Abstract
Background
Hepatocellular carcinoma (HCC) is one of the most common and most frequently lethal cancers worldwide. Although many advances have been made in the analysis of multistage hepatocarcinogenesis, we still lack information to guide adequate clinical management options for HCC. A large number of genetic alterations occur during hepatocarcinogenesis, and many genetic studies have indicated that one of the most frequently mutated oncogenes found in HCC is β-catenin.
Summary
Molecular subclassification of HCC based on gene expression signatures has identified a typical hepatocyte-like subclass of HCC harboring β-catenin mutations; this subclass is characterized by better histological differentiation and a less aggressive nature. We previously identified overexpression of the leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5), also known as GPR49, in HCC with β-catenin mutations. LGR5 has been indicated as one of the downstream target genes of the Wnt signaling pathway; however, the functional role of LGR5 in cancer is largely unknown. We demonstrated that HCC cells transfected with LGR5 exhibited higher colony forming activity and were more resistant to a cytotoxic drug than the control HCC cells were. Overexpression of LGR5 also retarded cell migration. LGR5-transfected HCC cells formed nodule-type tumors in the livers of immunodeficient mice, whereas control cells formed more invasive tumors. Results of our recent research suggest that aberrant expression of LGR5 could regulate the epithelial cell phenotype and promotes HCC cell survival. HCC cells overexpressing LGR5 seem to represent a typical phenotype of a less aggressive HCC.
Key messages
Recent efforts on the molecular classification of HCC have led us to new strategies for dealing with HCC. These specific signatures may predict the risk of recurrence or the patient survival rate, which affect the outlook and may suggest treatment strategies for HCC patients.
Key Words: HCC molecular subclassification, HCC typical phenotype, LGR5, LGR5-overexpressing cells, WNT/β-catenin
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer in men and the seventh in women, and because of high levels of mortality, it ranks as the third most common cause of death from cancer worldwide [1]. The steps involved in malignant transformation in HCC are well characterized: from chronic viral infection and liver cirrhosis to dysplastic nodules and then to early through to advanced HCC. Many studies have elucidated the molecular mechanisms involved in HCC's multistage hepatocarcinogenesis and helped to identify molecular markers in the development of HCC [2]. Although many advances have been made in the analysis of multistage hepatocarcinogenesis, we still lack information to guide adequate clinical management or indicate the most effective treatment options for HCC. Most HCC patients have underlying chronic liver diseases. Advanced liver cirrhosis resulting from either hepatitis B virus or hepatitis C virus infection is a contraindication for surgical liver resection, one of the potentially curative options in HCC treatment. Prognosis assessment and treatment decisions depend on the tumor staging; however, that depends on several different staging systems, and this fact can lead to difficulty in grading tumors uniformly [3]. Ideally, we need to standardize the histological molecular markers used in the diagnosis of HCC despite the diversity in clinical backgrounds and in the process of hepatocarcinogenesis. We also need to detail the molecular characteristics or histological patterns of HCC in order to establish accurate staging and improve treatment strategies. Here, we will discuss progress in molecular classification that has been made across different clinical settings in HCC patients worldwide and focus on the leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5) gene, one of the genes already identified as being overexpressed in HCC, which represents a typical subgroup of HCC.
Molecular Subclassification of HCC
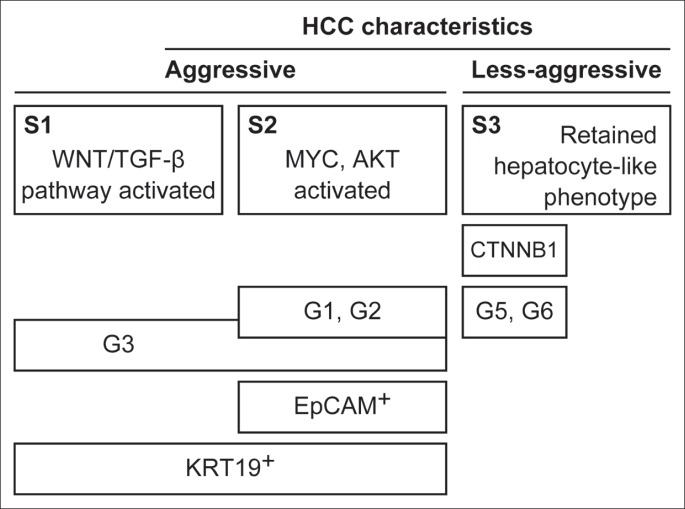
Some new approaches and methodologies have been developed to provide more credible histopathologic information that may be useful in predicting patient outcome and in assisting the clinical decision making process. Currently, specific genes and molecular pathways involved in the pathogenesis of HCC have been studied, and some progress has been made through genome-wide gene expression studies. Multiple studies have suggested that, despite highly variable clinical backgrounds and molecular heterogeneity, HCCs may share a similar gene-expression pattern [4]. A study by Boyault and colleagues in 2007 revealed six robust subgroups of HCC (designated G1-G6) by using microarray analysis and quantitative reverse transcription polymerase chain reaction (RT-PCR) data in a series of 120 HCCs and 3 hepatocellular adenomas. Subgroups G1-G3 were associated with high chromosomal instability, and the poorest prognosis was associated with subgroup G3, whereas better survival was evident in subgroups G4-G6 [5]. A subsequent study showed that the tumor-associated G3-proliferation signature and the poor-survival signature from adjacent nontumoral cirrhotic tissue were independent predictors of HCC recurrence, along with the presence of satellites [6]. A 2009 study by Hoshida and colleagues that involved a total of 603 HCC patients found three robust HCC subclasses (termed S1, S2, and S3): each subclass correlated with clinical parameters such as tumor size, the extent of cellular differentiation, and the serum α-fetoprotein (AFP) level. Subclasses S1 and S2 were associated with large tumors and poor histological differentiation, whereas subclass S3 was associated with a retained hepatocyte-like phenotype and a less-aggressive nature [7]. Some of the groups identified in these important studies had similar characteristics. Activations of Wnt/TGF-β, MYC, and AKT with inactivation of p53 were associated with subclasses S1 and S2, and this was in accordance with Boyoult's subgroups G1-G3. In addition, the less-aggressive S3 subclass resembled Boyault's G5 and G6 subgroups. Subgroups S3 and G5/G6 were strongly associated with β-catenin (CTNNB1) mutations that lead to Wnt pathway activation. As previously reported, a high prevalence of CTNNB1 mutations indicates alterations to canonical Wnt signaling in a subset of HCC [8]. Yamashita's classification of HCC subtypes with respect to expression levels of epithelial cell adhesion molecule (EpCAM) and AFP serum elevation was consistent with subclass S2, which also had an enriched EpCAM signature [7, 9]. EpCAM is known to be a hepatic stem cell marker, and EpCAM-positive HCC cells displayed a distinct molecular signature with features of hepatic progenitor cells [10]. In addition to EpCAM, KRT19 has also been identified as a gene associated with the hepatic progenitor cell-like signature and has already been described as a prognostic marker in HCC. Patients with HCCs that stain positive for KRT19 have a worse prognosis than those with KRT19-negative HCCs [11]. Subclasses S1 and S2 were associated with a KRT19-rich signature [4]. A simple global overview of HCC classification, modified from Hoshida's figure [4], is given in fig. 1. A recent study of 314 HCC samples from French patients identified a “five-gene score” for HCC (using a panel of five genes: TAF9, RAMP3, HN1, KRT19, and RAN) that was associated with outcome in HCC patients undergoing surgical resection [12]. This score was further validated in two groups of patients: one from Europe and the United States (most with hepatitis C virus and cirrhosis) and one from China (most with chronic hepatitis B virus infection). The five-gene score was significantly associated with disease-specific survival, independent of tumor stage, etiology, or the presence of cirrhosis. The cell cycle regulator (TAF9), the adrenomedullin receptor (RAMP3), the androgen (HN1), and the ras/raf/MAP kinase node (RAN) are new markers that have been linked to early tumor recurrence after surgery and to survival after recurrence [11, 13, 14]. Despite some additional validations being required before it is used as a robust predictive tool, the five-gene score was suggested to be superior in predicting patient prognosis than the previously established G3 HCC subgroup [12]. These novel strategies in HCC subclassification still need to undergo a long and challenging process of evaluation before they are applied in routine clinical practice. However, in the current era of advanced molecular medicine in which much effort has been expended on analyzing signaling pathways and mechanisms, a new approach to classifying typical HCC has been clearly indicated.
Fig. 1.
Simple HCC classification. HCC was divided into two groups: aggressive and less-aggressive types. The global subclasses of HCC (e.g., S1-S3 [7] and G1-G6 [5]) share some common characteristics. These specific signatures of HCC may enable us to predict patient outcome and lead to improved management and treatment.
LGR5 Expression in Cancer
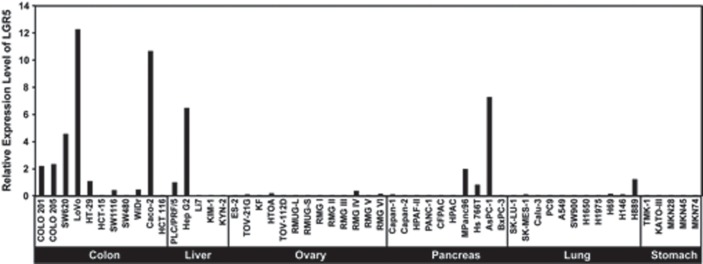
The Wnt/β-catenin signaling pathway is one of the major signaling pathways known to be involved in hepatocarcinogenesis and is also activated in Hoshida's S3 subclass and Boyault's G5/G6 subgroups. The Wnt signaling pathway is an evolutionarily conserved pathway that regulates crucial aspects of cell fate determination, cell migration, cell polarity, neural patterning, and organogenesis during embryonic development. It also plays a critical role in tumorigenesis. There are three main pathways related to Wnt signaling, and the canonical β-catenin-dependent pathway has been the most intensively studied. Mutations at sites that affect β-catenin phosphorylation can lead to dysregulation of the pathway [15,16]. We reported overexpression of LGR5, also known as G-protein-coupled receptor 49 (GPR49), in HCC carrying β-catenin mutations [17]. LGR5 is structurally related to members of the G-protein-coupled receptor (GPCR) family, which comprises proteins with seven transmembrane domains. GPCRs function as receptors for various classes of ligand, including peptide hormones and chemokines [18]. Some studies have suggested that LGR5 is a downstream target of the Wnt signaling pathway [19, 20]. It has been shown that LGR5 expression in a human colon cancer cell line was downregulated on the induced inhibition of canonical Wnt pathway activity [21]. LGR5 was also identified as a novel stem cell marker of the small intestine, colon, and the hair follicles of mice. In the gastrointestinal tract, LGR5 seems to mark the cycling crypt base columnar cells that are suggested to be true intestinal stem cells and can give rise to all cell types of the gut epithelium. LGR5 was restrictedly expressed in the population of actively cycling cells of the bulge of the telogen hair follicles. LGR5-positive hair follicles are self-renewing and multipotent, exhibiting the minimal properties of stem cells [22, 23]. The expression pattern of LGR5 suggests that it may represent a global marker of adult stem cells [21, 23]. Upregulation of LGR5 has also been reported in cancers other than HCC, and we previously reported LGR5 expression in a variety of cancer cell lines [24]. According to our data, a higher expression of LGR5 was observed in colon, liver, and pancreatic cancer cell lines than in ovarian, lung, and stomach cancer cell lines (fig. 2). LGR5 plays a significant role in tumor formation and cell proliferation in basal cell carcinoma, a common malignant skin tumor [25]. LGR5 is also involved from the early stages of colorectal tumorigenesis and is significantly correlated with lymphatic invasion in colorectal and gastric cancer [24, 26]. In intestinal-type gastric cancer, there is a correlation between expression of LGR5 and local tumor growth, nodal spread, and patient survival [27]. Also, in colon and ovarian carcinomas, overexpression of LGR5 has been identified, suggesting LGR5 as a new potential molecular target and biomarker in colon and ovarian tumors [28]. Although many studies have revealed upregulation of LGR5 along with increased Wntsignaling in cancer cells, indicating an important role of LGR5 in oncogenesis, the functional role of LGR5 in cancer cells is still poorly understood.
Fig. 2.
LGR5 expression in a variety of cancer cell lines. LGR5 expression was analyzed by quantitative RT-PCR. A higher expression of LGR5 was observed in colon, liver, and pancreatic cancer cell lines than in ovarian, lung, and stomach cancer cell lines.
LGR5 Expression in HCC
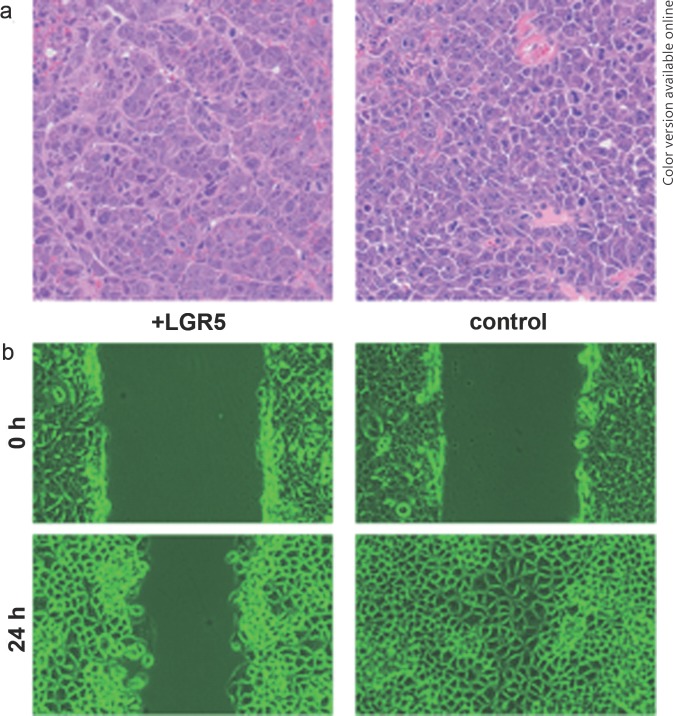
Continuing our findings of LGR5 overexpression in HCC, we analyzed LGR5 function using HCC cell lines. HepG2 and PLC/PRF/5 were used as cell lines with activated Wnt signaling and high levels of LGR5 mRNA expression, whereas KYN2 was used as a cell line with low expression of LGR5 mRNA [17, 24]. We established stable clones expressing high levels of LGR5. Cells transfected with LGR5 showed an ability to survive longer in overgrowth conditions and were more resistant to the cytocidal effects of puromycin than the control empty vector cells were. LGR5-overexpressing HCC cells formed nodular tumors with a tightly aggregated morphology, whereas downregulation of LGR5 transformed the cells into a loosely associated morphology allowing easy infiltration to contiguous tissue. Loss of LGR5 expression also increased cell motility: in the scratch wound motility assay, LGR5-overexpressing cells exhibited slow migration compared with that of the empty vector control cells (fig. 3) [16]. In the above study, we observed that high levels of LGR5 expression altered tumors from a diffuse to a more nodular phenotype and also from a metastatic to a less metastatic phenotype. These findings showed that LGR5 expression in HCC likely affects the morphology and the metastatic properties of the tumor. Another report using colorectal cancer cell lines indicated that loss of LGR5 may contribute to the invasive phenotype of colorectal carcinoma, suggesting that LGR5 is a negative regulator of tumorigenicity [29]. Our previous clinicopathological study showed that overexpression of LGR5 was frequently found in well to moderately differentiated HCC [16]. Also, in HCC cell lines, LGR5 was highly expressed in HepG2 and PLC/PRF/5, which are categorized as non-metastatic (fig. 2). Our findings regarding LGR5 function in HCC cells seem to be in agreement with Hoshida's S3 subclass and Boyault's G5/G6 subgroups, which are characterized by a less aggressive, more differentiated tumor and the retention of normal liver function. Therefore, we suggest that the typical morphological and biological features of LGR5 expression in HCC may characterize a subset of the typical phenotype of less-aggressive HCC.
Fig. 3.
Histological analysis and cell motility assay in LGR5-overexpressing cells. (a) Transplantation of LGR5-overexpressing cells (+LGR5) into the livers of NOG (NOD/Shi-scid/IL-2γ-/-) mice resulted in tumors with a trabecular pattern, whereas the control cells formed tumors with an ill-defined trabecular pattern. (b) In a scratch wound motility assay, retardation of cell migration was observed in LGR5-overexpressing cells compared to the control cells.
Conclusion
Recent studies on the molecular classification of HCC are shedding some light on progress in the management of HCC. In addition to other markers such as EpCAM and KRT19, LGR5 expression may reflect a subset of HCC classification. These specific signatures are associated with clinical outcomes and may suggest treatment strategies for HCC patients. Thus, combining the use of immunohistochemical markers and functional role studies with a systematic framework of HCC classification will provide a more accurate understanding of hepatocarcinogenesis and point the way toward the personalized treatment of HCC.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Sakamoto M, Effendi K, Masugi Y. Molecular diagnosis of multistage hepatocarcinogenesis. Jpn J Clin Oncol. 2010;40:891–896. doi: 10.1093/jjco/hyq099. [DOI] [PubMed] [Google Scholar]
- 3.Marrero JA, Fontana RJ, Barrat A, Askari F, Conjeevaram HS, Su GL, Lok AS. Prognosis of hepatocellular carcinoma: comparison of 7 staging systems in an American cohort. Hepatology. 2005;41:707–716. doi: 10.1002/hep.20636. [DOI] [PubMed] [Google Scholar]
- 4.Hoshida Y, Toffanin S, Lachenmayer A, Villanueva A, Minguez B, Llovet JM. Molecular classification and novel targets in hepatocellular carcinoma: recent advancements. Semin Liver Dis. 2010;30:35–51. doi: 10.1055/s-0030-1247131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyault S, Rickman DS, de Reyniès A, Balabaud C, Rebouissou S, Jeannot E, Hérault A, Saric J, Belghiti J, Franco D, Bioulac-Sage P, Laurent-Puig P, Zucman-Rossi J. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 6.Villanueva A, Hoshida Y, Battiston C, Tovar V, Sia D, Alsinet C, Cornella H, Liberzon A, Kobayashi M, Kumada H, Thung SN, Bruix J, Newell P, April C, Fan JB, Roayaie S, Mazzaferro V, Schwartz ME, Llovet JM. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140:1501–1512. doi: 10.1053/j.gastro.2011.02.006. e1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K, Hashimoto M, Watanabe G, Gabriel S, Friedman SL, Kumada H, Llovet JM, Golub TR. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, LeBlanc AC, Donovan DJ, Thung SN, Solé M, Tovar V, Alsinet C, Ramos AH, Barretina J, Roayaie S, Schwartz M, Waxman S, Bruix J, Mazzaferro V, Ligon AH, Najfeld V, Friedman SL, Sellers WR, Meyerson M, Llovet JM. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–6788. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, Budhu A, Zanetti KA, Chen Y, Qin LX, Tang ZY, Wang XW. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 10.Yamashita T, Budhu A, Forgues M, Wang XW. Activation of hepatic stem cell marker EpCAM by Wnt-beta-catenin signaling in hepatocellular carcinoma. Cancer Res. 2007;67:10831–10839. doi: 10.1158/0008-5472.CAN-07-0908. [DOI] [PubMed] [Google Scholar]
- 11.Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, Mikaelyan A, Roberts LR, Demetris AJ, Sun Z, Nevens F, Roskams T, Thorgeirsson SS. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 12.Nault JC, De Reyniès A, Villanueva A, Calderaro J, Rebouissou S, Couchy G, Decaens T, Franco D, Imbeaud S, Rousseau F, Azoulay D, Saric J, Blanc JF, Balabaud C, Bioulac-Sage P, Laurent A, Laurent-Puig P, Llovet JM, Zucman-Rossi J. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology. 2013;145:176–187. doi: 10.1053/j.gastro.2013.03.051. [DOI] [PubMed] [Google Scholar]
- 13.Nikitenko LL, Fox SB, Kehoe S, Rees MC, Bicknell R. Adrenomedullin and tumour angiogenesis. Br J Cancer. 2006;94:1–7. doi: 10.1038/sj.bjc.6602832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuen HF, Chan KK, Grills C, Murray JT, Platt-Higgins A, Eldin OS, O'Byrne K, Janne P, Fennell DA, Johnston PG, Rudland PS, El-Tanani M. Ran is a potential therapeutic target for cancer cells with molecular changes associated with activation of the PI3K/Akt/mTORC1 and Ras/MEK/ERK pathways. Clin Cancer Res. 2012;18:380–391. doi: 10.1158/1078-0432.CCR-11-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuma M, Tanese K, Effendi K, Yamazaki K, Masugi Y, Suda M, Sakamoto M. Leucine-rich repeat-containing G protein-coupled receptor 5 regulates epithelial cell phenotype and survival of hepatocellular carcinoma cells. Exp Cell Res. 2013;319:113–121. doi: 10.1016/j.yexcr.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto Y, Sakamoto M, Fujii G, Tsuiji H, Kenetaka K, Asaka M, Hirohashi S. Overexpression of orphan G-protein-coupled receptor, Gpr49, in human hepatocellular carcinomas with beta-catenin mutations. Hepatology. 2003;37:528–533. doi: 10.1053/jhep.2003.50029. [DOI] [PubMed] [Google Scholar]
- 18.Schöneberg T, Schultz G, Gudermann T. Structural basis of G protein-coupled receptor function. Mol Cell Endocrinol. 1999;151:181–193. doi: 10.1016/s0303-7207(99)00017-9. [DOI] [PubMed] [Google Scholar]
- 19.Garcia MI, Ghiani M, Lefort A, Libert F, Strollo S, Vassart G. LGR5 deficiency deregulates Wnt signaling and leads to precocious Paneth cell differentiation in the fetal intestine. Dev Biol. 2009;331:58–67. doi: 10.1016/j.ydbio.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 20.Carmon KS, Lin Q, Gong X, Thomas A, Liu Q. LGR5 interacts and cointernalizes with Wnt receptors to modulate Wnt/β-catenin signaling. Mol Cell Biol. 2012;32:2054–2064. doi: 10.1128/MCB.00272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 22.Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, Toftgård R. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 23.Haegebarth A, Clevers H. Wnt signaling, lgr5, and stem cells in the intestine and skin. Am J Pathol. 2009;174:715–721. doi: 10.2353/ajpath.2009.080758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uchida H, Yamazaki K, Fukuma M, Yamada T, Hayashida T, Hasegawa H, Kitajima M, Kitagawa Y, Sakamoto M. Overexpression of leucine-rich repeat-containing G protein-coupled receptor 5 in colorectal cancer. Cancer Sci. 2010;101:1731–1737. doi: 10.1111/j.1349-7006.2010.01571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanese K, Fukuma M, Yamada T, Mori T, Yoshikawa T, Watanabe W, Ishiko A, Amagai M, Nishikawa T, Sakamoto M. G-protein-coupled receptor GPR49 is up-regulated in basal cell carcinoma and promotes cell proliferation and tumor formation. Am J Pathol. 2008;173:835–843. doi: 10.2353/ajpath.2008.071091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamanoi K, Fukuma M, Uchida H, Kushima R, Yamazaki K, Katai H, Kanai Y, Sakamoto M. Overexpression of leucine-rich repeat-containing G protein-coupled receptor 5 in gastric cancer. Pathol Int. 2013;63:13–19. doi: 10.1111/pin.12013. [DOI] [PubMed] [Google Scholar]
- 27.Simon E, Petke D, Böger C, Behrens HM, Warneke V, Ebert M, Röcken C. The spatial distribution of LGR5+ cells correlates with gastric cancer progression. PLoS ONE. 2012;7:e35486. doi: 10.1371/journal.pone.0035486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McClanahan T, Koseoglu S, Smith K, Grein J, Gustafson E, Black S, Kirschmeier P, Samatar AA. Identification of overexpression of orphan G protein-coupled receptor GPR49 in human colon and ovarian primary tumors. Cancer Biol Ther. 2006;5:419–426. doi: 10.4161/cbt.5.4.2521. [DOI] [PubMed] [Google Scholar]
- 29.Walker F, Zhang HH, Odorizzi A, Burgess AW. LGR5 is a negative regulator of tumourigenicity, antagonizes Wnt signalling and regulates cell adhesion in colorectal cancer cell lines. PLoS ONE. 2011;6:e22733. doi: 10.1371/journal.pone.0022733. [DOI] [PMC free article] [PubMed] [Google Scholar]