Abstract
Background
Bacterial populations communicate through the cell density-dependent mechanism of quorum sensing (QS). Vibrio harveyi, one of the best studied model organisms for QS, was used to explore effects of the synthetic cannabinoid HU-210 on QS and different QS-regulated physiological processes in bacteria.
Results
Analysis of QS-regulated bioluminescence in wild-type and mutant strains of V. harveyi revealed that HU-210 affects the autoinducer-2 (AI-2) pathway, one of three known QS cascades of V. harveyi. Furthermore, QS-mediated biofilm formation and swimming motility in the mutant strain BB152 (AI-1−, AI-2+) were significantly reduced in the presence of HU-210. HU-210 inhibited QS-mediated virulence factor production without any inhibitory effect on bacterial growth. It also alters the expression of several genes, which are regulated by QS, specifically downregulating the genes of the AI-2 QS cascade.
Conclusion
First evidence is being provided for interference of bacterial signal-transduction systems by a synthetic cannabinoid. The effect of HU-210 was specific to the AI-2 cascade in V. harveyi. AI-2 is known as a "universal autoinducer" and interference with its activity opens a broad spectrum of applications for synthetic cannabinoids in future research as a potential anti-QS agent.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-015-0499-0) contains supplementary material, which is available to authorized users.
Background
Bacteria communicate and coordinate population behavior through the mechanism of quorum sensing (QS), which controls the expression of genes that affect a variety of bacterial processes [1, 2]. QS is based on small signaling molecules, termed autoinducers (AIs), which control factors such as bioluminescence, pigment production, motility and biofilm formation, among many others [3–5]. The QS, free-living marine bacterium Vibrio harveyi produces and responds to at least three distinct AIs [6]: HAI-1, an acyl homoserine lactone [7]; AI-2, a furanosyl-borate-diester [8]; and CAI-1, a long-chain amino ketone (Z)-3-aminoundec-2-en-4-one (Ea-C8-CAI-1) [9]. AI-2 is referred to "universal autoinducer" as it is found in numerous Gram-positive and Gram-negative bacteria [10, 11]. A better understanding of QS actions and novel ways to affect them is providing an opportunity to manipulate bacterial properties that are of major importance in industry, agriculture and medicine.
Plant and endogenous cannabinoids have been investigated extensively, particularly as agonists of the endocannabinoid signaling system and as therapeutically active substances [12, 13]. Both delta-9-tetrahydrocannabinol (THC), the major psychoactive constituent of cannabis [14] and cannabidiol (CBD), a non-psychoactive constituent, alter immune functions, mostly via suppression. The mechanism governing these activities has not been fully elucidated (for a recent review see Cabral et al. [15]). In most investigations, these phytocannabinoids have been found to enhance the susceptibility of mice to microorganisms (both bacteria and viruses). However, several reports have indicated that administration of THC leads to lower viral loads and moderation of disease progression [15].
The effect of cannabinoids on bacterial QS has never been investigated. We chose to study the activity of the synthetic cannabinoid HU-210 (Fig. 1), because its physical and chemical properties have been well defined. HU-210 was first synthesized from (1R,5S)-myrtenol [16]. It displays a multiplicity of biochemical, pharmacological, and behavioral effects, most of which depend on its potent agonistic activity at the cannabinoid receptors CB1 and CB2 [17].
Fig. 1.
Chemical structure of the synthetic cannabinoid HU-210, used in this study
Results
Effect of HU-210 on V. harveyi bioluminescence
To investigate the effect of HU-210 on the QS cascade of V. harveyi wild-type strain BB120 and QS-mutant strains (Table 1), bioluminescence was measured with and without HU-210 in the growth media. First, we determined whether addition of HU-210 to the growth media alters the bioluminescence of the wild-type V. harveyi strain. Results showed no significant difference in bioluminescence between samples with and without HU-210 (Fig. 2a). The experiments were repeated with other V. harveyi strains mutated in the QS cascade. Addition of HU-210 to the growth media resulted in an up to 98 % decrease in the bioluminescence of V. harveyi mutant BB152 (AI-1−, AI-2+), in a dose-dependent manner, whereas bioluminescence of the mutant strain MM30 (AI-1+, AI-2−) was not significantly affected (Fig. 2a). However growth of V. harveyi mutant BB152 (Fig. 2b) and other mutant strains (data not shown) was not affected by HU-210. Results indicated that a notable effect of HU-210 was specific to AI-2 in the QS cascade of V. harveyi, as this AI regulates QS of V. harveyi mutant strain BB152.
Table 1.
V. harveyi strains used in this study
| V. harveyi strain | Relative genotype | Relative phenotype |
|---|---|---|
| BB120 | Wild type | AI-1+, AI-2+; sensor-1+, sensor-2+ |
| BB152 | (BB7 or BB120) luxLM::Tn5 | AI-1−, AI-2+; sensor-1+, sensor-2+ |
| MM30 | (BB120) luxS::Tn5 | AI-1+, AI-2−; sensor-1+, sensor-2+ |
| MM77 | luxLM::Tn5, luxS::Tn5 | AI-1−, AI-2−; sensor-1+, sensor-2+ |
| BB170 | luxN::Tn5 | AI-1+, AI-2+; sensor-1−, sensor-2+ |
Fig. 2.
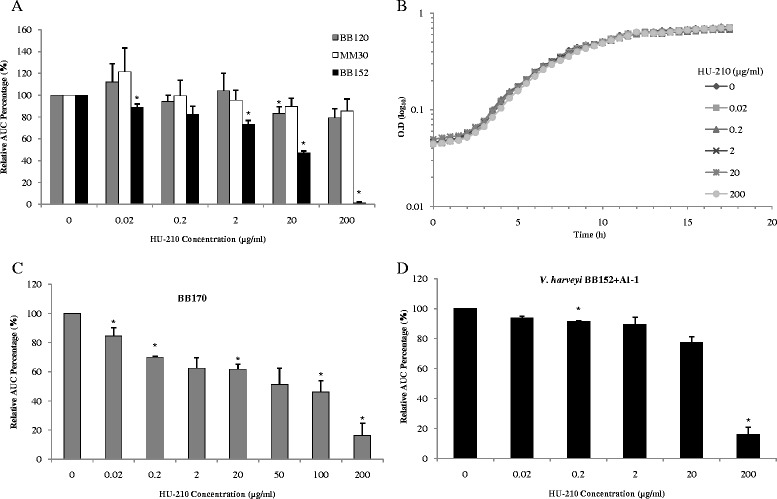
Effect of HU-210 on growth and bioluminescence production of V. harveyi. (a) Comparison of relative bioluminescence production by V. harveyi BB120 (wild type), MM30 (AI-1+, AI-2−) and BB152 (AI-1−, AI-2+) with different HU-210 concentrations, presented as area under the curve. (b) Growth curve of V. harveyi BB152 with different HU-210 concentrations. (c) Relative bioluminescence production by V. harveyi BB170 (Sensor-1−, Sensor-2+) with different HU-210 concentrations. (d) Relative bioluminescence production by V. harveyi BB152 with different HU-210 concentrations and simultaneous supplementation with exogenous AI-1 isolated from V. harveyi MM30 (AI-1+, AI-2−), presented as area under the curve. Presented data are means and SD of three independent experiments, each performed in triplicate.*P < 0.05 compared with control
To further analyze the mode of action of HU-210, another mutant strain, BB170 (Sensor-1−, Sensor-2+) was tested. Bioluminescence in V. harveyi BB170 is mainly controlled by AI-2, as this strain is not responsive to AI-1 stimulation due to the absence of sensor-1 [18]. In the presence of HU-210, bioluminescence production by mutant strain BB170 was decreased by up to 85 %, further confirming the specific action of HU-210 on AI-2 (Fig. 2c). To validate the molecular QS target of HU-210, another experiment was designed in which mutant strain MM77 (AI-1−, AI-2−), in the absence or presence of different doses of HU-210, was simultaneously incubated with 10 % (v/v) spent medium containing exogenous AI-1 and AI-2 isolated from V. harveyi MM30 (AI-1+, AI-2−) and BB152 (AI-1−,AI-2+) respectively. The inhibitory effect of HU-210 on bioluminescence production was more pronounced in samples to which exogenous AI-2 had been added (~95 % decrease), suggesting that the effect of HU-210 is not only on the production of AI-2 signal, but also on the reception of those signal molecules by hybrid sensor kinases present in mutant strain MM77 (Additional file 1: Figure S1).
Interestingly when mutant strain BB152 (AI-1−, AI-2+) was supplemented with exogenous AI-1 from MM30 (AI-1+, AI-2−) and simultaneously treated with different concentrations of HU-210, the latter's dose-dependent inhibitory effect could be reversed and bioluminescence level of this mutant strain restored to the similar level of the wild-type BB120, except in sample with highest HU-210 concentration (200 μg/ml) (Fig. 2d).
Effect of HU-210 on V. harveyi biofilm biomass and DNA quantity
The effect of HU-210 on other QS-controlled processes in V. harveyi, such as biofilm formation, was also tested. Biofilms were grown with and without different concentrations of HU-210 using different V. harveyi strains (Table 1). The biofilm biomass formed by the different strains was quantified using crystal violet (CV) staining. The effect of HU-210 on biofilm biomass formed by wild-type BB120 and mutant strain MM30 was minimal, as no significant decrease was observed with increasing concentrations of HU-210 (Additional file 1: Figure S2). Consistent with these results, an up to 80 % decrease was observed in biofilm biomass of mutant strain BB152 (AI-1−, AI-2+). Inhibition of biofilm biomass formation by HU-210 in mutant BB152 was found to be dose-dependent (Additional file 1: Figure S2).
This anti-biofilm effect was further confirmed using a DNA-quantification method which specifically quantified the DNA present in different V. harveyi biofilms. DNA quantification results showed the same trend of inhibitory action for HU-210: biofilm in the mutant strain BB152 was dose-dependently inhibited by up to 80 % (Fig. 3), whereas there was no significant decrease in the amount of DNA present in wild-type BB120 and mutant MM30 biofilms.
Fig. 3.
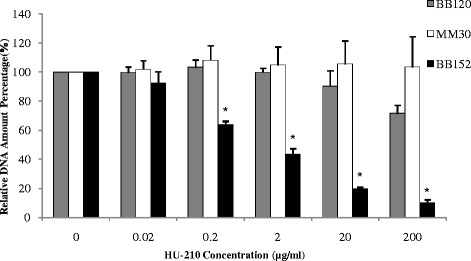
DNA quantification by qPCR. Comparison of quantified DNA of V. harveyi BB120 (wild type), MM30 (AI-1+, AI-2−) and BB152 (AI-1−, AI-2+) biofilm formed with or without different concentrations of HU-210. Presented data are means and SD of three independent experiments, each performed in triplicate.*P < 0.05 compared with control
Biofilm depth, bacterial viability and presence of extracellular polysaccharides (EPS)
To further evaluate the effect of HU-210 on biofilm formation by V. harveyi, we analyzed the morphology of the biofilms generated by the V. harveyi wild type BB120, and mutants MM30 (AI-1+, AI-2−) and BB152 (AI-1−, AI-2+) using confocal laser scanning microscopy (CLSM). The depth of the biofilms formed by the wild type and mutant strain MM30 was least affected by the addition of HU-210. However biofilms formed by mutant BB152 in control samples (without HU-210) were more confluent (95 μm) than those formed in the presence of HU-210 (55 μm), and showed a gradual decrease in average depth with increasing concentrations of HU-210 (Fig. 4a & b).
Fig. 4.
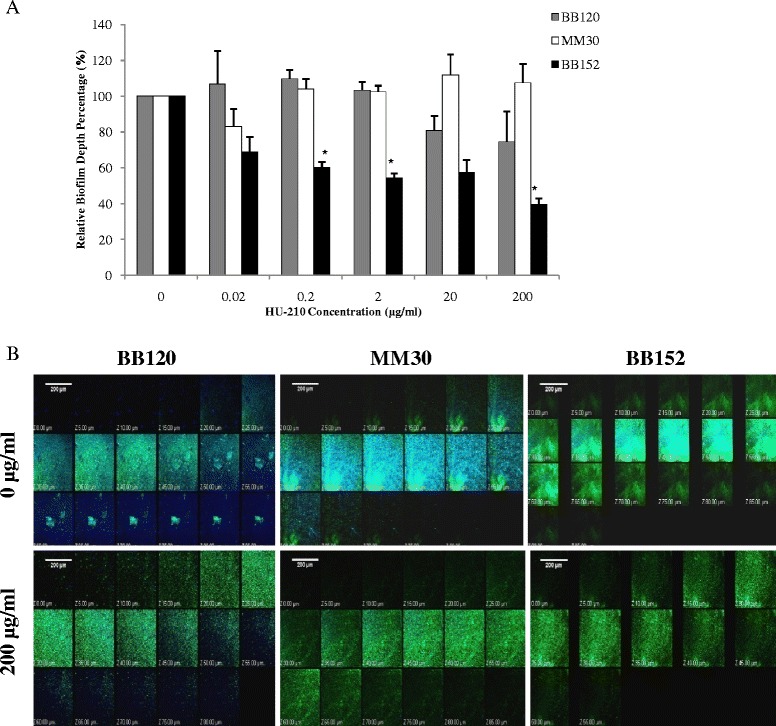
Analysis of V. harveyi biofilms by CLSM. (a) Comparison of biofilm depth of V. harveyi BB120 (wild type), MM30 (AI-1+, AI-2−) and BB152 (AI-1−, AI-2+) formed with or without different concentrations of HU-210. Presented data are means and SD of three independent experiments.*P < 0.05 compared with control. (b) V. harveyi BB120 (wild type), MM30 (AI-1+, AI-2−) and BB152 (AI-1−, AI-2+) biofilms grown with or without HU-210 (200 μg/ml) and stained with SYTO 9 dye (green), which stains live bacteria, and with concanavalin A–Alexa Fluor 647 conjugates (blue), which stain extracellular polysaccharides. Biofilm depth of V. harveyi BB120 (wild type), MM30 (AI-1+, AI-2−) and BB152 (AI-1−, AI-2+) biofilms in control (without HU-210) is 85 μm, 80 μm ,95 μm and in samples with HU-210 (200 μg/ml) is 80 μm, 85 μm, 55 μm respectively
The amount of EPS was normalized to live cells in the biofilm, and we observed less EPS in biofilms formed by all tested V. harveyi strains in the presence of HU-210 vs. controls. However, the decrease was only significant for mutant strain BB152, where up to 75 % less EPS was measured in the presence of 200 μg/ml HU-210 vs. controls. Bacterial cell viability in the biofilms was not significantly affected by HU-210 in any of the tested V. harveyi strains (Table 2).
Table 2.
Analysis of biofilms formed by V. harveyi strains in the presence/absence of HU-210
| V. harveyi strains | Flourescence Intensity | |
|---|---|---|
| with/without HU-210 | Live/Dead | Relative EPS/Live Percentage |
| BB120 | 2.40 ± 1.23 | 100 ± 0 |
| BB120 + 200 μg/ml HU-210 | 2.80 ± 1.13 | 45.98 ± 8.71 |
| MM30 | 1.60 ± 0.99 | 100 ± 0 |
| MM30 + 200 μg/ml HU-210 | 2.81 ± 1.15 | 88.75 ± 2.94 |
| BB152 | 1.89 ± 0.69 | 100 ± 0 |
| BB152 + 200 μg/ml HU-210 | 3.63 ± 0.03 | 25.50 ± 4.50* |
Based on data obtained from CLSM. Fluorescence intensity was measured using Image J software. Presented
data are means and SD of three independent experiments.*P < 0.05
Effect of HU-210 on swimming motility of V. harveyi
Swimming motility assays were performed with wild-type strain BB120 and mutant strains MM30 and BB152. A significant decrease (P < 0.05) in swimming motility was observed in all three strains in the presence of the highest tested concentration of HU-210 (200 μg/ml). This effect of HU-210 was most evident in mutant strain BB152 (AI-1−, AI-2+) as it interfered with swimming motility at lower concentrations as well (above 2 μg/ml) (Fig. 5). In both the wild-type strain BB120 and the mutant strain BB152, the observed reduction in swimming motility was dose-dependent (up to 80 % and 85 %, respectively), whereas in mutant strain MM30, up to 95 % inhibition was observed at the highest HU-210 concentration tested (200 μg/ml).
Fig. 5.
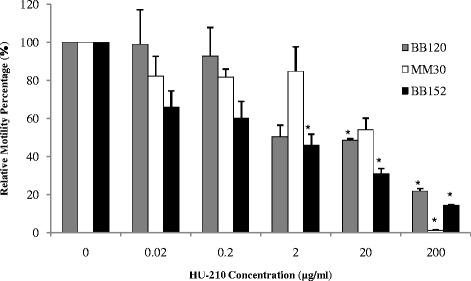
Comparison of swimming motility of V. harveyi BB120 (wild type), MM30 (AI-1+, AI-2−) and BB152 (AI-1−, AI-2+) with different concentrations of HU-210 quantified using Image J Software. Presented data are means and SD of two independent experiments (*P < 0.05)
Effect of HU-210 on mRNA levels of different genes involved in QS regulation
The molecular basis of the inhibitory action of HU-210 on QS and biofilm formation in V. harveyi mutant strain BB152 was investigated. We quantified mRNA levels of six genes which had been previously found to be regulated by the AI-1- and AI-2-dependent QS in Vibrio. Genes specific to the AI-2 QS cascade—luxS, luxP and luxQ—were downregulated in the presence of 2 μg/ml HU-210 in wild-type BB120 and mutant BB152, whereas genes specific to the AI-1 cascade—luxM and luxN—were upregulated in wild-type BB120 and mutant strain MM30. Significant downregulation of AI-2 synthase luxS and master regulator luxR was only observed in mutant strain BB152 treated with HU-210 (Table 3). Results suggested that HU-210 can inhibit QS through the AI-2 cascade by inhibiting the production of AI-2 signal molecules through luxS. Furthermore, decreased expression of luxP and luxQ in mutant strain BB152 also suggested that the mechanism of action proceeds through these hybrid sensor kinases. As an outcome, there was less expression of luxR—the master regulator of QS target genes—resulting in suppressed activity of the AI-2-mediated QS cascade present in mutant strain BB152. This could also explain the specific inhibitory effect of HU-210 on bioluminescence, biofilm-forming ability and swimming motility of mutant strain BB152 observed throughout the study.
Table 3.
Relative mRNA expression of different genes in V. harveyi
| Relative mRNA expression | BB120 | MM30 | BB152 |
|---|---|---|---|
| with HU-210 (2 μg/ml) | RQ value | ||
| luxS | 0.81 ± 0.03 | - | 0.42 ± 0.05* |
| luxP | 0.98 ± 0.45 | - | 0.37 ± 0.25 |
| luxQ | 1.1 ± 0.65 | - | 0.36 ± 0.11 |
| luxM | 3.05 ± 0.46 | 4.04 ± 1.42 | - |
| luxN | 1.31 ± 0.31 | 2.34 ± 0.87 | - |
| luxR | 0.45 ± 0.12 | 1.34 ± 0.19 | 0.43 ± 0.11* |
Relative expression of genes was measured using real-time RT-PCR as described in Materials and Methods. Fold change of the transcriptional level was calculated using ABI-Prism 7300 software v1.1 with RQ study 1.0, Applied Biosystems. The expression results represent mean ± SD of two independent experiments. *P < 0.05
Discussion
Bacteria communicate via QS using extracellular chemical signaling molecules. AI-2 used for QS signaling is found in both Gram-positive and Gram-negative bacteria. Recent studies have focused on finding small molecules analogous to AIs which can alter QS-regulated pathogenic behaviors and biofilm formation in bacteria [19]. The action of cannabinoids on microbes and viruses has been documented [15]. However their effect on bacterial QS pathways is unknown.
V. harveyi is one of the best-characterized model organisms for the study of QS [20, 21]. Therefore, we investigated the effect of the potent synthetic cannabinoid HU-210 on QS-regulated bioluminescence, biofilm formation and swimming motility of V. harveyi strains. Further research on cannabinoid–bacteria interactions will involve exploring more derivatives and their effects on other types of bacteria.
Our study focused mainly on the effect of HU-210 on QS-mediated pathways at concentrations below the minimum inhibitory concentration (MIC). Growth of V. harveyi strains was unaffected at all tested concentrations (0.2–200 μg/ml) of HU-210. In experiments with V. harveyi wild-type strain BB120 and mutant strains BB152 and MM30, HU-210 only significantly reduced bioluminescence of the mutant strain BB152. Strain BB152 is a HAI-1 synthase mutant and inhibition of its bioluminescence by HU-210 led us to postulate that this synthetic cannabinoid acts on the AI-2 mediated QS cascade. Using the bioluminescence reporter strain BB170 (Sensor-1−, Sensor-2+), we were able to address the specific inhibitory action of HU-210 on AI-2. The mutant BB152 (AI-1−, AI-2+) was incubated simultaneously with different doses of HU-210 and exogenous AI-1 and assessed for further effects of HU-210. Interestingly, exogenous AI-1 reversed HU-210 inhibition, indicating complementation by exogenous AI-1 for the dose-dependent effect of HU-210, except in samples with the highest tested HU-210 concentration (200 μg/ml). This could be because at the highest concentration (200 μg/ml), HU-210 strongly inhibited AI-2 activity (~95 % decrease) in BB152 and the observed bioluminescence production was due only to the excess exogenous AI-1 available at the time.
The plant kingdom is a well-known source of medicines, including numerous antimicrobial agents. Several other quorum-quenching molecules have been isolated from plants [22]. For example, the plant constituent curcumin was recently shown to interfere with QS-mediated bioluminescence production in V. harveyi [23]. Similarly, extracts of garlic have been shown to block QS of Pseudomonas aeruginosa [24], and vanilla extracts are capable of interfering with QS in Chromobacterium violaceum [25].
Previous studies have confirmed that biofilm formation is one of the primary attributes toward bacterial pathogenicity and that it is partially controlled by QS [26]. Results of the present study suggest that HU-210 at the tested concentrations significantly inhibited the biofilm-forming capability of V. harveyi mutant strain BB152 (AI-1−, AI-2+). The amount of total biomass in the biofilm, determined by CV staining and DNA quantification, decreased with increasing concentration of HU-210. Since the inhibitory action of HU-210 was more prominent in the mutant strain BB152 (AI-1−, AI-2+) than in the other tested V. harveyi strains, HU-210 seems to interfere with the AI-2-dependent QS pathway, resulting in less QS and ultimately less biofilm formation. Results of the current study are consistent with previous observations made in Vibrio spp. wherein a pytochemical, resveratrol significantly reduced total biofilm bimoass [27].
The anti-biofilm effect of HU-210 was further confirmed by CLSM observation of biofilm architecture, showing a significant decrease in the depth of the biofilm formed by mutant BB152; however, there was no such effect on biofilm formed by wild-type strain BB120 or mutant strain MM30. The average thickness of the biofilm formed by these strains remained almost the same in HU-210-treated and untreated samples. The substantial reduction in biofilm thickness for strain BB152 was comparable to the result of a study in which S6-15, an extract from the marine bacterium Bacillus pumilus, was shown to be effective against biofilm formation in a wide range of Gram-positive and Gram-negative bacteria [28].
Bacterial motility is an important virulence factor in many pathogens. Some Vibrio spp. (including V. harveyi) possess dual flagellar systems that are suited to movement under different conditions [29]. A recent study reported that QS enhances flagellar motility in pathogenic V. harveyi [30]. To further investigate the effect of HU-210 on QS, we checked its possible impact on swimming motility of V. harveyi. The results indicated an inhibitory effect of HU-210 on QS via its restrictive effect on the motility of the wild type as well as mutant strains BB152 and MM30. Observations from this study fall in line with another study in which curcumin was shown to affect swimming motility of V. harveyi [23].
Biofilm formation is a developmental process due to changes in the structural and regulatory genes required at various steps of its formation [31–33]. To unveil the molecular basis of V. harveyi BB152 bioluminescence and biofilm inhibition by HU-210, we measured differential expression of genes regulating QS-phenotypes in the presence and absence of HU-210. In V. harveyi, the AI-2-mediated QS system is composed of LuxS, LuxP and LuxQ. LuxS is responsible for the synthesis of AI-2 [8]. LuxP is a periplasmic protein that binds AI-2, and LuxQ is a hybrid sensor kinase. Lux R is the master regulator of all three parallel QS systems in V. harveyi [34]. We observed less expression of luxS, luxP, luxQ and luxR in mutant strain BB152 treated with HU-210, suggesting that this cannabinoid interferes with AI-2 production, as well as with reception of these QS signals by sensor kinases, resulting in lower activity of LuxR and less activation of QS target genes. This notion is in agreement with several other reports showing disruption of QS-regulated gene expression in V. harveyi by halogenated furanone [35, 36].
It has been reported that three QS systems work in parallel, based on a coincidence detection scheme, to regulate different genes' expression in V. harveyi [4]. We noticed that luxM (AI-1 producer) and luxN (hybrid sensor kinase which perceives AI-1) in wild-type BB120 as well as in mutant strain MM30 were upregulated in the presence of 2 μg/ml HU-210. It has been recently shown that in a growing wild-type V. harveyi culture, AI-2 synthesis is quickly followed by that of HAI-1 [21]. Based on this knowledge, it becomes evident that the observed limited effect of HU-210 on wild-type BB120 bioluminescence and biofilm formation is a result of the coincidence detection scheme in which AI-1 of BB120 (AI-1+, AI-2+) complements the QS system when AI-2 activity is interfered with by HU-210.
The present study is the first report on the effect of a cannabinoid on QS-mediated processes, namely bioluminescence, biofilm formation and swimming motility of a bacterial species. The inhibitory action of HU-210 was found to proceed through AI-2 and it was the result of altered expression of genes involved in the AI-2 QS cascade. AI-2 is known as the "universal autoinducer" and interference in its activity opens a broad spectrum of applications for synthetic cannabinoids in future research with prokaryotes. Further investigations need to be carried out to establish the molecular aspects and mode of action of cannabinoids' effects on QS.
Conclusions
The synthetic cannabinoid HU-210 has an inhibitory effect on QS and QS-dependent properties, such as bioluminescence, biofilm formation and swimming motility of V. harveyi, without affecting its growth. Furthermore, HU-210's effect was found to proceed through the AI-2-mediated QS cascade. Results of the present study suggest a novel mode of action for synthetic cannabinoids on bacteria via the bacterial communication system, and potential use of cannabinoids as anti-QS agents.
Methods
Bacterial strains and growth
V. harveyi wild-type and mutant strains (Table 1) were grown aerobically overnight at 30 °C in AB-medium. Bacterial strains were kindly provided by B. Bassler (Princeton University).
HU-210 preparation
HU-210 was synthesized as described previously [37]: m.p. 140–1 °C (from pentane), [α]D -227 (CHCl3), δ (CDCl3) 6.40, 6.25 (2 arom H's), 5.6 (C = CH), 4.09 (CH2–O). Stock solutions were prepared by dissolving 5 mg HU-210 in 250 μl of 100 % ethanol and stored at 4 °C until use. Working concentrations were prepared from the stock solutions by diluting with sterile AB-medium to final concentrations of 0.2–200 μg/ml.
MIC determination
V. harveyi wild-type strain BB120 was grown at 30 °C in the presence of HU-210 at concentrations ranging from 2–4 mg/ml in 96-well transparent plates (Nunc, Roskilde, Denmark). After 24 h of incubation, bacterial growth was determined by absorbance at 595 nm (OD595) (TECAN GENios reader, Schweiz, Austria). The MIC was recorded as the minimum concentration that exhibited visible growth inhibition and all further experiments were performed at sub-MIC [38].
Bioluminescence assay
QS regulates bioluminescence in V. harveyi [39]. For the bioluminescence assay, V. harveyi overnight culture (O.D595 ~ 0.7) was diluted 1:200 in fresh AB-medium along with different concentrations of HU-210. After dilution, 180 μl bacterial culture was transferred to 96-well white plates with an optic bottom (Nunc) and the readings were taken using the GENios reader at 30 °C. Luminescence was measured every 30 min in parallel with absorbance measurement (OD595) for 18 h. Relative luminescence unit (the amount of luminescence per unit of growth) was calculated as the quotient of luminescence value and growth for each well. The area under the curve was calculated for each sample, compared with a control sample (without HU-210) and reported as a percentage [38, 40].
Biofilm biomass quantification using the CV method
Biofilm biomass was quantified by CV staining as described previously [41] with slight modifications. Briefly, a 96-well polystyrene plate (Nunc) was seeded with 20 μl of V. harveyi overnight culture (O.D595 ~ 0.7) followed by addition of 160 μl AB-medium and 20 μl of HU-210 at different concentrations for biofilm growth. The plate was incubated for 24 h at 30 °C. Generated biofilms were washed carefully twice using 200 μl saline solution. The biofilms were stained with 200 μl of 0.1 % (w/v) CV (Merck, Darmstadt, Germany) solution for 20 min, then washed twice with saline and the stained biofilms were dried overnight at room temperature. Next, 33 % acetic acid was added to elute the CV for 60 min along with shaking at room temperature. The extract was transferred to a new 96-well plate and OD595 was measured using the GENios reader.
DNA quantification by quantitative (q) PCR
For biofilm growth, a polystyrene 48-well plate (Nunc) was seeded with 50 μl of V. harveyi overnight culture (O.D595 ~ 0.7) followed by addition of 400 μl AB-medium and 50 μl HU-210 at different concentrations. The plate was incubated for 24 h at 30 °C. Generated biofilms were washed carefully twice using 500 μl of saline solution.
DNA extraction and quantification were performed as described previously [42, 43]. Briefly, 160 μl NaOH (0.05 M) (Bio Lab Ltd., Jerusalem, Israel) and 40 μl DEPC-treated water (Bio Basic Canada Inc., Markham, Ontario, Canada) were added to each well. The plate was then immersed in a hot water bath for 1 h at 60 °C. Then, 18.5 μl Tris buffer (pH 7) (Eastman Kodak Company, Rochester, NY, USA) was added to each well and extracted DNA samples were stored at −20 °C until further use.
DNA samples from different V. harveyi biofilms were quantified by qPCR with specific primers for 16S rRNA using an ABI prism instrument (Applied Biosystems Prism 7300, Foster City, CA, USA). The amount of DNA was quantified according to the specific standard curve. Total genomic DNA was extracted from an overnight culture of V. harveyi BB120 using GenElute Bacterial Genomic DNA kit (Sigma Aldrich, St. Louis, MO, USA) as per the manufacturer’s instructions. Extracted genomic DNA for the standard curve analysis was stored at −20 °C.
Biofilm depth, cell viability and presence of EPS
Bacterial cell viability, presence of EPS and depth of the biofilms grown as described above were examined by CLSM (Olympus Fluoview 300, Tokyo, Japan) with a UPLSA 10X/0.4 lens. The biofilm was formed by seeding 20 μl V. harveyi overnight culture (O.D595 ~ 0.7), 160 μl AB- medium and 20 μl of HU-210 at different concentrations in a 96-well plate (Nunc). The plate was incubated for 24 h at 30 °C under aerobic conditions. Biofilms were then washed carefully with saline solution and stained for 20 min at room temperature with different stains. Biofilms were washed again with saline solution before visualization under the microscope.
To examine cell viability, biofilms were stained with LIVE/DEAD BacLight fluorescent dye (Life Technologies, Carlsbad, CA, USA). SYTO9 fluorescence was measured using 488 nm excitation and 515 nm emission filters, and propidium iodide (PI) fluorescence was measured using 543 nm excitation and 570 nm emission filters (Olympus). EPS were stained with concanavalin A–Alexa Fluor 647 conjugates (Invitrogen Molecular Probes, Carlsbad, CA, USA). The biofilm depth was determined by acquiring optical sections at 5-μm spacing using a previously described method [31]. Biofilm depth was examined at the center of each well and quantified as the distance between the highest and lowest section. Fluorescence intensity per unit area was calculated using Image J software (National Institutes of Health) for each color separately.
Swimming motility assay
The swimming motility assay was performed on soft agar plates using a previously described method [30] with some modifications. Briefly, AB-medium with 0.2 % (w/v) agar was prepared and autoclaved. After cooling to 60 °C, different concentrations of HU-210 were added and the medium was poured into small petri dishes. After solidification, 3 μl V. harveyi overnight culture (O.D595 ~ 0.5) was inoculated at the center of the agar. Agar plates without HU-210 served as controls. The plates were incubated at 30 °C for 15 h, then the area of the motility halo was measured using Image J software and compared with controls.
RNA extraction
V. harveyi wild-type BB120 and mutant strains MM30 and BB152 were grown overnight planktonically with and without (control) 2 μg/ml HU-210. V. harveyi overnight culture (200 μl; O.D595 ~ 0.7) was added in triplicate to a 12-well polystyrene plate (Nunc) along with 1.6 ml AB-medium and 200 μl HU-210 (2 μg/ml). The plate was incubated for 24 h at 30 °C under aerobic conditions. Following incubation, the bacterial culture in each well was mixed vigorously and transferred to 10-ml tubes. Further RNA extraction was performed using a previously described method [32]. Briefly, 2 ml of RNA Protect (Qiagen, Hilden, Germany) was added to each tube, mixed well and incubated at room temperature for 5 min. Cells were harvested and DNA-free RNA was isolated using the RNeasy MINI kit (Qiagen) according to the manufacturer’s instructions. RNA purity and quantity were determined using Nanodrop (Nanovue, GE Healthcare Life Sciences, Buckinghamshire, UK). RNA integrity was determined using a bioanalyzer (Agilent, Santa Clara, CA, USA) and the samples were stored at −80 °C for later use.
Reverse transcription (RT) and real-time PCR
Reverse transcription was performed with the SuperScript™ First-Strand Synthesis Kit (Invitrogen, Life Technologies) in accordance with the manufacturer’s instructions.
The synthesized cDNA was used for real-time PCR analysis with specific primers for the genes examined in this study. The RT-PCR was performed as described previously [44]. The PCR thermal profile included an initial denaturation at 95 °C for 10 min, followed by a 40-cycle amplification consisting of denaturation at 95 °C for 15 s and annealing and extension at 60 °C for 1 min.
As an additional control for each primer pair and each RNA sample, the cDNA synthesis reaction was performed in the absence of reverse transcriptase in order to check for contamination with residual genomic DNA. The expression levels of all genes tested by real-time PCR were normalized to the 16S rRNA gene of V. harveyi as an internal standard. There was no significant difference in the expression of the 16S rRNA gene for the two conditions and samples. Fold change of transcription level was calculated using ABI Prism 7300 SDS Software v1.1 with RQ Study 1.0 (Applied Biosystems) [43, 45].
Statistical analysis
Experiments were performed independently three times in triplicate and the data were analyzed statistically using Student's t test, with a P value of less than 0.05 considered significant.
Acknowledgements
We would like to thank members of the Steinberg laboratory, especially Danielle Assaf-Duanis, for helpful discussions.
Abbreviations
- QS
Quorum sensing
- AI
Autoinducer
- CB1
Cannabinoid receptor type-1
- CB2
Cannabinoid receptor type-2
- OD
Optical density
- RT
Reverse transcription
- PCR
Polymerase chain reaction
- qPCR
Quantitative real-time PCR
- CV
Crystal violet
Additional file
Comparison of relative bioluminescence production by V. harveyi MM77 (AI-1-, AI-2-, sensor-1+, sensor-2+) with different HU-210 concentrations when simultaneously supplemented with exogenous AI-1 and AI-2 from V. harveyi mutant strains MM30 (AI-1+, AI-2-) and BB152 (AI-1-, AI-2+) respectively, presented as area under the curve. Presented data are means and SD of three independent experiments, each performed in triplicate. *P < 0.05. Figure S2. Biofilm biomass quantification of V. harveyi wild type BB120, mutant strain MM30 and BB152 biofilms using CV staining. The staining strength is an indication of the amount of biofilm mass formed in AB –media with/without different concentrations of HU-210. Graph represents calculation of biofilm biomass relatively with control. Presented data are means and SD of three independent experiments, each performed in triplicate. *P < 0.05 (DOC 827 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DS performed all of the experimental work and wrote the manuscript. RS, AB and RM contributed reagents. *DS initiated and followed the study. All authors reviewed the manuscript. All authors read and approved the final manuscript.
Contributor Information
Divya Soni, Email: divyasony.11@gmail.com.
Reem Smoum, Email: reems@ekmd.huji.ac.il.
Aviva Breuer, Email: aviva.breuer@mail.huji.ac.il.
Raphael Mechoulam, Email: raphaelm@ekmd.huji.ac.il.
Doron Steinberg, Email: dorons@ekmd.huji.ac.il.
References
- 1.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176(2):269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 3.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50(1):101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 4.Henke JM, Bassler BL. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J Bacteriol. 2004;186(20):6902–6914. doi: 10.1128/JB.186.20.6902-6914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waters CM, Bassler BL. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev. 2006;20(19):2754–2767. doi: 10.1101/gad.1466506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 7.Cao JG, Meighen EA. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J Biol Chem. 1989;264(36):21670–21676. [PubMed] [Google Scholar]
- 8.Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415(6871):545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 9.Ng WL, Perez LJ, Wei Y, Kraml C, Semmelhack MF, Bassler BL. Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol Microbiol. 2011;79(6):1407–1417. doi: 10.1111/j.1365-2958.2011.07548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xavier KB, Bassler BL. LuxS quorum sensing: more than just a numbers game. Curr Opin Microbiol. 2003;6(2):191–197. doi: 10.1016/S1369-5274(03)00028-6. [DOI] [PubMed] [Google Scholar]
- 11.Schauder S, Bassler BL. The languages of bacteria. Genes Dev. 2001;15(12):1468–1480. doi: 10.1101/gad.899601. [DOI] [PubMed] [Google Scholar]
- 12.Mechoulam R. Cannabis--a valuable drug that deserves better treatment. Mayo Clin Proc. 2012;87(2):107–109. doi: 10.1016/j.mayocp.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pertwee RG. Elevating endocannabinoid levels: pharmacological strategies and potential therapeutic applications. Proc Nutr Soc. 2014;73(1):96–105. doi: 10.1017/S0029665113003649. [DOI] [PubMed] [Google Scholar]
- 14.Mechoulam R, Gaoni Y. Isolation, Structure, and Partial Synthesis of an Active Constituent of Hashish. J Am Chem Soc. 1964;86(8):1646–1647. doi: 10.1021/ja01062a046. [DOI] [Google Scholar]
- 15.Cabral GA, Raborn SE, Ferreira GA. Phytocannabinoids and the Immune System. First Edition, Chapter 14, Oxford University Press: Handbook of Cannabis 2014:261–79
- 16.Mechoulam R, Feigenbaum JJ, Lander N, Segal M, Jarbe TU, Hiltunen AJ, Consroe P. Enantiomeric cannabinoids: stereospecificity of psychotropic activity. Experientia. 1988;44(9):762–764. doi: 10.1007/BF01959156. [DOI] [PubMed] [Google Scholar]
- 17.Ottani A, Giuliani D. Hu 210: a potent tool for investigations of the cannabinoid system. CNS drug reviews. 2001;7(2):131–145. doi: 10.1111/j.1527-3458.2001.tb00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bassler BL, Wright M, Showalter RE, Silverman MR. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9(4):773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 19.Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2(11). doi:10.1101/cshperspect.a012427 [DOI] [PMC free article] [PubMed]
- 20.Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13(2):273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 21.Anetzberger C, Reiger M, Fekete A, Schell U, Stambrau N, Plener L, Kopka J, Schmitt-Kopplin P, Hilbi H, Jung K. Autoinducers act as biological timers in Vibrio harveyi. PloS one. 2012;7(10):e48310. doi: 10.1371/journal.pone.0048310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nazzaro F, Fratianni F, Coppola R. Quorum sensing and phytochemicals. Int J Mol Sci. 2013;14(6):12607–12619. doi: 10.3390/ijms140612607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Packiavathy IA, Sasikumar P, Pandian SK, Veera Ravi A. Prevention of quorum-sensing-mediated biofilm development and virulence factors production in Vibrio spp. by curcumin. Appl Microbiol Biotechnol. 2013;97(23):10177–10187. doi: 10.1007/s00253-013-4704-5. [DOI] [PubMed] [Google Scholar]
- 24.Bjarnsholt T, Jensen PO, Rasmussen TB, Christophersen L, Calum H, Hentzer M, Hougen HP, Rygaard J, Moser C, Eberl L, et al. Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infections. Microbiology. 2005;151(Pt 12):3873–3880. doi: 10.1099/mic.0.27955-0. [DOI] [PubMed] [Google Scholar]
- 25.Choo JH, Rukayadi Y, Hwang JK. Inhibition of bacterial quorum sensing by vanilla extract. Lett Appl Microbiol. 2006;42(6):637–641. doi: 10.1111/j.1472-765X.2006.01928.x. [DOI] [PubMed] [Google Scholar]
- 26.Defoirdt T, Boon N, Sorgeloos P, Verstraete W, Bossier P. Quorum sensing and quorum quenching in Vibrio harveyi: lessons learned from in vivo work. ISME J. 2008;2(1):19–26. doi: 10.1038/ismej.2007.92. [DOI] [PubMed] [Google Scholar]
- 27.Augustine N, Goel AK, Sivakumar KC, Kumar RA, Thomas S. Resveratrol--a potential inhibitor of biofilm formation in Vibrio cholerae. Phytomedicine. 2014;21(3):286–289. doi: 10.1016/j.phymed.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Nithya C, Devi MG, Karutha Pandian S. A novel compound from the marine bacterium Bacillus pumilus S6-15 inhibits biofilm formation in gram-positive and gram-negative species. Biofouling. 2011;27(5):519–528. doi: 10.1080/08927014.2011.586127. [DOI] [PubMed] [Google Scholar]
- 29.Kim YK, McCarter LL. Cross-regulation in Vibrio parahaemolyticus: compensatory activation of polar flagellar genes by the lateral flagellar regulator LafK. J Bacteriol. 2004;186(12):4014–4018. doi: 10.1128/JB.186.12.4014-4018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Q, Defoirdt T: Quorum sensing positively regulates flagellar motility in pathogenic Vibrio harveyi. Environ Microbiol. 2015;17(4)960-68 [DOI] [PubMed]
- 31.Shemesh M, Tam A, Aharoni R, Steinberg D. Genetic adaptation of Streptococcus mutans during biofilm formation on different types of surfaces. BMC Microbiol. 2010;10:51. doi: 10.1186/1471-2180-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sol A, Feuerstein O, Featherstone JD, Steinberg D. Effect of sublethal CO2 laser irradiation on gene expression of streptococcus mutans immobilized in a biofilm. Caries Res. 2011;45(4):361–369. doi: 10.1159/000329390. [DOI] [PubMed] [Google Scholar]
- 33.Shemesh M, Tam A, Feldman M, Steinberg D. Differential expression profiles of Streptococcus mutans ftf, gtf and vicR genes in the presence of dietary carbohydrates at early and late exponential growth phases. Carbohydr Res. 2006;341(12):2090–2097. doi: 10.1016/j.carres.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 34.Mok KC, Wingreen NS, Bassler BL. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J. 2003;22(4):870–881. doi: 10.1093/emboj/cdg085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Defoirdt T, Crab R, Wood TK, Sorgeloos P, Verstraete W, Bossier P. Quorum sensing-disrupting brominated furanones protect the gnotobiotic brine shrimp Artemia franciscana from pathogenic Vibrio harveyi, Vibrio campbellii, and Vibrio parahaemolyticus isolates. Appl Environ Microbiol. 2006;72(9):6419–6423. doi: 10.1128/AEM.00753-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manefield M, Harris L, Rice SA, de Nys R, Kjelleberg S. Inhibition of luminescence and virulence in the black tiger prawn (Penaeus monodon) pathogen Vibrio harveyi by intercellular signal antagonists. Appl Environ Microbiol. 2000;66(5):2079–2084. doi: 10.1128/AEM.66.5.2079-2084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mechoulam R, Lander N, Breuer A, Zahalka J. Synthesis of the individual, pharmacologically distinct, enantiomers of a tetra- hydrocannabinol derivative. Tetrahedron:Asymmetry. 1990;1(5):315–318. doi: 10.1016/S0957-4166(00)86322-3. [DOI] [Google Scholar]
- 38.Aharoni R, Bronstheyn M, Jabbour A, Zaks B, Srebnik M, Steinberg D. Oxazaborolidine derivatives inducing autoinducer-2 signal transduction in Vibrio harveyi. Bioorg Med Chem. 2008;16(4):1596–1604. doi: 10.1016/j.bmc.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 39.Niu C, Afre S, Gilbert ES. Subinhibitory concentrations of cinnamaldehyde interfere with quorum sensing. Lett Appl Microbiol. 2006;43(5):489–494. doi: 10.1111/j.1472-765X.2006.02001.x. [DOI] [PubMed] [Google Scholar]
- 40.Feldman M, Weiss EI, Ofek I, Steinberg D. Interference of cranberry constituents in cell-cell signaling system of Vibrio harveyi. Curr Microbiol. 2009;59(4):469–474. doi: 10.1007/s00284-009-9462-3. [DOI] [PubMed] [Google Scholar]
- 41.O'Toole GA. Microtiter dish biofilm formation assay. J Vis Exp. 2011;(47):2437. doi:10.3791/2437 [DOI] [PMC free article] [PubMed]
- 42.Periasamy S, Kolenbrander PE. Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel. J Bacteriol. 2009;191(22):6804–6811. doi: 10.1128/JB.01006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Assaf D, Steinberg D, Shemesh M. Lactose triggers biofilm formation by Streptococcus mutans. Int Dairy J. 2014;42:51–57. doi: 10.1016/j.idairyj.2014.10.008. [DOI] [Google Scholar]
- 44.Shemesh M, Tam A, Steinberg D. Expression of biofilm-associated genes of Streptococcus mutans in response to glucose and sucrose. J Med Microbiol. 2007;56(Pt 11):1528–1535. doi: 10.1099/jmm.0.47146-0. [DOI] [PubMed] [Google Scholar]
- 45.Feldman M, Al-Quntar A, Polacheck I, Friedman M, Steinberg D. Therapeutic potential of thiazolidinedione-8 as an antibiofilm agent against Candida albicans. PloS one. 2014;9(5):e93225. doi: 10.1371/journal.pone.0093225. [DOI] [PMC free article] [PubMed] [Google Scholar]


