 7). To establish their contribution to hydrolase activity, we replaced Glu-136 and Glu-222 individually with aspartate using site-directed mutagenesis (Figure 6A) and expressed the mutant proteins (E136Drec and E222Drec) in P. pastoris (Supplemental Figure 2). Hydrolase assays with the purified proteins showed that the xyloglucan-degrading endoglucanase activity was abolished in both mutant proteins (Table 1). To determine the relationship between hydrolase activity and cell death activity, purified E136Drec and E222Drec were infiltrated into leaves of N. benthamiana and soybean. Both mutated proteins could induce visible cell death in soybean and cell death response in N. benthamiana that was detectable by trypan blue staining 48 h after infiltration (Figures 6B and 6C).
Figure 6.
Cell Death-Inducing Activities of Wild-Type and Mutant Forms of XEG1.
(A) Diagram of XEG1 mutants.
(B) and (C) Representative soybean and N. benthamiana leaves infiltrated with the indicated proteins (300 nM). Soybean leaves were photographed 2 d postinfiltration. N. benthamiana leaves were photographed 2 d postinfiltration after staining with trypan blue. PEVC is the culture supernatant control.
(D) Representative N. benthamiana leaves 5 d after agroinfiltration using constructs encoding E136D and E222D XEG1 mutant proteins and indicated controls.
To further assess the elicitor activity of E136Drec and E222Drec, we transiently expressed the proteins in N. benthamiana and found that both triggered cell death to the same extent as XEG1 (Figure 6D). Production of the proteins was confirmed by immunoblot analysis (Supplemental Figure 3). The results of the protein infiltration and transient expression experiments indicate that the enzymatic activity of XEG1 is not required for cell death activity.
XEG1 Induces Disease Resistance in N. benthamiana and Soybean
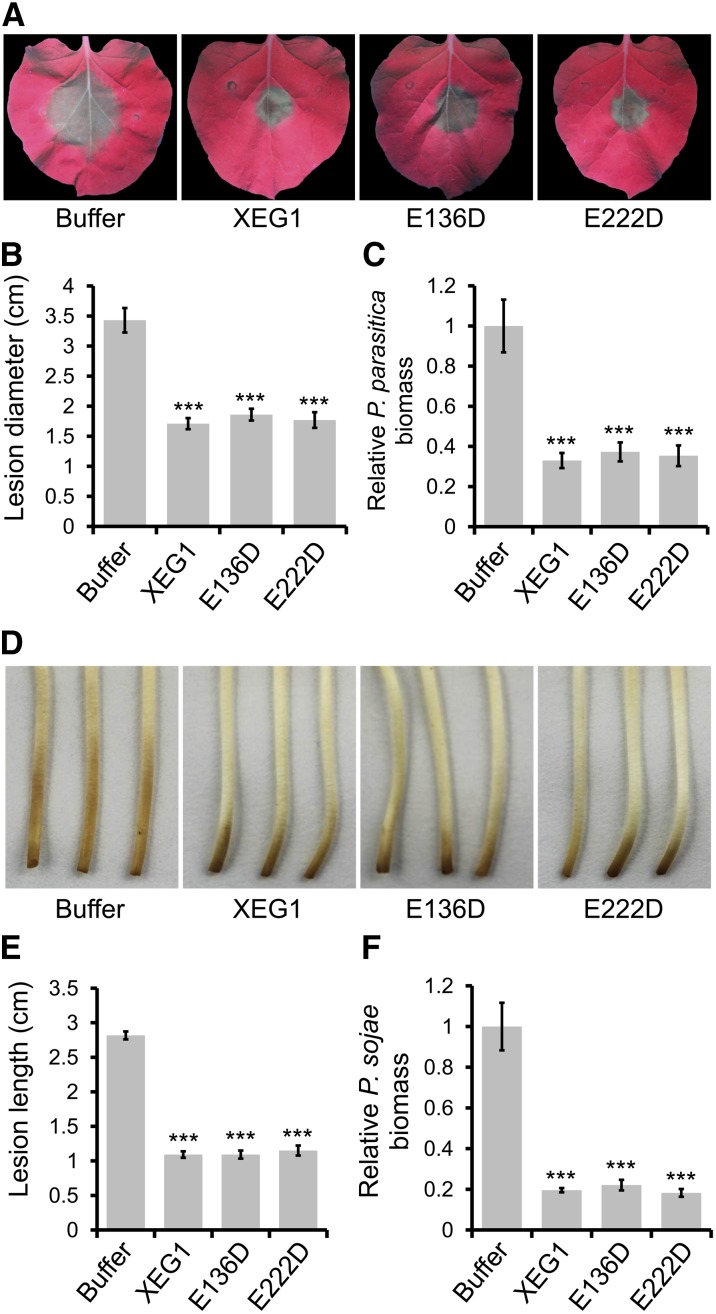
To assess whether XEG1 could elicit defense responses in addition to cell death, N. benthamiana leaves were treated separately with a low concentration of XEG1rec, E136D, or E222D (300 pM) or with a buffer control, and then 24 h later were inoculated with Phytophthora parasitica var nicotianae zoospores. Disease symptoms became visible 48 h after pathogen inoculation in N. benthamiana leaves that had been pretreated with buffer, whereas XEG1, E136D, or E222D-pretreated leaves were protected against disease development (Figure 7A). Determination of P. parasitica lesion diameter (Figure 7B) and biomass (Figure 7C) in the inoculated N. benthamiana plants by quantitative genomic DNA PCR revealed that XEG1, E136D, or E222D reduced P. parasitica growth significantly. In order to further evaluate the biological significance of XEG1’s activity on the cognate host, the cotyledon and root of etiolated soybean hypocotyl were cut off and were placed into 300 pM XEG1, E136D, or E222D protein solutions. Twelve hours later, the etiolated soybean hypocotyl was taken out from protein solution and inoculated with a piece of mycelium of P. sojae on the root side. Symptoms became visible after 36 h on plants pretreated with buffer, whereas XEG1, E136D, or E222D-pretreated plants were protected against disease development (Figure 7D). Determination of P. sojae lesion length and biomass in the inoculated soybean plants by quantitative genomic DNA PCR revealed that XEG1, E136D, or E222D significantly reduced P. sojae growth (Figures 7E and 7F). These findings suggest that XEG1 is an efficient elicitor of N. benthamiana and soybean defense mechanisms, and defense elicitation is independent of the enzymatic activity.
Figure 7.
XEG1 Induces Disease Resistance in N. benthamiana and Soybean.
(A) to (C) Disease resistance induced by pretreatment of N. benthamiana leaves with 300 pM XEG1 and then inoculated 24 h later with 100 zoospores of P. parasitica var nicotianae. The whole area of leave was injected and the P. sojae spores were applied at the center of leaves. The experiment was replicated six times with six plants per biological replicate and three leaves analyzed per plant. Lesions were assessed under UV light 48 hpi.
(A) Representative photographs.
(B) Average lesion diameters with standard errors.
(C) Relative biomass of P. parasitica with standard errors, as measured by genomic DNA qPCR and normalized to the buffer control. Means and standard errors from six biological replicates are shown.
(D) to (F) Induction of disease resistance to P. sojae induced by pretreatment of etiolated soybean hypocotyls with 300 pM XEG1. The cotyledons and roots of etiolated soybean hypocotyls were cut off and the remaining entire stem was then submerged under 300 pM XEG1, E136D, E222D protein solution, or buffer (1× PBS). Twelve hours later, the etiolated soybean hypocotyl was removed from the protein solution and then inoculated with a piece of mycelium of P. sojae on the root side. Lesions were assessed 36 hpi. The experiment was replicated six times with six plants per biological replicate.
(D) Representative photographs.
(E) Average lesion length with standard errors.
(F) Relative biomass of P. sojae in the infected etiolated soybean hypocotyls (at 36 hpi). DNA of P. sojae in infected soybean hypocotyls was measured by genomic DNA qPCR and was normalized to that treated with buffer. Means and standard errors from six biological replicates are shown.
In (B), (C), (E), and (F), asterisks indicate statistically significant differences to control based on Student’s t test (***P ≤ 0.001).
XEG1 Is Required for Virulence, but Also Triggers PAMP-Triggered Immunity during Infection
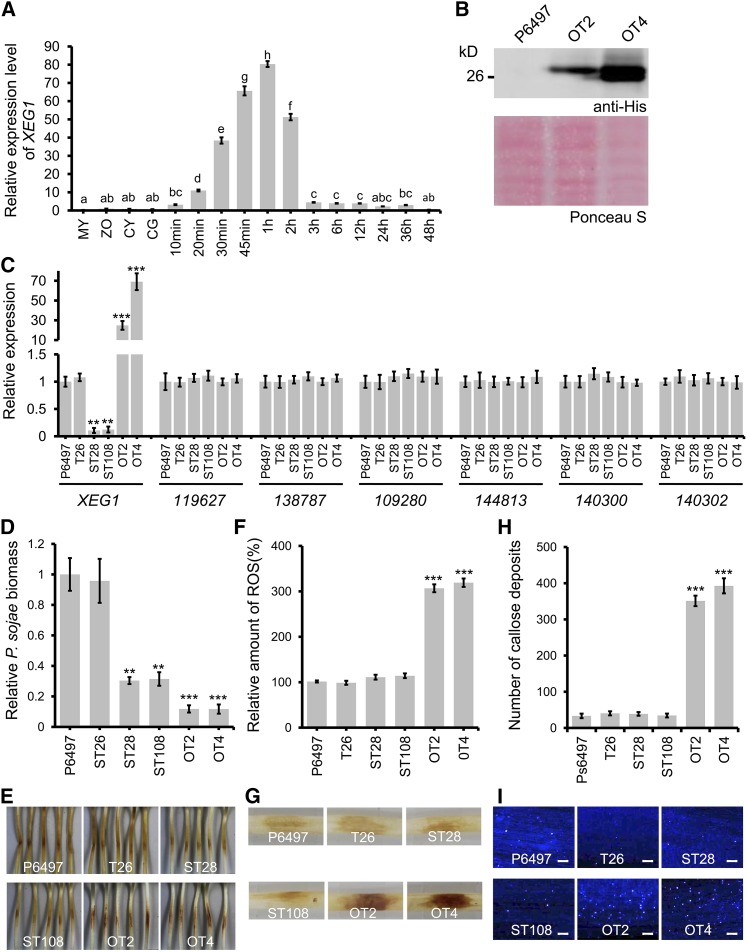
To explore the possible contribution of XEG1 to P. sojae virulence, we determined the expression patterns of XEG1 during different stages of development, including mycelia, zoospores, cysts, and germinating cyst and at 12 time points of infection following inoculation of etiolated soybean hypocotyls with zoospores. XEG1 was expressed at high levels during very early infection stages (20 min to 2 h) and then rapidly declined from 3 h onwards (Figure 8A). To more directly establish the role of XEG1 during infection, we silenced and overexpressed XEG1 by transformation with an antisense or sense construct, respectively (Whisson et al., 2007; Dou et al., 2008). XEG1 transcript levels were reduced to around 11% in two silenced lines (ST28 and ST108) and increased by 20- and 65-fold in two overexpression lines (OT2 and OT4) compared with the wild-type (P6497) strain (Figure 8C). We also checked the transcript levels of six other GH12 genes most similar to XEG1, in the silenced and overexpression lines. The transcript levels of these genes showed no significant differences compared with the wild-type strain (P6497) (Figure 8C). Immunoblot analysis using an α-His antibody demonstrated that XEG1 protein was elevated in the mycelia of the two overexpression transgenic lines (Figure 8B). A nonsilenced line (T26) expressing XEG1 at levels similar to those in the P6497 line was used as a negative transgenic control (Figure 8C). All transformants showed normal filamentous growth. To assay the virulence of the transformants, zoospores were inoculated onto etiolated seedlings of the susceptible soybean cultivar Williams. Both silencing and overexpression of XEG1 reduced P. sojae virulence (Figure 8E). Determination of P. sojae biomass in the inoculated soybean plants by quantitative genomic DNA PCR revealed that virulence in the OT2, OT4, ST28, and ST108 transgenic lines was severely attenuated compared with that in the P6497 and T26 lines (Figure 8D). Furthermore, the P. sojae biomass was even lower in etiolated seedlings infected with the OT2 and OT4 lines than with the ST28 and ST108 lines (Figure 8D).
Figure 8.
Virulence and Elicitor Functions of XEG1 during Infection.
(A) Transcript levels of XEG1 in different P. sojae tissues (My, nonsporulating mycelium grown in V8 broth; Zo, zoospores; Cy, cysts; GC, germinating cysts) or in P. sojae-infected Williams soybean (mpi, minutes postinoculation). Transcript levels measured by RT-qPCR were normalized to levels in zoospores using the Actin gene as an internal reference. Means and standard errors from three biological replicates are shown. Letters represent significant differences (P ≤ 0.05) as measured by Duncan’s multiple range test.
(B) Immunoblot analysis of proteins isolated from P. sojae transformants overexpressing XEG1-His (OT2 and OT4) and from control strain P6497. Upper panel, XEG1 proteins detected using anti-His antibody; lower panel, Ponceau S staining of total proteins.
(C) Transcript levels of XEG1 and its six closest paralogs in mycelia of P. sojae transformants, measured using RT-qPCR and normalized to the level in P6497 mycelia using Actin as the internal reference. The T26 line was a negative control transformant. Bars indicate standard errors from three biological replicates. Asterisks indicate significant differences with P6497 based on Student’s t test (**P ≤ 0.01 and ***P ≤ 0.001).
(D) Relative biomass of wild-type and transgenic P. sojae infecting etiolated soybean hypocotyls (at 48 hpi), as measured by genomic DNA quantitative PCR, and normalized to P6497. Means and standard errors from three biological replicates are shown. Asterisks indicate significant differences with P6497 based on Student’s t test (**P ≤ 0.01 and ***P ≤ 0.001).
(E) Visual symptoms 48 h after inoculation of etiolated soybean seedlings by P. sojae wild-type and transgenic lines. Representative photographs are shown.
(F) and (G) ROS accumulation at 6 hpi in hypocotyls infected with P. sojae transformants and controls, as detected by DAB staining.
(F) Quantification using ImageJ software. Means and standard errors from three independent replicates are shown. Significant differences with P6497 based on Student’s t test are indicated by the asterisks (***P ≤ 0.001).
(G) Representative photographs of visual symptoms.
(H) and (I) Callose deposition in etiolated soybean hypocotyl epidermal cells infected by P. sojae wild type and transgenic line zoospores.
(H) Number of callose deposits per microscopic field quantitated using ImageJ software. Means and standard errors from four independent replicates are shown. Asterisk indicate significant differences with P6497 based on Student’s t test (***P ≤ 0.001).
(I) Representative images of callose deposition. Bars = 50 μm.
Since XEG1 induced cell death in the soybean cultivar Williams, we determined reactive oxygen species (ROS) levels in etiolated soybean hypocotyl lesions 6 h after inoculation with zoospores of the transgenic silencing (ST28 and ST108) or overexpression lines (OT2 and OT4), or of the controls (Figures 8F and 8G). ROS accumulation in response to infection of the OT2 and OT4 lines was substantially enhanced compared with the ST28, ST108, T26, and P6497 lines (Figures 8F and 8G). We also examined callose deposition under the same conditions. At 6 h postinoculation (hpi), epidermal cells inoculated with OT2 or OT4 zoospores exhibited strong callose deposition compared with those inoculated with ST28, ST108, T26, and P6497, all of which exhibited low or undetectable levels of callose deposition (Figures 8H and 8I). These results indicate that XEG1 overexpression in P. sojae resulted in a strong defense response that could be responsible for limiting P. sojae infection. In contrast, ROS levels and callose deposition in soybean cells infected by ST28 and ST108 were quantitatively similar to those of the T26 and P6497 lines, suggesting that the loss of virulence of ST28 and ST108 is not associated with an obviously elevated plant defense response.
RXLR Effectors of P. sojae Can Suppress the XEG1-Induced Cell Death and PAMP-Triggered Immunity
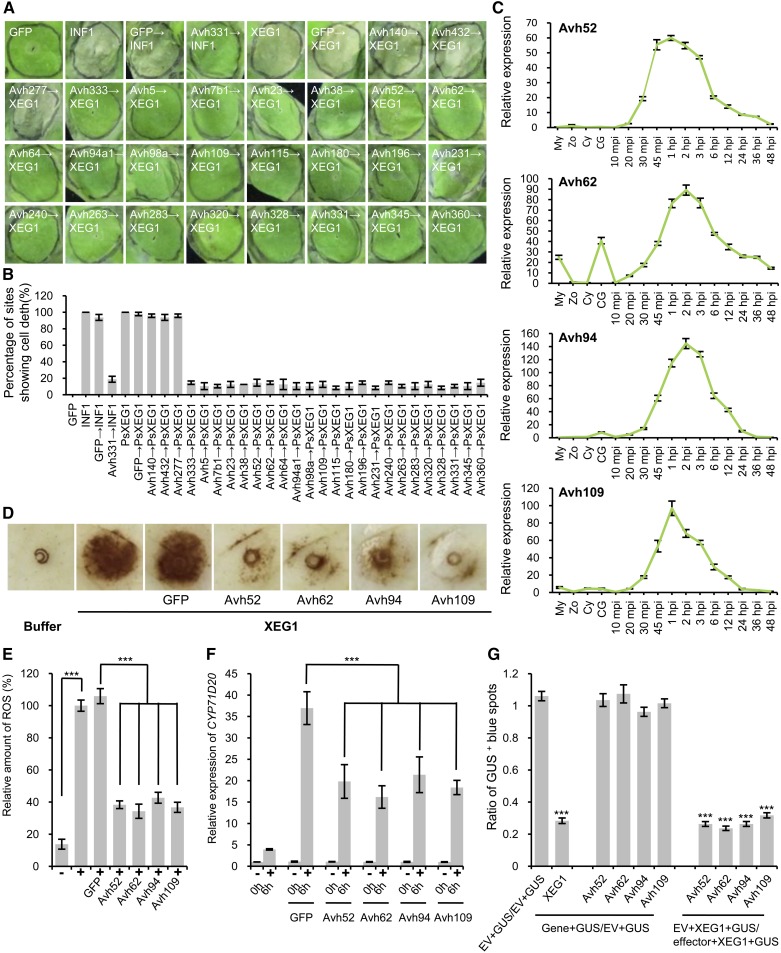
Although PAMPs can trigger strong defense responses when overexpressed in plants, many P. sojae RXLR effectors can suppress immune responses including PAMP-triggered cell death (Wang et al., 2011). Since P. sojae can successfully infect soybean, despite producing a PAMP (XEG1) that can trigger effective defense responses, we hypothesized that the defense responses triggered by XEG1, including cell death, might be suppressed by P. sojae RXLR effectors. To test this hypothesis, we selected a set of RXLR effectors previously shown to suppress cell death induced by the PAMP INF1 and by other effectors (Avh241 or Avh238) (Wang et al., 2011). We expressed XEG1 in N. benthamiana leaves 12 h after expressing the RXLR effectors or GFP (negative control) using agroinfiltration. All of the RXLR effectors that could suppress INF1-triggered cell death also suppressed cell death triggered by PsXEG1 (Figures 9A and 9B). Three RXLR effectors, Avh140, Avh432, and Avh277, which suppressed cell death induced by effectors, but not by INF1 (Wang et al., 2011), did not suppress PCD triggered by XEG1 (Figures 9A and 9B). These results suggested that cell death triggered by XEG1 could be effectively suppressed by P. sojae effectors in N. benthamiana. Several of the RXLR effectors, especially Avh52, Avh62, Avh94, and Avh109, were strongly expressed during very early infection, with similar dynamics to XEG1 (Figure 9C).
Figure 9.
Suppression of XEG1-Induced Cell Death and PTI by P. sojae RXLR Effectors.
(A) and (B) Suppression of PsXEG1-induced cell death in N. benthamiana leaves by agroinfiltration of P. sojae RXLR effector constructs 12 h beforehand. Experiments were performed three times with 16 sites for each RXLR effector or GFP and assessed 5 d after PsXEG1 agroinfiltration.
(A) Representative photographs.
(B) Mean percentages of cell death sites in infiltrated leaves with standard errors.
(C) Transcription patterns of four strongly expressed P. sojae RXLR effector genes measured as in Figure 9A, with zoospore levels set to 1.0. Bars represent standard errors from three independent biological replicates.
(D) and (E) P. sojae RXLR effectors can suppress XEG1-induced ROS in N. benthamiana. P. sojae RXLR effectors (Avh52, Avh62, Avh94, or Avh109) were transiently expressed in N. benthamiana leaves by agroinfiltration 48 h before infiltration of 300 nM XEG1 protein (+). GFP was used as an agroinfiltration control and buffer (−) was used as protein infiltration control. The infiltrated sites were stained with DAB.
(D) Representative photographs.
(E) Quantification of the DAB staining using ImageJ software. Means and standard errors from five independent replicates are shown. Significant differences based on Student’s t test are indicated by the asterisks (***P ≤ 0.001).
(F) Transcript level of CYP71D20 induced by 300 nM XEG1 or buffer in Agrobacterium-infiltrated N. benthamiana leaf tissue expressing GFP, Avh52, Avh62, Avh94, or Avh109, respectively. CYP71D20 gene expression was assessed at time 0 and 6 h after treatment (+, XEG1; −, buffer) using RT-qPCR. EF1α was used as an endogenous control. Means and standard errors from five independent replicates are shown. Significant differences based on Student’s t test are indicated by the asterisks (***P ≤ 0.001).
(G) Suppression of XEG1-triggered cell death in soybean by Avh52, Avh62, Avh94, or Avh109. Double-barreled particle bombardment of soybean Williams leaves with a GUS reporter gene was used to assay cell death. Leaves bombarded on one side with GUS plus empty vector (EV) and the other side with GUS plus DNA encoding XEG1 or effectors were used to test if XEG1 or effectors could induce cell death. The ratio of Gene+GUS/EV+GUS was measured to assess induction of cell death. The ratio of EV+GUS/EV+GUS was used as the control. A ratio similar to 1.0 indicates expression of the corresponding gene could not induce cell death, while a ratio significantly less than 1.0 indicates expression of the corresponding gene could induce cell death. The ratio of EV+XEG1+GUS/effector+XEG1+GUS was measured to assess the ability of effectors to suppress XEG1-induced cell death; a ratio similar to 1.0 indicates the corresponding effector could not suppress XEG1-induced cell death, while a ratio significantly less than 1.0, indicates the corresponding effector could suppress XEG1-induced cell death. Bars represent standard errors from four independent biological replicates. Significant differences based on Student’s t test are indicated by the asterisks (***P ≤ 0.001).
To further assess whether XEG1-induced defense responses other than cell death could also be suppressed by P. sojae effectors, we measured whether oxidative burst production and defense gene induction triggered by XEG1 could be suppressed in N. benthamiana by transiently expressing Avh52, Avh62, Avh94, or Avh109. We infiltrated XEG1rec into N. benthamiana leaves 48 h after expressing each RXLR effector or GFP (negative control) using agroinfiltration. Detection of ROS by staining of leaves 6 h after infiltration demonstrated that Avh52, Avh62, Avh94, and Avh109 suppressed XEG1-induced ROS compared with GFP (Figures 9D and 9E). Large sets of plant genes are upregulated in response to biotic stresses (Zipfel et al., 2006; Zhang et al., 2014). The tobacco Cyp71D20 gene, which serves as marker gene for responses to the flg22 N-terminal epitope of the flagellin protein was upregulated in N. benthamiana within 30 min of treatment with flg22 (Navarro et al., 2004). The expression level of this gene was determined in XEG1-treated N. benthamiana leaves transiently expressing Avh52, Avh62, Avh94, or Avh109. RT-qPCR analysis revealed that all four RXLR effectors could reduce the induction of CYP71D20 by XEG1 (Figure 9F). These results indicated that in addition to cell death, PAMP-triggered immunity (PTI) responses induced by XEG1 could also be suppressed by P. sojae RXLR effectors.
To validate that Avh52, Avh62, Avh94, and Avh109 could suppress XEG1-triggered cell death in soybean as well as in N. benthamiana, we used double-barrel particle bombardment experiments to test if coexpression of the effectors could suppress cell death triggered by XEG1. By testing the number of surviving GUS-positive blue patches, we inferred that Avh52, Avh62, Avh94, and Avh109 could all suppress the cell death triggered by XEG1 in soybean (Figure 9G; Supplemental Figure 4). Thus, the immune responses induced by XEG1 could be effectively suppressed by P. sojae effectors in both N. benthamiana and soybean.
DISCUSSION
In plant-pathogen interactions, the apoplastic space represents a complex battlefield involving many kinds of interactions in which attacking enzymes and counteracting inhibitors play an important role in determining the overall outcome of infection (Juge, 2006; Misas-Villamil and van der Hoorn, 2008; Doehlemann and Hemetsberger, 2013). Here, we identified a participant in this battlefield, the GH12 glycosyl hydrolase XEG1, which acts as a key virulence determinant of P. sojae but is at the same time recognized as a PAMP by the host plant soybean. The defense response triggered by XEG1 has the potential to block infection of soybean by P. sojae; however, numerous RXLR effectors secreted by P. sojae have the ability to suppress the XEG1-triggered defense response.
We initially identified XEG1 as a component of a protein fraction of the P. sojae culture supernatant that exhibited elicitor activity. XEG1 produced in planta has the ability to trigger cell death in N. benthamiana, while XEG1 purified from P. pastoris could trigger cell death in N. benthamiana, tomato, pepper, and soybean, but not in maize or cotton. XEG1 was identified as a GH12 glycosyl hydrolase with activity against both xyloglucans and β-glucans. Amino acid substitutions in two conserved glutamate residues produced the expected loss of xyloglucanase activity. The hydrolase activity was not required for the ability to trigger cell death. Triggering of cell death in N. benthamiana required secretion of XEG1 and also required BAK1 (Serk3), suggesting that XEG1 detection may be mediated by a cell surface pattern recognition receptor. XEG1 could still trigger cell death in Eix2-silenced tomato and in N. benthamiana, indicating that XEG1 functions independently of the perception system for fungal xylanases such as EIX. Silencing of XEG1 greatly reduced the virulence of P. sojae transformants. However, overexpression of XEG1 impaired P. sojae virulence even more strongly; soybean plants inoculated with these transformants exhibited a much more vigorous oxidative burst and callose deposition than plants inoculated with wild-type or XEG1-silenced P. sojae lines. Although XEG1 is normally induced strongly very early in infection (reaching a maximum level by 1 h), P. sojae produces many RXLR effectors that can suppress the XEG1 response. Several of those RXLR effectors are also expressed at very high levels during early infection stages with similar dynamics to XEG1, which may enable the pathogen to quickly suppress the plant’s responses to the rising levels of XEG1. On the other hand, when XEG1 was constitutively expressed in transformants, and thus prior to induction of expression of the RXLR effectors, infection failed.
Xyloglucan is an abundant hemicellulose in primary cell walls (Cosgrove, 2005). To penetrate and colonize plants, pathogens secrete a diverse array of CWDEs, including pectinases, polygalacturonases, cellulases, and hemicellulases such as xylanases (Walton, 1994; Juge, 2006). Plant pathogens also produce these enzymes to obtain nutrients from the cell wall hydrolysate (An et al., 2005; Hématy et al., 2009). Xylanases catalyze the initial breakdown of the xylan backbone of hemicelluloses (Collins et al., 2005).
Xylanases have been reported as virulence factors in several plant pathogens. For example, deletion of the xyn11A gene had a marked effect on the ability of B. cinerea to infect tomato leaves and grape berries (Brito et al., 2006), and a mutation in the xynB endoxylanase gene partially affected the virulence of X. oryzae pv oryzae (Rajeshwari et al., 2005). As in these examples from fungi and bacteria, XEG1 is an important virulence factor in P. sojae, as evidenced by its strong expression during infection and the severe effect of silencing the gene. Although XEG1 could trigger cell death, it was most strongly expressed very early during the biotrophic stage of infection when the pathogen actively suppresses cell death, with a strong peak at 1 h. Since P. sojae does not begin to form abundant haustoria until 2 to 4 h of infection (Enkerli et al., 1997), the very strong and early expression of XEG1 may indicate that it is a critical source of nutrition prior to the development of haustoria, in addition to the more obvious role of physically facilitating invasion of the host tissue.
Among plant-associated microbes, GH12 proteins are particularly abundant in Phytophthora species. Except for the three regions of sequence conservation, around the active site residues and at the N terminus, the Phytophthora GH12 sequences showed extensive diversification. Most subfamilies of oomycete GH12 sequences did not show consistent patterns of orthologs across Phytophthora species. For example, the XEG1 subfamily contained two members from P. sojae and P. parasitica, and one from Phytophthora capsici, but no members from Phytophthora infestans or H. arabidopsidis. Thus, the entire family of Phytophthora GH12 proteins may be under diversifying selective pressure.
The detection of PAMPs by pattern recognition receptors to trigger PTI is a major component of plant defense responses. A variety of proteins exhibiting hydrolytic activity, as well as various cell wall constituents that originate from host plants, have been identified as PAMPs (Beliën et al., 2006). For example, the EIX protein from Trichoderma viride induced a variety of plant defense responses, and the response was specific to particular plant species and/or varieties (Ron and Avni, 2004; Beliën et al., 2006). Fungal PGs can also be recognized as PAMPs by plants (Nürnberger et al., 2004; Zhang et al., 2014). Oligogalacturonides, which are released by degradation of pectin by PGs, also activate a variety of defense responses (Prade et al., 1999; D’Ovidio et al., 2004).
Our finding that not only XEG1, but also at least half of the GH12 proteins from oomycetes, and several from fungi, could trigger cell death in N. benthamiana suggests that this protein family represents a major new class of PAMPs. The ability to recognize XEG1 resulting in cell death was found in soybean, tomato, pepper, and N. benthamiana, but not in cotton or maize. Many other PAMPs exhibit restricted host ranges. For example, Arabidopsis and other Brassicaceae recognize EF-Tu peptides, but Nicotiana tabacum, sunflower (Helianthus annuus), soybean, and Medicago sativa do not (Kunze et al., 2004). Phytophthora elicitins are only recognized by Nicotiana species and some Raphanus sativus and Brassica campestris cultivars (Kamoun et al., 1993). Since we did not screen our collection of GH12 proteins for the ability to trigger non-cell-death PAMP responses, the number of GH12 proteins acting as PAMPs may be even larger than we have enumerated here. Similarly, the range of plant species responding to GH12 proteins may be larger than we detected. XEG1 does not require hydrolase activity to act as a PAMP, but we cannot rule out that other GH12 family members may act by releasing xyloglucan hydrolysis products; the ability of plants to detect these carbohydrate fragments needs further investigation.
The importance of XEG1 recognition by soybean is underlined by our observations that XEG1rec could protect etiolated soybean hypocotyls against subsequent inoculation with P. sojae and that constitutive overexpression of XEG1 severely limited the virulence of P. sojae transformants on soybean. Overexpression of XEG1 in P. sojae transformants resulted in the plants producing abundant ROS and callose at the site of infection. The constitutive level of expression of XEG1 in the transformants (20- to 65-fold over the levels in mycelia) was not markedly higher than the normal induction level of the gene during early infection (15- to 300-fold over the levels in mycelia and 10- to 85-fold over the levels in zoospores). However, it is much higher than the normal level at the immediate outset of infection (8- to 65-fold in mycelia or zoospores of the transformants compared with P6497). Therefore, we hypothesize that premature expression of XEG1 in these transformants resulted in an unusually fast and vigorous PAMP-triggered immune response that the pathogen was unable to overcome.
Despite the ability of plants such as soybean to recognize and respond to XEG1 and its homologs in other oomycete species, these pathogens have clearly evolved mechanisms to surmount the plants’ defense responses. Interference with PTI by effectors produced by pathogens has been reported in many systems (Jones and Dangl, 2006). P. sojae secretes a complex repertoire of RXLR effector proteins to manipulate host innate immunity, and most of the RXLR effectors expressed early during infection by P. sojae could suppress cell death induced in N. benthamiana by the PAMP INF1 (Wang et al., 2011). We showed here that 23 RXLR effectors encoded in the P. sojae genome could suppress cell death triggered by XEG1 in N. benthamiana. All of these effectors could also suppress cell death triggered by BAX, INF1, and two P. sojae effectors, suggesting that they all act on a common pathway that leads to cell death and defense. Several of these effectors, particularly Avh62, Avh52, Avh94, and Avh109, are expressed strongly during very early infection and show similar expression dynamics as XEG1, suggesting that these effectors may act preemptively to block the plants’ ability to respond to XEG1 and other PAMPs. Evidently, however, these effectors are unable to suppress PTI when XEG1 is already highly expressed at the moment of contact with the plant, prior to induction of the RXLR effectors, as in the overexpression transformants. The plants’ ability to respond to the presence of XEG1 thus places an upper limit on the level of expression of this virulence protein by the pathogen. It will be interesting to determine whether soybean and other plants produce specific inhibitors of GH12 hydrolases. The production of GH12 inhibitors would tend to place a lower limit on the level of GH12 hydrolases that would be effective in promoting virulence, thus tightly constraining the level of pathogen expression of these enzymes.
As an essential virulence factor, XEG1 functions for the pathogen as an apoplastic effector. Since it is also recognized by the plant as a PAMP, XEG1 further blurs the distinction between PTI and effector-triggered immunity (Thomma et al., 2011). XEG1 adds to a small but growing list of apoplastic effectors that, like XEG1, resemble PAMPs as they are widely distributed across diverse tax and/or are recognized by plasma membrane receptors. Other examples include necrosis- and ethylene-inducing peptide (Nep1)-like protein (NLP) toxins that distributed among bacteria, fungi, and oomycetes (Gijzen and Nürnberger, 2006) but also recognized as PAMPs (Böhm et al., 2014; Oome et al., 2014); fungal xylanases such as EIX recognized by tomato Eix1 and Eix2 (Ron and Avni, 2004); fungal PGs recognized by Arabidopsis RLP42 (Zhang et al., 2014); fungal LysM effectors such as Cladosporium Ecp6 (Thomma et al., 2011); and Avr3 from Fusarium oxysporum f.s. lycopersici that is required for virulence and is recognized by RLK I-3 (Catanzariti et al., 2015). Together with a substantial list of R genes that encode membrane receptors such as the tomato Cf genes (Joosten and de Wit, 1999) and Ve-1 (Fradin et al., 2009), rice (Oryza sativa) Pi-d2 (Chen et al., 2006) and Xa-21 (Song et al., 1995), canola (Brassica napus) Rlm2 (Larkan et al., 2015) and LepR3 (Larkan et al., 2013), and barley (Hordeum vulgare) Rpg1 (Brueggeman et al., 2002), PAMP-like effectors such as XEG1 suggest that apoplastic versus cytoplasmic immunity may represent a more natural distinction than the one between PTI and ETI.
METHODS
Microbial Cultures
Phytophthora sojae strains (P6497 and transformants) and Phytophthora parasitica strains (Pp025) were maintained on 2.5% vegetable (V8) juice medium at 25°C in the dark (Qutob et al., 2002). P. sojae mycelia and zoospores were prepared as previously described (Hua et al., 2008).
Preparation and Purification of Proteins Secreted from P. sojae
To produce large quantities of secreted proteins, P. sojae was cultivated in synthetic liquid medium (Kamoun et al., 1993). Ten-day-old P. sojae culture supernatants were collected and clarified by filtration through a 0.22-μm polyvinylidene fluoride membrane (Millipore). Proteins were precipitated overnight at 0°C by addition of 70 g (NH4)2SO4 per 100 mL culture filtrate. The precipitate was collected by centrifugation at 12,000g for 10 min at 4°C and then resuspended in 10 mM Tris-HCl (pH 7.5) and 1 mM EDTA (TE). The resuspended proteins were fractionated by gel permeation chromatography on Sephacryl S-300 in TE (GE Healthcare). Fractions corresponding to absorbance peaks at OD280 were collected. Selected fractions were further purified by ion-exchange chromatography on a Mono Q 5/50 GL column (GE Healthcare) equilibrated with TE and eluted with a linear sodium chloride gradient from 0.0 to 1.0 M in TE. Fractions corresponding to absorbance peaks were desalted and concentrated using 30-kD molecular mass centrifugal filter devices (Millipore). Fractions with cell death activity were analyzed by liquid chromatography-tandem mass spectrometry as described (Heese et al., 2007).
Immunoblotting
Protein extractions and immunoblots were performed as previously described (Yu et al., 2012). P. sojae transformants were assessed using an anti-His6-tag primary monoclonal antibody (Sigma-Aldrich). Transient protein expression in Nicotiana benthamiana was assessed using anti-HA-tag or anti-GFP tag primary monoclonal antibody (Sigma-Aldrich).
Plasmid Construction and Preparation
Plasmids used were constructed using standard techniques (Sambrook et al., 1989). We amplified full-length GH12 gene sequences from genomic DNA or from the cDNA of various species, depending on predicted introns, as listed in Supplemental Data Set 1, using high-fidelity PrimeSTAR HS DNA Polymerase (TaKaRa Bio) and using the primers and restriction enzymes listed in Supplemental Data Set 3. Full-length GH12 sequences were cloned into the PVX vector pGR107 (Jones et al., 1999). The INF1 gene was amplified from Phytophthora infestans isolate 88069 genomic DNA (Kamoun et al., 1998) and cloned into pGR107.
For XEG1 expression and purification, the sequence encoding mature XEG1 protein without the putative signal peptide was cloned into pPICZαA. For transient expression in N. benthamiana, sequences encoding a PR1 signal peptide-XEG1 fusion protein and XEG1 deletion mutants were cloned into pGR107. For particle bombardment assays, full-length XEG1 was cloned into pFF19. Constructs used for VIGS of BAK1 were generated in the pTRV2 vector as described (Heese et al., 2007). For P. sojae stable transformation, full-length XEG1 with a 6xHis tag fused to the C-terminal end was inserted in the forward direction into the pTOR vector for XEG1 overexpression, and full-length XEG1 was inserted in the reverse direction into the pTOR vector for XEG1 silencing (Whisson et al., 2007). All of the RXLR genes used to assay suppression of the cell death induced by XEG1 were cloned into pGR107 as described (Wang et al., 2011). All of the oligonucleotides used for plasmid construction are documented in Supplemental Table 2 and Supplemental Data Set 3. PCR products were digested with the appropriate enzymes, as listed in Supplemental Table 2. The constructs were further confirmed by sequencing (Invitrogen).
Expression and Purification of Recombinant XEG1 Protein
Pichia pastoris KM71H (Muts) (Invitrogen) was used as the expression host. Yeast cultures were maintained on yeast extract-peptone-dextrose (YEPD) medium. For growth and induction, BMGY (buffered glycerol-complex medium) and BMMY (buffered methanol-complex medium), respectively, were used, both at pH 6.5 (EasySelectPichia expression kit; Invitrogen). P. pastoris transformants were screened for protein induction in 24-well plates as described (Boettner et al., 2002). Induction of protein expression was performed according to the manufacturer’s instructions. Purification of recombinant XEG1 protein from the culture supernatant was performed by affinity chromatography using Ni-NTA Superflow resin (Qiagen).
Bioinformatic Analysis
The Conserved Domain Database was searched using XEG1 protein sequences, resulting in matches to the GH12 domain (pfam01670). XEG1 sequences lacking the signal peptide sequence were used as queries to search against the protein sequence databases listed in Supplemental Data Set 1 using reciprocal BLAST. After correction for redundancy, signal peptide prediction, and functional prediction, 68 protein sequences were identified (Supplemental Data Set 1). Signal peptide prediction was performed using the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/). The putative GH12 protein sequences were further confirmed using annotations in the NCBI database. The mature protein sequences were aligned using the ClustalW2 program, and phylogenetic dendrograms were constructed using PhyML implemented in SeaView (http://doua.prabi.fr/software/seaview) (Gouy et al., 2010) and graphically viewed using MEGA 5 (www.megasoftware.net) with maximum likelihood (Tamura et al., 2011).
VIGS Assays in N. benthamiana and Tomato
For VIGS assays, pTRV1, pTRV2:BAK1, or pTRV2:Eix2 plasmid constructs were introduced into Agrobacterium tumefaciens GV3101 by electroporation. The cultured Agrobacterium cells were harvested and resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES, pH 5.7, and 150 mM acetosyringone) to an OD of 0.4 and left at room temperature for 2 h. Agrobacterium strains harboring pTRV2-BAK1 or pTRV2:Eix2 vector combining with that harboring pTRV1 vector were mixed in a 1:1 ratio. The cocultures were then infiltrated into two primary leaves of a plant at the four-leaf stage. The effectiveness of the VIGS assay was evaluated using the PDS gene as described (Liu et al., 2002). The silencing efficiency of BAK1 or Eix2 was validated using RT-qPCR analysis of RNA from the leaves in the locations corresponding to the albino leaves.
Particle Bombardment Assays
Particle bombardment assays were performed using a double-barreled extension of the Bio-Rad He/1000 particle delivery system (Dou et al., 2008; Song et al., 2013). To quantitate induction of cell death by XEG1, construct carrying this gene (1.7 µg/shot) was cobombarded into soybean leaves with construct carrying a GUS reporter gene (1.7 µg/shot). The EV construct was cobombarded with the GUS reporter gene construct as a control. To test the cell death induction ability induced by XEG1 or effectors, for each pair of shots, the ratio of the number of blue spots produced by the PsXEG1 construct to the number of spots produced by the EV control construct was calculated. To test the suppression of XEG1-induced cell death by effector, the ratio of the number of blue spots produced by the XEG1 and EV control construct to the number of spots produced by the XEG1 and effector construct was calculated. Each assay consisted of six pairs of shots and was conducted three times. Statistical analysis employed the Wilcoxon rank sum test (Dou et al., 2008).
P. sojae Transformation and Virulence Assays in Soybean Seedlings
Gene-silenced and overexpression P. sojae transformants were generated using polyethylene glycol-mediated protoplast transformation (Dou et al., 2008). Putative transformants were screened by RT-qPCR to identify silenced or overexpression lines. Overexpression lines were confirmed by immunoblot. The pathogenicity phenotypes of P. sojae transformants were determined by inoculation of hypocotyls of etiolated soybean seedlings. Approximately 100 zoospores of each transformant and the wild-type strain were inoculated onto the hypocotyls of etiolated soybean seedlings (susceptible Williams cultivar). Disease symptoms were scored 48 h after infection. At 6 h after infection, the etiolated soybean seedlings were stained using 3,3′-diaminobenzidine (DAB) or aniline blue as described (Dong et al., 2011). At least three replications were performed for infected seedlings of each transformant. ROS accumulation and callose deposits were counted using Image J software. Significant differences were identified using Student’s t test.
Agrobacterium and Protein Infiltration Assays
Agrobacterium-mediated transient expression was performed as described (Wang et al., 2011). To assay suppression of XEG1-triggered cell death by RXLR effectors, Agrobacterium cells carrying each effector gene were infiltrated into leaves. The same infiltration site was challenged 12 h later with Agrobacterium carrying XEG1. Agrobacterium strains carrying XEG1 or eGFP alone were infiltrated in parallel as controls. Leaves were scored and photographed 6 d after initial inoculation. Each assay consisted of at least three plants inoculated on three leaves on at least two different dates.
To test the induction of cell death by recombinant proteins produced in P. pastoris, 300 pM to 3 μM protein solution were infiltrated into leaves of N. benthamiana, and 300 nM solutions of the proteins were infiltrated into leaves of soybean (Glycine max), pepper (Capsicum annum), tomato (Solanum lycopersicum; M82), cotton (Gossypium hirsutum; Xumian21), or maize (Zea mays; Zhongdan909). Leaves were photographed at 2, 2, 7, 8, 8, and 8 days postinfiltration, respectively. Each assay consisted of three or four plants inoculated on two or three leaves on at least two different dates. As a control, culture supernatants from empty vector control strains of P. pastoris were processed for protein purification.
To test the suppression of P. parasitica infection by XEG1, infiltrated leaves were detached 24 h after infiltration, maintained on half-strength Murashige and Skoog medium in a Petri dish, and then inoculated on the infiltrated regions with 100 P. parasitica strain 025 zoospores. Disease lesions were photographed, and the diameters were measured at 48 hpi.
To test the induction of plant defense-related gene expression by XEG1, RNA was extracted from transgenic N. benthamiana leaves expressing the constructs pBinPlus:Avh52, pBinPlus:Avh62, pBinPlus:Avh94, pBinPlus:Avh109, or pBinPlus:GFP at 0 and 6 h after infiltration with XEG1. SYBR green qRT-PCR assays were performed to determine CYP71D20 expression levels. Three independent biological replicates were conducted for each experiment.
Enzyme Activity Assays
Enzyme activities were determined as described (Grishutin et al., 2004) using xyloglucan and β-glucan (Megazyme) as substrates. Assays were performed in 96-well microplates.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: PsXEG1 (EGZ16757.1), Ps109281 (EGZ11358.1), Ps137930 (EGZ16993.1), and Ps159318 (EGZ09324.1). Accession numbers for GH12 genes from different microbes used in this article are listed in Supplemental Data Set 1.
Supplemental Data
Supplemental Figure 1. SDS-PAGE of Ps109681rec from P. pastoris stained with Coomassie blue.
Supplemental Figure 2. SDS-PAGE analysis of E136Drec and E222Drec protein stained with Coomassie blue.
Supplemental Figure 3. Immunoblot analysis of GFP, INF1, and XEG1 wild-type and mutant proteins produced in N. benthamiana during the experiments shown in Figure 6D.
Supplemental Figure 4. Representative soybean leaves for suppression of XEG1-triggered cell death in soybean by Avh52, Avh62, Avh94, or Avh109.
Supplemental Figure 5. Immunoblot analysis of GFP, Avh52, Avh62, Avh94, and Avh109 produced in N. benthamiana during the experiments shown in Figure 8.
Supplemental Figure 6. Classification of P. sojae effectors based on suppression of cell death triggered by BAX, RXLR effectors, or the PAMPs, INF1 and XEG1.
Supplemental Figure 7. ClustalW alignment of 68 GH12 protein sequences.
Supplemental Table 1. Identification of the proteins found in peak 3 (following ultrafiltration) by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Supplemental Table 2. List of primers used in this study.
Supplemental Table 3. Plasmid constructs used in the study.
Supplemental Data Set 1. List of all candidate GH12 genes from the genomes of various species.
Supplemental Data Set 2. Text file of alignment corresponding to the phylogenetic analysis in Figure 4.
Supplemental Data Set 3. GH12 proteins tested for induction of cell death in N. benthamiana by agroinfiltration.
Supplementary Material
Acknowledgments
We thank Francine Govers (Wageningen University) and Wenbo Ma (University of California, Riverside) for helpful suggestions. This work was supported in part by grants to Y.W. from China National Funds for Distinguished Young Scientists (31225022), Key Program of National Natural Science Foundation of China (31430073), Special Fund for Agro-scientific Research in the Public Interest (201303018), and the China Agriculture Research System (CARS-004-PS14), by a 111 International Cooperation grant (B07030) to Nanjing Agricultural University from the Chinese government, and by grants to B.M.T. from the National Research Initiative of the USDA National Institute of Food and Agriculture (Grants 2011-68004-30104 and 2010-65110-20764).
AUTHOR CONTRIBUTIONS
Y.C.W., B.M.T., and Z.M. designed the research. Z.M., T.S., and L.Z. performed the research. S.D., W.Y., and Y.W. contributed new tools. Y.C.W., B.M.T., Z.M., T.S., D.D., Z.Z., and X.Z. analyzed the data. B.M.T., Y.C.W., and Z.M. wrote the article. All authors commented on the article before submission.
Glossary
- PAMP
pathogen-associated molecular pattern
- CWDE
cell wall-degrading enzyme
- PG
endopolygalacturonase
- EIX
ethylene-inducing xylanase
- VIGS
virus-induced gene silencing
- RT-qPCR
reverse transcription-quantitative PCR
- qRT-PCR
quantitative RT-PCR
- ROS
reactive oxygen species
- hpi
hours postinoculation
- PTI
PAMP-triggered immunity
- DAB
3,3′-diaminobenzidine
Footnotes
Articles can be viewed online without a subscription.
References
- An H.J., Lurie S., Greve L.C., Rosenquist D., Kirmiz C., Labavitch J.M., Lebrilla C.B. (2005). Determination of pathogen-related enzyme action by mass spectrometry analysis of pectin breakdown products of plant cell walls. Anal. Biochem. 338: 71–82. [DOI] [PubMed] [Google Scholar]
- Beliën T., Van Campenhout S., Robben J., Volckaert G. (2006). Microbial endoxylanases: effective weapons to breach the plant cell-wall barrier or, rather, triggers of plant defense systems? Mol. Plant Microbe Interact. 19: 1072–1081. [DOI] [PubMed] [Google Scholar]
- Ben-Daniel B.H., Bar-Zvi D., Tsror Lahkim L. (2012). Pectate lyase affects pathogenicity in natural isolates of Colletotrichum coccodes and in pelA gene-disrupted and gene-overexpressing mutant lines. Mol. Plant Pathol. 13: 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner M., Prinz B., Holz C., Stahl U., Lang C. (2002). High-throughput screening for expression of heterologous proteins in the yeast Pichia pastoris. J. Biotechnol. 99: 51–62. [DOI] [PubMed] [Google Scholar]
- Böhm H., Albert I., Oome S., Raaymakers T.M., Van den Ackerveken G., Nürnberger T. (2014). A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PLoS Pathog. 10: e1004491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito N., Espino J.J., González C. (2006). The endo-beta-1,4-xylanase xyn11A is required for virulence in Botrytis cinerea. Mol. Plant Microbe Interact. 19: 25–32. [DOI] [PubMed] [Google Scholar]
- Brueggeman R., Rostoks N., Kudrna D., Kilian A., Han F., Chen J., Druka A., Steffenson B., Kleinhofs A. (2002). The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc. Natl. Acad. Sci. USA 99: 9328–9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner F., Rosahl S., Lee J., Rudd J.J., Geiler C., Kauppinen S., Rasmussen G., Scheel D., Nürnberger T. (2002). Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases. EMBO J. 21: 6681–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti A.M., Lim G.T., Jones D.A. (2015). The tomato I-3 gene: a novel gene for resistance to Fusarium wilt disease. New Phytol. 207: 106–118. [DOI] [PubMed] [Google Scholar]
- Chen X., et al. (2006). A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 46: 794–804. [DOI] [PubMed] [Google Scholar]
- Collins T., Gerday C., Feller G. (2005). Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 29: 3–23. [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6: 850–861. [DOI] [PubMed] [Google Scholar]
- Doehlemann G., Hemetsberger C. (2013). Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytol. 198: 1001–1016. [DOI] [PubMed] [Google Scholar]
- Dong S., et al. (2011). Phytophthora sojae avirulence effector Avr3b is a secreted NADH and ADP-ribose pyrophosphorylase that modulates plant immunity. PLoS Pathog. 7: e1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou D., et al. (2008). Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell 20: 1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ovidio R., Mattei B., Roberti S., Bellincampi D. (2004). Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant-pathogen interactions. Biochim. Biophys. Acta 1696: 237–244. [DOI] [PubMed] [Google Scholar]
- Enkerli J., Felix G., Boller T. (1999). The enzymatic activity of fungal xylanase is not necessary for its elicitor activity. Plant Physiol. 121: 391–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkerli K., Mims C.W., Hahn M.G. (1997). Ultrastructure of compatible and incompatible interactions of soybean roots infected with the plant pathogenic oomycete Phytophthora sojae. Can. J. Bot. 75: 1493–1508. [Google Scholar]
- Fradin E.F., Zhang Z., Juarez Ayala J.C., Castroverde C.D., Nazar R.N., Robb J., Liu C.M., Thomma B.P. (2009). Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150: 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulin E., et al. (2006). Cellulose binding domains of a Phytophthora cell wall protein are novel pathogen-associated molecular patterns. Plant Cell 18: 1766–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijzen M., Nürnberger T. (2006). Nep1-like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry 67: 1800–1807. [DOI] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. (2010). SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Grishutin S.G., Gusakov A.V., Markov A.V., Ustinov B.B., Semenova M.V., Sinitsyn A.P. (2004). Specific xyloglucanases as a new class of polysaccharide-degrading enzymes. Biochim. Biophys. Acta 1674: 268–281. [DOI] [PubMed] [Google Scholar]
- Heese A., Hann D.R., Gimenez-Ibanez S., Jones A.M.E., He K., Li J., Schroeder J.I., Peck S.C., Rathjen J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104: 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hématy K., Cherk C., Somerville S. (2009). Host-pathogen warfare at the plant cell wall. Curr. Opin. Plant Biol. 12: 406–413. [DOI] [PubMed] [Google Scholar]
- Hua C., Wang Y., Zheng X., Dou D., Zhang Z., Govers F., Wang Y. (2008). A Phytophthora sojae G-protein α subunit is involved in chemotaxis to soybean isoflavones. Eukaryot. Cell 7: 2133–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Jones L., Hamilton A.J., Voinnet O., Thomas C.L., Maule A.J., Baulcombe D.C. (1999). RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 11: 2291–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten M., de Wit P. (1999). The tomato-Cladosporium fulvum interaction: A versatile experimental system to study plant-pathogen interactions. Annu. Rev. Phytopathol. 37: 335–367. [DOI] [PubMed] [Google Scholar]
- Juge N. (2006). Plant protein inhibitors of cell wall degrading enzymes. Trends Plant Sci. 11: 359–367. [DOI] [PubMed] [Google Scholar]
- Kamoun S., van West P., Vleeshouwers V.G., de Groot K.E., Govers F. (1998). Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell 10: 1413–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S., Young M., Glascock C.B., Tyler B.M. (1993). Extracellular protein elicitors from Phytophthora: host-specificity and induction of resistance to bacterial and fungal phytopathogens. Mol. Plant Microbe Interact. 6: 15–25. [Google Scholar]
- Karlsson J., Siika-aho M., Tenkanen M., Tjerneld F. (2002). Enzymatic properties of the low molecular mass endoglucanases Cel12A (EG III) and Cel45A (EG V) of Trichoderma reesei. J. Biotechnol. 99: 63–78. [DOI] [PubMed] [Google Scholar]
- Kikot G.E., Hours R.A., Alconada T.M. (2009). Contribution of cell wall degrading enzymes to pathogenesis of Fusarium graminearum: a review. J. Basic Microbiol. 49: 231–241. [DOI] [PubMed] [Google Scholar]
- Klarzynski O., Plesse B., Joubert J.M., Yvin J.C., Kopp M., Kloareg B., Fritig B. (2000). Linear beta-1,3 glucans are elicitors of defense responses in tobacco. Plant Physiol. 124: 1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline J.N. (2009). Pathogen recognition and new insights into innate immunity. In Allergy Frontiers: Classification and Pathomechanisms, R. Pawankar, S.T. Holgate, and L.J. Rosenwasser, eds (Springer), pp. 19–30. [Google Scholar]
- Kunze G., Zipfel C., Robatzek S., Niehaus K., Boller T., Felix G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkan N.J., Lydiate D.J., Parkin I.A., Nelson M.N., Epp D.J., Cowling W.A., Rimmer S.R., Borhan M.H. (2013). The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 197: 595–605. [DOI] [PubMed] [Google Scholar]
- Larkan N.J., Ma L., Borhan M.H. (2015). The Brassica napus receptor-like protein RLM2 is encoded by a second allele of the LepR3/Rlm2 blackleg resistance locus. Plant Biotechnol. J., in press [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Marathe R., Dinesh-Kumar S.P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30: 415–429. [DOI] [PubMed] [Google Scholar]
- Master E.R., Zheng Y., Storms R., Tsang A., Powlowski J. (2008). A xyloglucan-specific family 12 glycosyl hydrolase from Aspergillus niger: recombinant expression, purification and characterization. Biochem. J. 411: 161–170. [DOI] [PubMed] [Google Scholar]
- Medzhitov R., Janeway C.A. Jr. (1997). Innate immunity: the virtues of a nonclonal system of recognition. Cell 91: 295–298. [DOI] [PubMed] [Google Scholar]
- Misas-Villamil J.C., van der Hoorn R.A. (2008). Enzyme-inhibitor interactions at the plant-pathogen interface. Curr. Opin. Plant Biol. 11: 380–388. [DOI] [PubMed] [Google Scholar]
- Morrissey J.P., Osbourn A.E. (1999). Fungal resistance to plant antibiotics as a mechanism of pathogenesis. Microbiol. Mol. Biol. Rev. 63: 708–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger T., Brunner F. (2002). Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 5: 318–324. [DOI] [PubMed] [Google Scholar]
- Navarro L., Zipfel C., Rowland O., Keller I., Robatzek S., Boller T., Jones J.D. (2004). The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135: 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M.-A., Sundelin T., Nielsen J.T., Erbs G. (2013). MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front. Plant Sci. 4: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger T., Brunner F., Kemmerling B., Piater L. (2004). Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198: 249–266. [DOI] [PubMed] [Google Scholar]
- Oome S., Raaymakers T.M., Cabral A., Samwel S., Böhm H., Albert I., Nürnberger T., Van den Ackerveken G. (2014). Nep1-like proteins from three kingdoms of life act as a microbe-associated molecular pattern in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 16955–16960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prade R.A., Zhan D., Ayoubi P., Mort A.J. (1999). Pectins, pectinases and plant-microbe interactions. Biotechnol. Genet. Eng. Rev. 16: 361–391. [DOI] [PubMed] [Google Scholar]
- Qutob D., Kamoun S., Gijzen M. (2002). Expression of a Phytophthora sojae necrosis-inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 32: 361–373. [DOI] [PubMed] [Google Scholar]
- Rajeshwari R., Jha G., Sonti R.V. (2005). Role of an in planta-expressed xylanase of Xanthomonas oryzae pv. oryzae in promoting virulence on rice. Mol. Plant Microbe Interact. 18: 830–837. [DOI] [PubMed] [Google Scholar]
- Ron M., Avni A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16: 1604–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald P.C., Beutler B. (2010). Plant and animal sensors of conserved microbial signatures. Science 330: 1061–1064. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Sandgren M., Shaw A., Ropp T.H., Wu S., Bott R., Cameron A.D., Ståhlberg J., Mitchinson C., Jones T.A. (2001). The X-ray crystal structure of the Trichoderma reesei family 12 endoglucanase 3, Cel12A, at 1.9 A resolution. J. Mol. Biol. 308: 295–310. [DOI] [PubMed] [Google Scholar]
- Sandgren M., Ståhlberg J., Mitchinson C. (2005). Structural and biochemical studies of GH family 12 cellulases: improved thermal stability, and ligand complexes. Prog. Biophys. Mol. Biol. 89: 246–291. [DOI] [PubMed] [Google Scholar]
- Sharp J.K., Valent B., Albersheim P. (1984). Purification and partial characterization of a beta-glucan fragment that elicits phytoalexin accumulation in soybean. J. Biol. Chem. 259: 11312–11320. [PubMed] [Google Scholar]
- Song T., Kale S.D., Arredondo F.D., Shen D., Su L., Liu L., Wu Y., Wang Y., Dou D., Tyler B.M. (2013). Two RxLR avirulence genes in Phytophthora sojae determine soybean Rps1k-mediated disease resistance. Mol. Plant Microbe Interact. 26: 711–720. [DOI] [PubMed] [Google Scholar]
- Song W.Y., Wang G.L., Chen L.L., Kim H.S., Pi L.Y., Holsten T., Gardner J., Wang B., Zhai W.X., Zhu L.H., Fauquet C., Ronald P. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804–1806. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B.P., Nürnberger T., Joosten M.H. (2011). Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23: 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton J.D. (1994). Deconstructing the cell wall. Plant Physiol. 104: 1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., et al. (2011). Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 23: 2064–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson S.C., et al. (2007). A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450: 115–118. [DOI] [PubMed] [Google Scholar]
- Yakoby N., Beno-Moualem D., Keen N.T., Dinoor A., Pines O., Prusky D. (2001). Colletotrichum gloeosporioides pelB is an important virulence factor in avocado fruit-fungus interaction. Mol. Plant Microbe Interact. 14: 988–995. [DOI] [PubMed] [Google Scholar]
- Yu L.M. (1995). Elicitins from Phytophthora and basic resistance in tobacco. Proc. Natl. Acad. Sci. USA 92: 4088–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Tang J., Wang Q., Ye W., Tao K., Duan S., Lu C., Yang X., Dong S., Zheng X., Wang Y. (2012). The RxLR effector Avh241 from Phytophthora sojae requires plasma membrane localization to induce plant cell death. New Phytol. 196: 247–260. [DOI] [PubMed] [Google Scholar]
- Zhang L., Kars I., Essenstam B., Liebrand T.W., Wagemakers L., Elberse J., Tagkalaki P., Tjoitang D., van den Ackerveken G., van Kan J.A. (2014). Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the Arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 164: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.