Lampard, G.R., Wengier, D.L., and Bergmann, D.C. (2014). Manipulation of mitogen-activated protein kinase kinase signaling in the Arabidopsis stomatal lineage reveals motifs that contribute to protein localization and signaling specificity. Plant Cell 26: 3358–3371.
The image in Figure 6E was inadvertently duplicated from Figure 6D. The corrected version of Figure 6 is shown below, wherein panel 6E has been replaced with the correct image representing SPCHp:CAMKK5ΔNINT. The figure legend and remaining figure panels are unaltered from the original.
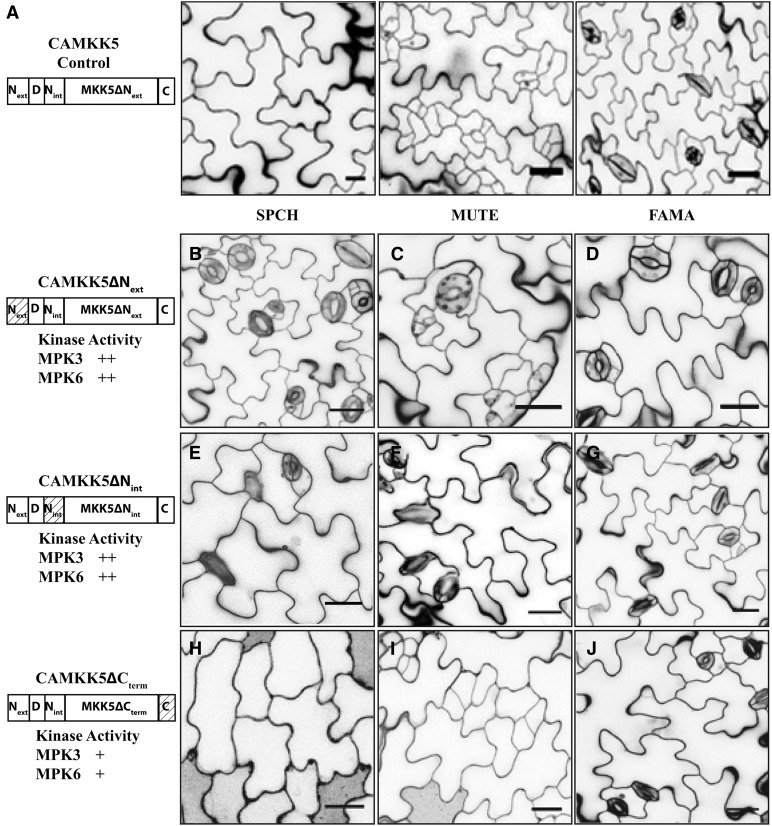
Figure 6.
N-Terminal Extensions in CAMKK4/5 Promote Stomatal Inhibition in the SPCH and MUTE Stages.
(A) Protein schematic (left) and images of phenotypes associated with expressing CAMKK5 in the SPCH, MUTE, and FAMA stages of stomatal development as indicated.
(B) to (D) The 10-DPG seedlings expressing CAMKK5∆Next in the SPCH (B), MUTE (C), and FAMA (D) stages show wild-type stomatal development patterns.
(E) to (G) The 10-DPG seedlings expressing CAMKK5∆Nint in the SPCH (E), MUTE (F), and FAMA (G) stages show wild-type stomatal development patterns. Thus, both Next and Nint have a positive function in inhibiting stomatal development but do not prevent CAMKK4/5 from inducing stomatal clustering in the FAMA stage.
(H) to (J) The 10-DPG seedlings expressing CAMKK5∆Cterm in the SPCH (H), MUTE (I), and FAMA (J) stages show the same stomatal development patterns as controls (A).
The relative activity of each kinase variant in in vitro kinase assays is depicted as follows (+, less active than full length; ++, comparable activity to the full-length CAMKK). Hashed boxes in schematic representations of kinase variants indicate the region(s) that were removed. All images are of 10-DPG abaxial cotyledons, and bars = 25 μm.
Editor’s note: the corrected figure and accompanying text were reviewed by members of The Plant Cell editorial board.